Rice pps is a heterochronic mutant that shows a prolonged juvenile phase by repressing GA biosynthetic genes and altering expression patterns of microRNA genes but is early flowering. Positional cloning revealed that PPS is the ortholog of Arabidopsis COP1. Although PPS has a role in photomorphogenesis as COP1 does, PPS has additional roles in vegetative phase change and flowering time regulation.
Abstract
Because plant reproductive development occurs only in adult plants, the juvenile-to-adult phase change is an indispensable part of the plant life cycle. We identified two allelic mutants, peter pan syndrome-1 (pps-1) and pps-2, that prolong the juvenile phase in rice (Oryza sativa) and showed that rice PPS is an ortholog of Arabidopsis thaliana CONSTITUTIVE PHOTOMORPHOGENIC1. The pps-1 mutant exhibits delayed expression of miR156 and miR172 and the suppression of GA biosynthetic genes, reducing the GA3 content in this mutant. In spite of its prolonged juvenile phase, the pps-1 mutant flowers early, and this is associated with derepression of RAP1B expression in pps-1 plants independently of the Hd1-Hd3a/RFT1 photoperiodic pathway. PPS is strongly expressed in the fourth and fifth leaves, suggesting that it regulates the onset of the adult phase downstream of MORI1 and upstream of miR156 and miR172. Its ability to regulate the vegetative phase change and the time of flowering suggests that rice PPS acquired novel functions during the evolution of rice/monocots.
INTRODUCTION
The life cycle of higher plants has three mutually distinct developmental stages: the embryogenetic, vegetative, and reproductive stages. The vegetative stage can be further divided into the juvenile and adult phases, which are distinguished by many morphological and physiological differences both in woody plants, such as English ivy (Hedera helix) and Eucalyptus occidentalis (Poethig, 1990; Jaya et al., 2010), which exhibit obvious heteroblasty, and in herbaceous plants, which exhibit different morphological and physiological characteristics in the juvenile and adult phases (Lawson and Poethig, 1995; Telfer et al., 1997; Asai et al., 2002). Importantly, because plants can initiate reproductive growth under appropriate environmental conditions only during the adult phase (Simpson et al., 1999; Poethig, 2003), the juvenile-to-adult phase change (also known as the vegetative phase change) has a critical role in plant development. Although the mechanisms underlying the vegetative phase change remain largely unknown, recent studies using heterochronic mutants have revealed that microRNAs (miRNAs) play a significant role in the phase change.
Arabidopsis thaliana rosette leaves are rounded with smooth leaf margins in the juvenile phase and are long, ovate, and serrated in the adult phase. The juvenile and adult leaves also differ conspicuously in their patterns of trichome distribution; trichomes are found only on the adaxial side of juvenile leaves but are found on both sides of adult leaves (Telfer et al., 1997). These phenotypic markers have been used to identify Arabidopsis mutations causing a precocious phase change, many of which were subsequently associated with small RNAs. Mutations in ZIPPY, SUPPRESSOR OF GENE SILENCING3, and RNA-DEPENDENT POLYMERASE6, which are required for posttranscriptional silencing, are associated with the presence of trichomes on the abaxial sides of juvenile leaves (Hunter et al., 2003; Peragine et al., 2004). In the squint mutant, which forms abaxial trichomes on early leaves, the activity of microRNA156 (miR156) is decreased because the ARGONAUTE1 protein is misfolded, resulting in the enhanced expression of the target genes, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family genes encoding transcription factors (Smith et al., 2009). Ten of the 16 SPL genes in Arabidopsis are downregulated by miR156b (Schwab et al., 2005), and the spl9 and spl15 T-DNA insertion mutants exhibit prolonged juvenile phases (Schwarz et al., 2008). Conversely, overexpression of SPL3/4/5 lacking the miR156 target site causes a precocious phase change (Wu and Poethig, 2006). These findings indicate that small RNAs play an important role in the vegetative phase change in Arabidopsis.
In maize (Zea mays), juvenile leaves have epicuticular wax and no epidermal hair, and adult leaves have epidermal hair but no wax (Lawson and Poethig, 1995). As in Arabidopsis, miRNAs are involved in the vegetative phase change. The dominant Corngrass1 (Cg1) mutant encodes miR156, leading to overexpression of miR156 and prolongation of the juvenile phase (Chuck et al., 2007). The glossy15 (gl15) mutant shortens the juvenile phase in the maize epidermis (Moose and Sisco, 1994); GL15 is an AP2-like gene harboring two AP2 domains (Moose and Sisco, 1996) that are targets of miR172 (Lauter et al., 2005). In the early vegetative stage, transcription of miR156 far exceeds that of miR172, whereas later in the vegetative stage, the inverse pattern is seen: transcription of miR172 far exceeds that of miR156 (Lauter et al., 2005; Chuck et al., 2007). Inverse miR156 and miR172 expression patterns are also observed in Arabidopsis (Aukerman and Sakai, 2003; Wu and Poethig, 2006).
In a recent Arabidopsis study (Wu et al., 2009), miR172 expression was shown to occur downstream of SPL9 and SPL10 expression, which is regulated by miR156. Thus, miR156 and miR172 may be key genes in the juvenile–adult phase change. The targets of miR156 and miR172 are known, but little is known about their upstream genes.
Gibberellin (GA) is a well-established regulatory phytohormone of the juvenile–adult phase change. In Arabidopsis, mutations in genes functioning in the biosynthesis of and response to GA prolong the juvenile phase (Telfer et al., 1997; Telfer and Poethig, 1998); the most severe mutation, ga1-3, causes the plant to fail to develop adult leaves (Telfer et al., 1997; Telfer and Poethig, 1998). Furthermore, GA treatment promotes the transition to adult phase in both Arabidopsis and maize (Evans and Poethig, 1995; Schwarz et al., 2008).
Although GA is known to promote the vegetative phase change, the other upstream and downstream factors controlling this phase change are largely unknown. However, because the juvenile–adult phase change affects a large number of traits, it is safe to assume that a large number of genes, including those encoding miRNAs, are involved in its regulation. A comprehensive understanding of the phase change will require the identification and functional characterization of more of the relevant genes.
To date, molecular genetic studies of the vegetative phase change have been concentrated mainly in two species, Arabidopsis and maize. However, rice (Oryza sativa) also has many morphological and physiological traits that differ between the juvenile and adult phases. These traits include the size of the shoot apical meristem (SAM), size and shape of leaf blades, presence or absence of midribs, vascular orientation in the stem, node–internode differentiation, and photosynthetic rate (Itoh et al., 2005). Only one mutation affecting the rice vegetative phase change, mori1, has been reported to date (Asai et al., 2002). Although this mutant has been characterized as perpetually maintaining the juvenile phase (Asai et al., 2002), the causal gene has not yet been cloned.
In this study, we attempted to identify additional rice mutants with altered vegetative phase changes. As a result, we isolated the peter pan syndrome (pps) mutant, which exhibits a prolonged juvenile phase and early flowering. Positional cloning revealed that PPS is an ortholog of Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1).
RESULTS
Vegetative Phenotypes of pps Mutant
We identified one recessive rice mutant with a dwarf, dark-green phenotype (Figure 1A). The dwarf phenotype was maintained until the flowering stage. Because a detailed examination of the juvenile/adult marker traits (described below) of this mutant revealed that it has a prolonged juvenile phase, we named this mutant peter pan syndrome-1 (pps-1). Subsequently, we identified an additional allelic mutant, pps-2, that exhibited an even more severe mutant phenotype. Most pps-2 seeds could not germinate, and only ~5% of seeds could germinate on nutrient medium containing sucrose but died in a week (Figure 1B). Thus, our analyses were performed primarily on pps-1.
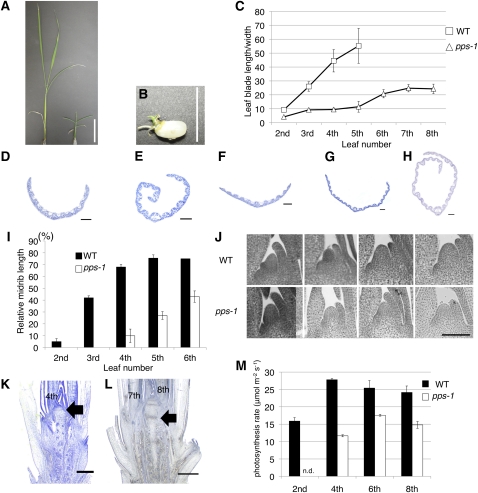
Figure 1.
Phenotypes of pps-1 Plants.
(A) Seedlings of 1-week-old wild type (left) and 1-week-old pps-1 (right).
(B) A rare germinated pps-2 seed.
(C) Comparison of the ratio of leaf blade length to width in the wild type (WT) versus pps-1.
(D) to (H) Cross sections of leaf blades.
(D) Cross section of wild-type second leaf blade cut at 10% of the distance from the base to the tip.
(E) Cross section of wild-type third leaf blade cut at 45% from the base.
(F) Cross section of pps-1 fourth leaf blade cut at 10% from the base.
(G) Cross section of pps-1 fifth leaf blade cut at 30% from the base.
(H) Cross section of pps-1 sixth leaf blade cut at 40% from the base.
(I) Comparison of relative midrib length in lead blades during development in the wild type versus pps-1. Midrib length is shown as a percentage of total blade length.
(J) Wild-type and pps-1 shoot apices showing temporal change of SAM size. From left to right, second leaf stage, third leaf stage, fourth leaf stage, and fifth leaf stage.
(K) Fourteen-day-old wild-type stem. Arrow indicates fourth leaf base node.
(L) Thirty-day-old pps-1 stem. Arrow indicates seventh leaf base node.
(M) Comparison of photosynthetic rates in the wild type and pps-1. Units = μmol CO2 m−2 s−1.
Data represent mean ± sd in (C) and (I) (n = 5) and in (M) (n = 3). Bars = 1 cm in (A), 5 cm in (B), 100 μm in (D) to (H), (K), and (L), and 50 μm in (J).
[See online article for color version of this figure.]
Of the considerable number of morphological and physiological traits known to differ between the juvenile and adult phases in rice (Itoh et al., 2005), we first examined leaf shape. We observed that the ratio of leaf blade length to width rapidly increases with elevation of leaf position (Figure 1C); the leaf-blade length:width ratio for the second leaf blade is ~10:1, whereas that of the fifth leaf blade, which is much more slender, is greater than 50:1. By contrast, no rapid increase of the length:width ratio with elevation of leaf position was observed in the pps-1 mutant; the ratio of even the eighth leaf blade of the mutant was comparable to that of the third leaf of wild-type plants (Figure 1C).
We also examined the leaf blades of the pps-1 mutant for the presence of a midrib, which is normally seen in adult leaves. In wild-type plants, midribs were rarely observed in second leaves (Figure 1D). In third and fourth leaves, midribs were observed to cover approximately one-half of the total length from leaf base to leaf tip (~40 and 60% of the total length, respectively; Figures 1E and 1I). In the higher leaves, midribs covered at least 75% of the length of the blade (Figure 1I). By contrast, second, third, and fourth pps-1 leaves exhibited almost no midrib (Figures 1F and 1I), and midrib formation in fifth leaf was ~20% of the total length (Figures 1G and 1I). Even in the sixth pps-1 leaf, midrib length was comparable to that of a wild-type third leaf (Figures 1H and 1I). Thus, second to fourth pps-1 leaves are structurally similar to wild-type second leaves, and fifth to sixth pps-1 leaves are comparable to wild-type third leaves. This suggests that the pps-1 sixth leaf is intermediate between juvenile and adult, suggesting that pps-1 has more juvenile leaves.
An examination of other morphological traits supported this characterization of the pps-1 mutant. Normally, SAMs enlarge gradually during development (Figure 1J), but the pps-1 SAM remained unchanged in size until at least the fifth-leaf stage (Figure 1J). Furthermore, the pps-1 mutant displayed different node–internode differentiation characteristics. Normally, below the insertion of the fourth leaf, the vascular bundles are irregular in orientation, and no node–internode differentiation is evident, whereas above the fourth leaf, the stem has obvious nodes (Figure 1K); in the mutant, on the other hand, the vascular orientation remained irregular up to the sixth leaf insertion, and no node differentiation was observable on the stem until the insertion of the seventh leaf (Figure 1L). Therefore, the stem structure of the pps-1 mutant becomes adult above the sixth or seventh leaf insertion and is also consistent with a prolonged juvenile phase.
The physiological traits associated with the phase change in the pps-1 mutant were also consistent with a prolonged juvenile phase. For example, in wild-type rice plants, photosynthesis is reduced in juvenile leaves (second leaves) and much more quickly in fourth and higher leaves (Figure 1M). By contrast, photosynthesis occurred slowly even in the fourth through eighth leaves of the pps-1 mutant; the rate was comparable to that in wild-type second leaves (Figure 1M). Thus, both morphological and physiological characteristics indicate that the juvenile phase extends to the sixth or eighth leaf in pps-1 plants. In addition, adult pps-1 plants were not normal (small stature and incomplete midrib formation). Thus, even in the late vegetative phase, juvenile characters coexist with adult characters.
Profile of miR156 and miR172 Expression
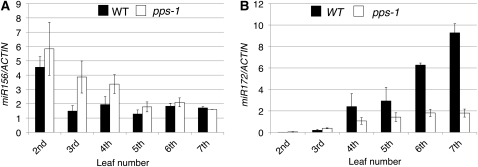
When we used real-time PCR to examine miR156 and miR172 expression in wild-type plants, we found that the level of miR156 expression was high in second leaves, but it decreased to less than half of this level in third leaves (Figure 2A). This low-level expression of miR156 was maintained through the seventh leaf (Figure 2A). The miR172 expression pattern was the inverse of that of miR156, as expected; the second and third leaves contained very little miR172 transcripts, and the amount of miR172 transcripts increased rapidly going up the stem to the seventh leaf (Figure 2B). Thus, expression of the miR156 and miR172 genes in wild-type rice is phase dependent and inversely regulated, as it is in maize and Arabidopsis.
Figure 2.
Expression of Two miRNAs in Wild-Type and pps-1 Leaves.
(A) Expression of miR156 in the second to seventh leaf blades. WT, wild type.
(B) Expression of miR172 in the second to seventh leaf blades. Expression levels are represented relative to ACTIN expression. Each value is the average of three independent real-time PCR assays.
Data in (A) and (B) represent means ± sd (n = 3).
In the pps-1 mutant, the miR156 expression level in third and fourth leaves was slightly lower than that in the second leaf but significantly higher than that in wild-type third and fourth leaves (Figure 2A). The miR156 expression levels in the pps-1 and wild-type plants did not became comparable until the sixth or seventh leaves (Figure 2A), indicating that pps-1 delays the downregulation of miR156 expression with elevation by two or three leaves. In addition, miR172 expression levels in the pps-1 mutant increased very slowly with development, so that the level in seventh pps-1 leaves was only 20% of that in wild-type seventh leaves (Figure 2B). This result is also consistent with a delay in the juvenile–adult phase change. Thus, the pps-1 mutation both affects the expression of miR156 and miR172 and prolongs the juvenile phase, providing strong evidence that miR156 and miR172 regulate the phase change downstream of PPS and that PPS promotes the transition to adult phase.
Flowering Time
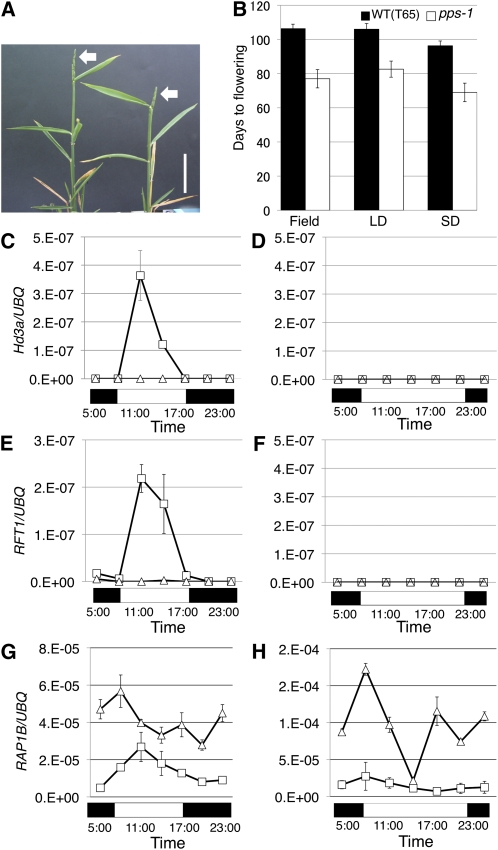
Notably, when about 10 leaves were formed, pps-1 plants flowered ~3 weeks early under field conditions despite their prolonged juvenile period (Figures 3A and 3B), and the early flowering was independent of daylength (Figure 3B). Thus, PPS appears not only to promote the juvenile–adult phase change but also to suppress the transition from vegetative to reproductive stage.
Figure 3.
Early Flowering Phenotypes in pps-1.
(A) Mature pps-1 plants. Arrows indicate panicles. Bar = 5 cm.
(B) Days to flowering in the wild type (WT) and pps-1 in field conditions, long day (LD) and short day (SD).
(C) Hd3a expression under short-day conditions.
(D) Hd3a expression under long-day conditions.
(E) RFT1 expression under short-day conditions.
(F) RFT1 expression under long-day conditions.
(G) RAP1B expression under long-day conditions.
(H) RAP1B expression under short-day conditions.
(C) to (H) White squares, the wild type; white triangles, pps-1. Black and white boxes under each panel represent periods of darkness and light, respectively. Data represent mean ± sd in (B) (n = 5) and in (C) to (H) (n = 3).
[See online article for color version of this figure.]
To examine the mechanism of early flowering in pps-1 plants, we first analyzed the expression of two florigen genes, Hd3a and RFT1, in plants with emerging sixth leaves. At this stage in typical photoperiodic cultivars of rice, the expression of the Hd3a and RFT1 genes in leaves is critically controlled by photoperiods (Itoh et al., 2010). In Taichung 65, the background cultivar of pps and the pps mutants, two major rice flowering-time genes, Hd1 (Heading date 1, a rice ortholog of Arabidopsis CO) and Ehd1 (Early heading date 1), were mutated (Doi et al., 2004). Therefore, expressions of these genes were only slightly induced in the sixth leaves of Taichung 65 plants under short-day conditions and completely suppressed under long-day conditions (Doi et al., 2004). Although we expected to see enhanced expressions of the florigen genes in pps-1 sixth leaves, they were completely suppressed (Figures 3C to 3F). This finding indicates that early flowering in pps-1 plants is not caused by precocious activation of the Hd1-Ehd1-Hd3a/RFT photoperiodic pathway.
Since the cop1 mutant exhibits a circadian clock phenotype (Millar et al., 1995), we further examined the diurnal expression of major circadian clock–related genes in rice (see Supplemental Figure 1 online). The results indicate that the amplitudes of Os GI and Os PRR1 in pps-1 were slightly reduced, but no critical changes were observed on phase setting of these genes. Similarly, no clear change was detected in Os LHY expression under short-day conditions. Together with the defects of Hd1 and Ehd1 in Taichung 65, which functions downstream of circadian clocks, it is unlikely that the early flowering phenotype of pps is caused by some defects in rice circadian clocks.
We next examined the expression of RAP1B/Os MADS14, which normally functions downstream of Hd3a/RFT1 in the positive regulation of floral homeotic genes (Komiya et al., 2008). RAP1B expression was higher in pps-1 plants than in wild-type plants under both short-day and long-day conditions (Figures 3G and 3H). Thus, the early flowering in pps-1 might result from derepression of of RAP1B, independently of the Hd3a/RFT1-mediated pathway. It is noted that overexpression of RAP1B caused a drastic early flowering (Jeon et al., 2000).
GA-Related Phenotypes
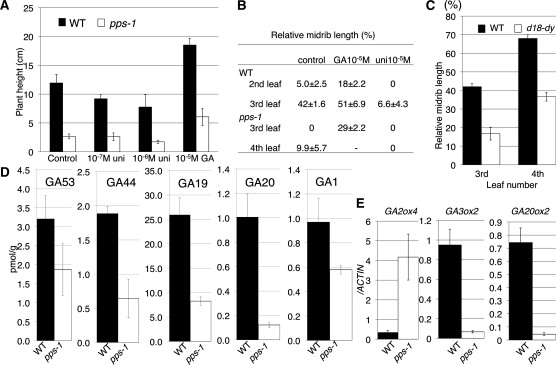
To confirm that GA promotes the juvenile–adult phase change in rice, as it does in Arabidopsis and maize (Evans and Poethig, 1995; Telfer et al., 1997; Telfer and Poethig, 1998), we treated wild-type plants with either GA3 or uniconazole, an inhibitor of GA biosynthesis. In addition to promoting rice plant growth (Figure 4A), as expected, GA3 treatment also induced midrib formation in the second leaf blades (Figure 4B). Uniconazole treatment suppressed both of these processes (Figures 4A and 4B). In addition, examination of the GA-deficient dwarf mutant d18-dy, which has a defect in the gene encoding GA3 OXIDASE2, revealed suppression of midrib formation in the third and fourth leaf blades (Figure 4C), consistent with a prolonged juvenile phase. These findings indicate that GA promotes the juvenile–adult phase change in rice.
Figure 4.
GA-Related Phenotypes in pps-1.
(A) Plant height of 1-week-old wild type (WT) and pps-1 germinated on medium containing GA3 or uniconazole.
(B) Relative length of midrib in the wild type and pps-1 germinated on medium containing GA3 or uniconazole. The values are shown as mean ± sd (n = 5).
(C) Relative length of midrib in third and fourth leaves of the wild type and d18-dy.
(D) Contents of GA53, GA44, GA19, GA20, and GA1 in 3-week-old wild-type and pps-1 seedlings.
(E) Real-time PCR analysis of GA metabolic gene expression in fourth leaves of the wild type and pps-1. Data represent mean ± sd in (A) to (C) (n = 5) and in (D) and (E) (n = 3).
Treatment of pps-1 plants with GA3 or uniconazole had effects that were similar to those seen in wild-type plants (Figures 4A and 4B), establishing that pps-1 plants have a normal sensitivity to GA. However, measurements of the amounts of active GA, GA1, and the intermediates GA53, GA44, GA19, and GA20 revealed a significant reduction in the levels of all of these species in pps-1 plants (Figure 4D). To determine the reason for the decreased GA content in pps-1 plants, we compared the expression of genes encoding GA metabolizing enzymes in pps-1 versus wild-type plants. Relative to the wild-type plants, the mutant plants expressed more GA2 OXIDASE4, which encodes a GA-catabolizing enzyme, with lower expression levels of GA3 OXIDASE2 and GA20 OXIDASE2, which encode enzymes involved in GA biosynthesis (Figure 4E). Accordingly, the low GA content in pps-1 plants appears to be caused by enhanced catabolism and suppressed anabolism of GA, indicating that PPS regulates the vegetative phase change upstream of GA.
Positional Cloning of the PPS Gene
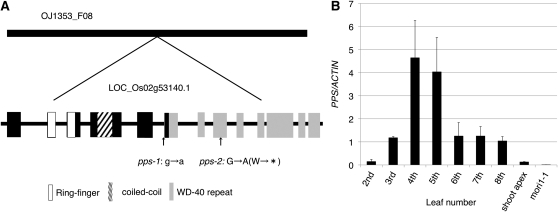
We identified the gene mutated in pps-1 using a map-based approach with the F2 and F3 hybrids of pps-1 heterozygotes and cv Kasalath (ssp indica). The PPS locus was mapped to ~139.5 centimorgans on the long arm of chromosome 2. This region of chromosome 2 harbors a putative Arabidopsis COP1 ortholog, Os02g53140.1 (Tsuge et al., 2001). Sequencing of the ortholog revealed mutations in this gene for both pps-1 and pps-2. The pps-1 mutation consists of a single base change (G→A) at the splicing acceptor site of the sixth exon of Os02g53140.1 on BAC clone OJ1353_F08. The pps-2 mutation consists of a single base change (G→A) in the eighth exon of the gene, generating a premature stop codon in the same gene (Figure 5A). In pps-1 plants, two types of COP1 ortholog transcripts were found; one of these transcripts contained an inserted intron, and the other lacked nine conserved amino acids in the sixth exon, the former being a little more abundant than the latter (see Supplemental Figure 2 online). Consistent with the mutant phenotypes, we presumed that pps-1 is a weaker allele than pps-2. Transformation of pps-1 plants with Os02g53140.1 cDNA under control of the ACTIN promoter restored the normal phenotype (see Supplemental Figure 3 online). We therefore concluded that Os02g53140.1 is the PPS gene.
Figure 5.
Molecular Characterization of the PPS Gene.
(A) PPS gene structure. White boxes, RING-FINGER domain; slanted line box, coiled-coil domain; gray box, WD-40 repeat. Mutation sites of two pps alleles are indicated by vertical arrows.
(B) Expression pattern of PPS (relative to ACTIN-1) in second to eighth wild-type leaf blades, 2-week-old wild-type shoot apex, and mori1-1 seedling. Data represent means ± sd (n = 3).
The PPS gene is composed of 13 exons comprising 2058 nucleotides encoding a 685–amino acid polypeptide (Figure 5A). Os02g53140.1 is known to be an ortholog of Arabidopsis COP1 (Tsuge et al., 2001). The PPS/COP1 protein includes three well-conserved domains (ring-finger, coiled-coil, and WD-40 repeat domains) that play roles in protein–protein interactions (Figure 5A). The amino acid sequences of PPS and COP1 are 73% identical.
Pattern of PPS Expression
Rice PPS was previously reported (as a COP1 ortholog) to be expressed in almost all tissues, including roots, calli, and leaves (Tsuge et al., 2001). In an RT-PCR analysis, we found low-level expression of PPS in shoot apices and young panicles (see Supplemental Figure 4 online). To elucidate the developmental regulation of PPS expression in leaves, we used real-time PCR to measure PPS mRNA levels in the second through eighth leaf blades. Expression of PPS was minimal in second leaves, began to increase in third leaves, reached a peak in fourth and fifth leaves, and declined in higher leaves (Figure 5B). Since the rice juvenile phase is limited to the second leaf, and the juvenile–adult transition occurs in the third to fifth leaves, this expression pattern indicates that PPS is expressed primarily during the transition stage. This expression pattern also provides a straightforward explanation for the delayed adult phase initiation phenotype of pps-1 (i.e., PPS is involved in the initiation of the phase transition). Consistent with the hypothesis that leaves play a major role in the phase change, the amount of PPS mRNA found in leaves of wild-type plants far exceeded that found in the shoot apices, including the SAMs and a few leaf primordia (Figure 5B).
The mori1 mutant is the only heterochronic rice mutant reported to date with a perpetual juvenile phase (Asai et al., 2002), suggesting that MORI1 is a master switch for the juvenile–adult phase change in rice. To examine the functional relationship between MORI1 and PPS, we constructed a mori1 pps-1 double mutant. This mutant exhibited a mori1 phenotype (data not shown), showing that MORI1 is epistatic to PPS. When we examined PPS expression in the mori1 mutant background, we found that PPS expression was completely suppressed (Figure 5B), indicating that MORI1 positively regulates PPS expression.
Comparison of pps-1 and cop1 Phenotypes
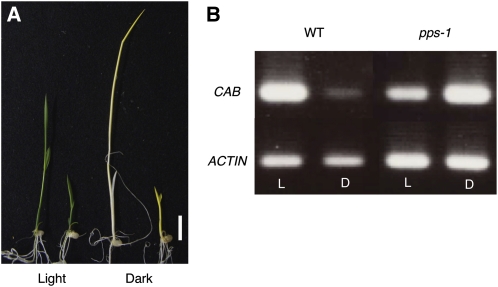
Because the Arabidopsis cop1 mutant exhibits photomorphogenesis in the dark, we expected the pps-1 mutant to also exhibit photomorphogenesis in the dark. Indeed, when we grew wild-type and pps-1 plants in dark conditions, the wild-type plants were etiolated and showed active internode elongation, whereas internode elongation was suppressed in the pps-1 plants, which were yellow (Figure 6A). In addition, the light-inducible CHLOROPHYLL A/B BINDING PROTEIN (CAB) gene was strongly expressed in dark-grown pps-1 plants but almost completely suppressed in the dark-grown wild-type plants (Figure 6B). These results indicate that PPS is functionally equivalent to COP1 in photomorphogenesis/scotomorphogenesis.
Figure 6.
Photomorphogenesis of pps-1 in the Dark.
(A) Seedlings grown in the light or in the dark. From left to right, 7-d-old wild type in the light, 7-d-old pps-1 in the light, 7-d-old wild type in the dark, and 7-d-old pps-1 in the dark. Bar = 1 cm.
(B) Expression of the CAB gene in the light (L) and dark (D). WT, wild type.
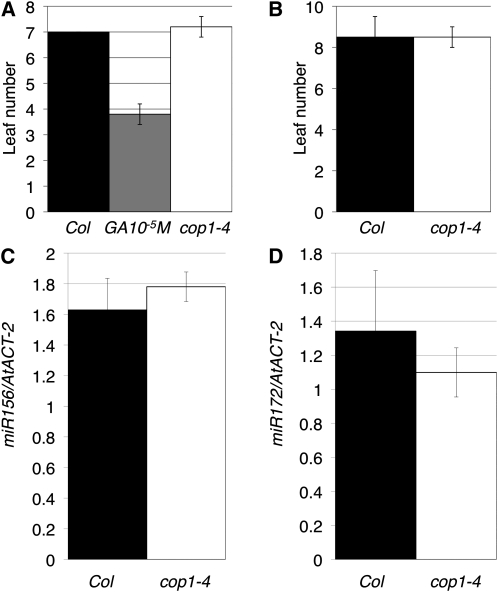
To investigate the effect of the cop1 mutation on the vegetative phase change in Arabidopsis that has not been previously reported, we examined trichome distribution in early leaves of Arabidopsis plants harboring cop1-4, a weak allele that allows development through the reproductive stage. The number of the leaf on which abaxial trichomes first appeared was the same in wild-type and cop1-4 plants (Figures 7A and 7B). In addition, expression levels of miR156 and miR172 genes in 10-d-old cop1-4 seedling were comparable with those of the wild type (Figures 7C and 7D). Thus, COP1, unlike PPS, is not involved in the juvenile–adult phase change.
Figure 7.
Vegetative Phase Change in Arabidopsis cop1-4.
(A) and (B) The leaf number on which abaxial trichomes first appeared in Columbia (Col), Col treated with 10−5 M GA3, and cop1-4 under long-day (A) and short-day (B) conditions.
(C) Real-time PCR analysis of miR156 in 10-d-old Columbia and cop1-4 seedlings.
(D) Real-time PCR analysis of miR172 in 10-d-old Columbia and cop1-4 seedlings.
Data represent mean ± sd (n = 3).
DISCUSSION
Our analyses indicated that the PPS gene temporally regulates the onset of the adult phase, acting downstream of MORI1 and upstream of miRNAs and GA-related genes. In addition, the pps-1 mutant flowered early possibly due to precocious activation of RAP1B, independently of the Hd3a/RFT pathway. Positional cloning revealed that PPS is an ortholog of Arabidopsis COP1. Like the cop1 Arabidopsis mutant, the pps-1 rice mutant exhibited photomorphogenesis in the dark. Thus, PPS appears to have acquired novel functions associated with phase change regulation during the evolution of rice/monocots, as COP1 does not appear to regulate the vegetative phase change or the time of flowering.
Our findings suggest that PPS promotes the juvenile–adult phase change in rice by regulating the expression of miR156, miR172, and GA-related genes. In maize, the dominant Cg1 mutant exhibits a prolonged juvenile phase that results from overexpression of miR156 (Chuck et al., 2007), and the glossy15 mutant exhibits a shortened juvenile phase that results from a deficiency in the AP2 domain–containing gene target of miR172. In Arabidopsis, miR156 and miR172 have been reported to regulate the juvenile–adult phase change (Wu and Poethig, 2006; Schwarz et al., 2008; Wu et al., 2009). We found that the rice pps-1 mutant exhibits a temporal delay in the downregulation of miR156 expression and the upregulation of miR172 expression during development. Thus, miR156 and miR172 might be ubiquitous regulators of the vegetative phase change in plants.
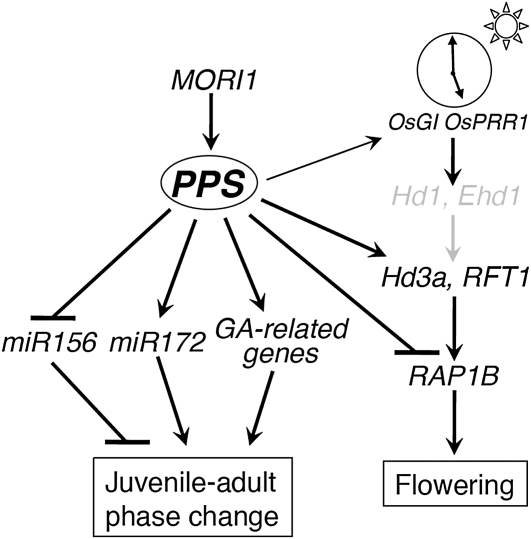
To comprehensively understand the genetic network regulating the juvenile–adult phase change, we must know which genes act upstream or downstream of miR156 and miR172. Maize GL15, which is a target of miR172 as described above, is exclusively associated with leaf epidermal identity (Moose and Sisco, 1994, 1996). On the other hand, there have been no reports of heterochronic gene(s) acting upstream of these miRNAs or of GA, although such genes would be expected to affect more traits than the miRNAs. Since PPS affects the expression of miR156, miR172, and GA metabolic genes and alters the temporal characteristics of a large number of traits, PPS appears to be positioned upstream of miR156 and miR172. mori1 is the first heterochronic rice mutant that remains perpetually in the juvenile phase and never flowers (Asai et al., 2002). On the other hand, pps-1 exhibits adult characters later and flowers. These phenotypic differences imply that PPS is downstream of MORI1. In fact, PPS is not expressed in mori1 mutants, suggesting that MORI1 is a master switch for the juvenile–adult phase change. Since many of the same traits are affected by the mori1 and pps mutations, PPS probably integrates genes acting immediately or almost immediately downstream of MORI1 (Figure 8).
Figure 8.
Schematic Representation of Gene Cascades Involved in Phase Changes in Rice.
Hd1 and EHD1 correspond to CO in Arabidopsis, and Hd3a and RFT to FT in Arabidopsis. Note that Hd1 and Ehd1 are written in gray because Taichung 65, the background cultivar of pps, harbors mutant alleles of Hd1 and Ehd1. Otherwise, they are written in black.
Our results also suggest that PPS regulates the biosynthesis of GA in rice and that GA promotes the vegetative phase change in rice, as it does in Arabidopsis and maize (Evans and Poethig, 1995; Telfer et al., 1997), by promoting the transition to adult phase. GA1 and the other intermediates of the GA biosynthetic pathway were present in lower concentrations in pps-1 plants than in wild-type plants. Additionally, GA catabolic and biosynthetic genes were significantly upregulated and downregulated, respectively, in pps-1 plants. These findings show that PPS is a positive regulator of GA synthesis.
Although close links between GA and photomorphogenesis have been demonstrated in Arabidopsis (Alabadí et al., 2008), whether COP1 directly regulates GA biosynthesis has not been established. LIP1, a pea (Pisum sativum) ortholog of COP1, was recently reported to positively regulate GA biosynthesis through negative regulation of LONG1, an ortholog of the COP1 target HY5, which negatively regulates GA biosynthesis (Weller et al., 2009). This case is comparable with that of PPS. Although the details of the mechanism by which PPS regulates GA biosynthesis remain to be determined, PPS acts upstream of GA metabolic genes, and the pleiotropic phenotypes in pps-1 plants can be at least partially explained by GA deficiency.
The decreased GA content in the pps-1 mutant appears to affect the vegetative phase change. Certainly, GA3 application promotes the vegetative phase change in this mutant, although it does not completely restore the normal phenotype. Accordingly, PPS seems to partially regulate the phase change by affecting the amount of GA, but it also controls phase change–specific pathways, such as those regulating miR156/miR172 expression, independently of GA.
In spite of its prolonged juvenile phase, the pps-1 mutant flowers early, indicating that its adult phase is shortened dramatically. This characteristic contrasts notably with the many heterochronic mutants of maize (Dudley and Poethig, 1993; Moose and Sisco, 1994; Evans and Poethig, 1995; Vega et al., 2002; Chuck et al., 2007), such as Teopod1 (Tp1), Tp2, Cg1, and dwarf1 (d1), that have a prolonged juvenile phase together with late flowering (Dudley and Poethig, 1993; Evans and Poethig, 1995; Chuck et al., 2007). In addition, the gl15 and early phase change maize mutants, which have a shortened juvenile phase, exhibit flowering times comparable to or earlier than that of the wild-type plant (Moose and Sisco, 1994; Vega et al., 2002). Thus, the duration of the juvenile phase does not seem to greatly affect the duration of the adult phase in these maize mutants. In this context, pps-1 is a unique heterochronic mutant.
The mechanism of early flowering in pps-1 is interesting. Although most early flowering rice mutants or cultivars exhibit precocious activation of Hd3a/RFT1 expression (Izawa et al., 2002; Kim et al., 2008; Andrés et al., 2009), Hd3a/RFT1 expression is almost completely suppressed in the pps-1 mutant, regardless of daylength. Instead, the mutant exhibits derepressed expression of RAP1B, which normally acts downstream of Hd3a/RFT1 and may function in floral meristem and determine floral organ identity. It has been reported that ectopic expression of RAP1B resulted in a drastic early flowering (Jeon et al., 2000). Therefore, PPS regulates flowering time by suppressing the expression of RAP1B (and possibly of other floral meristem identity genes) independent of the Hd1-Ehd1-Hd3a/RFT1 pathway (Figure 8; Itoh et al., 2010).
One of the major functions of Arabidopsis COP1 is repressing photomorphogenesis in the dark. In darkness, wild-type plants show elongated hypocotyls and unopened cotyledons, and mRNA levels of light-regulated genes such as CAB are strongly repressed (Deng et al., 1991). Conversely, dark-grown cop1-1 exhibits light-grown characteristics at both morphological and mRNA levels (Deng et al., 1991). Similar phenotypes were observed in pps-1 plants. The introduction of rice COP1/PPS into a cop1 mutant complements the photomorphogenesis phenotype in the dark (Tsuge et al., 2001). Thus, PPS and Arabidopsis COP1 have a conserved function: the repression of photomorphogenesis in the dark. This conclusion is supported by the fact that the pps-1 mutant exhibits photomorphogenesis in dark conditions.
Although PPS is similar to Arabidopsis COP1 in its photomorphogenesis-repressing function, PPS also appears to have functions that have not been reported for COP1. First, PPS promotes the vegetative phase change by regulating GA biosynthesis and miR156/miR172 expression, whereas the effect of COP1 on the vegetative phase change has not been reported. In fact, our analysis suggests that the timing of the juvenile–adult phase change is similar in wild-type and cop1-4 Arabidopsis plants. Second, PPS can control flowering time via the Hd1-independent pathway, since Hd1 was mutated in both Taichung 65 and pps mutants. The cop1 mutant exhibits a flowering time comparable to that of wild-type plants under long-day conditions and earlier than that of wild-type plants under short-day conditions (McNellis et al., 1994), and COP1affects flowering time via the CO-FT pathway, mainly due to the control of CO protein degradation via COP1/SPA1 system (Valverde et al., 2004; Laubinger et al., 2006; Jang et al., 2008; Liu et al., 2008; Chen et al., 2010). Thus, the mechanism by which PPS regulates flowering time is a unique one.
PPS has acquired important and opposing functions in two independent transitions: it promotes the juvenile–adult phase change and represses the vegetative–reproductive stage transition. By regulating several different genetic pathways, PPS performs an important biological function in plant development: it integrates the phase transitions. Although the functions of Arabidopsis COP1 in vegetative development in the light have not yet been intensively analyzed, our results underscore the divergence of the downstream events controlled by PPS/COP1 in different species.
We also characterized the temporal dynamics and sites of PPS expression. PPS expression is suppressed in juvenile leaves (i.e., leaves 1 and 2), begins to increase in third leaves, and reaches a maximum in fourth and fifth leaves, which are intermediate between the juvenile and adult leaves. This pattern indicates that PPS primarily regulates the onset of adult phase. The period of maximum PPS expression (Figure 5) coincides with the period of rapid change in miR156 and miR172 expression (Figure 2). Thus, we assume that PPS plays a role in triggering adult phase development downstream of MORI1 and upstream of GA-related genes and miRNAs.
PPS is expressed at a much lower level in the shoot apex than in leaves, indicating that the juvenile–adult phase change is initiated primarily in the leaves and not in the shoot apex. This aspect of PPS expression implies that certain factors determining adult phase identity may initiate in the leaves. These factors must be translocated to the rest of the plant, as adult phase traits are observed not only in leaves but also in the SAM and stem. Similarly, the results of clonal analysis and apex culture of maize have suggested that the vegetative phase identity of a leaf is determined after leaf initiation and that the factors initiating phase change do not originate in the SAM (Orkwiszewski and Poethig, 2000). A recent study also revealed that leaf-derived signal represses the miR156 transcription to regulate vegetative phase change in Arabidopsis and Nicotiana benthamiana (Yang et al., 2011). The vegetative–reproductive phase change is regulated by the FT/Hd3a protein, known as florigen, which is produced in leaves and translocates to the SAM to transform it into an inflorescence meristem (Tamaki et al., 2007). If the first event of the vegetative phase change takes place in leaves, some signal must be transmitted from the leaves to the SAM and stem, as SAM size, node–internode differentiation, and structural phenotypes are phase specific. Further studies of PPS-dependent phase regulation might result in the identification of a florigen-like factor mediating the vegetative phase change in rice.
This study was conducted to reveal the role of the PPS gene in the cascade associated with the vegetative phase change in rice. We conclude that PPS triggers adult development downstream of MORI1 and regulates the expression of downstream genes. It also regulates flowering time and thus might be a key gene, one that integrates phase changes during rice development (Figure 8).
METHODS
Plant Materials
Two single recessive allelic mutants, pps-1 and pps-2, of rice (Oryza sativa) showing small seedlings with small leaves were identified from an M2 population of cv Taichung 65 mutagenized with N-methyl-N-nitrosourea. Taichung 65 was used as the wild type. We also used the mori1-1 mutant reported previously (Asai et al., 2002), which perpetuates the juvenile phase and fails to become adult and also d18-dy, which is a dwarf mutant defective in the GA biosynthestic gene encoding GA3 OXIDASE2. Mutants and wild-type plants were grown in paddy fields or in pots under natural field conditions.
The Arabidopsis thaliana cop1-4 mutant (McNellis et al.,1994) and its background ecotype Columbia were grown in a growth chamber under daily cycles of long-day (14 h light/10 h dark) conditions at 23°C.
Paraffin Sectioning
Leaves and shoot apices were fixed with FAA (formalin:acetic acid:50% ethanol, 1:1:18) for 24 h at 4°C. They were dehydrated in a graded ethanol series and embedded in Paraplast plus (McCormick Scientific). Microtome sections (8 μm thick) were stained with Delafield’s hematoxylin (Muto Pure Chemicals) and observed with a light microscope.
Measurement of Photosynthetic Rate
For measuring photosynthesis, fully expanded second, fourth, sixth, eighth, and 10th leaves of the wild type and fourth, sixth, and eighth leaves of pps-1 mutant were used. Rates of photosynthetic CO2 assimilation were measured using a portable gas exchange system (LI-6400; Li-Cor). Measurements were made on intact leaf blades between 9 am and noon and replicated for three plants. Light was provided by an LED source (red/blue, 6400-02 LED source; Li-Cor). During the measurement of photosynthetic CO2 assimilation rates, the photon flux density was 1300 μmol photons m−2 s−1, leaf temperature was 25°C, and the reference CO2 concentration was 370 ppm.
Gene Expression Profiling
Total RNA was extracted using TRIzol reagent (Invitrogen) from 10-d-old wild-type and pps-1 seedlings under the light and dark conditions for CAB expression. For quantifying the CAB expression relative to ACT1 expression, RT-PCR was performed using Super Script III (Invitrogen). The primers for CAB and ACT1 are listed in Supplemental Table 1 online. We performed RT-PCR for appropriate cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s.
To monitor PPS expression, total RNA was extracted using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) from wild-type second, third, fourth, fifth, sixth, and seventh leaves. To quantify the PPS expression relative to ACT1 expression, PCR was performed using the TaqMan Fast Universal PCR Master Mix, FAM-labeled TaqMan probes for each gene (Applied Biosystems), and the StepOnePlus real-time PCR system (Applied Biosystem). The expression level of each sample was normalized against that of an internal control, ACT1. The primers and probes for ACT1 and PPS are listed in Supplemental Table 1 online. All TaqMan probes except ACT1 probe include FAM dye at the 5′-end and TAMRA at the 3′-end. ACT1 probe includes FAM dye at the 5′-end and BHQ at the 3′-end.
The real-time PCR for miR156 and miR172 was performed using TaqMan MicroRNA assay (Applied Biosystems). We used the same wild-type leaves as used for PPS expression. In addition, RNA was also isolated from pps-1 second, third, fourth, fifth, sixth, and seventh leaves. For miR156 and miR172 expression assay in Arabidopsis, total RNA was extracted using TRIzol reagent (Invitrogen) from 10-d-old Columbia and cop1-4 whole seedlings. The real-time PCR for miR156 and miR172 was performed using TaqMan MicroRNA assay. We used ACTIN-2 as an internal control, and quantitative PCR was conducted using SYBR green master mix (Applied Biosystems). The primers for ACTIN-2 are listed in Supplemental Table 1 online. For GA metabolic gene expression, total RNA was isolated from wild-type and pps-1 fourth leaves. The primers and probes for GA2ox4, GA3ox2, and GA20ox2 are listed in Supplemental Table 1 online.
For expression analysis of flowering time genes (Hd3a, RFT, and RAP1B/Os MADS14), wild-type and pps-1 seeds were imbibed in darkness (48 h at 30°C) and then sown in soil. Plants were grown in a growth chamber at 70% humidity under daily cycles of short-day (10 h light/14 h dark) or long-day (14.5 h light/9.5 h dark) conditions at 28°C in the light and 24°C in the dark. Light was applied by a metal halide lamp (photosynthetic photon flux density 450 μmol per m2 per s). Flowering time was counted as the time when the first panicle emerged. Total RNA was extracted from leaves of 14-d-old seedlings using an RNeasy Plant Mini Kit (Qiagen) in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized from 5 mg of total RNA using Superscript II reverse transcriptase (Invitrogen). Real-time quantitative RT-PCR was performed using the Taq-Man PCR method on an ABI PRISM 7900 sequence detection system, as described previously (Ogiso et al., 2010). Gene-specific primes and probes for Hd3a, RFT, RAP1B, and UBQ are listed in Supplemental Table 1 online.
Measurement of GA Content and GA3 Application
For measuring GA content, sampling of ~100 mg of 3-week-old seedlings from more than five plants was repeated three times for pps-1 and the wild type, respectively. Extraction and determination of GA for each sample were performed using a liquid chromatography–tandem mass spectrometry system (AQUITY UPLC/Quattro Premier) as described previously (Kojima et al., 2009).
For observing responses to GA, sterilized seeds of pps-1 and the wild type were plated on Murashige and Skoog (1962) medium containing 10−5 M GA3 (Sigma-Aldrich) or 10−5 M uniconazole P (Wako). Plants were grown in a growth chamber under the continuous light at 28°C.
Map-Based Cloning
To map the PPS locus, pps-1 heterozygous plant was crossed with cv Kasalath (ssp indica). Using mutant plants segregated in the F2 population, PPS locus was roughly mapped on the long arm of chromosome 2. Further mapping limited the PPS locus around 139.5 centimorgans between one cleaved-amplified polymorphic sequence marker and one sequence-tagged sites marker. These two closest markers are listed in Supplemental Table 1 online. The genomic sequence of PPS (LOC_Os02g53140 in TIGR) in pps-1 was amplified by PCR using ExTaq DNA polymerase (TaKaRa). The amplified PCR products were directly sequenced using a dye terminator cycle sequencing kit and an ABI PRISM 310 sequencer (Applied Biosystems).
Expression of Circadian Clock–Related Genes, GI, PRR1, and LHY
Wild-type and pps-1 plants were entrained under short-day conditions for 14 d. Then, samples were collected at every 3 h. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). cDNA was synthesized using SuperscriptII RTase (Invitrogen). Real-time quantitative PCR was performed by Taq-Man PCR method on an ABI PRISM 7900 sequence detection system. Primers and probes for rice (Os) GI, PRR1, and LHY are listed in Supplemental Table 1 online.
Detection of PPS Transcripts and Amino Acid Alignment of COP1 Orthologs
Total RNA was extracted using TRIzol reagent (Invitrogen) from wild-type and pps-1 1-week-old seedlings. RT-PCR was performed using Super Script III (Invitrogen). The primers are listed in Supplemental Table 1 online. We performed RT-PCR for appropriate cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s. The amino acid sequences of two genes that have the highest homology with PPS in maize and Arabidopsis were selected by BLAST research (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/).
Complementation Test
PPS cDNA was inserted into a binary vector containing the rice ACTIN promoter and nos terminator (pACT:PPS) and was introduced into the pps-1 homozygous calli using the Agrobacterium tumefaciens–mediated transformation method (Hiei et al., 1994). The primers for full-length PPS cDNA amplification are listed in Supplemental Table 1 online.
Tissue-Specific Expression Pattern of PPS
To examine PPS expression, total RNA was extracted using TRIzol reagent (Invitrogen) from wild-type fourth leaf of 1-week-old shoot apex, 3-week-old shoot apex, 5-week-old shoot apex, young panicle at primary rachis branch differentiation stage, young panicle at secondary rachis branch differentiation stage, and young panicle at floral organ differentiation stage. RT-PCR was performed using Super Script III (Invitrogen). The primers are listed in Supplemental Table 1 online. We performed quantitative RT-PCR for appropriate cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: AB634500 (PPS), AK058305 (CAB), AK058421 (ACT1), AK101713 (GA2ox4) AB056518 (GA3ox2), AB077025 (GA20ox2), AB052943 (Hd3a), AB062676 (RFT1), AK121171 (RAP1B), AK101547 (UBQ), AK072166 (Os GI), AK111828 (Os PRR1), AK111893 (Os LHY1), NP_188508 (ACT2), and NM 128855 (COP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Circadian Clock–Related Gene Expression under Short-Day Conditions.
Supplemental Figure 2. Detection of PPS Transcripts.
Supplemental Figure 3. Complementation Test of pps-1.
Supplemental Figure 4. RT-PCR Analysis of PPS Expression.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank T. Tsuge (Kyoto University) for kind gift of cop1-4 seeds. We also thank R. Soga and K. Ichikawa (University of Tokyo) for their assistance in cultivating rice plants at the Experimental Farm of the University of Tokyo. This work is supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20248001 and 23658005 to Y.N.).
AUTHOR CONTRIBUTIONS
N.T., J.-I.I., and Y.N. designed the experiments. N.T. performed most experiments. M.K. and H.S. measured GA content. N.S. measured photosynthetic rate. H.I. and T.I. performed experiments on flowering time–related gene expression. N.T., J.-I.I., and Y.N. wrote the article.
References
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Andrés F., Galbraith D.W., Talón M., Domingo C. (2009). Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. 151: 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K., Satoh N., Sasaki H., Satoh H., Nagato Y. (2002). A rice heterochronic mutant, mori1, is defective in the juvenile-adult phase change. Development 129: 265–273 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley M., Poethig R.S. (1993). The heterochronic Teopod1 and Teopod2 mutations of maize are expressed non-cell-autonomously. Genetics 133: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.M., Poethig R.S. (1995). Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 108: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Genes Dev. 18: 2368–2379 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaya E., Kubien D.S., Jameson P.E., Clemens J. (2010). Vegetative phase change and photosynthesis in Eucalyptus occidentalis: Architectural simplification prolongs juvenile traits. Tree Physiol. 30: 393–403 [DOI] [PubMed] [Google Scholar]
- Jeon J.-S., Lee S., Jung K.-H., Yang W.-S., Yi G.-H., Oh B.-G., An G. (2000). Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol. Breed. 6: 581–592 [Google Scholar]
- Kim S.K., Yun C.H., Lee J.H., Jang Y.H., Park H.Y., Kim J.K. (2008). OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228: 355–365 [DOI] [PubMed] [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., Ashikari M., Ueguchi-Tanaka M., Matsuoka M., Suzuki K., Sakakibara H. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Marchal V., Le Gourrierec J., Wenkel S., Adrian J., Jang S., Kulajta C., Braun H., Coupland G., Hoecker U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 Erratum. Development 133: 4608 [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102: 9412–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson E.J., Poethig R.S. (1995). Shoot development in plants: Time for a change. Trends Genet. 11: 263–268 [DOI] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.J., Straume M., Chory J., Chua N.H., Kay S.A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Moose S.P., Sisco P.H. (1994). Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell 6: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose S.P., Sisco P.H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10: 3018–3027 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Ogiso E., Takahashi Y., Sasaki T., Yano M., Izawa T. (2010). The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 152: 808–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkwiszewski J.A., Poethig R.S. (2000). Phase identity of the maize leaf is determined after leaf initiation. Proc. Natl. Acad. Sci. USA 97: 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Gendall A.R., Dean C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 15: 519–550 [DOI] [PubMed] [Google Scholar]
- Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Möller B., Weijers D., Poethig R.S. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman K.M., Poethig R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Telfer A., Poethig R.S. (1998). HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125: 1889–1898 [DOI] [PubMed] [Google Scholar]
- Tsuge T., Inagaki N., Yoshizumi T., Shimada H., Kawamoto T., Matsuki R., Yamamoto N., Matsui M. (2001). Phytochrome-mediated control of COP1 gene expression in rice plants. Mol. Genet. Genomics 265: 43–50 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vega S.H., Sauer M., Orkwiszewski J.A., Poethig R.S. (2002). The early phase change gene in maize. Plant Cell 14: 133–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., Hecht V., Vander Schoor J.K., Davidson S.E., Ross J.J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell 21: 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Conway S.R., Poethig R.S. (2011). Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 138: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.