This work examines downstream targets of MYB80 and shows that an aspartic protease regulated by MYB80 is involved in the timing of programmed cell death of the tapetum in the Arabidopsis anther.
Abstract
Arabidopsis thaliana MYB80 (formerly MYB103) is expressed in the tapetum and microspores between anther developmental stages 6 and 10. MYB80 encodes a MYB transcription factor that is essential for tapetal and pollen development. Using microarray analysis of anther mRNA, we identified 404 genes differentially expressed in the myb80 mutant. Employing the glucocorticoid receptor system, the expression of 79 genes was changed when MYB80 function was restored in the myb80 mutant following induction by dexamethasone. Thirty-two genes were analyzed using chromatin immunoprecipitation, and three were identified as direct targets of MYB80. The genes encode a glyoxal oxidase (GLOX1), a pectin methylesterase (VANGUARD1), and an A1 aspartic protease (UNDEAD). All three genes are expressed in the tapetum and microspores. Electrophoretic mobility shift assays confirmed that MYB80 binds to all three target promoters, with the preferential binding site containing the CCAACC motif. TUNEL assays showed that when UNDEAD expression was silenced using small interfering RNA, premature tapetal and pollen programmed cell death occurred, resembling the myb80 mutant phenotype. UNDEAD possesses a mitochondrial targeting signal and may hydrolyze an apoptosis-inducing protein(s) in mitochondria. The timing of tapetal programmed cell death is critical for pollen development, and the MYB80/UNDEAD system may regulate that timing.
INTRODUCTION
The tapetum forms a cell layer surrounding developing microspores within the anther locule. In Arabidopsis thaliana, tapetal cells are derived from primary sporogenous cell lineages during anther developmental stage 4 (Sanders et al., 1999). During microspore development, the tapetum supplies necessary nutrients and structural components. As the pollen matures, the tapetum undergoes programmed cell death (PCD), releasing tapetal remnants, including elaioplasts and tapetosomes, which are incorporated into the coat of mature pollen grains (Wu et al., 1997; Hsieh and Huang, 2007; Parish and Li, 2010). Tapetal PCD is a highly orchestrated event that occurs synchronously with pollen mitotic division and formation of the exine coat (Sanders et al., 1999). Tapetal degeneration begins at stage 10 and is completed by stage 11. Tapetal PCD appears to be apoptosis like, as it is relatively rapid and possesses characteristic features of apoptosis, such as chromatin condensation, DNA fragmentation, and mitochondrial and cytoskeletal disintegration (Papini et al., 1999; Love et al., 2008).
Transcription factors known to be involved in tapetal development include DYSFUNCTIONAL TAPETUM1 (DYT1) (Zhang et al., 2006), MYB33/MYB65 (Millar and Gubler, 2005), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1) (Zhu et al., 2008), ABORTED MICROSPORES (AMS) (Sorensen et al., 2003), MALE STERILITY1 (MS1) (Wilson et al., 2001; Ito and Shinozaki, 2002) and MYB80 (formerly MYB103) (Higginson et al., 2003; Li et al., 2007; Zhang et al., 2007). DYT1 and AMS both encode basic-helix-loop-helix proteins, whereas TDF1 and MS1 encode MYB and PHD transcription factors, respectively. The dyt1, tdf1, and ams mutants exhibit a similar tapetal defect, namely, increased vacuolation and enlargement of the tapetum resulting in arrested microspore development. Transcript levels of AMS, MYB80, and MS1 are significantly reduced in the dyt1 mutant, suggesting that they act downstream of DYT1 (Zhang et al., 2006). Based on analysis of transcript levels within tdf1 and ams mutants, Zhu et al. (2008) suggested that TDF1 functions upstream of AMS and that AMS is upstream of MYB80. Xu et al. (2010) identified 13 genes as direct targets of AMS using chromatin immoprecipitation, but MYB80 was not among them. Transcript levels of MS1, MS2, and A6 are downregulated in the myb80 mutant, suggesting they act downstream of MYB80 (Zhang et al., 2007). It is not known if the three genes are directly or indirectly regulated by MYB80.
MYB80 was isolated from an Arabidopsis genomic library using degenerate primers covering a conserved region within the third repeat of several MYB genes (Li et al., 1999). MYB80 expression is clearly detected in the tapetum and microspores of anthers containing tetrads (stage 7), and expression persists until stage 10 when the tapetum begins to degenerate (Higginson et al., 2003; Li et al., 2007). Functional disruption of MYB80 using antisense (Higginson et al., 2003), T-DNA knockout (Li et al., 2007), or point mutation (Zhang et al., 2007) resulted in a partial (in the case of the antisense lines) or complete male sterility. The myb80 mutant exhibits signs of premature tapetal degeneration in stage 6 anthers, which becomes more pronounced at stage 7, where an increase in tapetal vacuolation occurs. At stage 7, the tapetal cell wall does not degrade in the mutant, suggesting that the transition to a secretory tapetum fails to occur. The release of tetrads, which requires callose dissolution by enzymes secreted from the tapetum, is significantly reduced in the myb80 mutant. Expression of A6, a gene encoding for a putative callase resembling β1,3-glucanases, is reduced in the mutant (Zhang et al., 2007).
Here, microarray analysis identified 404 genes showing differential expression in young anthers of the myb80 mutant compared with wild-type anthers. To narrow down the number of candidate target genes, an inducible system using dexamethasone as the inducer of an AtMYB80:glucocorticoid receptor (GR) construct was used to restore fertility in the myb80 mutant. After 24 h of DEX treatment, the expression levels of 79 genes were significantly changed in anthers. Thirty-two were selected for chromatin immunoprecipitation (ChIP) analysis and three found to be directly regulated by MYB80. The in vitro DNA binding specificity and anther expression patterns were examined. Two of the genes, encoding a pectin methylesterase and a glyoxal oxidase, appeared to be downregulated by MYB80, while the third gene, encoding an A1 aspartic protease, was upregulated. When the latter was silenced using small interfering RNA (siRNA), TUNEL assays showed that premature tapetal and microspore cell death occurred, a phenotype resembling the myb80 mutant. Consequently, we named the aspartic protease gene UNDEAD. UNDEAD has a mitochondrial targeting signal, and the MYB80/UNDEAD system may regulate the timing of tapetal PCD.
RESULTS
Identification of MYB80 Downstream Genes Using Microarray Analysis of myb80 versus Wild-Type Anthers
In order to identify the genes regulated by MYB80, microarray technology was employed to analyze the expression levels of genes that were differentially regulated in the myb80 mutant when compared with wild-type anthers. Approximately 1000 anthers at stages 5 to 8 were dissected from the wild type and the myb80 mutant for each biological replicate for subsequent RNA isolation, labeling, and hybridization to Affymetrix Arabidopsis ATH1 genome arrays. In the microarray analysis, 404 genes were differentially expressed by >2-fold in the myb80 mutant compared with the wild type and possessed a P value of <0.02. The data set included 297 genes that were downregulated in myb80 and 107 genes that were upregulated, with differential gene expression ranging from 2- to 17-fold.
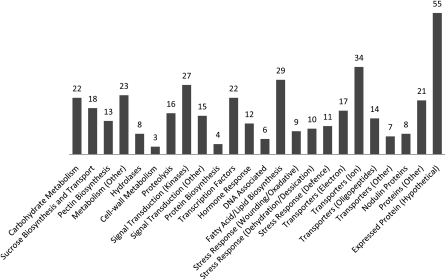
Genes were categorized according to known or putative function based on gene ontology annotations obtained from Affymetrix. Differentially expressed genes included those involved in a variety of biological processes, such as signal transduction, carbohydrate metabolism, transport, lipid and fatty acid metabolism, and Suc metabolism along with several transcription factors and genes involved in ion transport (Figure 1). The complete list is presented in Supplemental Data Set 1 online.
Figure 1.
Genes That Are Differentially Expressed in myb80 Mutant Anthers.
The genes are categorized into groups based on known and predicted functions. The data consist of genes that have a fold change between 2 and 17 and a P value <0.02. The most highly represented biological function groups includes transporters (19%), metabolism (12%), signal transduction components (11%), fatty acid/lipid biosynthesis (7%), and transcription factors (5%). The complete list is presented in Supplemental Data Set 1 online.
Eight genes were selected from the data set for verification of the microarray data. These genes were selected on the basis of high differential expression and low P values. RT-PCR analysis correlated with results obtained in the microarray experiment (see Supplemental Figure 1 online).
Ten genes were selected for further analysis based on substantial changes in gene expression in the myb80 mutant and low P values. T-DNA insertion mutants were obtained for each of the selected genes as summarized in Supplemental Table 1 online. Plants homozygous for the T-DNA were examined for any phenotypic changes in pollen morphology and tested for pollen viability by Alexander’s staining. There were no distinguishable phenotypic changes in the homozygous mutants when compared with the wild type, and male fertility was unaffected. This indicates that the genes examined are redundant or are not direct targets of MYB80.
The myb80 T-DNA mutant is characterized by premature degradation of the tapetum and pollen grains. Hence, it is probable that many of the genes observed in the microarray data set were differentially expressed as a consequence of tapetal breakdown rather than a direct result of eliminating functional MYB80 protein.
Microarray Analysis of AtMYB80:GR-Inducible Lines
To facilitate the identification of MYB80 target genes, an inducible system was employed. The GR domain was fused to the C terminus of MYB80 and the construct driven by the endogenous MYB80 promoter (Figure 2). This construct was transformed into myb80 mutant plants. Following application of dexamethasone (DEX), the AtMYB80:GR fusion protein is able to enter the nucleus, resulting in complete restoration of male fertility (Li et al., 2007). Comparison of gene expression levels just prior to and following application of DEX was expected to enable identification of MYB80 target genes.
Figure 2.
Schematic Diagram of the AtMYB80:GR-Inducible Construct.
The construct consists of 1104 bp of MYB80 promoter sequence and 1471 bp of coding sequence fused to 1.5 kb of the GR gene in the pCambia 1380 vector. Restriction enzymes used in the cloning process are indicated above the construct.
Postfunctional MYB80 protein induction was examined using microarray analysis. Treatments included pre-DEX (noninduced) and 3 to 5 h, 8 to 10 h, and 24 h post-DEX induction. Each time course consisted of four biological replicates. Three biological replicates of myb80 mutants at 8 to 10 h post-DEX (MT) were included as a control. Each replicate contained RNA extracted from ~1000 developmental stage 5 to 8 anthers. Analysis of data obtained from the 3 to 5 h and 8 to 10 h post-DEX samples when compared with pre-DEX and/or the myb80 8 to 10 h control samples revealed only minor differential gene expression changes, with all the genes showing a change of <2-fold. This suggests that 3 to 5 h and 8 to 10 h DEX exposure is insufficient for downstream gene induction to occur.
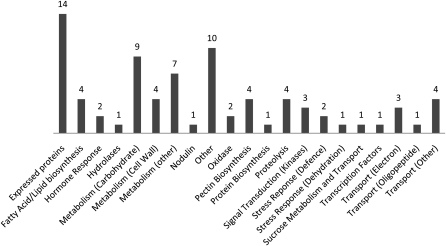
The 24 h post-DEX induction microarray data sets were separately analyzed as two groups to identify overlap. Group A consisted of the pre-DEX and myb80 mutant (MT) data sets versus 24-h post-DEX (Dex24). Group B consisted of wild-type versus pre-DEX and myb80 (MT) mutant data sets. Group A reflects gene expression changes seen in anthers as they transition from male sterility to full fertility after MYB80 induction. Group B represents differences in gene expression between the wild type and the pre-DEX (noninduced) and myb80 mutant. The genes shared by the two comparisons reflect the subset of genes most likely to be directly regulated by MYB80 (Figure 3). The overlapping data set consists of 79 genes that were differentially expressed by 2- to 87-fold with corresponding P values of <0.02 (see Supplemental Table 2 online).
Figure 3.
Genes That Are Differentially Expressed 24 h Postfunctional MYB80 Induction.
The genes were categorized into groups based on known and predicted functions. The data consist of genes that have a fold change between 2 and 87 and a P value <0.02. The highest represented biological function groups include metabolism (25%), transporters (10%), fatty acid/lipid biosynthesis (5%), pectin biosynthesis (5%), and proteolysis (5%). The complete data set is presented in Supplemental Table 2 online.
In order to verify the microarray data and identify genes directly downstream of MYB80, RT-PCR analysis of nine genes selected on the basis of low P values was conducted over a time course including pre-DEX plus 10, 24, 48, and 72 h post-DEX (see Supplemental Figure 2 online). All RNA samples were extracted from young anthers at developmental stages 5 to 8. RT-PCR analysis showed that the transcript levels of four genes, including At3g11980, At1g01280, At4g22080, and At1g66460, were induced in young anthers 24 h after the application of DEX. Expression levels peaked at either 48 or 72 h. Low levels of At1g02050 and At4g12920 transcripts were observed pre-DEX. Expression increased at 24 h post-DEX and peaked at 48 or 72 h post-DEX for At4g12920 and At1g02050, respectively. These results are consistent with the microarray data analysis. Twelve additional genes were analyzed using T-DNA insertion mutants; however, all exhibited wild-type pollen development and seed set (see Supplemental Table 1 online).
ChIP Identifies Three MYB80 Direct Target Genes
To identify directly regulated downstream genes, ChIP was employed. Saleh et al. (2008) successfully achieved enrichment of specific targets using ~0.2 g of plant material. Hence, we performed ChIP using this protocol with some modifications, in particular, the use of young floral buds and dissected anthers rather than whole seedlings. To enrich for the AtMYB80:GR complex, the anti-GR antibody PA1-516 was used. Twenty-eight genes were selected from the 24 h postinduction microarray data set for the ChIP assay (Table 1). The criteria for selection included stamen-specific expression, although gene function was the key determinant. Three of these genes are known to be involved in male fertility, namely, VANGUARD1 (VGD1) (At2g47040), MALE STERILITY2 (At3g11980), and CYTOCHROME P450 (At1g01280) (Aarts et al., 1993; Jiang et al., 2005; Morant et al., 2007). In addition, genes downstream of MYB80, such as MALE STERILITY1 and A6 (Zhang et al., 2007), were selected. Tandem genes, such as At4g30030 (aspartic protease) and At2g47050 (invertase/pectin methylesterase inhibitor), were also examined. The remaining genes on the list are proteases or participate in structural component biosynthesis. Proteases may function in tapetal and pollen development as many proteases were found to be downregulated in previous ms1 and ams gene expression studies (Ito et al., 2007; Yang et al., 2007; Xu et al., 2010).
Table 1.
A Subset of Genes from the Inducible MYB80 Microarray Data Set Analyzed by ChIP
| Gene Locus ID | Name or Annotation | Predicted MYB Binding Motifs | Dex24 | ChIP |
| At4g12920 | Aspartic protease/UNDEAD | MYB1AT x8, MYBST1 x3, MYB1LEPR x2, MYBGAHV | 14.67 | + |
| At1g67290 | Glyoxal oxidase/GLOX1 | MYB1AT, MYBPZM | −2.91 | + |
| At2g47040 | PME/VANGUARD1 | MYB1AT x4, MYBCORE, MYBGAHV, MYBST1 | −29.19 | + |
| At1g01280 | Cytochrome p450 | MYBCORE x2, MYBGAHV x3, MYB1LEPR | 87.68 | − |
| At1g02790 | Polygalacturonase 4 | MYBCORE, MYBGAHV x2 | −21.22 | − |
| At1g02930 | Glutathione S-transferase, putative | MYB1AT x2, MYBGAHV, MYBST1, MYB1LEPR, MYBCOREATCYCB1 | −2.62 | − |
| At1g29270 | Expressed protein | MYB1AT | 13.14 | − |
| At1g54540 | late embryogenesis abundant (LEA) | MYB1AT, MYBCORE | 10.16 | − |
| At1g78140 | Methyltransferase | MYB1AT x3, MYBST1 | 2.08 | − |
| At2g02810 | UDP-galatose transporter 1 | MYB1AT x3, MYBCOREATCYCB1 | 2.53 | − |
| At2g03740 | LEA | MYB1AT, MYBGAHV x2, MYB1LEPR | 2.15 | − |
| At2g42570 | Expressed protein | MYB1AT x3 | 2.90 | − |
| At2g43520 | Trypsin inhibitor, putative | MYB1AT x2, MYBGAHV | 2.16 | − |
| At2g46660 | Cytochrome p450, putative | MYB1AT, MYBST1, MYBCOREATCYCB1 | 2.6 | − |
| At2g47050 | Invertase/PME inhibitor | MYB1AT x2, MYBCORE | −8.85 | − |
| At3g01700 | Arabinogalactan protein 11 | MYBST1 | −3.41 | − |
| At3g01290 | Band 7 family protein | MYB1AT, MYBST1 | −2.13 | − |
| At3g02480 | LEA, abscisic acid–responsive protein | MYB1AT, MYBCORE x2, MYBST1, MYBGAHV | −5.27 | − |
| At3g11980 | Male Sterility2 (MS2) | MYB1AT x3, MYBGAHV x2, MYB1LEPR | 24.85 | − |
| At3g23770 | Glycosyl hydrolase | MYB1AT, MYBCORE, MYB1LEPR, MYBPZM | 37.49 | − |
| At3g62180 | PME | MYBCORE, MYBGAHV, MYBCOREATCYCB1 x2, MYBPZM | −2.25 | − |
| At4g11760 | Pollen coat protein | MYB1AT x2, MYBCORE x2, MYBGAHV | −11.75 | − |
| At4g14080 | A6, putative callase/MEE48 | MYBPZM | <2 | − |
| At4g16160 | Inner membrane translocase | MYBCORE, MYBST1, MYBCOREATCYCB1 x2 | −4.27 | − |
| At4g22080 | Pectate lyase | 14.7 | − | |
| At4g30030 | Aspartic protease | MYB1AT, MYBCORE x2 | N/A | |
| At4g30040 | Aspartic protease | MYB1AT, MYBST1 | 9.85 | − |
| At5g09550 | Rab GDP dissociation inhibitor | MYB1AT, MYBCORE x2, MYBCOREATCYCB1 | −7.65 | − |
| At5g52160 | Protease inhibitor | MYBST1 | 5.97 | − |
| At5g22260 | Male Sterility 1 (MS1) | N/A | − | |
| At5g48880 | Acetyl-CoA C-acyltransferase 1 | MYBCORE, MYBST1 x2 | 3.23 | − |
| At5g56110 | MYB80/MYB103 | MYBST1, MYBGAHV x2, MYBPZM | <2 | − |
Predicted MYB binding motifs present in the promoters (up to −600 bp from the ATG) were obtained using cis-PLACE analysis. ChIP identified UNDEAD, GLOX1, and VGD1 as direct targets of MYB80. The aspartic protease At4g30030 was examined by ChIP as it occurs in tandem with At4g0040. MS1 is absent from the Affymetrix ATH1 array. A6 and MYB80 transcript difference was <2-fold. Dex24, differential gene expression (fold change) 24 h postfunctional MYB80 induction. MYB1AT, A/TAACCA (Abe et al., 2003); MYBCORE, CNGTTA/G (Urao et al., 1993); MYBST1, GGATA (Baranowskij et al., 1994); MYBGAHV, TAACAAA (Gubler et al., 1999); MYB1LEPR, GTTAGTT (Chakravarthy et al., 2003); MYBCOREATCYCB1, AACGG (Planchais et al., 2002); MYBPZM, CCA/TACC (Grotewold et al., 1994). N/A, not present on the ATH1 array. Primers are presented in Supplemental Table 5 online.
The promoters of the selected genes were analyzed for the presence of putative MYB binding elements immediately upstream of the TATA box transcription start site using the cis-PLACE (http://www.dna.affrc.go.jp/PLACE) database. PCR primers were designed to flank MYB binding elements to generate an amplicon of ~300 to 400 bp. All primers were tested by PCR using wild-type genomic DNA and sonicated ChIP samples as template.
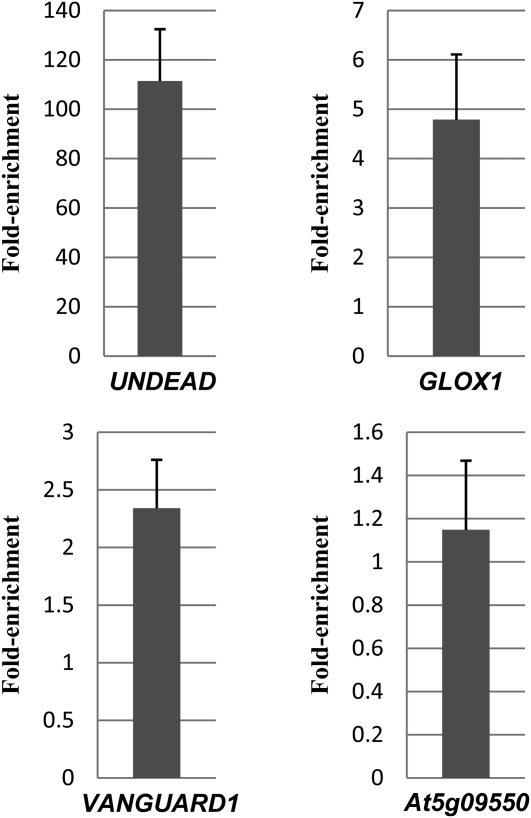
Three out of the 32 genes analyzed were positively enriched in the +AB (antibody) compared with the –AB samples (Figure 4). These genes encode for VGD1 (At2g47040), a glyoxal oxidase (At1g67290, named GLOX1), and an A1 aspartic protease (At4g12920, named UNDEAD). The remaining genes did not exhibit enrichment in the +AB samples and so are unlikely to be direct targets of MYB80 (see Supplemental Figures 3 and 4 online). MS2, A6, and MS1 were not enriched. Hence, their downregulation in the myb80 mutant (Zhang et al., 2007) may be a consequence of tapetal degeneration. Alternatively, the genes may be indirectly activated by MYB80. MYB80 does not appear to bind to its own promoter even though there are several MYB cis-elements present in the region immediately upstream of the TATA box.
Figure 4.
qChIP-PCR Analysis of Enrichment of MYB80 Target Genes in Floral Buds.
Fold enrichment represents the fold change in +AB (antibody) compared with −AB samples. qPCR data were gathered from two biological and three technical replicates. Error bars represent sd. qPCR data correlate with standard ChIP-PCR with UNDEAD strongly enriched, followed by moderate enrichment of GLOX1 and weak enrichment of VGD1 (see Supplemental Figures 3 and 4 online). At5g09550 levels were not affected by the anti-GR PA-516 antibody, suggesting it is not a direct target.
MYB80 Directly Binds to MYB Binding cis-Elements Present in the Promoters of VGD1, GLOX1, and UNDEAD
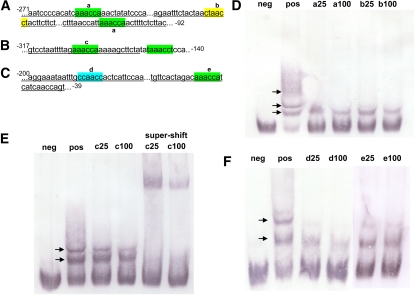
To complement the ChIP results, electrophoretic mobility shift assays (EMSAs) were used to confirm binding to the target promoters by MYB80 and determine if any of the cis-elements were preferentially bound. Recombinant full-length and truncated MYB80 protein was expressed and purified from Escherichia coli (see Supplemental Figure 5 online). The full-length MYB80 protein was expressed at very low levels and so the truncated protein was used. The truncated MYB80 protein consists of the entire MYB domain and sequences up to the amino acids LLTKKRV. Based on our work and other previous work, truncation of MYB proteins beyond the MYB domain does not appear to affect specificity (Ramsay et al., 1992; Li and Parish, 1995). Probes were generated using PCR to incorporate digoxigenin-labeled dUTP into the amplified product. As the consensus MYB80 binding motif was unknown, the probes were designed based on the promoter sequence targeted by the ChIP analysis and were ~170 to 180 bp in length containing one or more potential MYB binding motifs (Figures 5A to 5C). Sequence analysis identified three types of MYB binding motifs present in the probes, namely, MYBPZM (CCA/TACC), MYB1AT (A/TAACCA), and MYB1LEPR (GTTAGTT) (Grotewold et al., 1994; Abe et al., 2003; Chakravarthy et al., 2003). To identify the preferred binding sequence of MYB80, 30-bp unlabeled probes were generated to span the individual cis-elements and used as competitors.
Figure 5.
EMSA Shows MYB80 Is Able to Bind to the Promoter of All Three Target Genes at Several Different MYB Binding cis-Elements.
(A) The UNDEAD probe contains three MYB binding cis-elements. Underlined sequences represent unlabeled competitors “a” and “b.”
(B) The VGD1 probe contains two MYB binding cis-elements with a single unlabeled competitor “c” underlined.
(C) The GLOX1 probe contains two MYB binding cis-elements. Underlined sequences are unlabeled competitors “d” and “e.” Numbers denote location relative to the ATG translational start site. Highlighted colors represent the motifs MYB1AT (green), MYB1LEPR (yellow), and MYBPZM (blue).
(D) MYB80 is able to bind to all three cis-elements present in the UNDEAD promoter. Nonlabeled competitors are able to reduce the visible shift significantly (arrows), resulting in an increase in free probe.
(E) MYB80 is able to bind to both MYB1AT cis-elements present in the VGD1 promoter. The unlabeled “c” competitor even at 100-fold does not significantly reduce the second visible shift, suggesting GAAACCA is not the preferred motif. The reduction is more prominent in the supershift using a T7 antibody against the fusion MYB80 protein.
(F) MYB80 preferentially binds to the MYBPZM cis-element (CCAACC) in the GLOX1 promoter as “d” is the most effective competitor at reducing the visible shifts. neg, free labeled probe (no MYB80 protein); pos, labeled probe and MYB80; a-e/25/100, pos + unlabeled competitors at 25- and 100-fold compared with labeled probe.
MYB80 appears able to bind all three target gene promoters (Figures 5D to 5F); however, there are multiple shifts, which indicates either dimer formation or the binding of more than one protein molecule to the target probe. The latter is the more likely explanation as it has been demonstrated that c-Myb is able to bind multiple times to probes that contain more than one MYB binding site (Ramsay et al., 1992). The addition of competitor probes also reduced or abolished the multiple shifts, supporting multiple protein/DNA interactions rather than dimer formation. The MYB binding cis-elements present in the promoters of UNDEAD, GLOX1, and VGD1 all share the same core sequence of AACC with differences occurring in the adjacent nucleotides. AACC forms the core of the MYB1AT cis-element bound by MYB2 with a consensus sequence of CTAACCA (Abe et al., 1997). The least effective competitor contains the 5′ nucleotide G adjacent to the core motif (Figures 5B and 5E). The most effective competitor probe contains a single MYBPZM cis-element (Figures 5C and 5F). MYB80 appears to have the highest binding affinity for the sequence CCAACCA, which is also the preferential binding site of the maize (Zea mays) P and C1 MYB proteins (Grotewold et al., 1994; Sainz et al., 1997).
Expression Analysis of the Target Genes
A positively regulated target gene should possess a similar expression pattern to MYB80, namely, expression in the tapetum and/or microspores from anther developmental stage 6 until stage 10. MYB80 expression is absent in stage 11 anthers (Higginson et al., 2003; Li et al., 2007). Promoter:GUS (β-glucuronidase) constructs were created, and quantitative RT-PCR (qRT-PCR) analysis was performed to detect transcript levels of the three target genes and MYB80 (see Supplemental Figure 6 online) in young floral buds (corresponding with anther stages 9 and below) and mature floral buds (anther stages 10 to 12).
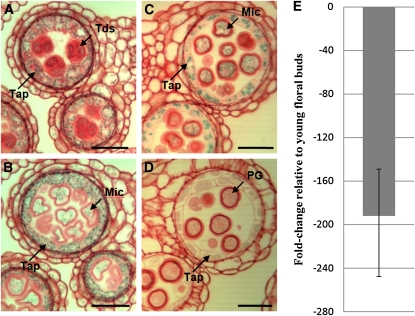
The GUS staining pattern shows that the UNDEAD promoter is active specifically in anthers of young floral buds, with weak expression present in stage 6 increasing at stage 7 to 9 anthers (see Supplemental Figure 7 online). Sections reveal strong GUS staining in the tapetal cell layer and in the developing microspores (Figures 6A to 6D). No GUS staining was observed in stage 11 and beyond, mirroring the expression pattern of MYB80. qRT-PCR detected high levels of UNDEAD transcript in young floral buds, but levels were significantly reduced in mature floral buds (Figure 6E).
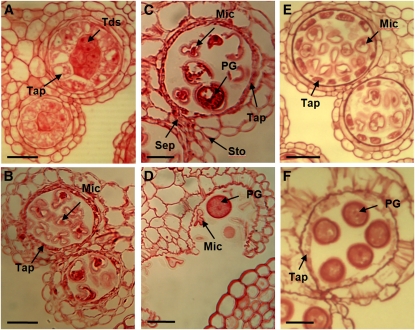
Figure 6.
ASPARTIC PROTEASE/UNDEAD Expression Analysis.
(A) to (D) Sections (3 μm) of UNDEAD promoter:GUS anthers stained with safranin. Light microscopy of anthers at stages 7 (A), 8 (B), 9 (C), and early 10 (D) shows tapetum- and microspore-specific expression. Weak GUS activity is present in stage 10 pollen grains. No GUS activity was detected in stage 11 and beyond.
(E) Comparative qRT-PCR analysis of UNDEAD transcript levels in wild-type mature (anther stages 10 to 12) versus young (anther stages ≤ 9) floral buds. The UNDEAD transcript level is lower in mature floral buds. Error bar represents sd. Tap, tapetum; Tds, tetrads; Mic, microspores; PG, pollen grain. Bars = 25 μm.
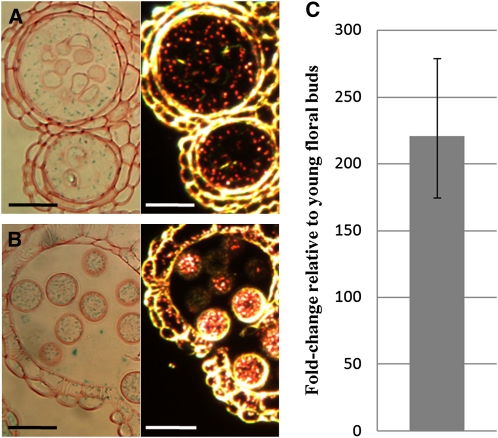
The GLOX1 promoter drives GUS expression within anthers of floral buds from stage 8 onwards with very strong GUS staining present in mature and released pollen grains (see Supplemental Figure 7 online). GUS staining was strongest in stage 10 anthers and persisted at high levels up to stage 14. Sections show strong GUS staining in the tapetum, developing microspores, and mature pollen grains from late stage 8 (Figures 7A and 7B). qRT-PCR detected low levels of GLOX1 transcript in young floral buds but levels were significantly increased in mature floral buds (Figure 7C).
Figure 7.
GLOX1 Expression Analysis.
(A) and (B) Sections (3 μm) of GLOX1 promoter:GUS anthers stained with safranin. Light and dark-field microscopy (GUS activity is visualized as red crystals) of a late stage 8 progressing to 9 (A) and stage 12 (B) anther. GUS crystals are present in the tapetum and developing pollen grains.
(C) Comparative qRT-PCR analysis of GLOX1 transcript levels in wild-type mature (anther stages 10 to 12) versus young (anther stages ≤ 9) floral buds. The GLOX1 transcript level is higher in mature floral buds. Error bar represents sd. Bars = 25 μm.
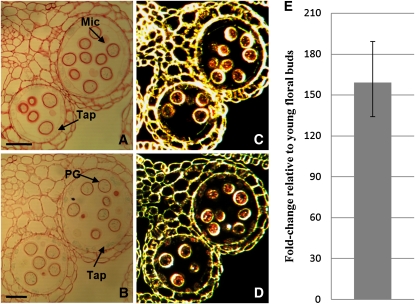
The microarray analysis suggests VGD1 expression is strongly repressed by MYB80. Therefore, VGD1 should not be expressed at high levels during early anther developmental stages where MYB80 is most active. Previously, Jiang et al. (2005) showed VGD1 expression is specific to mature pollen grains and pollen tubes; however, there was no examination or mention of the tapetum. GUS staining shows the promoter of VGD1 is active within the late tapetum and mature pollen grains. GUS staining was faint but present in stage 9 anthers and became progressively stronger in stage 10 anthers and beyond. Sections identified tapetum-specific expression occurring in stage 10 to 11 anthers where the tapetum is undergoing PCD but activity was predominantly in mature pollen grains (Figures 8A to 8D). qRT-PCR detected low levels of VGD1 transcript in young floral buds but levels were significantly increased in mature floral buds (Figure 8E).
Figure 8.
VGD1 Expression Analysis.
(A) to (D) Sections (3 μm) of VGD1 promoter:GUS anthers stained with safranin. Light microscopy of anthers at stages 10 (A) and 11 (B). Blue crystals are weakly present within the tapetum and developing pollen grains. Dark-field microscopy of anthers at stages 10 (C) and 11 (D). GUS activity is visualized as red crystals. GUS activity was not detected in stage 8 or earlier stages.
(E) Comparative qRT-PCR analysis of VGD1 transcript levels in wild-type mature (anther stages 10 to 12) versus young (anther stages ≤ 9) floral buds. The VGD1 transcript level is higher in mature floral buds. Error bar represents sd. Mic, microspores; Tap, tapetum; PG, pollen grain. Bars = 25 μm.
Functional Analysis of the Target Genes
The function of VGD1, encoding a pectin methylesterase, has been studied by Jiang et al. (2005). The vgd1 mutant was isolated from enhancer-trap dissociation (Ds) insertion lines in Arabidopsis ecotype Landsberg erecta (Sundaresan et al., 1995). The homozygous mutant has smaller and shorter siliques with fewer seeds, and pollen tube growth in the style and transmitting tract is severely retarded. Tapetal cell development in the mutant was not examined, and vgd1 pollen appeared morphologically normal (Jiang et al., 2005).
SALK_000947 seeds containing a T-DNA insertion in the first exon of GLOX1 (Scholl et al., 2000; European Arabidopsis Stock Centre) were obtained and germinated on kanamycin germination medium. PCR confirmed the presence of the T-DNA in the 23 lines that survived selection. However, no homozygous glox1 lines could be identified.
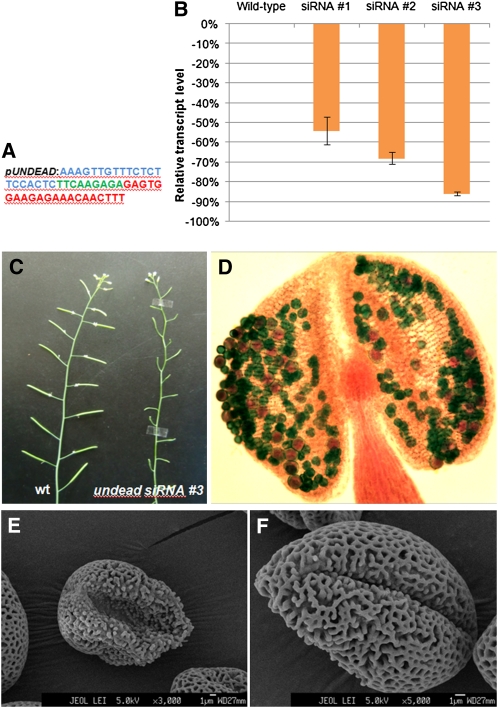
Gene silencing was used to downregulate UNDEAD transcript levels as no suitable insertion mutants are available. Three constructs were created using the endogenous UNDEAD promoter to drive two siRNAs and an artificial microRNA (amiRNA). The two siRNAs consisted of a short 21-nucleotide hairpin targeting the UNDEAD transcript at the nucleotide positions 121 and 297 (Figure 9A). The amiRNA (MIR319a backbone) also targeted the UNDEAD transcript at nucleotide position 121. The silencing vectors were transformed into wild-type Arabidopsis (Columbia-0), and over 30 transgenic lines were obtained for each construct. The majority (>70%) of transgenic siRNA-121 lines exhibited partial male sterility with reduced silique elongation and seed set. Both the siRNA-297 and amiRNA transgenic lines also displayed partial male sterility, but the majority (~75%) of lines were identical to the wild type.
Figure 9.
Functional Characterization of UNDEAD via siRNA-Mediated Gene Silencing.
(A) The siRNA construct consists of 21 nucleotides targeting the UNDEAD transcript at nucleotide position 121 (blue), a short 9-nucleotide loop (green), and the reverse compliment 21 nucleotides (red). The siRNA was driven by the endogenous UNDEAD promoter.
(B) Real-time qPCR analysis of three different undead siRNA lines identified varying levels of transcript silencing. Error bars represent sd. Line #3 exhibited the greatest reduction of UNDEAD transcript.
(C) Silencing of UNDEAD transcript resulted in severe male sterility with line #3 displaying a near complete lack of silique elongation.
(D) Alexander’s stain of an undead stage 12 anther shows a reduction in the number of pollen grains, the majority of which are collapsed and lacking viable cytoplasm (green staining).
(E) and (F) Scanning electron micrographs of undead pollen grains reveal irregularly shaped and collapsed grains with abnormal exine pattern formation. Wild-type pollen grain scanning electron micrograms are presented in Supplemental Figure 8 online.
Real-time qPCR was performed on young floral buds at stages 6 to 10 from three siRNA-121 lines selected for their varying degrees of partial male sterility (Figure 9B). Line #1 had less than half of the siliques elongating and had a full complement of seeds, whereas line #3 was severely male sterile with almost no seed set (Figure 9C). The endogenous UNDEAD promoter was effective in driving the siRNA as a reduction in UNDEAD transcript occurred in all three lines tested. Line #3 had the highest level of UNDEAD transcript silencing, with the mRNA level reduced by ~85% compared with the wild type. The qRT-PCR data indicated a correlation between the level of UNDEAD transcript and the severity of the male sterility phenotype.
Alexander’s stain was used to determine pollen cytoplasmic viability (Figure 9D), with viable pollen staining red and pollen grains lacking cytoplasm staining green. Most of the siRNA-121 undead stage 10 and beyond anthers examined possessed reduced numbers of pollen grains when compared with the wild type, and the majority of pollen grains stained green. Mature siRNA-121 undead stage 13 anthers released only a few pollen grains. Scanning electron microscopy showed that of the few pollen grains released, the majority were morphologically abnormal and collapsed, the exine coat appearing deformed and irregular (Figures 9E and 9F) compared with the wild type (see Supplemental Figure 8 online).
Sections of undead floral buds were examined to identify changes in tapetal and microspore development (Figure 10). At stage 7, the tapetal cell layer in undead exhibited increased vacuolation, similar to changes seen in the myb80 mutant (Li et al., 2007; Zhang et al., 2007). Stage 10 undead anthers had high levels of aborted microspores (Figure 10C), again resembling the myb80 mutant. At anther stages 12 and 13, the septum and stomium cell layers break down as normal. However, the majority of pollen grains are collapsed, clump together, and are not released (Figure 10D).
Figure 10.
The undead Mutant Exhibits Premature Tapetal Degeneration.
Sections (3 μm) of undead and wild-type anthers stained with safranin. MC, meiotic cell; Tap, tapetum; Tds, tetrads; Mic, microspores; PG, pollen grain; Sep, septum; Sto, stomium. Bars = 20 μm.
(A) Stage 7, tetrads are formed. The tapetum exhibits increased vacuolation similar to the myb80 T-DNA mutant (Li et al., 2007).
(B) Stage 8, the tapetum appears sparse, exhibiting advanced degeneration commonly seen in wild-type stage 10 anthers.
(C) Stage 10, most of the microspores are aborted. A thin tapetal layer still remains.
(D) Stage 13, anther dehiscence and the breakage of both septum and stomium layers occur as normal. The collapsed pollen grains clump together and are not able to be released.
(E) Wild-type stage 8.
(F) Wild-type stage 11.
To determine the possible localization of UNDEAD, the peptide sequence was analyzed using TargetP (http://www.cbs.dtu.dk/services/TargetP/) and iPSORT (http://ipsort.hgc.jp/), which are known to be ~85% accurate in signal peptide prediction (Emanuelsson and von Heijne, 2001). Both analyses identified the presence of a signal peptide (KTTMNFVFLFF) targeting UNDEAD to the mitochondria, in agreement with a previous study by Beers et al. (2004).
Apoptosis-Like PCD Occurs Prematurely in myb80 and undead Tapetum and Pollen
PCD is believed to be responsible for tapetal degeneration (Parish and Li, 2010). This process is characterized by cellular condensation, mitochondria and cytoskeleton degeneration, nuclear condensation, and internucleosomal cleavage of chromosomal DNA. To investigate the nature of the tapetal breakdown in the myb80 and undead mutants, TUNEL assays were performed on 6-μm transverse sections of paraffin-embedded wild-type, myb80, and siRNA #3 undead anthers. The TUNEL assay detects in situ DNA cleavage, a hallmark feature of apoptosis-like PCD, by enzymatically incorporating fluorescein-12-dUTP into the 3′-OH ends of fragmented DNA. Anther staging was based primarily on floral bud size and stages; however, key anther morphology traits that are unaffected in the myb80 mutant, such as tetrad formation corresponding to anther stage 7 and tetrad release at stage 8, were also used as an indicator of anther developmental stages (Smyth et al., 1990; Bowman et al., 1991; Peirson et al., 1996; Sanders et al., 1999). In the myb80 mutant, tapetal and microspore degeneration commences at stage 9, resulting in distorted morphology. In the determination of anther stages 9 to 11, floral bud size was a key indicator.
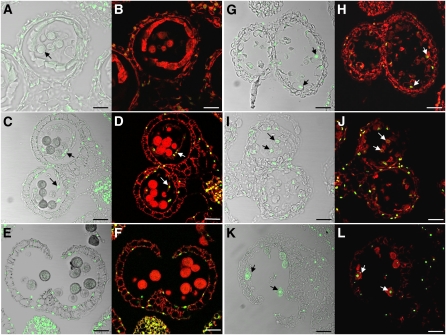
Analysis of the wild type at anther stage 9 where microspores first develop the pollen wall (Figure 11A, arrow) failed to detect TUNEL signals in tapetal cells and microspores. However, TUNEL-positive signals were detected in the myb80 tapetum at this stage (Figures 11G and 11H). Wild-type tapetal cells start to degenerate rapidly at anther stage 10 and are no longer detected at anther stage 12 (Sanders et al., 1999). During wild-type anther stage 10, the first observations of TUNEL signal in the tapetum were made, consistent with the findings of Vizcay-Barrena and Wilson (2006) (Figures 11C and 11D). TUNEL-positive signals in the myb80 stage 10 anther became more prominent in the tapetal layer and also appeared in collapsing microspores (Figures 11I and 11J).
Figure 11.
Tapetal PCD Occurs Prematurely in the myb80 Mutant.
Confocal microscopy of DNA fragmentation detected using the TUNEL assay in 6-μm sections of wild-type ([A] to [F]) and myb80 ([G] to [L]) anthers. TUNEL-positive signal is indicated by the green fluorescence of fluorescein, and nuclei fluoresce deep red due to the counterstain propidium iodide. Bars = 25 μm.
(A) and (B) Wild-type anther stage 9 with the first appearance of the pollen coat (arrow) was TUNEL-negative.
(C) and (D) TUNEL signal is first observed in the tapetum of wild-type anthers at stage 10 (arrows), which correlates with the onset of tapetal cell breakdown.
(E) and (F) At stage 13, the tapetum has degenerated completely, and TUNEL signal is visible only in the anther epidermis and endothecium.
(G) and (H) myb80 anthers corresponding to stage 9 exhibit TUNEL signal in the tapetal layer but not in microspores (arrows).
(I) and (J) At stage 10 in myb80, TUNEL-positive signals appeared in the collapsing microspores (arrows).
(K) and (L) At stage 13 in myb80, TUNEL-positive nuclei are observed in the collapsed pollen (arrows).
Anther dehiscence, characterized by breakage of the stomium, occurs during stage 13 of anther development. Pollen grains have reached maturity with the completion of pollen sexine development and mitotic divisions to generate the tricellular pollen. In the myb80 mutant (Figures 11K and 11L), TUNEL signal is clearly evident in the collapsed pollen and possibly in remnants of the tapetum among the cellular debris, whereas the wild-type pollen remains TUNEL negative (Figures 11E and 11F).
Premature PCD also occurs in the undead mutant anthers. Positive TUNEL signals first appear in stage 9 tapetum (Figures 12A and 12D). However, no TUNEL signal was detected in collapsed or the few morphologically normal microspores, consistent with the myb80 TUNEL data for this stage. At stage 11, the remaining pollen grains appeared collapsed, and intense TUNEL signals were present in both the remnant tapetal cells and pollen grains (Figures 12B and 12E). At stage 12, the undead anther locule contains collapsed pollen grains also undergoing PCD (Figures 12C and 12F).
Figure 12.
Tapetal PCD Occurs Prematurely in the undead Mutant.
Confocal microscopy of DNA fragmentation detected using the TUNEL assay in 8-μm sections of siRNA undead anthers. TUNEL-positive signal is indicated by the green fluorescence of fluorescein. Grayfield ([A], [C], and [E]) and propidium iodide counterstain ([B], [D], and [F]) of stage 9 ([A] and [B]), 11 ([C] and [D]), and 12 anthers ([E] and [F]). TUNEL-positive signals are strongly present in stage 9 undead tapetum, suggesting that advanced PCD degeneration has occurred (arrows). TUNEL signal is present in collapsing microspores at stages 11 and 12. Bars = 25 μm.
DISCUSSION
MYB80 is essential for pollen development. In the myb80 mutant, tapetal cell vacuolation and breakdown are premature and the small oil bodies, plastids, and vesicles are absent (Higginson et al., 2003; Li et al., 2007; Zhang et al., 2007). Tetrad callose dissolution is greatly reduced and the few microspores produced lack exine (Zhang et al., 2007). The AMS gene, which is expressed in the tapetum post microspore meiosis, does not appear to be part of the MYB80 pathway (Xu et al., 2010). Microarray analysis of the ms1 and ams buds in comparison to the wild type identified 260 and 549 differential expressed genes, respectively (Yang et al., 2007; Xu et al., 2010). Our transcriptome analysis identified 404 differentially expressed genes in young anthers of the myb80 mutant compared with the wild type. The myb80 mutant microarray shares 30 and 66 genes with the ms1 and ams microarrays, respectively (see Supplemental Tables 3 and 4 online). Based on transcript analysis, Zhang et al. (2007) suggested that MS1, MS2, and the putative callase A6 act downstream of MYB80. However, our ChIP analysis does not support direct regulation. In the ams mutant, vacuolation increases in the tapetal cells, which enlarge (hypertrophy) until they fill the vacuole (Sorensen et al., 2003). In the ms1 mutant, abnormal vacuolation of tapetal cells also occurs. However, no DNA fragmentation is detected in the tapetum (Vizcay-Barrena and Wilson, 2006). The A6 gene plays a role in dissolution of the tetrad callose wall (Hird et al., 1993). Hence, blocking expression of the three genes previously shown to be downstream of MYB80 does not result in phenotypes that fully resemble the myb80 mutant. In particular, tapetal PCD is not premature.
The differential expression of genes in the myb80 mutant might simply be a consequence of the premature tapetal degeneration. Consequently, we used the GR system to restore MYB80 function in the myb80 mutant. Ito et al. (2007) used the same approach to study genes regulated by MS1. We selected 12 of the resulting differentially expressed genes based on their reported anther expression and obtained insertion mutants for them, but all anther phenotypes were identical to the wild type.
An additional 32 genes were then selected for ChIP analysis, again based on stamen-specific expression. The genes encoding the pectin methylesterase VGD1, GLOX1, and an A1 aspartic protease (UNDEAD) were positively enriched. EMSA confirmed that MYB80 binds to the promoters of all three genes with the motif CCAACC as the preferred binding site. MS2, CYTOCHROME P450 (At1g01280), A6, and MS1, all genes involved in pollen development and expressed in the tapetum, were not enriched. Hence, they are not direct targets of MYB80.
VGD1 expression is absent or very weak in the tapetum when MYB80 is most active in anther stages 6 to 9, and the increase in VGD1 expression correlates with the downregulation of MYB80 expression at stage 10. VGD1 encodes a pectin methylesterase homologous (PME) protein consisting of a PME inhibitor homologous domain, a pectinesterase homologous domain, and a secretion-related transmembrane domain (Jiang et al., 2005). PMEs are a group of cell wall–modifying enzymes that catalyze the demethylesterification of cell wall pectin (Jiang et al., 2005). The vgd1 mutant exhibits partial male sterility, seed production being restricted to the top section of the mature siliques. Due to the specific expression in pollen and growing pollen tubes, VGD1 may play a key role in strengthening the pollen tube cell walls, thereby increasing the stability of the structure during growth (Jiang et al., 2005). Alternately, VGD1 could be involved in modifying material released from the degenerating tapetum to form part of the pollen coat.
MYB80 may partially repress GLOX1 transcription up to anther stages 8 to 9, and other transcription factors activate expression during the latter stages of pollen development. Overexpression of a Vitus pseudoreticulata GLOX can suppress powdery mildew hyphal development, possibly due to the generation of reactive oxygen species (ROS) (Guan et al., 2011). The authors suggest Vp-GLOX regulates cellular H2O2 levels since H2O2 can severely inhibit the enzyme. Vp-GLOX is a homolog of an Arabidopsis GLOX (At3g53950) whose function is unknown. The changes in and role of ROS during tapetum and pollen development are unclear. In plants, ROS may be a signaling molecule responsible for opening the permeability transition pore in mitochondria leading to PCD (Reape and McCabe, 2010). A homozygous Arabidopsis glox1 insertion mutant could not be obtained for further study.
It appears that At-MYB80 acts as both repressor and activator in the same tissue. Bifunctional transcription factors have been described previously in plants (Mena et al., 2002; Bossi et al., 2009; Ikeda et al., 2009). The nature of the promoter region of a gene and the binding of other factors may be involved in the conversion from repressor to activator.
The expression pattern of UNDEAD mirrors that of MYB80 (Higginson et al., 2003; Li et al., 2007). A suitable insertion mutant was unavailable, so siRNA and amiRNA were used to downregulate UNDEAD expression, and the degree of male sterility observed correlated with the reduction in UNDEAD transcript levels. Changes in the tapetal cells in anthers when the UNDEAD transcript levels were low resembled the myb80 mutant, including increased tapetal vacuolation and premature cell death. myb80 pollen grains appear to lack the exine (outer) layer (Zhang et al., 2007). Exine formation was also abnormal in the undead mutant but not to the extent of the myb80 pollen. This is presumably because many of the genes downstream of MYB80 are involved in lipid and sporopollenin synthesis (Zhang et al., 2007), and this is reflected by our myb80 microarray analysis, as a large cluster of lipid synthesis and transport genes are affected.
DNA fragmentation is indicative of apoptosis-like PCD (Papini et al., 1999; Balk and Leaver, 2001; Love et al., 2008; Parish and Li, 2010). Using the TUNEL assay, we found that DNA fragmentation commences at stage 10 of wild-type tapetal cells. However, in the myb80 mutant, TUNEL signal was first detected at anther stage 9 in tapetal cells. In the siRNA undead mutants, TUNEL signal also appeared in tapetal cells of anther stage 9. At stage 11, the signal was detected in remnant tapetal cells and collapsed pollen grains. The results are consistent with MYB80 upregulation of UNDEAD, thereby delaying tapetal PCD, the program being initiated once MYB80 and hence UNDEAD expression ceases. This differs from the ms1 mutant where microspore and tapetal cells also collapse and degenerate; however, no DNA fragmentation is detected in the tapetum, suggesting necrotic breakdown rather than PCD is occurring (Vizcay-Barrena and Wilson, 2006).
Although UNDEAD gene expression ceases in the microspores of wild-type anthers at stage 11 as a consequence of a decrease in MYB80 expression, unlike tapetal cells, the microspores do not undergo PCD. The PCD-inducing protein(s) hydrolyzed by UNDEAD may no longer be expressed in microspores at this stage or perhaps UNDEAD is replaced by another protease. Alternately, the UNDEAD protein may persist in mature pollen grains while transcript has dissipated.
We conclude that MYB80 is delaying PCD by activating transcription of the UNDEAD gene. Proteases might be expected to activate rather than suppress PCD (Parish and Li, 2010); however, the Arabidopsis PROMOTION OF CELL SURVIVAL1 (PCS1) gene also encodes an aspartic protease (Ge et al., 2005). PCS1 is expressed in developing flowers and young siliques. Promoter:GUS analysis identified activity in anthers and pollen, but no information about tapetal expression was provided. Male and female gametophyte degeneration and excessive embryo apoptotic cell death prior to the torpedo stage occurred with a loss-of-function mutation. Anther dehiscence was blocked when PCS1 was ectopically expressed using the 35S cauliflower mosaic virus promoter. The death of stomium and septum cells was prevented and leaf senescence was delayed.
Both UNDEAD and PCS1 belong to the A1 family of Arabidopsis pepsin-like aspartic proteases (Beers et al., 2004). At the amino acid level, PCS1 is only 27% similar to UNDEAD. Despite the differences, both PCS1 and UNDEAD appear to regulate the timing of PCD. PCS1 and UNDEAD both possess signal sequences; however, PCS1 localization remains unclear, whereas the UNDEAD signal sequence indicates the protein is directed to the mitochondria. Changes in mitochondrial outer membrane permeabilization and subsequent activation of cytoplasmic caspase proteases are responsible for apoptotic cell death in mammals (Tait and Green, 2010), but no caspase homologs have been found in plants (Sanmartín et al., 2005; Parish and Li, 2010). Release of apoptosis-inducing factor as a result of mitochondrial outer membrane permeabilization may contribute to caspase-independent cell death in mammals (Tait and Green, 2010). Vaux (2011) argues that cytochrome c is the only protein released from mitochondria for which there is strong evidence it is involved in mammalian cell death. Other proteins are released, some secondarily from the outer membrane by caspase activity, but evidence for a major role in cell death is lacking. A role for plant mitochondria has not yet been established. Cytochrome c is released from plant mitochondria by a variety of PCD-inducing stimuli, such as heat shock, d-mannose, menadione, and ceramide (Balk et al., 1999; Stein and Hansen, 1999; Sun et al., 1999; Reape and McCabe, 2008). The partial release of cytochrome c through transient pores in the mitochondrial membrane correlates with cytological abnormalities in tapetal cells of the Petiolaris-cytoplasmic male sterile (PET1-CMS) sunflower (Helianthus annuus; Balk and Leaver, 2001). However, purified cytochrome c added to plant mitochondria did not induce DNA fragmentation (Balk et al., 2003). Nonetheless, the possible mitochondrial location of UNDEAD indicates that the organelle may be involved in plant PCD.
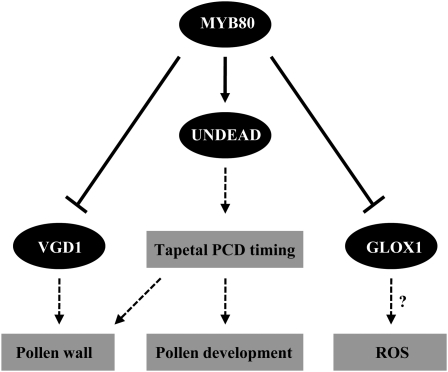
UNDEAD may hydrolyze an apoptosis-inducing protein in the mitochondria that participates in PCD, either by being released into the cytoplasm or modifying mitochondrial outer membrane permeability. One function of MYB80 could be to induce transcription of the nuclear UNDEAD gene to ensure mitochondrial damage was prevented in tapetal cells. The downregulation of MYB80 expression at stage 10 would shut down UNDEAD transcription and allow PCD to proceed. Such a model assumes a short half-life for the UNDEAD protein in mitochondria. A proposed model of MYB80 function is presented in Figure 13. The timing of tapetal PCD is critical for pollen development, and any delay or inhibition results in male sterility (Kawanabe et al., 2006). The MYB80/UNDEAD system may play a central role in this timing.
Figure 13.
A Proposed Model of MYB80 Function in Tapetal and Pollen Development.
Solid lines represent direct gene regulation (arrows represent positive regulation and closed arrows represent negative regulation, respectively). Dashed lines represent function. ?, Hypothesized function. For a comprehensive model, see Wilson and Zhang (2009) and Parish and Li (2010).
METHODS
Plant Materials and Growth
Arabidopsis thaliana accession Columbia (Col-0) was used for all gene transfer experiments and wild-type controls. Plants were grown on soil at 22°C under constant illumination or on germination medium containing the appropriate selective antibiotic. T-DNA insertion mutant lines were obtained from GABI-Kat (Max Planck Institute for Plant Breeding Research), the European Arabidopsis Stock Centre, and the ABRC. The insertion mutant lines used in this research are presented in Supplemental Table 1 online. Transgenic plants were transformed with Agrobacterium tumefaciens strain GV3101 by dripping of the Agrobacterium solution (40 mL of a 2-d culture resuspended in 20 mL of infiltration medium: 1 g sucrose, 6 μL Silwet per 20 mL) onto each floret. One week later, the dripping procedure was repeated.
Plasmid Construction
Promoter:GUS constructs were generated by PCR amplification from genomic DNA. Fragments were cloned into the pENTR/D-TOPO Gateway vector (Invitrogen) and then cloned into pkGWFS7 using the LR clonase reaction. The AtMYB80:GR construct previously described by Li et al. (2007) was generated by PCR amplification of 1104 bp of the At-MYB80 promoter sequence and 1471 bp of the At-MYB80 coding sequence. This fragment was transcriptionally fused to 1.5 kb of the rat GR ligand gene using the restriction sites PstI and SpeI then cloned into the pCambia 1380 vector (CAMBIA) using SalI and SpeI restriction enzyme sites. The siRNA constructs were created by cloning the UNDEAD promoter into pCambia 1380 using BamHI and HindIII restriction enzyme sites. Primer pairs (forward and reverse complement primers) were designed for each construct to incorporate 21 nucleotides targeting the UNDEAD transcript, a short 9-nucleotide loop, and the reverse complementary sequence of the 21-nucleotide target region. The primers also incorporated HindIII and SpeI restriction enzyme sites for cloning into pCambia 1380 containing the UNDEAD promoter. The At-MYB80 protein was expressed using MYB80 cDNA cloned into the pRSETB (Invitrogen) vector at the KpnI and HindIII sites. Primers are listed in Supplemental Table 5 online.
RT-PCR and qRT-PCR Analysis
Flower bud length was measured and staged according to Peirson et al. (1996). The anthers at stages 5 to 8 (0.4 to 0.9 mm) were dissected in RNAlater solution (Ambion). Total RNA was extracted from the isolated anthers using an RNeasy plant kit (Qiagen). First-strand cDNA synthesis was performed according to the manufacturer’s instructions (Invitrogen Superscript III reverse transcriptase and reagents). The conditions for PCR amplification of cDNA were as follows: first cycle, 94°C for 3 min; second cycle, 94°C for 30 s; 55°C for 30 s and 72°C for 50 s; third cycle, 72°C for 10 min. PCR products were visualized by running on a 1% agarose gel stained with ethidium bromide and captured digitally using a UV video capture system. qRT-PCR was performed using the iQ SYBR Green Supermix (Bio-Rad) on the MyiQ iCycler (Bio-Rad). The PCR conditions were as follows: 94°C for 3 min; 36 cycles of 94°C for 30 s; 51 to 56°C for 30 s; 72°C for 20 s; one cycle at 72°C for 5 min. Data were analyzed using the iQ5 (Bio-Rad) software, and differences in gene expression were calculated using the 2^(−deltaCT) analysis method. Gene-specific primers are listed in Supplemental Table 5 online.
Sectioning of Resin-Embedded Floral Buds
Florets were fixed, embedded, and sectioned as described by Li et al. (2007).
Histochemical Assay of Transformed Arabidopsis Plants
Plant tissue was incubated in X-gluc solution at 37°C for 16 h. The chlorophyll was leached from the plant tissue with 70% ethanol. GUS staining was examined under a dissecting microscope. Determination of anther stage was based on bud measurements and stages of anther development described by Peirson et al. (1996) and reconciled with anther stages described by Sanders et al. (1999). Anthers were stained with Alexander’s stain (Alexander, 1969) and examined microscopically.
Microarray Analysis
Approximately 1000 anthers at stages 5 to 8 were dissected from the wild type, AtMYB80:GR transgenic plants, and the myb80 mutant for each biological replicate for subsequent RNA isolation. Each sample contained four biological replicates. To induce functional MYB80, plants were sprayed with 20 μM DEX at the appropriate time prior to anther dissection. Total RNA was extracted from each collection of 1000 anthers using the RNeasy plant kit (Qiagen). Microarray analysis was performed using 300 ng of total RNA per sample as described in the Genechip Expression Analysis Technical Manual (Affymetrix). Following biotin labeling of cRNA and fragmentation, samples were hybridized to Affymetrix Arabidopsis ATH1 genome arrays, washed using a Genechip fluidics station, and scanned using the Genechip Scanner 3000. Microarray data were processed using the ArrayAssist 5.5.1 software provided by Stratagene and Strand Life Sciences. The CEL files were analyzed with the PLIER algorithm, which provides background subtraction, normalization, and probe summarization. Variance stabilization was performed to add a fixed quantity (16) to all linear scale signal values, which suppresses noise at log signal values. Linear scale data were converted into log scale, where logs are taken to base 2. The log data set was then subjected to significance analysis (unpaired t test) to obtain a P value, a fold change, and a direction of change (up or down) for each gene. The Benjamini-Hochberg false discovery rate method was used to obtain P values corrected for multiple testing.
TUNEL Assay
Whole inflorescences were fixed in 4% (v/v) paraformaldehyde in PBS, 0.1% (v/v) Triton X-100, and 0.1% (v/v) Tween 20 for 1 h at room temperature under vacuum and then incubated in fresh fixative overnight at 4°C. Samples were washed with PBS, dehydrated in a graded ethanol series, and cleared in ethanol/histoclear (2:1, 1:1, and 1:2) for 1 h each and three times in 100% histoclear for 1 h each. Inflorescences were embedded in paraffin wax. Sections of 6 μm were cut using a Jung rotary microtome and attached to silane-coated slides. For the TUNEL assay, sections were deparaffinized with 100% xylene, dehydrated in a graded ethanol series, and permeabilized in proteinase K. Nick-end labeling of fragmented DNA was performed using the DeadEnd Fluorometric TUNEL system (Promega) according to the manufacturer’s instructions. Slides were counterstained with 1 μg/mL of propidium iodide and mounted with SlowFade Gold antifade reagent (Invitrogen). Cover slips were sealed with clear nail varnish and slides stored at 4°C with a desiccant. Samples were analyzed under a fluorescence scanning confocal microscope (Leica TCS SP2) using excitation at 488 nm and emission at 509 nm to view the green fluorescence of fluorescein and a 538/617-nm excitation/emission spectrum to view the red fluorescence of propidium iodide. Merged images were generated using the ImageJ program (National Institutes of Health).
ChIP and Promoter Sequence Analysis
To use ChIP, a specific antibody to the target protein of interest is required. We decided not to create an MYB80 antibody as it may not be specific given the high sequence similarity within the MYB family or the epitope may not be accessible in planta. The anti-GR PA-516 (Upsate) antibody was used to enrich for the AtMYB80:GR protein/DNA complex. The ChIP experiment was performed using the protocol described by Saleh et al. (2008). myb80 mutant plants containing the AtMYB80:GR transgene were sprayed with 20 μM DEX. Anthers or floral buds were harvested 24 h after DEX induction. PCR was performed using a standard GoTaq (Promega), 50 μL total reaction volume, and the following cycling parameters: first cycle, 94°C for 2 min; 36 cycles of 94°C for 30 s, 51°C for 30 s and 72°C for 40 s; and one cycle of 72°C for 2 min. To identify putative MYB binding motifs, ~600 bp of the 5′ untranslated promoter region was analyzed using cis-PLACE (www.dna.affrc.go.jp).
EMSA
Recombinant MYB80 protein was purified using Ni-NTA Superflow columns (Qiagen) according to the manufacturer’s protocol. EMSA was performed using a nonradioactive protocol available on the website of the Hammer Lab (University of Michigan; http://www.med.umich.edu/hammerlab/) and the protocol described by Li and Parish (1995). Probes were labeled with digoxigenin via PCR using digoxigenin-labeled dUTP (Roche). Primers were designed to amplify a probe of 150 to 200 bp in length based on the promoter region, which was the target of the ChIP analysis. Protein-probe binding was performed using 400 ng total protein, 3 μL of 5× binding buffer (20 mM NaCl, 5 mM MgCl2, 20 mM Tris, pH 8, 10% glycerol, 0.5 mM EDTA, and 0.5 mM DTT), 1 μL of poly d(I-C), and 1 to 4 μL of unlabeled competitor probe.
Scanning Electron Micrograph
Scanning electron microscopy was performed as described by Li et al. (2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases and are presented in Table 1 (gene ID/locus) or Supplemental Table 1 online (T-DNA insertions).
AUTHOR CONTRIBUTIONS
H.A.P. designed the research, performed the research, analyzed the data, and wrote the article. S.I. designed the research, performed the research, and wrote the article. S.F.L. designed the research and analyzed the data. R.W.P. designed the research and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Analysis to Validate the Microarray Data.
Supplemental Figure 2. RT-PCR Analysis to Validate the Inducible Microarray Data.
Supplemental Figure 3. ChIP Assay Using Floral Buds as Input Material.
Supplemental Figure 4. ChIP Assay Using Dissected Anthers (Stages 5 to 8) as Input Material.
Supplemental Figure 5. Recombinant MYB80 Protein Expression and Purification.
Supplemental Figure 6. Comparative qRT-PCR Analysis of MYB80 Transcript Levels in Wild-Type Mature (Anther Stages 10 to 12) versus Young (Anther Stages ≤ 9) Floral Buds.
Supplemental Figure 7. Promoter:GUS Expression Analysis.
Supplemental Figure 8. Scanning Electron Micrograph of Wild-Type Arabidopsis Pollen Grain.
Supplemental Table 1. Summary of T-DNA Insertion Mutants Analyzed for Phenotypic Changes in Male Fertility.
Supplemental Table 2. Genes Identified to Be Differentially Expressed 24 h after Dexamethasone Induction of Functional MYB80 Compared with Noninduced and myb80 Mutant.
Supplemental Table 3. List of Overlapping Genes (30) Present in Both the ms1 (Yang et al., 2007) and myb80 Microarray Data Set.
Supplemental Table 4. List of Overlapping Genes (66) Present in Both the ams (Xu et al., 2010) and myb80 Microarray Data Set.
Supplemental Table 5. Primer Sequences Used in This Article.
Supplemental Data Set 1. Microarray Analysis of myb80 versus Wild-Type Anthers at Stages 5 to 8.
Supplementary Material
Acknowledgments
We thank Edgar Sakers (La Trobe University) for his hand-sectioning skills and technical help, Tracie Webster and Nga Nguyen (Victorian AgriBiosciences Center) for their assistance with the Microarray Fluidics Station/Scanner, and Alexander Fink and Tim Brown (La Trobe University) for the introductory lesson on using the scanning electron and confocal microscopes. Part of this research was funded by an Australian Research Council Linkage Grant and a Grains Research and Development Corporation Support Grant to R.W.P.
References
- Aarts M.G., Dirkse W.G., Stiekema W.J., Pereira A. (1993). Transposon tagging of a male sterility gene in Arabidopsis. Nature 363: 715–717 [DOI] [PubMed] [Google Scholar]
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Balk J., Chew S.K., Leaver C.J., McCabe P.F. (2003). The intermembrane space of plant mitochondria contains a DNase activity that may be involved in programmed cell death. Plant J. 34: 573–583 [DOI] [PubMed] [Google Scholar]
- Balk J., Leaver C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Leaver C.J., McCabe P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463: 151–154 [DOI] [PubMed] [Google Scholar]
- Baranowskij N., Frohberg C., Prat S., Willmitzer L. (1994). A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers E.P., Jones A.M., Dickerman A.W. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65: 43–58 [DOI] [PubMed] [Google Scholar]
- Bossi F., Cordoba E., Dupré P., Mendoza M.S., Román C.S., León P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S., Tuori R.P., D’Ascenzo M.D., Fobert P.R., Despres C., Martin G.B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15: 3033–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., von Heijne G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541: 114–119 [DOI] [PubMed] [Google Scholar]
- Ge X., Dietrich C., Matsuno M., Li G., Berg H., Xia Y. (2005). An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep. 6: 282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond B.J., Bowen B., Peterson T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Gubler F., Raventos D., Keys M., Watts R., Mundy J., Jacobsen J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Guan X., Zhao H., Xu Y., Wang Y. (2011). Transient expression of glyoxal oxidase from the Chinese wild grape Vitis pseudoreticulata can suppress powdery mildew in a susceptible genotype. Protoplasma 248: 415–423 [DOI] [PubMed] [Google Scholar]
- Higginson T., Li S.F., Parish R.W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35: 177–192 [DOI] [PubMed] [Google Scholar]
- Hird D.L., Worrall D., Hodge R., Smartt S., Paul W., Scott R. (1993). The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to beta-1,3-glucanases. Plant J. 4: 1023–1033 [DOI] [PubMed] [Google Scholar]
- Hsieh K., Huang A.H.C. (2007). Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Nagata N., Yoshiba Y., Ohme-Takagi M., Ma H., Shinozaki K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Shinozaki K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43: 1285–1292 [DOI] [PubMed] [Google Scholar]
- Jiang L., Yang S.L., Xie L.F., Puah C.S., Zhang X.Q., Yang W.C., Sundaresan V., Ye D. (2005). VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T., Ariizumi T., Kawai-Yamada M., Uchimiya H., Toriyama K. (2006). Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 47: 784–787 [DOI] [PubMed] [Google Scholar]
- Li S.F., Higginson T., Parish R.W. (1999). A novel MYB-related gene from Arabidopsis thaliana expressed in developing anthers. Plant Cell Physiol. 40: 343–347 [DOI] [PubMed] [Google Scholar]
- Li S.F., Iacuone S., Parish R.W. (2007). Suppression and restoration of male fertility using a transcription factor. Plant Biotechnol. J. 5: 297–312 [DOI] [PubMed] [Google Scholar]
- Li S.F., Parish R.W. (1995). Isolation of two novel myb-like genes from Arabidopsis and studies on the DNA-binding properties of their products. Plant J. 8: 963–972 [DOI] [PubMed] [Google Scholar]
- Love A.J., Milner J.J., Sadanandom A. (2008). Timing is everything: Regulatory overlap in plant cell death. Trends Plant Sci. 13: 589–595 [DOI] [PubMed] [Google Scholar]
- Mena M., Cejudo F.J., Isabel-Lamoneda I., Carbonero P. (2002). A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 130: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Gubler F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini A., Mosti S., Brighigna L. (1999). Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 207: 213–221 [Google Scholar]
- Parish R.W., Li S.F. (2010). Death of a tapetum: A programme of developmental altruism. Plant Sci. 178: 73–89 [Google Scholar]
- Peirson B.N., Owen H.A., Feldmann K.A., Makaroff C.A. (1996). Characterization of three male-sterile mutants of Arabidopsis thaliana exhibiting alterations in meiosis. Sex Plant Reprod. 9: 1–16 [Google Scholar]
- Planchais S., Perennes C., Glab N., Mironov V., Inzé D., Bergounioux C. (2002). Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath;CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol. Biol. 50: 111–127 [DOI] [PubMed] [Google Scholar]
- Ramsay R.G., Ishii S., Gonda T.J. (1992). Interaction of the Myb protein with specific DNA binding sites. J. Biol. Chem. 267: 5656–5662 [PubMed] [Google Scholar]
- Reape T.J., McCabe P.F. (2008). Apoptotic-like programmed cell death in plants. New Phytol. 180: 13–26 [DOI] [PubMed] [Google Scholar]
- Reape T.J., McCabe P.F. (2010). Apoptotic-like regulation of programmed cell death in plants. Apoptosis 15: 249–256 [DOI] [PubMed] [Google Scholar]
- Sainz M.B., Grotewold E., Chandler V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Sanmartín M., Jaroszewski L., Raikhel N.V., Rojo E. (2005). Caspases. Regulating death since the origin of life. Plant Physiol. 137: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl R.L., May S.T., Ware D.H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A.M., Kröber S., Unte U.S., Huijser P., Dekker K., Saedler H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Stein J.C., Hansen G. (1999). Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol. 121: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.L., Zhao Y., Hong X., Zhai Z.H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462: 317–321 [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tait S.W., Green D.R. (2010). Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11: 621–632 [DOI] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux D.L. (2011). Apoptogenic factors released from mitochondria. Biochim. Biophys. Acta 1813: 546–550 [DOI] [PubMed] [Google Scholar]
- Vizcay-Barrena G., Wilson Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57: 2709–2717 [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Morroll S.M., Dawson J., Swarup R., Tighe P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Zhang D.B. (2009). From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wu S.S., Platt K.A., Ratnayake C., Wang T.W., Ting J.T., Huang A.H. (1997). Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc. Natl. Acad. Sci. USA 94: 12711–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang C., Yuan Z., Zhang D., Gondwe M.Y., Ding Z., Liang W., Zhang D., Wilson Z.A. (2010). The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Vizcay-Barrena G., Conner K., Wilson Z.A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang Z.B., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhu J., Chen H., Li H., Gao J.F., Jiang H., Wang C., Guan Y.F., Yang Z.N. (2008). Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55: 266–277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.