This work shows that a mutation of the PIN1 ortholog in Medicago truncatula, SLM1, caused multiple terminal leaflets and a reduction of lateral leaflets. Characterization of the mutant reveals that the terminal leaflet primordium in M. truncatula has a unique developmental mechanism and the elaboration of compound leaves is context dependent and tightly correlated with the auxin/SLM1 module.
Abstract
Compound leaf development requires highly regulated cell proliferation, differentiation, and expansion patterns. We identified loss-of-function alleles at the SMOOTH LEAF MARGIN1 (SLM1) locus in Medicago truncatula, a model legume species with trifoliate adult leaves. SLM1 encodes an auxin efflux carrier protein and is the ortholog of Arabidopsis thaliana PIN-FORMED1 (PIN1). Auxin distribution is impaired in the slm1 mutant, resulting in pleiotropic phenotypes in different organs. The most striking change in slm1 is the increase in the number of terminal leaflets and a simultaneous reduction in the number of lateral leaflets, accompanied by reduced expression of SINGLE LEAFLET1 (SGL1), an ortholog of LEAFY. Characterization of the mutant indicates that distinct developmental domains exist in the formation of terminal and lateral leaflets. In contrast with the pinnate compound leaves in the wild type, the slm1 sgl1 double mutant shows nonpeltately palmate leaves, suggesting that the terminal leaflet primordium in M. truncatula has a unique developmental mechanism. Further investigations on the development of leaf serrations reveal different ontogenies between distal serration and marginal serration formation as well as between serration and leaflet formation. These data suggest that regulation of the elaboration of compound leaves and serrations is context dependent and tightly correlated with the auxin/SLM1 module in M. truncatula.
INTRODUCTION
Leaves are the main photosynthetic organs of flowering plants and show considerable diversity in shape and size. Diverse leaf forms can be categorized into two major types: simple leaves and compound leaves. Simple leaves often have a single unit of undivided blade. Compound leaves consist of multiple discontinuous blade subunits, termed leaflets, that are attached to a rachis and display different forms such as pinnate and palmate compound leaves (Kim et al., 2003a). Simple and compound leaf morphology can be further characterized based on leaf margins, such as entire, serrated, or lobed (Goliber et al., 1999).
Leaves are derived from a pluripotent cell population named the shoot apical meristem (SAM). The leaf founder cells at the flanks of SAM are specified and grow into leaf primordia. Leaf development proceeds through primary morphogenesis, during which leaflets and serrations are produced by cell division, and secondary morphogenesis, during which final leaf size and shape are determined by cell expansion. The distal portion normally displays secondary morphogenesis earlier than the proximal portion in a developing leaf. As a result, different developmental stages can be observed at the same time in a leaf (Hagemann and Gleissberg, 1996; Ori et al., 2007). In simple leaf species, such as Arabidopsis thaliana, the blade expands from a region at the edge of the leaf primordium termed the marginal blastozone, which maintains morphogenetic activity (Hagemann and Gleissberg, 1996). In compound-leafed species, such as tomato (Solanum lycopersicum), leaflet primordia that are marked by rapid cell division can also initiate from the marginal blastozone, resulting in discrete leaflets (Hagemann and Gleissberg, 1996; Koenig et al., 2009).
The plant hormone auxin is known to regulate the initiation of organs from the SAM, the formation of leaf serrations, and the patterning of leaf veins (Benková et al., 2003; Reinhardt et al., 2003; DeMason and Chawla, 2004; Hay et al., 2006; Scarpella et al., 2006; Barkoulas et al., 2008; DeMason and Polowick, 2009; Koenig et al., 2009; Bilsborough et al., 2011). Auxin distribution follows a polar gradient with the actions of influx and efflux transporters. Previous work on PIN-FORMED1 (PIN1) has shown that PIN1 actively directs auxin efflux in Arabidopsis (Benková et al., 2003; Reinhardt et al., 2003). An auxin maximum can be generated in the L1 surface layer of meristem via PIN1 localization toward the auxin convergence point at the center of the incipient primordium (Benková et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005; Hay et al., 2006). Therefore, an auxin maximum is the earliest marker of a new lateral organ primordium (Heisler et al., 2005; Barkoulas et al., 2008; Bayer et al., 2009; DeMason and Polowick, 2009; Koenig et al., 2009). Loss of function of PIN1 leads to defects in initiation and separation of lateral organs, such as fused cotyledons and leaves, pin-like inflorescences, and abnormal branches (Vernoux et al., 2000; Reinhardt et al., 2003). In addition, the auxin/PIN1 module that triggers initiation of the leaf primordium at the flanks of the SAM is probably redeployed in leaves to regulate leaf shape (Scarpella et al., 2010; Bilsborough et al., 2011). The PIN1 convergence points in the epidermis are associated with auxin activity maxima at the tips forming serrations, and the sites of lateral vein formation are defined by internalizing auxin through the center of the serrations (Hay et al., 2006; Scarpella et al., 2006; Kawamura et al., 2010). As a result, the pin1 mutant has a smooth leaf margin (Hay et al., 2006). A recent study shows that two feedback loops are involved in Arabidopsis leaf margin development. The first one relates to the transport of auxin regulated by its own distribution via PIN1. In the second loop, CUP-SHAPED COTYLEDON2 (CUC2) promotes the generation of auxin activity maxima while auxin represses CUC2 expression (Bilsborough et al., 2011). In addition, CUC3 also plays a role in sculpting leaf margin serrations (Hasson et al., 2011).
In compound-leafed species, such as tomato, hairy bittercress (Cardamine hirsuta), and pea (Pisum sativum), the initiations of leaflet primordia are correlated with local peaks of auxin response. Perturbation of auxin transport by 1-N-naphthylphthalamic acid (NPA) or inactivation of PIN1 orthologs inhibited the formation of leaflets in tomato and C. hirsuta (Barkoulas et al., 2008; Koenig et al., 2009). In addition, differential auxin distribution is capable of delineating the initiation of lobes and patterning blade outgrowth in tomato (Koenig et al., 2009). Adult leaves of pea possess both leaflets and tendrils and the tendril is probably an abaxialized leaflet (Hofer et al., 2009). Auxin is tightly associated with the initiation of pinna primordia during compound leaf development in pea (DeMason and Polowick, 2009). In NPA-treated plants, terminal tendrils were converted to leaflets in some cases, and the number of lateral pinna pairs was reduced (DeMason and Chawla, 2004; DeMason and Hirsch, 2006). Furthermore, the development of axillary meristem and the outgrowth of axillary buds require auxin synthesis and transport (Reinhardt et al., 2003; Ongaro and Leyser, 2008; Balla et al., 2011). The recently described model for pea bud outgrowth indicates that auxin is involved in the determination of plant architecture (Balla et al., 2011). These results demonstrate that auxin distribution and auxin response are central to the regulation of plant growth.
Much effort has been devoted to the identification of regulators for compound leaf development. Several mechanisms have been shown to be involved in the developmental window to elaborate leaf formation (Braybrook and Kuhlemeier, 2010; Efroni et al., 2010). As the first homeodomain factors identified in plants, Class I KNOTTED1-like homeobox (KNOX1) genes are essential for the regulation of indeterminacy of SAM, but their expression is excluded from incipient leaf primordia in both simple-leafed and compound-leafed plants (Hake et al., 2004). The repression of KNOX1 genes persists during leaf formation in simple-leafed plants, such as Arabidopsis (Byrne et al., 2000; Ori et al., 2000; Hay and Tsiantis, 2006; Uchida et al., 2007). In some compound-leafed plants, KNOX1 expression is reestablished later in developing primordia (Hareven et al., 1996; Bharathan et al., 2002; Kim et al., 2003b; Uchida et al., 2007; Shani et al., 2009). For example, in tomato, KNOX1 is expressed in developing leaf primordia rather than only in the SAM (Hareven et al., 1996). In C. hirsuta, an Arabidopsis relative with dissected leaves, transgenic lines with reduced expression of KNOX1 have fewer leaflets, and ectopic KNOX1 expression leads to increased leaflet number, suggesting that KNOX1 proteins are required for leaflet formation in this species (Hay and Tsiantis, 2006). Furthermore, leaflet formation in C. hirsuta involves auxin activity maxima accompanied by downregulation of KNOX1 gene expression, implying a manner similar to the leaf initiation process at the SAM (Barkoulas et al., 2008).
Genetic regulation of compound leaf development is complex in various compound-leafed species. The FLORICAULA (FLO)/LEAFY (LFY) putative orthologs, such as UNIFOLIATA/SINGLE LEAFLET1 (SGL1) in some leguminous plants belonging to the inverted repeat lacking clade (IRLC), including pea, alfalfa (Medicago sativa), and Medicago truncatula, may function in place of KNOX1 to regulate compound leaf development (Hofer et al., 1997; Wojciechowski et al., 2004; Champagne et al., 2007; Wang et al., 2008). The leaves of the pea uni mutant have one to three leaflets, which are simpler than wild-type leaves. Neither rachises nor tendrils are formed (Hofer et al., 1997). In M. truncatula, leaves of the sgl1 mutant turn into a simple form (Wang et al., 2008). In these IRLC species, the expression of KNOX1 genes is not associated with compound leaves, although overexpression of KNOX1 in alfalfa results in an increase in leaflet number (Hofer et al., 2001; Champagne et al., 2007). On the other hand, downregulation of the expression of the FLO/LFY gene in the non-IRLC legumes, such as soybean (Glycine max) and Lotus japonicus, leads to moderate simplifications of compound leaves (Dong et al., 2005; Champagne et al., 2007). These data suggest that the FLO/LFY putative orthologs in IRLC species play an important role in compound leaf development.
Independent studies in different species shed light on the elaboration mechanisms of branches, leaves, leaflets, lobes, and serrations, in which the auxin/PIN1 module is extensively involved. However, thus far, all mutants that have been found to affect lateral leaflet development do not affect the initiation of the terminal leaflet (Efroni et al., 2010). Furthermore, although it has been reported that PIN1 is involved in the regulation of LFY via local accumulation of auxin in Arabidopsis (Vernoux et al., 2000), it is not clear how the putative orthologs of PIN1 and LFY interact in compound-leafed species. M. truncatula is a model legume species whose adult leaves are trifoliates with serrations on the leaf margin. The mechanism of leaf development in this species is largely unknown, and only a few genes have been identified (Wang et al., 2008; Chen et al., 2010). In this study, a mutant with a smooth leaf margin, smooth leaf margin1 (slm1), was isolated from a Tnt1 retrotransposon-tagged mutant population of M. truncatula. SLM1 was identified by thermal asymmetric interlaced-PCR and association analysis. Molecular analysis shows that SLM1 encodes an auxin efflux carrier protein Mt PIN10 in M. truncatula (Schnabel and Frugoli, 2004) and is the ortholog of Arabidopsis PIN1. SLM1 loss of function causes diffuse auxin distribution, ultimately resulting in pleiotropic phenotypes in different regions, such as leaves, leaf margins, and flowers. The most striking feature of the slm1 mutant is the formation of multiple terminal leaflets and a simultaneous reduction in the number of lateral leaflets accompanied with reduced SGL1 expression. In the slm1 sgl1 double mutant, only the formation of lateral leaflets was affected, suggesting that distinct developmental domains exist in the initiation of lateral and terminal leaflets. In addition, different ontogenies were observed between distal serration and marginal serration formation, as well as leaflet formation. We present a possible model for the regulation of elaboration of compound leaves in M. truncatula, which is context dependent and tightly correlated with the auxin/SLM1 module.
RESULTS
SLM1 Is Required for Lateral Organ Development at the Vegetative Stage
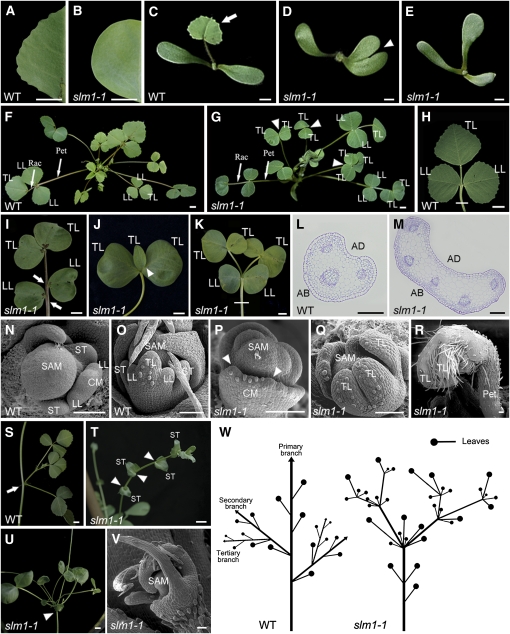
To identify mutants with defects in compound leaf development, ~10,000 independent lines of Tnt1 retrotransposon-tagged M. truncatula populations were screened. Three mutant lines with obvious changes in leaf margin were identified. In contrast with the wild type, which exhibits serrations on the leaf margin (Figure 1A), these mutants showed an obvious smooth leaf margin phenotype (Figure 1B). The mutants were named slm1 (slm1-1, slm1-2, and slm1-3).
Figure 1.
The slm1-1 Mutant of M. truncatula Shows Developmental Defects at the Vegetative Stage.
(A) and (B) Leaf margin of the wild type (WT) (A) and slm1-1 (B).
(C) to (E) Four-day-old seedlings of the wild type (C) and slm1-1 ([D] and [E]). The arrow points to the first true leaf in the wild type (C). Note that the development of the first true leaf was abolished in slm1-1 ([D] and [E]). The arrowhead points to a cotyledon fusion (D).
(F) and (G) Five-week-old plants of the wild type (F) and slm1-1 (G). Arrowheads point to three adult leaves of slm1-1. Two of the adult leaves have double terminal leaflets developed at the distal end of petiole, and one has three terminal leaflets (G). No lateral leaflets developed in all three marked adult leaves (G). Rac, rachis; Pet, petiole; TL, terminal leaflet; LL, lateral leaflet.
(H) to (K) Adult leaves of the wild type (H) and slm1-1 ([I] to [K]). Note that three terminal leaflets developed on the petiole and no lateral leaflets were produced (J). Petiole fusion could be observed in (K). Two terminal leaflets developed on the distal end of each petiole, respectively (K). Arrows indicate asymmetric lateral leaflets on the petiole (I). Arrowhead indicates two fused terminal leaflets (J). TL, terminal leaflet; LL, lateral leaflet.
(L) and (M) Transverse sections of petioles in the wild type (L) and slm1-1 (M); the sectioning regions are shown in (H) and (K) by white lines, respectively. AD, adaxial side; AB, abaxial side.
(N) to (R) Scanning electron micrographs of leaf primordia in the wild type at stage 3 (N) and stage 5 (O) and in slm1-1 at stage 3 (P) and stage 5 (Q), and the developing leaf in slm1-1 at stage 9 (R). Arrowheads point out that at least two terminal leaflet primordia initiated from a common leaf primordium (P). CM, common leaf primordium; TL, terminal leaflet primordium; LL, lateral leaflet primordium; ST, stipule primordium; Pet, petiole.
(S) to (V) Development of branches in the wild type (S) and slm1-1 ([T] to [V]). Arrow points to the node that bears one trifoliate and a higher-order branch in the wild type (S). Arrowheads point to nodes without branches and leaves in (T) and to the distal portion of stem with radial multiple leaves and branches in (U). Scanning electron micrograph shows the SAM with radial lateral organs in slm1-1 (V). ST, stipule.
(W) A schematic illustration of branch arrangement in the wild type (left) and slm1-1 (right) at the vegetative stage.
Bars = 5 mm in (A) to (K) and (S) to (U), 200 μm in (L) and (M), and 50 μm in (N) to (R) and (V).
Alterations in SLM1 activity not only affected leaf margin but also dramatically affected the formation of lateral organs, such as cotyledons, leaves, flowers, and branches, and showed increased indeterminacy throughout plant growth. slm1-1 seedlings showed abnormal cotyledons, in which 24% displayed fused cotyledons and 11% displayed triple cotyledons (Figures 1C to 1E, n = 50), suggesting that SLM1 affects the initiation of cotyledons or the partitioning of the embryo apical domain. In addition, the elaboration of veins was abnormal in cotyledons of slm1-1, indicating that SLM1 is required for vascular patterning in cotyledons (see Supplemental Figures 1A to 1C online).
In the wild type, the juvenile leaf (first true leaf) has a simple leaf morphology, and all other adult leaves are in trifoliate form (Figures 1C and 1F; see Supplemental Figure 1D online). In adult leaves of the wild type, a single terminal leaflet develops on the distal end of the petiole/rachis and a pair of lateral leaflets develops on the sides of the petiole (Figure 1H). The epicotyl length of slm1-1 was increased compared with that of the wild type (see Supplemental Figures 1G to 1K online). In the slm1-1 mutant, the juvenile leaf did not develop in most cases (Figures 1D and 1E; see Supplemental Figure 1L online). However, adult leaves in slm1-1 could be produced continuously (see Supplemental Figure 1E online). In addition, the first adult leaf developed at almost the same time in the wild type and slm1-1. Therefore, the initiation of the juvenile leaf in slm1-1 was abolished instead of being skipped. This observation suggests that the formation of the juvenile leaf is more sensitive than that of adult leaves in slm1-1 and implies that loss of function of SLM1 has a greater impact on the juvenile stage than adult stage.
A striking change in slm1-1 is the development of multiple terminal leaflets at the distal portion of rachis, while the lateral leaflet number was reduced (Figures 1G, 1I, to 1K; see Supplemental Figure 1M online). In the slm1-1 mutant, 42% of adult leaves (n = 100) did not produce any lateral leaflets, and 45% of adult leaves (n = 100) developed more than one terminal leaflet. Additionally, the terminal leaflet length of the mutant was reduced (Table 1). Fused leaflets were observed in rare cases (3%, n = 100) in slm1-1, resulting in a malformed leaflet (Figure 1J, arrowhead). Fusion between petioles, which still show distinct domains of adaxial and abaxial sides, was frequently observed in slm1-1 and was confirmed by anatomical analysis (Figures 1L and 1M).
Table 1.
Phenotypic Characterization of Wild-Type and slm1 Plants of M. truncatula
| Genotype | Terminal Leaflet Length (cm) | Terminal Leaflet Width (cm) | Petiole Length (cm) | Rachis Length (cm) |
| Wild type | 1.80 ± 0.20 | 2.13 ± 0.12 | 6.53 ± 0.58 | 0.95 ± 0.13 |
| slm1-1 | 1.47 ± 0.17* | 2.24 ± 0.15 | 7.41 ± 0.43* | 1.64 ± 0.15* |
| slm1-2 | 1.40 ± 0.18* | 2.10 ± 0.19 | 7.44 ± 0.55* | 1.61 ± 0.17* |
| slm1-3 | 1.51 ± 0.26* | 2.17 ± 0.16 | 7.38 ± 0.73* | 1.42 ± 0.37* |
Leaflet length was measured from tip to base of the terminal leaflet. Leaflet width was measured from margin to margin of the terminal leaflet. Petiole and rachis length were measured on the first fully expanded trifoliate of 8-week-old plants. Numbers are presented as mean ± sd. The number of observations in each mean is 35. Asterisks indicate that the differences between the wild type and slm1 are statistically significant at P < 0.05.
To better characterize compound leaf defects in slm1-1, scanning electron microscopy analysis of leaf development was performed. In M. truncatula, lateral leaflet primordia and the terminal leaflet primordium do not develop at the same stage (Wang et al., 2008). A common leaf primordium (CM) that has the potential to differentiate into leaflet primordium developed first (stages 1 and 2) (Wang et al., 2008). Then, a pair of lateral leaflet primordia (LL) emerged at the proximal end of the common leaf primordium at stage 3 (Figure 1N). Last, the common leaf primordium differentiated into a single terminal leaflet primordium (TL) at stage 4 (Wang et al., 2008). The ontogeny of compound leaf development probably implies that the developmental identities between lateral leaflet primordia and the terminal leaflet primordium are different in M. truncatula. At stage 5, the wild type showed a single terminal leaflet primordium, two lateral leaflet primordia, and two stipule primordia (ST) (Figure 1O). In slm1-1, at least two terminal leaflet primordia were initiated from a common leaf primordium during the development of some adult leaves (Figure 1P). In some cases, three terminal leaflet primordia were developed and no lateral leaflet primordium was formed at stage 5 (Figure 1Q), resulting in a compound leaf with three terminal leaflets and without lateral leaflets (Figure 1R). Accompanying these changes was an increase of the petiole and rachis length of mature leaves in 8-week-old slm1-1 plants, suggesting that the proximal-distal axis of slm1-1 was also altered (Table 1).
Shoot branching of slm1-1 was also altered. In the wild type, one node bears one trifoliate and one higher order branch (Figures 1S and 1W). By contrast, the development of higher-order branches and leaves on some nodes of slm1-1 was abolished, suggesting defects in the initiation of axillary meristems and/or in the outgrowth of axillary buds (Figure 1T). Moreover, multiple leaves and branches frequently developed at the distal portion of stem in slm1-1 (Figures 1U to 1W). These observations indicate that SLM1 is required for the determinacy of shoot branching in M. truncatula. As a result, the architecture of the slm1-1 plant was affected (Figure 1W) and displayed a semidwarf phenotype at the reproductive stage (see Supplemental Figure 1F online). In addition, all three alleles of SLM1 exhibited the same defects in leaves with variation of leaflet number and showed a semidwarf phenotype at the vegetative stage (Table 1; see Supplemental Figure 1N online).
SLM1 Is Required for Flower Development
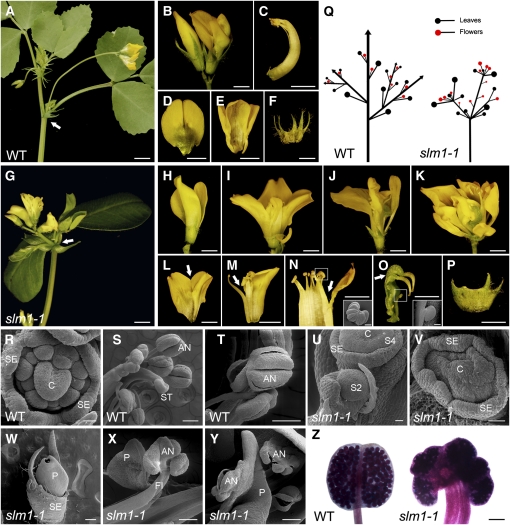
Wild-type M. truncatula enters the reproductive stage after ~60 d of growth. At the reproductive stage, a node in the wild type bears one to three open flowers besides shoot branches and leaves (Figure 2A). The wild-type flowers are comprised of a central carpel enclosed by a stamina tube, vexillum, sepal, and fused alae and keel (Figures 2B to 2F). By contrast, the lesion in SLM1 resulted in abnormal flowers with mild, moderate, and severe phenotypes in slm1-1 (Figures 2G to 2K), but flowering time of the mutant was not affected. Moreover, malformed floral organs and fused floral organs were frequently observed (Figures 2L to 2P), suggesting that SLM1 regulates the initiation and separation of floral organs. In addition, similar to shoot branching at the vegetative stage, flower arrangement was severely affected in slm1-1, indicating that branching is also abnormal at the reproductive stage in slm1-1 (Figure 2Q). To further investigate the defects in floral organs in slm1-1, the developmental processes of flowers between the wild type and slm1-1 were compared by scanning electron microscopy. At stage 6 (Benlloch et al., 2003), floral organ primordia were completely differentiated and carpel suture became visible in the wild type (Figure 2R). As for mutants, slm1-1 exhibited defects in the separation of floral primordia, which could be seen as early as at stage 4 (Figure 2U). At stage 6, fused floral primordia were more obvious (Figure 2V), resulting in fused floral organs (Figures 2W to 2Y), which are distinct from those of the wild type (Figure 2S). At the late stage of flower development, anthers of slm1-1 dehisced normally, similar to those of the wild type (Figures 2T and 2Y), and their pollen was viable, as revealed by pollen staining (Figure 2Z). The three alleles of SLM1 showed the same defective phenotype of flowers. However, variation in fertility among the three slm1 alleles was observed. slm1-1 was infertile, while slm1-2 and slm1-3 could occasionally produce seedpods and seeds with normal germination ability (see Supplemental Figure 2 online).
Figure 2.
The slm1-1 Mutant of M. truncatula Shows Developmental Defects at the Reproductive Stage.
(A) Flower development in the wild type. Arrow indicates a node that bears two open flowers and one fully expanded trifoliate.
(B) Flower phenotype in the wild type. The flowers of the wild type show bilateral symmetry along the dorsal-ventral axis.
(C) to (F) Dissected floral organs of the wild type. The side view of the central carpel (C), top view of vexillum (D), alae and keel (E), and sepal (F).
(G) Flower development in slm1-1. Arrow indicates that flowers and leaves develop radially at the distal portion of stem.
(H) to (K) Flower phenotype in slm1-1 with mild (H), moderate ([I] and [J]), and severe (K) alterations.
(L) to (P) Dissected floral organs of slm1-1. Fusions between floral organs were frequently observed; for example, the fusion between vexillums (L), between stamen and petal (M), between anthers (N), and between pistils (O). The sepal is also abnormal (P). The insets in (N) and (O) show fused anthers and exposed ovules by scanning electron microscopy, respectively. Arrows in (L) to (O) indicate the fusion of floral organs.
(Q) A schematic illustration of branch arrangement of the wild type (left) and slm1-1 (right) at the reproductive stage.
(R) to (T) Scanning electron microscopy analysis of floral organs in the wild type. Representative images show floral primordia at stage 6 (R), anthers and stigma (S), and dehiscing anthers (T) in a mature flower.
(U) to (Y) Scanning electron microscopy analysis of floral organs in slm1-1. Representative images show floral primordia at stage 2 ([U], S2), stage 4 ([U], S4), and stage 6 (V). At the late stage of floral development, fully fused petals (W), fused filament and petal (X), and dehiscing anther (Y) were observed.
(Z) Pollen staining in the wild type (left) and slm1-1 (right).
C, carpel; SE, sepal; P, petal; ST, stigma; AN, anther; FI, filament; S2, stage 2; S4, stage 4; WT, wild type. Bars = 5 mm in (A) and (G), 2 mm in (B) to (F) and (H) to (P), 200 μm in (S), (T), (W) to (Y), and the insets in (N) and (O), 100 μm in (Z), and 50 μm in (R), (U), and (V).
Molecular Cloning of SLM1
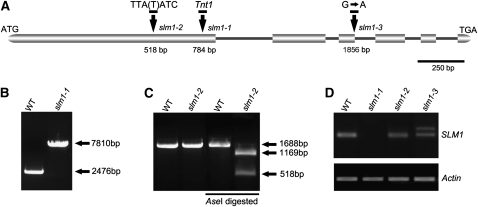
To identify the gene responsible for the developmental defects, thermal asymmetric interlaced-PCR was performed to recover the flanking sequences from the three mutant lines. Surprisingly, except for slm1-1, the flanking sequences recovered from either slm1-2 or slm1-3 could not be associated with the mutant phenotype. Further analysis with the slm1-1 mutant identified 19 flanking sequences, and one was confirmed to be associated with the mutation. BLAST analysis using this flanking sequence was performed against the M. truncatula genome from the National Center for Biotechnology Information. A full-length genomic sequence of 2476 nucleotides was obtained. The full-length coding sequence (CDS) of SLM1 was cloned by RT-PCR and found to contain 1776 nucleotides. Alignment between the cDNA and genomic sequences of SLM1 revealed that SLM1 consists of six exons and five introns (Figure 3A).
Figure 3.
Molecular Characterization of SLM1 in M. truncatula.
(A) Schematic diagram of the gene structure of SLM1. The positions of the ATG start and TGA stop codons are shown. Vertical arrows mark the nucleotide changes in various slm1 alleles. Numbers indicate nucleotide positions of the site of mutations. Boxes represent exons and lines represent introns. A single base, T (thymine), was deleted in slm1-2.
(B) PCR amplification of SLM1 from the wild type (WT) and slm1-1. A single insertion of the tobacco Tnt1 retrotransposon (~5.3 kb) was detected in slm1-1.
(C) Transcripts of SLM1 from the wild type and slm1-2 were amplified by RT-PCR and digested by AseI, resulting in length polymorphism because of a single-base-pair deletion mutation in slm1-2. Three technical replicates were performed.
(D) RT-PCR analysis of SLM1 transcripts in the wild type and slm1 alleles. Altered splicing of transcript in slm1-3 is shown. Actin was used as a loading control. Three technical replicates were performed.
PCR amplification of the SLM1 genomic sequence from the three mutant lines and the wild type revealed that only slm1-1 carried a single 5.3-kb Tnt1 retrotransposon insertion, resulting in the interrupted expression of SLM1 (Figures 3B and 3D). Amplification of the full-length CDS of SLM1 from slm1-2 and slm1-3 and sequence comparison revealed that both slm1-2 and slm1-3 contained lesions in this gene, which were not due to the Tnt1 insertion. slm1-2 carried a single-base-pair deletion mutation in the first exon, which caused a shift in the reading frame and resulted in premature termination of the encoded protein (Figure 3A). Moreover, this change introduced an additional AseI restriction site. This allowed us to amplify the sequence spanning the mutation site and generate a cleaved polymorphic sequence marker, which yielded products of 1169 and 518 bp after AseI digestion of the PCR product in slm1-2 but not in the wild type (Figure 3C). The other mutant, slm1-3, has a single-base-pair substitution (G to A) in the intron splicing site, resulting in altered mRNA splicing (Figure 3A). To verify this, the CDS of SLM1 in slm1-3 was amplified. Longer mRNA molecules were indeed amplified in slm1-3, confirming the altered splicing of SLM1 transcripts in this allele (Figure 3D).
SLM1 Complements the Mutant Phenotype of slm1
As Tnt1 retrotransposon-tagged lines of M. truncatula generally contain 20 to 50 insertions (Tadege et al., 2008), two backcrosses were performed to obtain a segregation population of slm1-1 to confirm that the mutant phenotype was caused by Tnt1 insertion in a single gene. The mutants and wild-type-like plants showed a segregation ratio of 1:3, suggesting that the mutant phenotype was associated with a single recessive locus (see Supplemental Figure 3A online). To further confirm that the mutant phenotype was caused by the mutation of this gene, a construct carrying a 5.2-kb genomic fragment containing the promoter region and SLM1 open reading frame was transformed into slm1-1 plants. Phenotypic observation confirmed that complementary SLM1 expression fully rescued leaf and floral defects in slm1-1 at different developmental stages (see Supplemental Figure 3B online).
SLM1 Is an Ortholog of Arabidopsis PIN1
BLASTX analysis using the SLM1 CDS revealed several hits belonging to the auxin efflux carrier protein family. Most known members of this family are PIN components of auxin efflux facilitators in plants. These carriers are auxin specific and localized to the basal ends of auxin transport-competent cells (Kramer, 2004; Blakeslee et al., 2005). Phylogenetic analyses with 18 members of the PIN family from Arabidopsis (At), C. hirsuta (Ch), P. sativum (Ps), Triticum aestivum (Ta), Oryza sativa (Os), and Brassica juncea (Bj) revealed that SLM1 was evolutionarily closer to the PIN1 family and showed 65% identity with At PIN1, suggesting a possible effect of SLM1 on polar auxin transport (see Supplemental Figure 4 and Supplemental Data Set 1 online). Sequence alignment was also performed among At PIN1, Ch PIN1, Ps PIN1, Ta PIN1, Os PIN1, Bj PIN1, and SLM1. SLM1 shared high sequence similarity with PIN1 proteins at the conserved C- and N-terminal domains (see Supplemental Figure 5 online). Based on these results, SLM1 is identified as a putative ortholog of Arabidopsis PIN1.
To assess the function of SLM1 as an auxin transporter, the Arabidopsis pin1 mutant was transformed with a construct containing the SLM1 CDS driven by the Arabidopsis PIN1 promoter. This construct was capable of fully rescuing the defects of the pin1 mutant, suggesting that SLM1 is a functional auxin efflux transporter (see Supplemental Figure 6 online). To examine the ability of Arabidopsis PIN1 to suppress the loss-of-function phenotype seen in slm1-1, the Arabidopsis ProPIN1:PIN1:GFP (green fluorescent protein) construct was introduced into the slm1-1 mutant. It has been shown that this construct was sufficient to rescue the C. hirsuta pin1 mutant (Barkoulas et al., 2008). The defects of slm1-1 were fully suppressed in eight transgenic plants and partially suppressed in seven other transgenic plants, suggesting functional conservation between SLM1 and PIN1.
Expression Pattern of SLM1
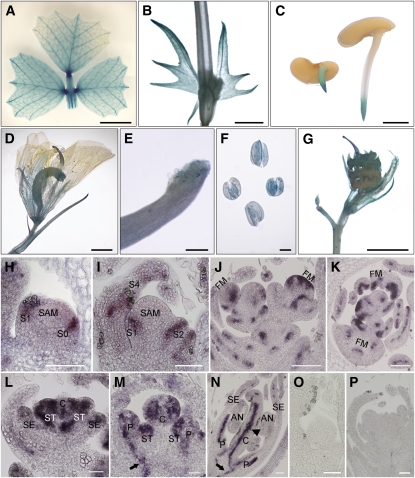
The expression pattern of SLM1 in different tissues and organs was analyzed using the M. truncatula Gene Expression Atlas (http://mtgea.noble.org/v2). The relative expression level of SLM1 was obtained using the probe set Mtr.47942.1.S1, which represented SLM1 in the microarray chip. The data revealed that SLM1 was expressed in almost all tissues. The expression of SLM1 was relatively high in the vegetative bud, root tip, and developing nodule (see Supplemental Figure 7 online). To determinate the expression pattern comprehensively, an SLM1 promoter-β-glucuronidase (GUS) construct was introduced into wild-type M. truncatula, and GUS activity was examined in transgenic plants. The transgenic plants showed GUS expression in leaf veins, the basal region of leaflets (Figure 4A), the stem, stipule (Figure 4B), and root tip of germinating seeds (Figure 4C). GUS expression was also detected in the basal region of the flower (Figure 4D), stigma (Figure 4E), anther (Figure 4F), and young seedpod (Figure 4G).
Figure 4.
Expression Pattern of SLM1 in M. truncatula.
(A) to (G) Promoter-GUS fusion studies of SLM1 expression in transgenic M. truncatula. SLM1 promoter driven GUS is expressed in the adult leaf (A), stem and stipule (B), root tip of germinating seeds (C), flower (D), stigma (E), anther (F), and 5-d-old seedpod (G).
(H) to (P) In situ hybridization analysis of SLM1 mRNA in vegetative and reproductive apices of the wild type.
(H) and (I) Longitudinal sections of the SAM at stage 1 (S1; [H]) and stage 4 (S4; [I]).
(J) and (K) Longitudinal section (J) and transverse section (K) of floral apices at stage 2.
(L) to (N) Longitudinal sections of the floral apical meristem at stage 3 (L), stage 5 (M), and stage 7 (N).
(O) and (P) The sense probe was hybridized and used as control. Arrows indicate vascular bundles. Arrowhead indicates the inside of the carpel.
FM, floral meristem; SE, sepal; P, petal; C, carpel; ST, stamen; AN, anther. Bars = 5 mm in (A), 2 mm in (B) to (D) and (G), 200 μm in (E) and (F), and 50 μm in (H) to (P).
The spatial and temporal localizations of SLM1 at the vegetative and reproductive stages were examined further by in situ hybridization analysis in the wild type. SLM1 mRNA was detected in the cells that give rise to leaf primordia at the SAM and in the developing leaf primordia (Figures 4H and 4I). Strong SLM1 expression was observed in floral meristems and restricted to the site that would give rise to floral organ primordia (Figures 4J and 4K). In M. truncatula, sepal primordia initiate first in floral meristems. Then, common primordia that produce petal and stamen primordia develop at stage 3 (Benlloch et al., 2003). High SLM1 expression was detected in the developing common primordia at stage 3 (Figure 4L) and in the developing petals, stigma, and stamens at stage 5 (Figure 4M). At stage 7, SLM1 mainly accumulated in petals and inside the carpel, where ovules were under development (Figure 4N). In addition, SLM1 mRNA was detected in the vascular bundles (Figures 4M and 4N, arrows).
Local Auxin Activity Maxima Facilitate the Initiation of Leaf and Floral Primordia
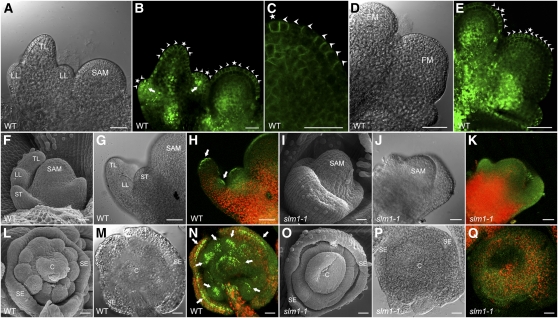
To investigate whether the initiation of leaf and floral primordia in M. truncatula is related to auxin transportation and accumulation, PIN1 localization was examined in M. truncatula plants transformed with the Arabidopsis ProPIN1:PIN1:GFP reporter (Benková et al., 2003). The results showed that PIN1 is apically localized at the epidermal cells of the leaf and floral meristem and mark the site of incipient primordia formation at the meristem flank toward primordia tips (Figures 5A to 5E). In addition, GFP expression was also upregulated in initiating lateral leaflet primordia (Figure 5B, arrows). These observations suggest that the local auxin activity maxima generated by PIN1/SLM1 probably facilitate the formation of both leaves and floral primordia. To verify this hypothesis, the DR5rev:GFP auxin response reporter was transformed into the wild type and slm1-1, respectively. A gradient of DR5 activity, with a maximum at the tips of leaves and floral organ primordia, was detected by GFP signal in the wild type, indicating that auxin maxima are required for the proper development of primordia (Figures 5F to 5H and 5L to 5N). However, auxin distribution was disturbed in slm1-1, which was defective in the positioning and separation of lateral organ primordia (Figures 5I to 5K and 5O to 5Q). These results demonstrate that the defects of slm1-1 are caused by disorders of auxin transportation and distribution, which are tightly correlated with SLM1.
Figure 5.
PIN1/SLM1-Dependent Auxin Gradients in Leaf and Floral Organ Formation in M. truncatula.
(A) Leaf primordia of the wild type (WT) at stage 4.
(B) and (C) Distribution of the ProPIN1:PIN1:GFP marker (green signal) in leaf primordia (B) and a close view of the localization of ProPIN1:PIN1:GFP marker in the SAM (C). Arrowheads mark the direction of PIN1 polarization. Asterisks indicate the auxin convergence points that mark the site of incipient primordium initiation.
(D) Floral primordia of the wild type at stage 2. FM, floral meristem.
(E) Distribution of the ProPIN1:PIN1:GFP marker in floral primordia. Arrowheads point to the direction of PIN1 polarization. Asterisks indicate the auxin convergence points.
(F) to (K) Leaf primordia of the wild type ([F] to [H]) and slm1-1 ([I] to [K]). Leaf primordia harboring the auxin response marker DR5 (DR5rev:GFP) were observed by scanning electron microscopy ([F] and [I]), light-field microscopy ([G] and [J]), and confocal microscopy ([H] and [K]). Arrows point to auxin accumulation at the tip of lateral and terminal leaflet primordia.
(L) to (Q) Floral primordia of wild-type ([L] to [N]) and slm1-1 ([O] to [Q]). Arrows point to auxin accumulation at the tip of floral organ primordia.
TL, terminal leaflet primordium; LL, lateral leaflet primordium; ST, stipule; C, carpel; SE, sepal; WT, wild type. Bars = 25 μm.
SLM1 Regulates Leaf Margin and Leaf Marginal Cell Development
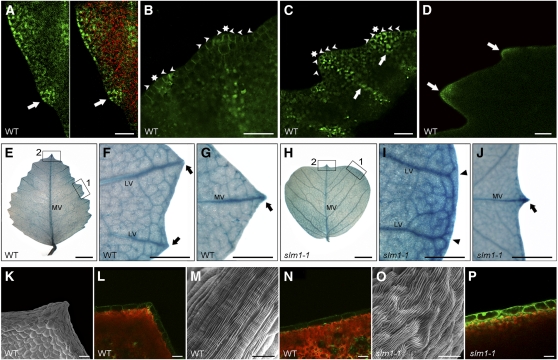
One prominent phenotype of slm1-1 was the conversion of the serrated leaf margin to smooth leaf margin. To understand how the leaf serrations are formed in M. truncatula, the DR5rev:GFP auxin response reporter and the ProPIN1:PIN1:GFP reporter were transformed into wild-type plants. DR5rev:GFP expression was detected in the tips of initiating serrations (Figure 6A). To examine whether auxin accumulation is generated by PIN1/SLM1-directed auxin efflux, localization of the ProPIN1:PIN1:GFP reporter was examined. Polar expression of ProPIN1:PIN1:GFP was observed in the epidermal cells, predicting that the flow of auxin converged to the site of serration initiation (Figure 6B). As the leaf serrations were expanding, ProPIN1:PIN1:GFP expression displayed evidence that the direction of auxin flux was toward the tips of serrations (Figure 6C) where auxin accumulated (Figure 6D). These observations demonstrate that auxin transportation and activity gradients are important for the formation of leaf marginal serrations.
Figure 6.
Involvement of SLM1 in Leaf Margin Development in M. truncatula.
(A) DR5rev:GFP expression maximum at the site of serration initiation of the leaf margin (green signal, left) and an overlay image with chlorophyll autofluorescence (red signal, right) in the wild type (WT). Arrows point to the site of serration initiation.
(B) and (C) PIN1:PIN1-GFP expression during the development of leaf serrations. The localization of ProPIN1:PIN1-GFP reporter is polar at the site of serration initiation (B) and developing serrations (C). Asterisks indicate auxin flow converging at the tip of a serration. Arrowheads indicate the orientation of auxin flow predicated by PIN1/SLM1. Arrows point to the location of lateral vein formation.
(D) DR5rev:GFP expression in developing leaf serrations. Arrows indicate auxin accumulation at the tip of serrations.
(E) to (G) DR5:GUS expression in the fully expanded terminal leaflet of the wild type (E). Close views of marginal serration (empty box 1) and distal serration (empty box 2) are shown in (F) and (G), respectively. Arrows mark auxin accumulation at the tip of serrations. MV, midvein; LV, lateral vein.
(H) to (J) DR5:GUS expression in a fully expanded terminal leaflet of slm1-1 (H). Close views of leaf margin (empty box 1) and distal serration (empty box 2) are shown in (I) and (J), respectively. Arrowheads point to lateral veins, which do not terminate at the margins. Arrow indicates auxin accumulation at the tip of the distal serration. MV, midvein; LV, lateral vein.
(K) to (P) Observation of marginal cells in the wild type ([K] to [N]) and slm1-1 ([O] and [P]). Scanning electron microscopy analysis of the surface of marginal cells at the tip (K) and the side (M) of serrations in the wild type and the surface of marginal cells in slm1-1 (O). DR5rev:GFP expression is shown in the marginal cells at the same location in the wild type ([L] and [N]) and slm1-1 (P).
Bar = 25 μm in (A) to (C), 5 mm in (E) and (H), 1 mm in (F), (G), (I), and (J), 150 μm in (D), 50 μm in (K), (L), (N), and (P), and 20 μm in (M) and (O).
To further elucidate the role of auxin in leaf margin morphogenesis, the DR5:GUS auxin response reporter (Ulmasov et al., 1997) was introduced into wild-type and slm1-1 plants to reflect relative auxin levels. Terminal leaflets were used to compare the configuration of the leaf margin and veins of both the wild type and slm1-1, since the terminal leaflet and lateral leaflet showed similar DR5:GUS expression patterns. In the wild type, auxin accumulated in the midvein and at the tips of serrations (Figure 6E). Furthermore, local auxin maxima at the tips of serrations were tightly associated with the positioning of lateral veins. Among the lateral veins, the auxin level gradually decreased from the tips of serrations to the midvein (Figures 6E and 6F). Auxin accumulation was also observed in the midvein of slm1-1. However, auxin distribution in lateral veins was diffuse. In contrast with the correlation between lateral veins and serrations in the wild type, higher-order and free-ending veins developed at the distal end of lateral veins in slm1-1 (Figures 6H and 6I). These observations indicate that SLM1 regulates the elaboration of leaf shape and the pattern of leaf venation by directing auxin distribution. Moreover, a smooth leaf margin was also observed in the NPA-treated plants, confirming that SLM1 is involved in polar auxin transport, which is correlated with the formation of the leaf margin (see Figure 8K). In addition, we noticed that the distal serration associated with the midvein was intact in slm1-1, similar to as in the wild type, implying that different developmental mechanisms exist between distal serration and marginal serrations (Figures 6G and 6J). On the other hand, the surface of marginal cells changed in the fully expanded leaves of slm1-1. The ridge-like structure was distorted due to the loss of function of SLM1, compared with that of the wild type (Figures 6K, 6M, and 6O). Furthermore, auxin accumulation was observed within marginal cells of both the wild type and slm1-1 by assaying DR5rev:GFP expression (Figures 6L, 6N, and 6P). GFP expression level was higher in slm1-1, suggesting that more auxin accumulated in the marginal cells of these plants. These observations imply that the development of marginal cells also requires proper auxin activity gradients.
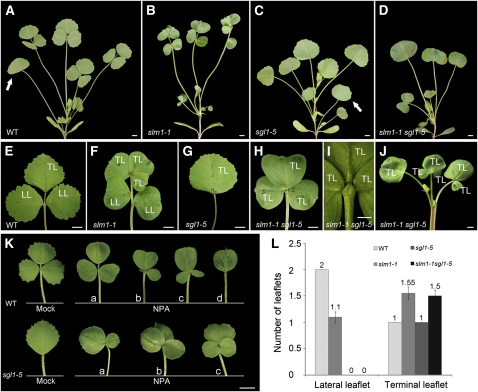
Figure 8.
SLM1 and SGL1 Regulate Compound Leaf Development in M. truncatula.
(A) to (D) Four-week-old plants of the wild type (WT) (A), slm1-1 (B), sgl1-5 (C), and slm1-1 sgl1-5 (D). Arrows indicate the juvenile leaf in (A) and (C). Note that the juvenile leaf did not develop in (B) and (D).
(E) to (J) Adult leaves of the wild type (E), slm1-1 (F), sgl1-5 (G), and slm1-1 sgl1-5 ([H] and [I]). Close view of the basal region of terminal leaflets of slm1-1 sgl1-5 (I). Note that three terminal leaflets were developed on the distal end of petiole (I). Radial multiple leaves developed at the distal portion of the stem in slm1-1 sgl1-5 (J). TL, terminal leaflet; LL, lateral leaflet.
(K) Adult leaf phenotype of wild-type (top) and sgl1-5 (bottom) plants grown on MS medium supplemented with 50 μM NPA. Control plants were grown on MS medium supplemented with the same concentration of DMSO. The letters a to d indicate variations of compound leaf forms in the wild type and sgl1-5 under NPA treatment.
(L) Number of lateral leaflets and terminal leaflets in the wild type and the mutants. Fifty-day-old plants were used for calculating the leaflet numbers of adult leaves. Means ± se are shown (n = 100).
Bars = 5 mm in (A) to (H), (J), and (K), and 2 mm in (I).
SGL1 Is Partially Involved in Lateral Leaflet Defects in slm1
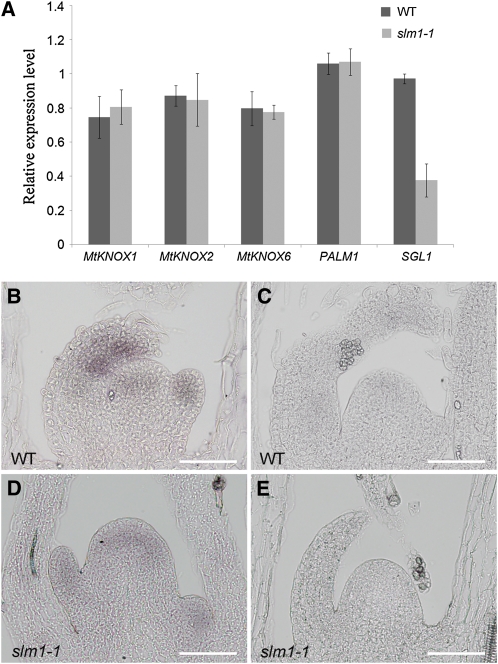
Defects in compound leaf development in slm1-1 suggest that SLM1 is required for the correct formation of both the lateral leaflet and terminal leaflet. Previous studies indicate that multiple genes are involved in leaf development (Champagne et al., 2007; Wang et al., 2008; Chen et al., 2010). Quantitative RT-PCR (qRT-PCR) analysis was performed to determine the expression of M. truncatula genes that have been proposed to regulate this process. These genes included M. truncatula homologs of the Class I KNOX1 homeobox gene family, Mt KNOX1 (SHOOT MERISTEMLESS-like), Mt KNOX6 (SHOOT MERISTEMLESS-like), Mt KNOX2 (KNAT1/BREVIPEDICELLUS-like) (Di Giacomo et al., 2008), M. truncatula PALMATE-LIKE PENTAFOLIATA1 (PALM1) (Chen et al., 2010), and SGL1 (Wang et al., 2008). The expression of SGL1 was suppressed in slm1-1, whereas the expression of other genes remained essentially unchanged (Figure 7A). The spatial localization of SGL1 in slm1-1 during leaf development was further examined by in situ hybridization analysis. mRNA expression of SGL1 was detected in the SAM and young leaf primordia in the wild type (Figure 7B). In slm1-1, the reduction in SGL1 expression supported the qRT-PCR results, illustrating that endogenous SGL1 expression was downregulated (Figure 7D). As a negative control, a sense probe did not give any hybridization signal (Figures 7C and 7E). Since SGL1 regulates lateral leaflet development, these observations indicate that downregulated expression of SGL1 probably contributes to the reduced lateral leaflet number in slm1-1.
Figure 7.
Expression Analysis of Genes Related to Compound Leaf Development in M. truncatula.
(A) Transcript levels of the M. truncatula KNOX1, PALM1, and SGL1 genes in the wild type (WT) and slm1-1. Transcript levels were measured by qRT-PCR using leaf meristems from 6-week-old plants. Means ± se are shown (n = 3).
(B) to (E) In situ hybridization and expression patterns of SGL1 in leaf primordia of the wild type (B) and slm1-1 (D). SGL1 sense probes were used as a negative control in the wild type (C) and slm1-1 (E). Bars = 50 μm.
The Development of the Terminal Leaflet Is Regulated by SLM1 Independently of SGL1
The role of SLM1 in promoting leaflet development was further examined by generating double mutants with the sgl1 mutant (Figures 8A to 8J; see Supplemental Figure 8 online). In slm1-1, the number of lateral leaflets decreased, but the number of terminal leaflets increased (Figures 8B, 8F, and 8L). In sgl1-5, all adult leaves were simple and only terminal leaflets were preserved (Figures 8C and 8G). The slm1-1 sgl1-5 double mutant did not produce lateral leaflets but developed multiple terminal leaflets whose number was similar to that of slm1-1 (Figure 8L). To examine whether local auxin activity gradients are required for the development of leaflets, the gradients were perturbed by growing wild-type and sgl1-5 plants on medium containing 50 μM NPA. The results showed that the phenotype of wild-type plants treated with NPA mimicked the slm1-1 phenotype. The leaf margin of all adult leaves became smooth and various leaflet numbers were noticed (Figure 8K, top panel, a to d). The following leaflet variations were observed: one lateral leaflet degenerated (Figure 8K, top panel, b); three terminal leaflets developed (Figure 8K, top panel, c), and a simple leaf formed (Figure 8K, top panel, d). In addition, the sgl1-5 mutants grown on the same medium developed two (Figure 8K, bottom panel, a) or three (Figure 8K, bottom panel, b and c) terminal leaflets. Taken together, these observations reveal that the local gradients of auxin activity, generated by SLM1, are differentially required for the development of lateral and terminal leaflets in M. truncatula. They also demonstrate that the development of terminal leaflets is independent of SGL1 activity.
DISCUSSION
SLM1 Is the M. truncatula Putative Ortholog of Arabidopsis PIN1
slm1, identified as a recessive mutant by segregation analysis, is defective in leaf and floral development. In this study, three independent SLM1 alleles were found. They showed the same defects in the development of compound leaves and flowers except for fertility. slm1-1 is infertile, and the expression of SLM1 in this allele is completely interrupted by a Tnt1 insertion, indicating that slm1-1 is a null allele. slm1-2 and slm1-3 are point mutations and retain low fertility. The maintenance of partial fertility in slm1-2 and slm1-3 is probably because the SLM1 proteins in the mutants contain partially conserved N-terminal domains (see Supplemental Figure 5 online).
SLM1 is identified as the M. truncatula putative ortholog of Arabidopsis PIN1 by the following lines of evidence. First, some of the defects in slm1 were similar to the classical Arabidopsis pin1 phenotypes, such as triple cotyledons, fused lateral organs, abnormal branching, and smooth leaf margin (Gälweiler et al., 1998; Vernoux et al., 2000; Reinhardt et al., 2003; Hay et al., 2006). Second, the expression patterns of SLM1 revealed by the SLM1 promoter-GUS reporter and in situ hybridization are similar to those of PIN1 in Arabidopsis (Palme and Gälweiler, 1999; Vernoux et al., 2000). Third, Arabidopsis PIN1 is an auxin efflux carrier required for polar auxin transport. Auxin distribution at the meristem of pin1 or NPA-grown plants is diffuse (Benková et al., 2003), which is similar to the auxin distribution pattern in slm1. Fourth, the Arabidopsis ProPIN1:PIN1:GFP construct has been used for PIN1 localization in different species to investigate the initiation of leaf/leaflet primordia and the development of the leaf margin (Benková et al., 2003; Barkoulas et al., 2008; Koenig et al., 2009). The Arabidopsis ProPIN1:PIN1:GFP construct is capable of fully rescuing the slm1 mutant, suggesting conserved function between PIN1 and SLM1. Cross-species complementation of PIN1 was also found between Arabidopsis and C. hirsuta (Barkoulas et al., 2008), indicating that the promoter of PIN1 can be trans-activated in both M. truncatula and C. hirsuta. Fifth, the SLM1 CDS driven by the Arabidopsis PIN1 promoter could complement the pin1 mutant phenotype, suggesting that SLM1 is a functional auxin efflux transporter and can restore polar auxin transport in a heterologous system.
Developmental Domains in the Elaboration of Lateral and Terminal Leaflets in M. truncatula
The development of compound leaves has been documented in several species such as tomato, C. hirsuta, pea, and M. truncatula (Hareven et al., 1996; Hofer et al., 1997; DeMason and Chawla, 2004; Hay and Tsiantis, 2006; Champagne et al., 2007; Barkoulas et al., 2008; Blein et al., 2008; Wang et al., 2008; DeMason and Polowick, 2009; Koenig et al., 2009; Shani et al., 2009; Chen et al., 2010). Several key genes were uncovered by analyses of various mutants with defects in compound leaf formation and development. However, to date, all the mutants identified to be defective in the initiation of lateral leaflets do not affect the formation of the terminal leaflet (Efroni et al., 2010). In our experiments, the mutation in SLM1 reveals a novel phenotype of increased terminal leaflet number, suggesting that a unique mechanism is involved in compound leaf development in M. truncatula.
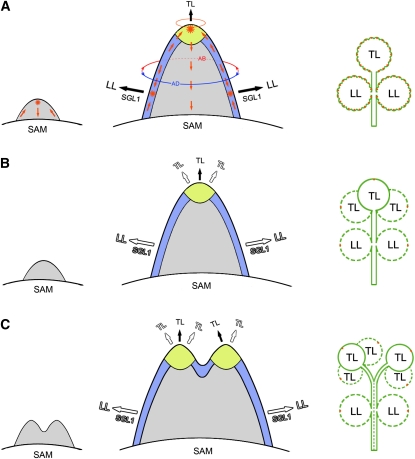
In compound-leafed species, lateral leaflets are considered to be formed from a region at the primordium margin named the marginal blastozone, which has meristematic potential (Hagemann and Gleissberg, 1996; Dengler and Tsukaya, 2001). In addition, the initiation of lateral leaflet primordia is associated with local peaks of auxin response (Barkoulas et al., 2008; DeMason and Polowick, 2009; Koenig et al., 2009). In pea, auxin peaks are also tightly associated with the initiation of pinna primordia that will differentiate into leaflets or tendrils (DeMason and Polowick, 2009). In accordance with these findings, we found that local auxin activity gradients generated by SLM1 facilitate the initiation of lateral leaflets in M. truncatula. In our experiments, auxin activity maxima were also observed at the apex of the common leaf primordium (Figure 5H). Here, we name the apex of the common leaf primordium the terminal zone, in reference to the concept of the marginal blastozone. In slm1, the number of terminal leaflets increased, while the number of lateral leaflets decreased. This observation suggests that the developmental characteristics of lateral leaflet primordia and the terminal leaflet primordium are probably different, implying that distinct developmental domains exist in the elaboration of lateral and terminal leaflets in M. truncatula. This hypothesis is also supported by the ontogenic analysis that the lateral leaflet and terminal leaflet have their own ontogenies with distinct developmental status in M. truncatula.
Previous studies showed that the leaf common primordium developed from an existing radial prepattern of SAM accompanied by the establishment of dorsiventral polarity (Hagemann and Gleissberg, 1996). Then, lateral leaflet primordia initiate from a common leaf primordium that has an existing dorsiventral prepattern (Efroni et al., 2010). By clonal analysis and examination of auxin maxima, a recent study showed that only one to four founder cells of the marginal cell files are involved in lateral leaflet initiation in C. hirsuta (Barkoulas et al., 2008; Efroni et al., 2010). Based on our data and previous research, we propose a model to explain possible differences between the marginal blastozone and terminal zone in M. truncatula. The marginal blastozone has existing dorsiventral polarity, although it appears to function in a manner that is mechanistically similar to SAM. Local auxin maxima mark the founder cells to initiate lateral leaflets in an SGL1-dependent manner (Figure 9A). In slm1, the founder cells cannot be identified in the marginal blastozone without auxin activity maxima, resulting in reduced lateral leaflet number (Figures 9B and 9C). On the other hand, a terminal zone is located at the apex of a common primordium and is probably more likely to resemble the SAM with a radial prepattern than to have dorsiventral polarity. In the wild type, the terminal zone is competent for the formation of multiple terminal leaflet primordia but is prevented from doing so by drainage of auxin into the tip of the terminal zone, which results in a single terminal leaflet primordium (Figure 9A). By contrast, multiple terminal leaf primordia can initiate from the terminal zone in slm1 due to its diffuse auxin distribution, resembling leaf primordia initiated from SAM and resulting in multiple terminal leaflets (Figures 9B and 9C).
Figure 9.
A Proposed Model for Compound Leaf Development Regulated by Auxin Polarity in M. truncatula.
(A) In the wild type, an incipient primordium is initiated at the flanks of the SAM (left). The convergence of epidermal auxin flow (red arrows) forms a maximum of auxin activity (asterisk) at the tip of the primordium and then the auxin is drained through the center of the primordium. During the development of the leaf primordium (middle), dorsiventral polarity (the part of lower circle with blue color: adaxial side; the part of lower circle with red color: abaxial side) is established and the pseudomeristematic region termed blastozone at the margin of the primordium is formed (blue). The auxin flow converges again to form the maxima of auxin activity (asterisk), marking the sites of incipient lateral leaflet (LL) primordia formation at the blastozone. Auxin activity maxima are also formed at the tip of the terminal zone (yellow), which gives rise to the terminal leaflet (TL) primordium. The terminal zone is probably more likely to resemble the SAM with a radial prepattern (the upper circle with orange color) than to develop dorsiventral polarity (middle). The initiation of lateral leaflet primordia is in an SGL1-dependent manner, but terminal leaflet development does not depend on SGL1. The formation of serrations on the leaf margin also correlates with auxin activity maxima (orange spots) at the tip of serrations (right).
(B) and (C) A developmental model of compound leaves in the slm1 mutant. As a result of diffuse auxin distribution in the slm1 mutant, the incipient primordia are able to initiate ([B], left), but fused leaf primordia initiate in some cases ([C], left), resulting in the formation of double terminal zones ([C], middle) and fused petioles ([C], right, broken line). Compared with the wild type, fewer lateral leaflet primordia (0 to 2) develop at the blastozone in an SGL1-dependent manner (empty fonts). However, the terminal zone has the potential to develop one to three terminal leaflet primordia ([B], middle; [C], middle). In addition, the leaf margin of slm1, except the distal serration (orange spots), becomes entire due to the abolished local auxin gradient activity ([B], right; [C], right). The broken line circle in (B) and (C) represents potential leaflets in slm1.
Different Molecular Mechanisms in the Marginal Blastozone and Terminal Zone in IRLC Legumes
Class I KNOX1 genes are expressed in the SAM and involved in acquiring and maintaining SAM activity (Hay and Tsiantis, 2009). The auxin and AS1 pathway repress the expression of the KNOX gene BREVIPEDICELLUS to promote leaf fate (Hay et al., 2006). KNOX1 proteins are involved in compound leaf patterning in a number of species (Bharathan et al., 2002; Hay and Tsiantis, 2006; Barkoulas et al., 2008; Shani et al., 2009) but excluded from leaflet formation in IRLC legumes (Champagne et al., 2007). The expression level of M. truncatula homologs of KNOX1 genes remained essentially unchanged in slm1, indicating that these genes are not involved in the defects in leaf development.
A reduction in LFY expression in the inflorescence apices of the pin1 mutant was reported in Arabidopsis previously, indicating that PIN1 probably regulates LFY expression indirectly via local accumulation of auxin (Vernoux et al., 2000). SGL1, the putative ortholog of LFY in M. truncatula, is required for the initiation of lateral leaflet primordia (Wang et al., 2008). It has been proposed that FLO/LFY may function in place of KNOX1 genes in the regulation of compound leaf development in IRLC legumes (Champagne et al., 2007). Thus, the reduced SGL1 expression in slm1 implies that SGL1 is likely partially responsible for the defects in lateral leaflet development. In addition, our data demonstrate that the downregulated expression of SGL1 is not caused by PALM1, which is a repressor of SGL1 (Chen et al., 2010), since the expression level of PALM1 did not change in slm1. These findings suggest that the expression of SGL1 is sensitive to local auxin activity gradients generated by SLM1 in compound leaf development and also imply that the change in SGL1 expression is probably a secondary effect.
As mentioned above, the marginal blastozone and terminal zone are associated with the formation of lateral leaflets and terminal leaflets, respectively. In the slm1 sgl1 double mutant, the formation of lateral leaflets was fully repressed, but the multiple terminal leaflets were unaffected. This phenotype was also confirmed by the ectopic formation of terminal leaflets in the sgl1 mutant, where auxin transport was perturbed by treatment with auxin transport inhibitors. The expression of SGL1 can be detected in the common leaf primordia at the early stage (this study) and throughout the developing leaflet primordia (Wang et al., 2008). These results suggest that although SGL1 is expressed in both the marginal blastozone and terminal zone, it may function with different interactors in the two domains, resulting in different developmental events. On the other hand, only a few fused leaflets are observed in slm1, implying that the genes that specify leaflet boundaries, such as the M. truncatula homologs of CUC (Blein et al., 2008; Berger et al., 2009), may be employed in both the marginal blastozone and terminal zone for leaflet separation. Taken together, these findings indicate that different molecular mechanisms are involved in these two distinct developmental domains.
Distinct Ontogenies between Distal Serration and Marginal Serration and between Serration and Leaflet
Recent studies have found that regulation of auxin gradients modulates leaf shape in both simple and compound leaf species (Hay et al., 2006; Barkoulas et al., 2008; Koenig et al., 2009; Bilsborough et al., 2011). In our experiments, the investigations on serration formation in the wild type and the observations of abolished serrations in slm1 as well as in NPA-treated plants support the theory that local auxin activity maxima are required for the elaboration of leaf serrations (Hay and Tsiantis, 2006; Nikovics et al., 2006; Scarpella et al., 2006; Bilsborough et al., 2011). Auxin maxima were detected in the tips of initiating serrations in Arabidopsis and C. hirsuta (Hay et al., 2006; Barkoulas et al., 2008). Similar auxin distribution was observed in the development of marginal serrations in M. truncatula. In addition, DR5rev:GFP expression was also detected throughout the leaf margin (Figures 6A and 6D), implying that auxin, not only in the tips of serration, but also in the leaf marginal cells, is probably required for the formation of a proper leaf margin in M. truncatula. Furthermore, the distal serration of leaflets in slm1 and NPA-treated plants was normal (Figures 6E and 6H). Auxin accumulation was also detected at the tip of the distal serration, as assayed by DR5:GUS (Figure 6J), which resembled that of the wild type (Figure 6G). These observations suggest that the developmental processes underlying distal serration and marginal serration are different. The defects were observed in both leaf serrations and leaflets in slm1, suggesting that similar developmental mechanisms may be involved in the formation of the leaf margin and leaflet. However, marginal serrations are completely abolished, while lateral leaflets still can be developed somehow, indicating the different ontogenies between them. Therefore, these observations provide evidence that the development of leaflets is different from that of serrations, although they may share common genetic components (Efroni et al., 2010).
Roles of Auxin in Branching and Phyllotaxy
The elaboration of branches generally comprises two different steps: the initiation of axillary meristems in the leaf axils and the outgrowth of axillary buds, resulting in the formation of shoot branches and inflorescence branches or flowers (Shimizu-Sato and Mori, 2001). The location and timing of axillary meristem initiation is one of the major determinants of plant architecture. Mutants that have defects in initiation of the axillary meristem have been identified in Arabidopsis (Greb et al., 2003), tomato (Schumacher et al., 1999; Schmitz et al., 2002), rice (Komatsu et al., 2003; Li et al., 2003), and maize (Zea mays; Gallavotti et al., 2004, 2010; Satoh-Nagasawa et al., 2006). In addition, vascular differentiation is also related to the formation of the axillary meristem (Schmitz and Theres, 1999; McHale and Koning, 2004; Schmitz and Theres, 2005). For example, inflorescence branches or flowers often fail to develop in the revolute mutant that exhibits defects in the vascular system (Otsuga et al., 2001).
Phyllotaxy refers to the relative arrangements of leaves or inflorescences along the stem following a regular pattern (Reinhardt, 2005). In Arabidopsis, PIN1 responds to the phyllotactic signal (auxin) and creates a phyllotactic pattern (Reinhardt et al., 2003). It has been reported that auxin synthesis or transport is required for the formation of branches (Reinhardt et al., 2003; Ongaro and Leyser, 2008; Balla et al., 2011). The phyllotactic patterning of the inflorescence in Arabidopsis cannot form independently of auxin (Reinhardt et al., 2003). Auxin is also required for the formation of paired spikelets in maize (Gallavotti et al., 2004; Wu and McSteen, 2007). Therefore, as expected, branching at both vegetative and reproductive stages was severely affected in slm1 (Figures 1W and 2Q). The defects of branching in slm1 can be categorized into two types. The first type is the lack of branches in some nodes of slm1. The second is the development of multiple shoot branches/flowers at the distal portions of the stem. Accompanying branching defects, leaf arrangement along the stem is also abnormal in slm1. Similar to the defects of branches, the development of leaves was also abolished in some nodes and multiple leaves developed at the tip of the stem. The similarity of defects in branching and phyllotaxy suggest that similar developmental regulators, such as the auxin/SLM1 module, are involved in these formation processes, even though branches develop from the axillary meristem and leaves develop from the SAM. Previous investigations show that leaves of seed plants likely evolved from branched shoots of early vascular plants (Floyd and Bowman, 2010). Therefore, the similar defects in branches and leaves induced by loss of function of SLM1 may be taken as evidence that leaves show branched shoot-like attributes in M. truncatula, even though leaves are considered to differ from shoots based on the prepattern paradigm (Hagemann and Gleissberg, 1996).
Morphological Defects in slm1 Are Context Dependent
Pleiotropic phenotypes in different organs were observed in the slm1 mutant. Our results show distinct morphological defects in the terminal leaflet and lateral leaflet in slm1. These observations suggest that primordia initiation differs between the lateral leaflet and terminal leaflet in M. truncatula. We propose that the different developmental identities between the lateral leaflet and terminal leaflet are the result of context-specific effects, even though common molecular mechanisms are shared in compound leaf development. The context-specific effects are further emphasized by the observation that, within a single leaflet in slm1, the distal serration is normal, but the marginal serrations are abolished. Such context-dependent effects of gene function are evident in different species. For example, KNOX1 genes are responsible for the leaf shape in naturally lobed Arabidopsis species (Piazza et al., 2010) and for leaflet formation in C. hirsuta (Hay and Tsiantis, 2006). The context-specific trans-activity of KNOX1 is also observed in leaves of Arabidopsis and tomato (Shani et al., 2009). The auxin/PIN1 module was considered to be the conserved mechanism for elaboration of leaves, leaflets, serrations, and branches in plants. In contrast with the ectopic terminal leaflets in slm1, the number of terminal leaflets in the C. hirsuta pin1 mutant did not change, with only a single terminal leaflet being present (Barkoulas et al., 2008). These observations indicate that the terminal leaflet primordium in M. truncatula has a unique developmental mechanism and that the multiple terminal leaflets in slm1 are context-dependent morphological outcomes.
In our results, the petioles of slm1 still have well-defined adaxial and abaxial domains, although the petioles are frequently fused to each other (Figures 1L and 1M). Therefore, the pinnate compound leaves in the wild type turn into nonpeltately palmate leaves instead of peltately palmate leaves in the slm1 sgl1 double mutant (Figures 8H and 8I). To gain a complete picture of compound leaf development, it will be useful to explore this context-specific mechanism for terminal leaflet development. Furthermore, it will be interesting to determine whether various compound leaf forms, including pinnate, nonpeltately palmate, and peltately palmate leaves, in other species are also context dependent and correlated with the auxin/PIN1 module.
In summary, we identified slm1 mutants with pleiotropic phenotypes in different organs that coincided with the loss of auxin response maxima. The functions of SLM1 were characterized and shown to regulate auxin polar transportation and distribution. The data suggest that SLM1 promotes the initiation and separation of aerial organs in a conserved mechanism. However, the phenotype of the terminal leaflet in slm1 mutants is different from that of any other mutants previously identified. This finding implies that the terminal leaflet primordium has its own domain and a unique developmental mechanism exists in M. truncatula. In addition, the development of distal serration and marginal serration occurs in a context-dependent manner. This study expands our knowledge of compound leaf development, especially of the distinct ontogenies that are tightly correlated with the auxin/SLM1 module during plant development.
METHODS
Plant Materials and Growth Conditions
Medicago truncatula ecotype R108 was used for all experiments described in this study. NF3969 (slm1-1), NF1349 (slm1-2), and NF6630 (slm1-3) alleles were identified from a tobacco (Nicotiana tabacum) Tnt1 retrotransposon-tagged mutant collection of M. truncatula (Tadege et al., 2008). A new allele of the sgl1 mutant NF5229 (sgl1-5) that has the Tnt1 insert in the third exon was identified from the same mutant population and confirmed by PCR and RT-PCR. Plants were grown in MetroMix 350 soil mix at 22°C day/20°C night temperature, 16-h-day/8-h-night photoperiod, 70 to 80% relative humidity, and 150 μmol/m2/s light intensity.
Arabidopsis thaliana plants were grown in a growth chamber at 20°C and a daylength of 18 h. The Arabidopsis pin1 allele (GK-051A10-012139, ecotype Columbia-0) was obtained from GABI-Kat.
Plasmids and Plant Transformation
To obtain the SLM1 genomic clone for functional complementation of the slm1 mutant, the 2.3-kb promoter sequence plus 2.9-kb SLM1 genomic sequence was amplified using primers gSLM1-F/gSLM1-R (see Supplemental Table 1 online) and transferred into the pEarleyGate 301 vector (Earley et al., 2006) using the Gateway LR reaction (Invitrogen). To generate the SLM1:GUS construct, a 2.3-kb promoter region of SLM1 was amplified using primers pSLM1-F/pSLM1-R and transferred into the pHGWFS7 vector (Karimi et al., 2002) for gene expression pattern analysis. To clone the SLM1 cDNA for genetic complementation of the Arabidopsis pin1 mutant, the SLM1 CDS from the start codon to stop codon was amplified, and NcoI and BstEII sites were introduced using primers cSLM1-F/cSLM1-R. The PCR product was digested with NcoI and BstEII and ligated into the corresponding site of vector pCAMBIA3301, resulting in pCAMBIA3301-SLM1. Then, the AtPIN1 promoter was amplified from the ProPIN1:PIN1:GFP clone (Benková et al., 2003) using primers pAtPIN1-F/pAtPIN1-R (see Supplemental Table 1 online), which introduced EcoRI and NcoI sites, and cloned into the same sites of pCAMBIA3301-SLM1. To generate the DR5:GUS construct, the DR5 promoter plus GUS gene was amplified from DR5 in the pUC19 construct (Ulmasov et al., 1997) using primers DR5GUS-F/DR5GUS-R (see Supplemental Table 1 online) and transferred into pEarleyGate 301 (Earley et al., 2006) using the Gateway LR reaction (Invitrogen).
The resulting constructs were introduced into the disarmed Agrobacterium tumefaciens EHA105 strain. For stable transformation, leaves of wild-type and slm1-1 were transformed with EHA105 harboring various vectors (Cosson et al., 2006; Crane et al., 2006). The strain EHA105 was also used for Arabidopsis transformation using the floral dip method (Clough and Bent, 1998).
Phylogenetic Analysis
Alignments were performed using ClustalW with default parameters. The phylogenetic tree was constructed using the MEGA2 program (http://www.megasoftware.net/) with 1000 bootstrap replicates.
RNA Extraction, RT-PCR, and qRT-PCR
The shoot meristem tissue of 4-week-old wild-type and mutants was collected for RNA isolation, and qRT-PCR was used to examine the gene expression level. Total RNA isolation, RNA purification, RT-PCR, and qRT-PCR were performed as described by Pang et al. (2009). The primers used for PCR are listed in Supplemental Table1 online. qPCR was performed with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems). SYBR Green was used as the reporter dye. Data were analyzed using the SDS 2.2.1 software (Applied Biosystems).
GUS Staining and NPA Treatment
GUS activities were histochemically detected as described by Jefferson et al. (1987). Images are representative of at least 25 viewed samples stained in three independent experiments. NPA (Sigma-Aldrich) was dissolved in DMSO (Sigma-Aldrich) to a stock concentration of 100 mM and added to Murashige and Skoog (MS) medium to a final concentration of 50 μM. MS medium with the same concentration of DMSO was used as control. Seeds of the wild type and the mutant were germinated on MS medium and then transferred onto MS medium with NPA and DMSO, respectively. The adult leaves were collected for photographing after 40 d of culture.
Histology and Pollen Staining
The petioles of the wild type and slm1 were fixed in glutaraldehyde in phosphate buffer. Dehydrated samples were embedded in LR white resin (London Resin), sectioned into 10- to 20-μm-thick sections using a Leica RM 2255 microtome (Leica Microsystems), and then stained with toluidine blue-O (Sigma-Aldrich) for observation.
To determine pollen viability, flowers of the wild type and slm1 were collected and fixed by Carnoy's fixative for 2 h at room temperature and then stained with Alexander’s solution for 2 h at room temperature (Alexander, 1969). The samples were destained in 10% glycerol for 45 min prior to observation.
Microscopy and Photography
For observation of venation patterns of cotyledons, dissected cotyledons of the wild type and slm1 mutant were fixed and then mounted in chloral hydrate solution according to published protocols (Tsugeki et al., 2009).
For scanning electron microscopy analysis, leaves, floral apical meristems, and organs at different stages were collected, dehydrated, and dried. Scanning electron microscopy analysis was performed as described by Zhao et al. (2010).
Light microscopy was performed using a Nikon SMZ 1500 stereomicroscope (Nikon). For fluorescent imaging, the microscope was equipped with a Leica TCS SP2 AOBS confocal laser scanning microscope using the 488-nm line of an argon laser for the GFP signal, and emission was detected at 510 nm (Leica Microsystems). All experiments are representative of at least 15 observed samples from three independent experiments. Other digital photographs were taken with the Nikon D300 camera.
In Situ Hybridization Analysis
The fragments of 624-bp SLM1 CDS and 427-bp SGL1 CDS were amplified using primer pairs prbSLM1-F/prbSLM1-R and prbSGL1-F/prbSGL1-R (see Supplemental Table 1 online), respectively. The PCR products were labeled with digoxigenin (Digoxigenin-11-UTP; Roche Diagnosis). Fixation/dehydration/paraffin embedding in tissue preparation was performed according to Long’s protocol (http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf). Prehybiridization, hybridization, and washing were conducted on the robotic GenePaint system (Tecan) following the manufacturer’s instructions. Sections were imaged with an Olympus BX 41 microscope using bright-field optics. Images were captured using an Olympus DP71 digital camera and Olympus DP Controller software.
Double Mutant Generation
The slm1-1 and sgl1-5 heterozygous plants were used as parents and crossed with each other to generate F1 plants. F1 plants were genotyped for slm1-1 and sgl1-5 by PCR to identify heterozygotes, which were then selfed to generate F2 plants. The novel slm1-1 sgl1-5 double phenotype was identified in a segregating population and was confirmed by PCR.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SLM1/MtPIN10, AAT48630; AtPIN1, AF089085; AtPIN2, AF086907; AtPIN3, AF087818; AtPIN4, AF087016; AtPIN5, AB005242; AtPIN6, AF087819; AtPIN7, AF087820; AtPIN8, AL391146; ChPIN1, ACH91863; PsPIN1, AY222857; PsPIN2, AB112364; TaPIN1, AY496058; OsPIN1, Q5SMQ9; OsPIN1b, 85542141; OsPIN1c, 75116026; BjPIN1, AJ132363; BjPIN2, AJ249297 and BjPIN3, AJ249298; MtKNOX1, EF128056; MtKNOX2, EF128057; MtKNOX6, EF128061; PALM1, HM038482; and SGL1, AY928184.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Developmental Defects in the slm1-1 Mutant of M. truncatula.
Supplemental Figure 2. Loss of Function in SLM1 Leads to Defects in Fertility.
Supplemental Figure 3. Genetic Segregation Analysis and Genetic Complementation of the slm1-1 Mutant.
Supplemental Figure 4. Phylogenetic Analysis of SLM1/Mt PIN10 and PIN.
Supplemental Figure 5. Alignment of Amino Acid Sequences of At PIN1, Ch PIN1, Ps PIN1, Ta PIN1, Os PIN1, Bj PIN1, and SLM1.
Supplemental Figure 6. Genetic Complementation of the Arabidopsis pin1 Mutant.
Supplemental Figure 7. Expression Profiling of the SLM1 Transcript.
Supplemental Figure 8. Flower Phenotype of the 70-d-Old Plants of the Wild Type and the Mutants Analyzed.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Protein Sequences of SLM1/Mt PIN10 and PIN for Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Qiao Zhao, Frank Hardin, and Amy Mason (all of the Samuel Roberts Noble Foundation) for helpful comments on the manuscript. We thank Jiří Friml (Ghent University, Belgium) for the gift of the plasmid ProPIN1:PIN1:GFP, Thomas Guilfoyle (University of Missouri) for the gift of the plasmid DR5:GUS, and Elison Blancaflor (Noble Foundation) for the gift of the plasmid DR5rev:GFP. We thank Xiaofei Cheng and Jiangqi Wen (both of the Noble Foundation) for mutant screening; Junying Ma and Yuhong Tang (both of the Noble Foundation) for in situ hybridization; Jin Nakashima (Noble Foundation) for help with histology analysis and confocal microscopy; and Li Quan (Noble Foundation) for help with pollen staining. We also thank GABI-Kat (Bielefeld University, Germany) for Arabidopsis pin1 mutant seed stocks. We are grateful to the greenhouse staff for assistance with plant growth and to the Samuel Roberts Noble Electron Microscopy Laboratory (Oklahoma University) for supporting the scanning electron microscopy work. We also thank the National Science Foundation Plant Genome Program (DBI 0703285) for supporting the generation of M. truncatula Tnt1 mutants and the National Science Foundation equipment grant (DBI 0400580) for the confocal microscope. This work was supported by the Samuel Roberts Noble Foundation and the BioEnergy Science Center. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Balla J., Kalousek P., Reinöhl V., Friml J., Procházka S. (2011). Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 65: 571–577 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Smith R.S., Mandel T., Nakayama N., Sauer M., Prusinkiewicz P., Kuhlemeier C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Benlloch R., Navarro C., Beltran J.P., Canas L.A. (2003). Floral development of the model legume Medicago truncatula: Ontogeny studies as a tool to better characterize homeotic mutations. Sex. Plant Reprod. 15: 231–241 [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J.P., Zinder M., Samach A., Eshed Y., Ori N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Bilsborough G.D., Runions A., Barkoulas M., Jenkins H.W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P., Tsiantis M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee J.J., Peer W.A., Murphy A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8: 494–500 [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Kuhlemeier C. (2010). How a plant builds leaves. Plant Cell 22: 1006–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Champagne C.E., Goliber T.E., Wojciechowski M.F., Mei R.W., Townsley B.T., Wang K., Paz M.M., Geeta R., Sinha N.R. (2007). Compound leaf development and evolution in the legumes. Plant Cell 19: 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 107: 10754–10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosson V., Durand P., d’Erfurth I., Kondorosi A., Ratet P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343: 115–127 [DOI] [PubMed] [Google Scholar]
- Crane C., Wright E., Dixon R.A., Wang Z.-Y. (2006). Transgenic Medicago truncatula plants obtained from Agrobacterium tumefaciens-transformed roots and Agrobacterium rhizogenes-transformed hairy roots. Planta 223: 1344–1354 [DOI] [PubMed] [Google Scholar]
- DeMason D.A., Chawla R. (2004). Roles for auxin during morphogenesis of the compound leaves of pea ( Pisum sativum). Planta 218: 435–448 [DOI] [PubMed] [Google Scholar]
- DeMason D.A., Hirsch A.M. (2006). Auxin/gibberellin interactions in pea leaf morphogenesis. Bot. J. Linn. Soc. 150: 45–59 [Google Scholar]
- DeMason D., Polowick P.L. (2009). Patterns of DR5:GUS expression in organs of pea (Pisum sativum). Int. J. Plant Sci. 170: 1–11 [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: Current issues. Int. J. Plant Sci. 162: 459–464 [Google Scholar]
- Di Giacomo E., Sestili F., Iannelli M.A., Testone G., Mariotti D., Frugis G. (2008). Characterization of KNOX genes in Medicago truncatula. Plant Mol. Biol. 67: 135–150 [DOI] [PubMed] [Google Scholar]
- Dong Z.C., Zhao Z., Liu C.W., Luo J.H., Yang J., Huang W.H., Hu X.H., Wang T.L., Luo D. (2005). Floral patterning in Lotus japonicus. Plant Physiol. 137: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S.K., Bowman J.L. (2010). Gene expression patterns in seed plant shoot meristems and leaves: Homoplasy or homology? J. Plant Res. 123: 43–55 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Long J.A., Stanfield S., Yang X., Jackson D., Vollbrecht E., Schmidt R.J. (2010). The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R.B., Ritter M.K., Doebley J.F., Pè M.E., Schmidt R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Goliber T., Kessler S., Chen J.J., Bharathan G., Sinha N. (1999). Genetic, molecular, and morphological analysis of compound leaf development. Curr. Top. Dev. Biol. 43: 259–290 [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152 [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hasson A., Plessis A., Blein T., Adroher B., Grigg S., Tsiantis M., Boudaoud A., Damerval C., Laufs P. (2011). Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2009). A KNOX family TALE. Curr. Opin. Plant Biol. 12: 593–598 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hofer J., Gourlay C., Michael A., Ellis T.H. (2001). Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol. Biol. 45: 387–398 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., Ellis N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Moreau C., Ambrose M., Isaac P., Butcher S., Weller J., Dupin A., Dalmais M., Le Signor C., Bendahmane A., Ellis N. (2009). Tendril-less regulates tendril formation in pea leaves. Plant Cell 21: 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62: 429–441 [DOI] [PubMed] [Google Scholar]
- Kim M., McCormick S., Timmermans M., Sinha N. (2003a). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424: 438–443 [DOI] [PubMed] [Google Scholar]
- Kim M., Pham T., Hamidi A., McCormick S., Kuzoff R.K., Sinha N. (2003b). Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- Koenig D., Bayer E., Kang J., Kuhlemeier C., Sinha N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M. (2004). PIN and AUX/LAX proteins: Their role in auxin accumulation. Trends Plant Sci. 9: 578–582 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- McHale N.A., Koning R.E. (2004). MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16: 1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V., Leyser O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Palme K., Gälweiler L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2: 375–381 [DOI] [PubMed] [Google Scholar]
- Pang Y., et al. (2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 151: 1114–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20: 2223–2228 [DOI] [PubMed] [Google Scholar]
- Reinhardt D. (2005). Regulation of phyllotaxis. Int. J. Dev. Biol. 49: 539–546 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D. (2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Barkoulas M., Tsiantis M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G., Theres K. (1999). Genetic control of branching in Arabidopsis and tomato. Curr. Opin. Plant Biol. 2: 51–55 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Theres K. (2005). Shoot and inflorescence branching. Curr. Opin. Plant Biol. 8: 506–511 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Tillmann E., Carriero F., Fiore C., Cellini F., Theres K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E.L., Frugoli J. (2004). The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol. Genet. Genomics 272: 420–432 [DOI] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Mori H. (2001). Control of outgrowth and dormancy in axillary buds. Plant Physiol. 127: 1405–1413 [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tsugeki R., Ditengou F.A., Sumi Y., Teale W., Palme K., Okada K. (2009). NO VEIN mediates auxin-dependent specification and patterning in the Arabidopsis embryo, shoot, and root. Plant Cell 21: 3133–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Townsley B., Chung K.H., Sinha N. (2007). Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc. Natl. Acad. Sci. USA 104: 15953–15958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Kronenberger J., Grandjean O., Laufs P., Traas J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127: 5157–5165 [DOI] [PubMed] [Google Scholar]
- Wang H., Chen J., Wen J., Tadege M., Li G., Liu Y., Mysore K.S., Ratet P., Chen R. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146: 1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M., Lavin M., Sanderson M. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Wu X.T., McSteen P. (2007). The role of auxin transport during inflorescence development in maize (Zea mays, Poaceae). Am. J. Bot. 94: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Gallego-Giraldo L., Wang H., Zeng Y., Ding S.Y., Chen F., Dixon R.A. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63: 100–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.