Abstract
Much of our knowledge about mammalian evolution comes from examination of dental fossils, because the highly calcified enamel that covers teeth causes them to be among the best-preserved organs. As mammals entered new ecological niches, many changes in tooth number occurred, presumably as adaptations to new diets. For example, in contrast to humans, who have two incisors in each dental quadrant, rodents only have one incisor per quadrant. The rodent incisor, because of its unusual morphogenesis and remarkable stem cell-based continuous growth, presents a quandary for evolutionary biologists, as its origin in the fossil record is difficult to trace, and the genetic regulation of incisor number remains a largely open question. Here, we studied a series of mice carrying mutations in sprouty genes, the protein products of which are antagonists of receptor-tyrosine kinase signaling. In sprouty loss-of-function mutants, splitting of gene expression domains and reduced apoptosis was associated with subdivision of the incisor primordium and a multiplication of its stem cell-containing regions. Interestingly, changes in sprouty gene dosage led to a graded change in incisor number, with progressive decreases in sprouty dosage leading to increasing numbers of teeth. Moreover, the independent development of two incisors in mutants with large decreases in sprouty dosage mimicked the likely condition of rodent ancestors. Together, our findings indicate that altering genetic dosage of an antagonist can recapitulate ancestral dental characters, and that tooth number can be progressively regulated by changing levels of activity of a single signal transduction pathway.
Keywords: FGF signaling, Sprouty genes, Incisor, Mouse
INTRODUCTION
Tooth number is highly variable among mammalian species because of adaptive changes that were selected during evolution. In many cases, evolution led to a reduction of tooth number in extant species compared with their ancestral forms. The laboratory mouse in particular has one of the most reduced mammalian dentitions. Like other rodents, mice have a single pair of ever-growing incisors in the upper and lower jaws (Fig. 1A-C). The ancestral dental formula of placental mammals includes three incisors, indicating that rodents have lost two pairs of upper and lower incisors during evolution. Unlike humans, mice do not replace their teeth, and the mouse incisor is considered to be an unreplaced deciduous tooth (Luckett, 1985; Moss-Salentijn, 1978). In contrast to rodents, the Lagomorpha (rabbits and pika) have a pair of ever-growing incisors as well as a second pair of upper incisors located lingual (nearer the tongue) to the first pair.
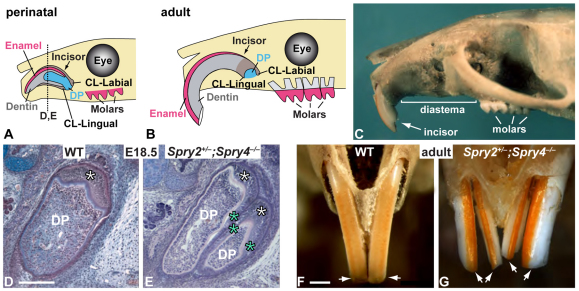
Fig. 1.
Incisor duplication in Spry2+/–;Spry4–/– mice. (A) Schematic of incisor in a perinatal mouse; dotted vertical black line indicates plane of section for D and E. (B) Schematic of incisor in an adult mouse. (C) Adult mouse skull showing the incisor, toothless diastema and molars. (D) Section of wild-type (WT) incisor at E18.5. (E) Section of Spry2+/–;Spry4–/–incisor at E18.5. (F) Adult wild-type incisors in frontal view. (G) Incisor duplication in adult Spry2+/–;Spry4–/– mutant mice. CL, cervical loop; DP, dental papilla. In D and E, white asterisks indicate labial ameloblasts and blue asterisks indicate ectopic ameloblasts. Arrows in F and G point to the incisors. Scale bars: 500 μm.
The mechanism by which the number of incisors decreased during evolution is poorly understood because of a lack of transitional fossils. However, the study of incisor development in wild-type and mutant mice can potentially shed light on these evolutionary events. Mice carrying mutations in Sostdc1 (wise, ectodin) (Munne et al., 2009; Murashima-Suginami et al., 2007), Lrp4 (Ohazama et al., 2008) or Di (Danforth, 1958) have supernumerary upper or lower incisors that are located lingually to the normal incisor, in a configuration that is similar to Lagomorpha upper incisors. The detailed study of incisor development in Sostdc1 mutants indicated that the supernumerary incisor corresponded to a replacement tooth (Munne et al., 2009). Splitting of the incisor placode has been observed in vivo in Sostdc1-follistatin double-null mice, in which there is a partial splitting of the placode leading to bifid incisors. In vitro studies using mandible explants have also shown that the main incisor placode is able to split and give rise to two incisors after activin or noggin treatments (Munne et al., 2010).
The normal mouse upper incisor, which is relatively large compared with the molars, has been documented to form through the fusion of six small primordia at early embryonic stages (Peterkova et al., 1993; Peterkova et al., 2006). Thus, previous studies suggest several potential mechanisms by which supernumerary incisors might arise in mice: failure of integration of the ancestral dental primordia (Hovorakova et al., 2011), development of a replacement tooth (Munne et al., 2009), splitting of a large placode into smaller elements (Munne et al., 2010) or development of a supernumerary germ (Sofaer, 1969).
Mammalian tooth morphogenesis is controlled by interactions between the oral epithelium and the neural crest-derived ectomesenchyme, and these interactions are mediated by signaling pathways, including the receptor-tyrosine kinase (RTK) pathway initiated by the secreted fibroblast growth factors (FGFs) (Pispa and Thesleff, 2003; Cobourne and Sharp, 2010). The sprouty (Spry) family of genes encodes proteins that are intracellular inhibitors of RTK signaling (Guy et al., 2003; Kim and Bar-Sagi, 2004). We have previously reported that mice carrying mutations in sprouty genes have dental abnormalities; in particular, Spry2+/–;Spry4–/– mice have abnormal enamel deposition on the lingual surface of the incisors in their lower jaws (Klein et al., 2006; Klein et al., 2008; Boran et al., 2009; Peterkova et al., 2009; Caton et al., 2009). Here, we focus on the upper jaws of Spry2+/–;Spry4–/– mice and show that loss of sprouty genes can increase the number of incisors by a subdivision of the single embryonic incisor anlage preceded by the subdivision of its gene expression domains. We also studied a dosage series of sprouty mutants to assess how fine-tuning levels of signaling affects the development of incisors and found that progressive changes in incisor number can occur as sprouty gene dosage is changed. The phenotype in sprouty loss-of-function mutants corresponds to the morphology of rodent ancestors, leading us to propose a new model for modification of incisor development during evolution.
MATERIALS AND METHODS
Mice
Mouse lines carrying mutant alleles of Spry1 (Basson et al., 2005), Spry2 (Shim et al., 2005), Spry4 (Klein et al., 2008) and Fgf10 (Min et al., 1998) were maintained on a mixed genetic background and genotyped as reported. For the study of dental development, we used age-matched CD1-K14-EGFP animals (Vaezi et al., 2002). In all other experiments, CD1 mice were used as wild-type controls. Noon of the day when a vaginal plug was detected was considered as embryonic day (E) 0.5. Because the Spry2+/–;Spry4–/– incisor duplication was not completely penetrant, we examined a large number of embryos (at least 20 mutants and 20 wild types between E12.5 and E14.5, and at least 12 mutants and 12 wild types for each time point between E15.5 and E17.5), and we selected the most representative for the figures on the study of dental development. For all other genotypes, at least five embryos per genotype were examined.
The pregnant mice were killed by carbon dioxide exposure followed by cervical dislocation. For 3D reconstructions, wet body weight of the fetuses was determined immediately after their removal from the uterus to be able to correctly compare age- and weight-matched embryos (Peterka et al., 2002).
The pK14-Spry4 construct was made by replacing the wise cDNA of pK14-wise (Ahn et al., 2010) with the mouse Spry4 cDNA (904 bases). The 3.8-kb K14-Spry4 fragment was gel purified and injected into a CBA/J × C57BL/10J one-cell zygote. A total of 40 K14-Spry4 embryos between E14.5 and E18.5 were examined.
Epithelial-mesenchymal dissociation
Fresh embryos were dissected in Hanks' medium, and the developing incisor was isolated and incubated in dispase at 37°C for 30 minutes to 7 hours depending on the age of the embryo; the longest incubation periods were used only for late embryos with mineralized tissues. After dispase treatment, which disrupts the adhesion between epithelium and mesenchyme, the samples were transferred to PBS and the epithelium was carefully pulled out of the mesenchyme. The epithelium was then fixed in 4% paraformaldehyde. We have previously used this method, which enables direct visualization of the tooth germ from all sides, and have found that the morphologies observed with this method were consistent with 3D reconstructions (Prochazka et al., 2010).
3D reconstructions of the dental and adjacent epithelium
Mouse embryonic heads at E12.5-18.5 were fixed in Bouin-Hollande fluid and embedded in paraffin. Sixty heads were cut in series of 7 μm frontal sections. The sections were stained with Hematoxylin-Eosin and Alcian Blue.
The specimens were ranked according to the chronological age, refined by body weight, of embryos. A succession of gradual stages of incisor development was determined on histological sections. For 3D reconstructions of the epithelium of the developing incisor, we selected ten representative series of routinely stained frontal paraffin sections of Spry2+/–;Spry4–/– mouse heads at E12.5-17.5 and seven series of wild-type mouse heads. In all these specimens, the epithelium of the right and/or left upper incisor area was reconstructed.
Contours of the dental and adjacent oral epithelium were drawn from frontal histological sections using a Leica DMLB microscope (Leica Microsystems, Wetzlar, Germany) equipped with a drawing chamber at a magnification of ×320. Apoptotic cells and bodies in the epithelium were recorded in the drawings. The digitalization of the serial drawings and correlation of successive images have previously been described (Lesot et al., 1996). Three-dimensional images were generated using VG Studio Max 2.0 software (VG Studio Max, Heidelberg, Germany).
Quantitative evaluation of proliferation and apoptosis
The area of dental epithelium was measured on each fifth frontal serial section using ImageJ (http://rsb.info.nih.gov/ij/). The measured area was delimited by the basement membrane, the oral surface of the epithelium, and the places where the thickness of the dental epithelium decreased to the thickness of the medially and laterally adjacent oral epithelium. The volume of dental epithelium was calculated as the mean of the area of dental epithelium multiplied by the length of the dental epithelium (corresponding to the number of sections with the thickened incisor epithelium multiplied by the section thickness 7 μm). The mitotic index was calculated as the percentage of cells in mitosis (from early metaphase to early telophase). The apoptotic rate was calculated as the number of observed apoptotic cells and bodies per 100 cells. For details, see Peterkova et al. (Peterkova et al., 2009).
In situ hybridization
After harvesting, embryos were fixed overnight in 4% paraformaldehyde. RNA in situ hybridizations were performed either on whole-mount embryos or on paraffin sections using standard protocols (Klein et al., 2006; Klein et al., 2008). Antisense, digoxigenin-labeled RNA probes were generated from full length Fgf4 and Shh cDNAs (kind gifts from Drs Gail Martin, UCSF, San Francisco, CA, USA, and Andrew McMahon, Harvard University, Cambridge, MA, USA, respectively).
Quantitative RT-PCR
RNA was isolated from the incisor developing area at E12.5 and E13.5 using a Qiagen RNeasy Mini Kit. Expression was assessed quantitatively using GoTaq qPCR Master Mix (Promega) in a Mastercycler Realplex (Eppendorf), and transcript levels were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Results are expressed as normalized expression values (equal to 2–ΔCt). Primer sequences used for RT-qPCR are listed in Table S1 in the supplementary material.
Study of label-retaining cells
We crossed Spry2+/–;Spry4–/– mice with K5tTA;H2B-GFP mice, in which expression of a doxycycline-repressible H2B-GFP transgene is controlled in the epithelium by the keratin 5 (K5; Krt5 – Mouse Genome Informatics) promoter. All epithelial cells were uniformly labeled by GFP expression before doxycycline was added. In the presence of doxycycline, expression of H2B-GFP was repressed and, subsequently, the H2B-GFP label was diluted out in proliferating cells (Tumbar et al., 2004), resulting in isolated populations of label-retaining cells (LRCs; see Fig. S2 in the supplementary material). Mice carrying both K5tTA (Diamond et al., 2000) and H2B-GFP (Tumbar et al., 2004) alleles were genotyped by observation of GFP-fluorescent skin using a fluorescence dissecting microscope. Adult mice were fed a doxycycline-containing diet for 6-8 weeks. Dissection of the incisor and observations were made under a Leica MZ16F dissecting microscope equipped with a Leica EL600 light source.
Scanning electron microscopy
A Hitachi S-4300SE/N environmental scanning electron microscope (ESEM; Hitachi America, Pleasanton, CA, USA) operating at 20 kV under high-resolution image mode was used to image incisors. Prior to mounting on a metal stub for imaging, samples were embedded in epoxy resin and sectioned using an ultra-microtome (Leica, LLC).
RESULTS
Spry2+/–;Spry4–/– mice have supernumerary incisors
The perinatal incisor (Fig. 1A), which appears as a tooth bud around embryonic day (E) 12, is unerupted. After eruption in the postnatal period, the incisor continues to grow for the rest of the animal's life because of the presence of stem cells in the cervical loops (CLs), the epithelial structures at the apical end of the incisors (Fig. 1B). Whereas the wild-type mouse always has a single upper incisor on each side of the jaw arch (Fig. 1C,D,F), approximately half of the Spry2+/–;Spry4–/– mice had two upper incisors per half arch instead of one (Fig. 1E,G). The presence of two incisors per quadrant, each with enamel on both lingual and labial surfaces, is reminiscent of the incisor configuration of most non-rodent mammals.
The duplicated incisors in the mutant were located side by side and shared the same bone socket, suggesting that the development of the two incisors was not independent. Examination of embryonic sections at E18.5 allowed us to look at the relationship between the dental epithelium, called the enamel organ, and the ensheathed dental mesenchyme (papilla). In the Spry2+/–;Spry4–/– mutants, we observed that the two incisors shared the same enamel organ split by an epithelial septum, each part accommodating a distinct dental papilla (prospective pulp). The septum was formed by a duplication of the layer of inner dental epithelium with interposed stratum intermedium cells, which participate in ameloblast differentiation.
In wild-type incisors, enamel-producing ameloblasts were only located on the labial (nearer the lip) side (white asterisk, Fig. 1D). In the mutant, the layer of ameloblasts also extended to the septum between the two incisors and to the lateral aspect of the lateral incisor (blue asterisks, Fig. 1E), although this was not reflected in the adult enamel pattern (Fig. 1G). We have previously reported that in lower incisors of Spry2+/–;Spry4–/– mice, enamel is abnormally present on the lingual surface (Klein et al., 2008). We performed scanning electron microscopy (SEM) and found that Spry2+/–;Spry4–/– mice also had lingual enamel in their upper incisors, and this enamel was normally organized (see Fig. S1 in the supplementary material).
Incisor growth is associated with the presence of stem cell niches in the CLs (Harada et al., 1999; Seidel et al., 2010; Parsa et al., 2010). We next set out to study the stem cell niches in the upper incisors of Spry2+/–;Spry4–/– mutants, in order to determine whether the duplicated incisors in the mutants grow from the same stem cell population. We took advantage of label retention, which is a property of many stem cell populations and can serve as an indication of the presence of slow-cycling stem cells (Fuchs, 2009); we have previously used this approach to identify slowly cycling stem cells in the wild-type incisor (Seidel et al., 2010). We crossed Spry2+/–;Spry4–/– mice with K5tTA;H2B-GFP mice and obtained LRCs as described previously (Seidel et al., 2010). In the wild-type adult mouse, the lingual CL contained one population of LRCs (see Fig. S2 in the supplementary material). In the mutant, the lingual part of the adult incisors contained a total of three stem cell niches (see Fig. S2 in the supplementary material). This result indicates that the maintenance of continuous growth of the two incisors was supported by multiple stem cell niches.
Incisor duplications in sprouty mutants arise following subdivision of the epithelium
To understand the origins of the incisor duplication, we investigated the development of the wild-type and Spry2+/–;Spry4–/– mutant upper incisors between E12.5 and E17.5 using a combined analysis of dissociated epithelia, histological sections and 3D reconstructions. Tooth germs from closely staged embryos were dissected and the incisor dental epithelium was removed using dispase treatment (Fig. 2A) in order to enable visualization of the epithelium from all sides in a large sample of specimens. The 3D reconstructions documented detailed morphology of the incisor epithelium at different developmental stages.
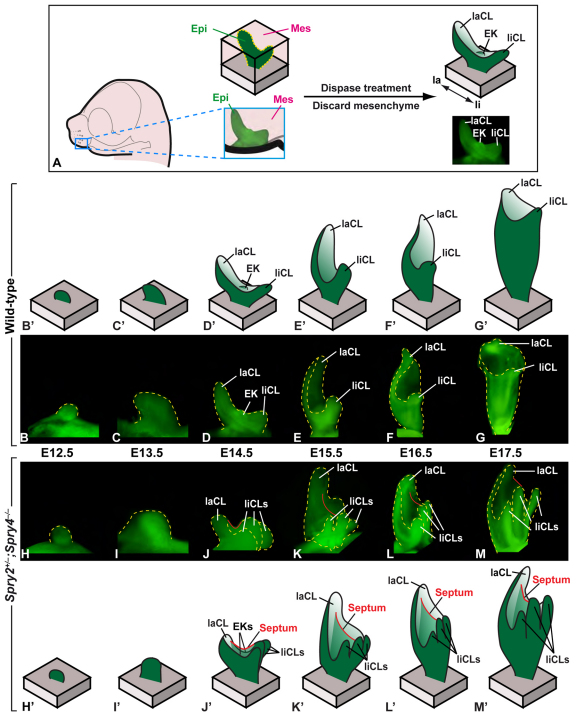
Fig. 2.
Upper incisor development in wild-type and Spry2+/–;Spry4–/– embryos. (A) Schematic of the epithelium-mesenchyme dissociation method. (B-G) Photographs of dissociated incisor epithelia in wild-type embryos from E12.5 to E17.5. (B′-G′) Schematic of upper incisor development in wild-type embryos. (H-M) Photographs of dissociated incisor epithelia in Spry2+/–;Spry4–/– embryos from E12.5 to E17.5. (H′-M′) Schematic representation of upper incisor development in Spry2+/–;Spry4–/– embryos. Yellow dashed lines outline the incisor epithelium; red line indicates position of septum. EK, enamel knot; Epi, epithelium; Mes, mesenchyme; la, labial; li, lingual; laCL, labial cervical loop; liCL, lingual cervical loop.
At E12.5, the upper incisors of both wild-type and mutant embryos were at the early bud stage (Fig. 2B,H), and a well-formed bud was present at E13.5 (Fig. 2C,I). The cap stage started one day later, at E14.5 (Fig. 2D,J). The wild-type incisor cap normally contains the labial and lingual CLs and one enamel knot (Fig. 2D) (Kieffer et al., 1999); the latter is a transient signaling center that controls the morphology of the tooth by directing the growth of surrounding epithelium and mesenchyme (Vaahtokari et al., 1996). In contrast to the wild type, the Spry2+/–;Spry4–/– incisor epithelium displayed a multiplication of the lingual CLs, such that three CLs were typically observed in the mutants (Fig. 2J). Additionally, in Spry2+/–;Spry4–/– embryos we observed two enamel knots instead of one, as well as a septum running from the labial CL to the middle CL located on the lingual side (Fig. 2J). This septum (red line in Fig. 2J) resulted from the splitting of the epithelium into two compartments, each one with its own enamel knot. At E15.5, the dental epithelium grew deeper into the mesenchyme, leading to a more elongated structure with the CLs on the top. The septum observed at E14.5 persisted as an epithelial demarcation between the two mesenchymal compartments at E15.5-17.5 (Fig. 2E-G,K-M).
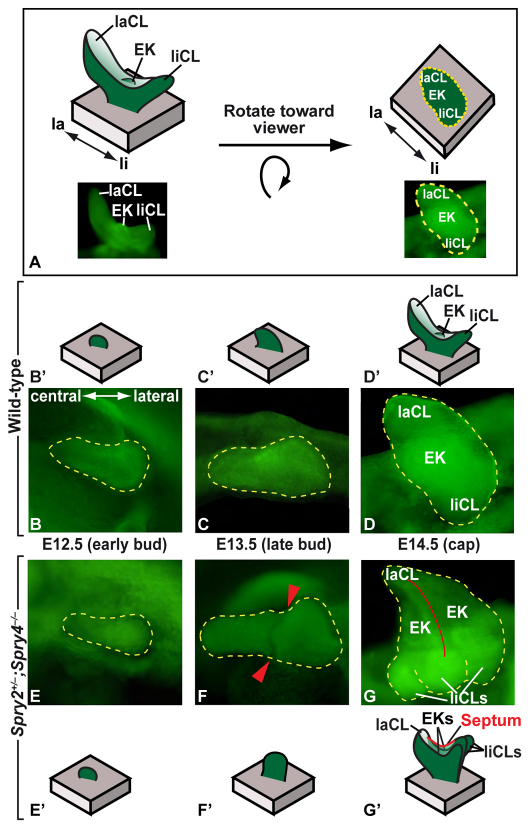
Further examination by rotating the dissected epithelia (Fig. 3A) and analyzing 3D reconstructions (Fig. 4A-F) revealed the morphological origins of the incisor duplication. At E12.5, the wild-type and mutant incisor epithelia appeared similar (Fig. 3B,E, Fig. 4A,D). The first obvious morphological differences were observed at E13.5, at which point the late bud stage wild-type epithelium had a smooth-appearing surface (Fig. 3C, Fig. 4B), whereas a notch could be seen in mutant epithelium (Fig. 3F, Fig. 4E). A histological section through the incisor bud at E13.5 showed that this notch was associated with an interruption of the regular arrangement of the inner epithelial cells that suggested an internal boundary between the cell population in the medial and lateral parts of the incisor bud (see Fig. S3 in the supplementary material). The location of the notch at E13.5 was the same as the future location of the septum, which was first visible at E14.5 (Fig. 3G) and persisted at later stages, dividing the papilla into two compartments (see Fig. S3 in the supplementary material). Thus, the earliest morphological change in the mutant embryos was a midline notch, first detectable at E13.5, and the two incisors observed in the jaw half of adult Spry2+/–;Spry4–/– mice resulted from the development of two compartments within the budding dental epithelium, followed by formation of an epithelial septum that split the mesenchyme inside the enamel organ into two papillae.
Fig. 3.
Septum formation during incisor development in Spry2+/–;Spry4–/– embryos. (A) Schematic of epithelium re-orientation. (B-D) Photographs of epithelia after re-orientation of wild-type embryos at E12.5, E13.5 and E14.5. (E-G) Photographs of epithelia of Spry2+/–;Spry4–/– embryos at E12.5, E13.5 and E14.5. (B′-G′) Schematics of epithelial development. Yellow dashed lines outline the incisor; red arrowheads in F point to notch; red line in G indicates septum. la, labial; li, lingual; EK, enamel knot; laCL, labial cervical loop; liCL, lingual cervical loop.
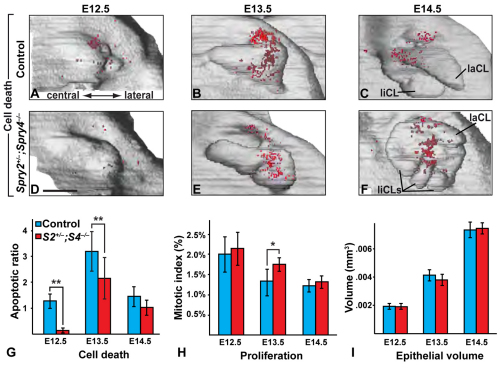
Fig. 4.
Cell death and proliferation in incisors of control and Spry2+/–;Spry4–/– embryos. (A-F) Three-dimensional reconstructions viewed from mesenchymal aspect and location of apoptotic elements (red dots) in epithelia of control (A-C) and Spry2+/–;Spry4–/– embryos (D-F) at E12.5 (A,D), E13.5 (B,E) and E14.5 (C,F). Scale bar: 100 μm. (G-I) Apoptotic rate (G), mitotic index (H) and epithelial volume (I) in control and Spry2+/–;Spry4–/– embryos. Mean±s.e.m. are plotted. *P<0.05, **P<0.01.
Decreased cell death is associated with abnormal subdivision of the epithelium
To understand the cellular basis for the incisor duplication, we next analyzed epithelial cell proliferation, cell death and tissue volume. Apoptotic cells and bodies were labeled in drawings of the dental epithelium realized from frontal histological sections, and their distribution was represented on 3D reconstructions of the dental epithelium (Fig. 4A-F). The proportion of apoptotic elements and proliferative cells was calculated from sections. Although the size of wild-type and Spry2+/–;Spry4–/– incisors did not differ (Fig. 4I) at E12.5, very little to no cell death, distributed throughout the epithelium, was detected in the mutant, representing a significant decrease from the wild type, which had apoptotic elements mostly present antero-labially in the dental epithelium (Fig. 4A,D,G). At E13.5, apoptotic elements were present in the central region in Spry2+/–;Spry4–/– incisors as in the wild type, but the number of apoptotic elements was still significantly lower in mutants compared with wild-type embryos (Fig. 4B,E,G). At E14.5, the amount of cell death was similar in wild-type and Spry2+/–;Spry4–/– mice (Fig. 4G), but the location of dying cells differed, as apoptosis was specifically concentrated along the central axis of the mutant incisor (Fig. 4C,F), corresponding to the area where the epithelial septum between the two duplicated papillae appears. This concentration along the midline was similar to the distribution of apoptotic cells one day earlier in wild-type mice and might reflect a failed attempt by the left and right parts of the incisor to fuse in the Spry2+/–;Spry4–/– embryos. Interestingly, the only difference in proliferation was a small but significant increase in the mutant compared with wild type at E13.5 (Fig. 4H). Although the overall size of the incisor epithelium was similar at all three time points examined (Fig. 4I), the mutant epithelium was larger when viewed from the mesenchymal aspect, whereas the wild-type epithelium was submerged more deeply into the mesenchyme. Thus, the septum that separated the two dental papillae was correlated with a delay in the onset of cell death from E12.5 to E13.5, increased proliferation at E13.5 and a change in the location of apoptotic cells at E14.5.
Splitting of gene expression domains foreshadows the incisor duplication
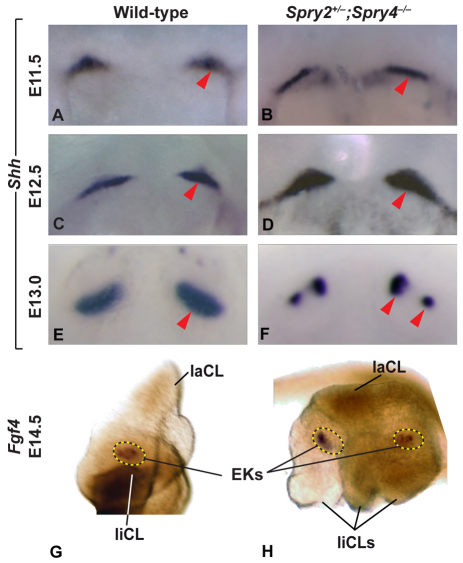
To determine how loss of sprouty genes might affect incisor bud development, we assessed the expression pattern of sonic hedgehog (Shh), an early marker of tooth development. Initially, Shh is expressed in a single domain in the incisor area (Pispa and Thesleff, 2003). At E11.5, Shh expression appeared similar in wild-type and mutant specimens (Fig. 5A,B). At E12.5, the Shh expression domain was similar in wild-type and mutant embryos (Fig. 5C,D), but the Shh expression staining was more intense in the mutants, and this finding was confirmed by qPCR analyses (see Fig. S4 in the supplementary material). At E13.0, a time point at which morphological differences were not detected in the mutant, the domain of Shh expression was already widely split in the mutant mice (Fig. 5F). In wild-type embryos, a transient splitting of the early Shh expression domain can be observed in the lower as well as upper mouse incisor (Hovorakova et al., 2011), but this is followed by the presence of a single Shh domain at E13.5. However, in the present study, the split domain of Shh expression was identified in mutants but not controls at E13.5 (data not shown). These two separate domains of Shh expression in the mutant suggest that the decision to split into two teeth occurs at or before this time point.
Fig. 5.
In situ hybridization in incisors of wild-type and Spry2+/–;Spry4–/– embryos. (A-F) Whole-mount RNA in situ hybridization for Shh in wild-type (A,C,E) and Spry2+/–;Spry4–/– (B,D,F) embryos at E11.5, E12.5 and E13.0. Red arrowheads point to the left incisor expression domain. (G,H) Whole-mount RNA in situ hybridization for Fgf4 in dissected incisor epithelia viewed from mesenchymal side of wild-type (G) and Spry2+/–;Spry4–/– (H) embryos at E14.5. EK, enamel knot; laCL, labial cervical loop; liCL, lingual cervical loop.
In our morphological analysis, we identified two raised structures as the likely enamel knots at E14.5 (Fig. 3G). We next assessed the expression of Fgf4 and Shh, which are markers of enamel knots, to determine whether each compartment in the mutant has a bona fide, distinct enamel knot. In situ hybridization on dissected epithelia showed one domain of Fgf4 and Shh expression in the wild type, located in the center of the incisor, whereas in the mutant, two Fgf4 and Shh expression domains representing the two enamel knots were present, one on each side of the septum (Fig. 5G,H; data not shown). Thus, the mutant incisor had two distinct enamel knots at E14.5.
Loss of sprouty genes causes hypersensitivity to Fgf10 in the dental epithelium
A previous study suggested that the lack of sprouty function leads to hypersensitivity of dental epithelial buds in the molar region to FGF signaling from the mesenchyme (Klein et al., 2006). To test for such hypersensitivity in the incisor of Spry2+/–;Spry4–/– embryos, we measured the relative expression of Etv5, a marker of the FGF pathway (Roehl and Nüsslein-Volhard, 2001). The observed relative increase of Etv5 at E12.5 and E13.5 indicated an upregulation of FGF signaling in the Spry2+/–;Spry4–/– mutants (see Fig. S4 in the supplementary material). We have previously shown that, in both the early incisor and molar, Spry1 is expressed in epithelium and mesenchyme, Spry2 in epithelium and Spry4 exclusively in the mesenchyme (Klein et al., 2006; Klein et al., 2008). qPCR analysis confirmed that Spry2+/–;Spry4–/– mutants had half the dosage of Spry2 and no expression of Spry4 (see Fig. S4A,B in the supplementary material).
Fgf10 is expressed during incisor development from early stages to adulthood (Kettunen et al., 2000). Its expression around CLs regulates epithelial stem cell survival, which is essential for continuous incisor growth (Harada et al., 2002; Yokohama-Tamaki et al., 2006). We therefore produced Spry2+/–;Spry4–/– mice lacking one Fgf10 allele to test whether decreased FGF dosage would rescue the incisor duplication phenotype. Whereas an incisor duplication was present in 43% of Spry2+/–;Spry4–/– mice, compound mutants with a Fgf10 null allele had a duplicated incisor in only 17% of specimens (see Fig. S5 in the supplementary material). Thus, a decrease in Fgf10 dosage caused a decreased penetrance of the duplication, suggesting that the incisor duplication observed in Spry2+/–;Spry4–/– mice resulted from hypersensitivity to FGF signaling. Interestingly, upregulation of Fgf10 was detected by qPCR at E12.5 but not at E13.5 in the mutant, whereas Fgf9 levels were unaltered (see Fig. S4A,B in the supplementary material), illustrating the importance of the proper dose and timing of activity of FGF signaling. This finding was reminiscent of the upregulation in Fgf10 expression that we had previously observed in incisors from the lower mandibles of Spry2+/–;Spry4–/– embryos (Klein et al., 2008).
Modulation of sprouty levels can lead to a phenotypic gradation in tooth number
The molecular regulation of tooth number is an important question in evolutionary biology (reviewed by Cobourne and Sharpe, 2010), and the phenotype in the Spry2+/–;Spry4–/– mutant, which lacks three sprouty alleles, was particularly intriguing in that the incisor duplication was present in the upper but not lower jaw. This suggested to us that a subtle dosage response might exist between the level of FGF signaling and the number of teeth. Therefore, we produced a mouse with increased sprouty dosage in which Spry4 was ectopically expressed in the epithelium under the control of the human keratin 14 promoter (K14-Spry4GOF). There was significant variability among these embryos (see Fig. S6 in the supplementary material), but it was interesting to note that in one K14-Spry4GOF embryo at E15.5, we observed fusion of left and right lower incisors along the midline (Fig. 6A-A′); we also observed in these mutants a case of lower incisor duplications (see Fig. S6 in the supplementary material). These data indicate that, in certain cases, increasing the expression of a signaling antagonist can lead to decreases in the number of teeth.
Fig. 6.
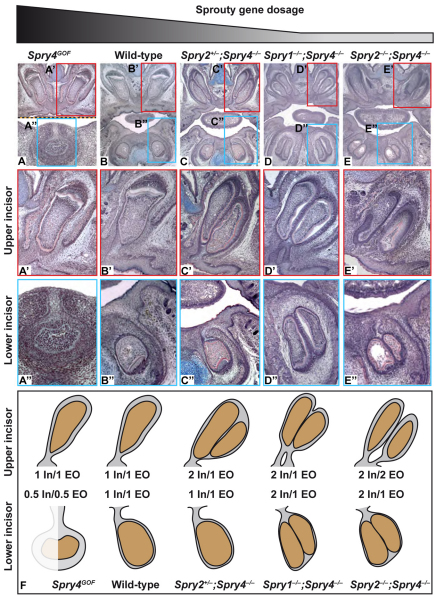
Incisor phenotypes in a Sprouty gene dosage series. (A-E) Frontal histological sections of embryonic head at E15.5 (A) and E18.5 (B-E) showing the phenotype of upper and lower incisors in K14-Spry4 (Spry4GOF) (A), wild-type (B), Spry2+/–;Spry4–/– (C), Spry1–/–;Spry4–/– (D) and Spry2–/–;Spry4–/– (E) embryos. (A′-E′) High magnification of upper incisor. (A′-E′) High magnification of lower incisor. (F) Schematic with indication of the number of incisors and enamel organs for each genotype. EO, enamel organ(s); In, incisor(s). Dotted line in A indicates that this composite image is composed of upper and lower jaw images from different sections.
We next produced a series of mice lacking different combinations of sprouty genes in order to progressively change the level of RTK signaling in the developing tooth. As described above, wild-type embryos have a single incisor in each dental quadrant (Fig. 6B-B′), and Spry2+/–;Spry4–/– embryos had two upper incisors and one lower incisor (Fig. 6C-C′). Further decreases in sprouty dosage in Spry1–/–;Spry4–/– and Spry2–/–;Spry4–/– embryos led to two upper and two lower incisors in each quadrant (Fig. 6D-E′). Interestingly, the phenotypic gradation observed with the decrease of sprouty dosage involved not only the number of incisors but also their inter-relationship. In Spry2+/–;Spry4–/– embryos, the two upper incisors were still in the same enamel organ (Fig. 6C′). In Spry1–/–;Spry4–/– embryos, the two upper incisors were in the same enamel organ, but the enamel organ had two stalks joining the enamel organ with the oral epithelium (Fig. 6D′). In Spry2–/–;Spry4–/– embryos, we identified specimens in which each duplicated upper incisor developed in its own enamel organ (Fig. 6E′; data summarized in Fig. 6F). A similar gradation from one to two teeth was seen in the lower incisors (Fig. 6B′,D′,E′), but in the case of the lower jaw, even in the double-null embryos the two incisors still developed in the same enamel organ. These data indicate that the tendency to increase tooth number in the lower jaw is less sensitive to sprouty dosage than in the upper jaw, as it occurred in the lower jaw only when four sprouty alleles were deleted versus three for the upper jaw (Fig. 6F). Overall, in both upper and lower incisors, the decrease of sprouty dosage led to progressively more independent teeth in both upper and lower jaws.
DISCUSSION
We report that modification of sprouty gene dosage regulates the number of incisors. As sprouty gene dosage was increased or decreased, a spectrum of phenotypes was observed. These ranged from one central incisor to two separate incisors in two different enamel organs located side-by-side, which resembled the dentition of most non-rodent mammals. Our study of dental development in Spry2+/–;Spry4–/– embryos indicates that sprouty genes control the integrity of the Shh expression domain. In these mutants, the duplication resulted from subdivision of the incisor enamel organ, which initially had a similar size to that of the control embryos, by an epithelial septum invading and splitting the mesenchyme into two papillae. This morphological splitting was associated with the persistent splitting of the Shh expression domain, with a delay in the onset of cell death at early stages and a later change in the location of apoptosis and increased proliferation. We also found that the sprouty genes antagonize the FGF pathway during regulation of incisor number by producing Spry2+/–;Spry4–/– mutants in which one allele of Fgf10 was inactivated.
The duplicated incisors belong to the same generation and are not replacement teeth
The supernumerary incisors reported in mutant mice have most often been located on the lingual side of the normal incisor (Danforth, 1958; Murashima-Suginami et al., 2007; Ohazama et al., 2008; Munne et al., 2009). The detailed analysis of one of these mutants (Sostdc1) showed that the supernumerary incisor developed on the lingual side of the normal one, and this tooth was considered to belong to a different tooth generation, corresponding to the revival of either the replacement incisor (Munne et al., 2009) or of a rudimentary lacteal incisor (Murashima-Suginami et al., 2007). The supernumerary incisor of Lrp4-null mice has been considered as having the same origin as the supernumerary incisor of Sostdc1 mutants (Ohazama et al., 2010). In the sprouty mutants reported here, the concomitant developmental origin and the side-by-side organization of the two incisors indicate that they must be considered as the same tooth generation. To our knowledge, the only adult mutants reported to have an incisor phenotype similar to the Spry2+/–;Spry4–/– mice are Eda-tabby (Sofaer, 1969) and the Pax6-Sey (small eye) homozygous mutants (Kaufman et al., 1995). In Eda mutants, only very rare cases of side-by-side upper incisor duplication have been reported (Sofaer, 1969); we did not detect this phenotype during our observation of 60 heterozygous and 80 null Eda mutants in previous studies (Charles et al., 2009a; Charles et al., 2009b). Pax6 mutant rats, as Pax6-Sey mice, also exhibit supernumerary incisors, and it has been suggested that these incisors result from the total lack of fusion between two components of the epithelial incisor anlage separated by a cleft lip (Kriangkrai et al., 2006) and not from the secondary splitting of the incisor anlage as we have observed. Splitting of the incisor anlage might occur in nestin-cre/K5-lacZfl-smad7 mice, but the splitting seemed to be a transient event, as the two incisors were fused at their base (Klopcic et al., 2007). The β-catΔPrx/lacZ mice have two incisors belonging to the same generation, but in these mice only the lower incisors have been reported to be affected (Fujimori et al., 2010). Thus, the Spry2+/–;Spry4–/– mice are the first in which supernumerary incisor development has been clearly shown in vivo to result from the secondary splitting of the incisor primordium and Shh expression domain after normal development at initial stages.
Splitting of the incisor primordium precedes the incisor duplication
In the sprouty dosage series reported here, we showed that the splitting of an anlage does not always occur as a sudden change but rather can also develop as a progressive process. The stable development of two incisors in the same enamel organ and the two independent incisors seen in Spry2+/–;Spry4–/– and Spry2–/–;Spry4–/– mice, respectively, might be facilitated by a varying degree of re-individualization of the ancestral primordia.
Etv5, which is known to be a downstream target of the FGF pathway (Roehl and Nüsslein-Volhard, 2001; Zhang et al., 2009), was upregulated in Spry2+/–;Spry4–/– embryos (see Fig. S4 in the supplementary material), indicating increased FGF signaling in these mutants. Because Shh is necessary for the survival of tooth bud epithelial cells (Cobourne et al., 2001), the lower cell death observed in the Spry2+/–;Spry4–/– embryos at E12.5 might have resulted from the higher levels of Shh expression (see Fig. S4 in the supplementary material). Future studies will be needed to test our hypothesis that the splitting of the Shh expression domain in the mutant at E13.0 creates a Shh-free region in the placode; this might correspond to the region in which cell death is subsequently concentrated and where the septum appears, leading to the morphological splitting of the incisor.
We have previously reported that inactivation of sprouty genes leads to decreased apoptosis in a rudimentary diastema tooth bud (Peterkova et al., 2009), indicating that these genes perform similar functions in the incisor and molar regions. The transient splitting of some gene expression domains in the budding dental epithelium during a very short time period has also recently been shown in the early incisor region in both lower and upper jaws of wild-type embryos without any tooth anomalies (Hovorakova et al., 2011; Nakatomi et al., 2010). This splitting of the early Shh domain was followed by the presence of a single Shh expression domain at the incisor bud at E13.5 (Hovorakova et al., 2011). However, splitting of the Shh expression domain leading to the development of a supernumerary lower incisor has been recently reported in β-catΔPrx/lacZ mice (Fujimori et al., 2010). In these embryos, the Shh expression domain splitting occurred at E13.0 and the first morphological evidence of the incisor duplication was noticed at E13.5. Together with our results, this study emphasizes that the integrity of Shh expression domain at the bud stage is crucial for the normal development of the mouse incisor. The developmental relationship between the early and later Shh expression domains (Hovorakova et al., 2011) and formation of the supernumerary incisors will require further investigation.
The absence of variability in the number of incisors in wild rodents suggests that incisor development is highly canalized. However, during our observation of wild-type embryos at E14.5 and E15.5, we found eight out of 55 specimens with a notch in the lingual CL, and two specimens even had a duplication of this CL (see Fig. S7 in the supplementary material). These specimens indicate that incisor development is not as highly canalized as it might seem when looking at adult mice, reflecting a degree of developmental plasticity that might explain the incomplete penetrance of the incisor duplication phenotype in Spry2+/–;Spry4–/– mice.
Activator-inhibitor balance allows a range of re-individualization of the ancestral tooth primordia
The fusion or separation of tooth primordia resulting in their respective joint or individual development has been proposed to depend on a balance between activators and inhibitors (Peterkova et al., 2002). In vitro modification of this balance by decreased concentration of noggin (considered to be an inhibitor) or increased concentration of activin (an activator) can lead to the development of two or three small incisors instead of the normal single thick incisor (Munne et al., 2010). In the present study, the decrease of sprouty gene dosage resulted in the splitting of the epithelial incisor anlage, providing an in vivo analog of the in vitro splitting (Munne et al., 2010). Previous studies on postnatal development of incisors in Fgf3–/–;Fgf10–/– mice showed that decreased FGF levels led to a single small incisor (Wang, X.-P., et al., 2007), which might develop from a smaller primordium. We can thus extrapolate the existence of a phenotypic gradation involving FGF signaling and sprouty genes, with a small incisor anlage in Fgf3–/–;Fgf10–/– embryos, a larger anlage in the wild type and a split incisor primordium in Spry2+/–;Spry4–/– embryos, which are hypersensitive to FGF10 signaling. The sprouty mutant series demonstrated such a gradation, from fusion of left and right lower incisors in one K14-Spry4GOF specimen to two incisors in two enamel organs in Spry1–/–;Spry4–/– mice. The fusion of the left and right incisors as seen in the fused K14-Spry4GOF specimen was also reported previously in mouse embryos with hypervitaminosis A (Knudsen, 1965). These mice showed a gradation of phenotypes ranging from normal to complete fusion of the two enamel organs (Knudsen, 1965), and similar observations were made in p53 mutant embryos (Kaufman et al., 1997). In a study of Bmp4hfΔPrx1/ΔlacZ embryos, the mandibular Shh expression domains were fused in the midline, whereas in β-catΔPrx/lacZ embryos, two incisors developed, pointing to important differences in the roles of BMP and WNT/β-catenin signaling in the regulation of incisor number in the mandible (Fujimori et al., 2010). In all these cases, we presume that the levels of activator and inhibitor of incisor development were modified, suggesting that in the sprouty series the incisor splitting was dosage dependant, with progressive splitting of the epithelial anlage as sprouty dosage was decreased.
The sprouty gene dosage series leads to the ancestral Glires phenotype
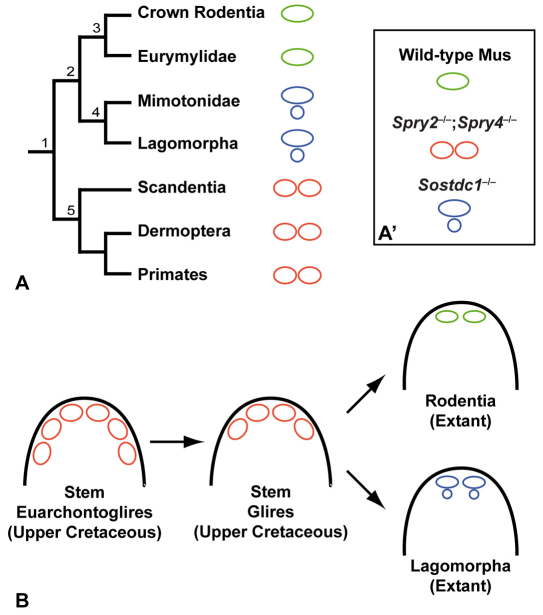
The number of upper incisors per quadrant varies from zero to five among extant mammals, and the genetic mechanisms underlying this variation are poorly understood. The progressive change in tooth number in the sprouty mutant series enabled us to explore the similarities between the incisor dental formulas (i.e. the number of incisors per quadrant) of sprouty mutants and wild Euarchontoglires, the group including rodents, rabbits and primates (Fig. 7A, node 1). Among Glires, the group that comprises rabbits and rodents (Fig. 7A, node 2), the number of incisors is reduced. This is particularly the case for rodents and Eurymilidae, a fossil family of rodent-related species, which together are called Simplicidentata (Fig. 7A, node 3) because they only possess one enlarged evergrowing incisor, considered to be the second deciduous incisor (dI2). Lagomorphs (rabbits and pikas) and the Mimotonidae family of Lagomorph-related fossil species are called Duplicidentata because they have two upper incisors (Fig. 7A, node 4): the enlarged evergrowing incisor typical of all Glires and a second, small tooth located behind the main incisor and considered to be the third permanent incisor (I3) (Luckett, 1985; Ooe, 1980). Almost all living Scandentia (treeshrews), Dermoptera (colugos) and primates, together named Euarchonta (Fig. 7A, node 5), have two upper incisors. From the fossil record, it appears that the stem Euarchontoglires (e.g. Purgatorius) had at least three incisors (Fig. 7B), indicating that all extant species in this group have lost at least one incisor during evolution. The incisors in Sostdc1 mutants (Fig. 7A′) have many similarities with those of the Lagomorphs and Mimotonidae. In both cases, the incisors are placed one behind the other and appear to arise from two different dental generations (Munne et al., 2010), suggesting that Sostdc1 might have played a role in the suppression of the permanent upper incisor in the dentition of rodents and Eurymilidae.
Fig. 7.
Upper incisor evolution in Euarchontoglires. (A) Phylogenetic tree of Euarchontoglires and schematic of the general upper incisor pattern of each taxon. Three patterns are indicated by color coding: one pair of incisors in Simplicidentata (green), two pairs of incisors (labial and lingual, each pair in tandem) in Duplicidentata (blue) and two pairs of incisors (frontals, each pair side by side) in Euarchonta (red). 1, Euarchontoglires; 2, Glires; 3, Simplicidentata; 4, Duplicidentata; 5. Euarchonta. (A′) Upper incisor phenotypes of mutants discussed in the text. (B) Schematic of incisor evolution in early Euarchontoglires showing the appearance of the three main incisor patterns.
The two upper incisors of sprouty mutants belong to the same dental generation and are situated one next to the other, as is the case in most mammals. It is known that basal placental mammals (ancestors of Euarchontoglires) had up to five upper incisors per quadrant (Ji et al., 2002). How the number of incisors decreased during evolution is currently an open question. However, the studies of incisor development in wild-type embryos have produced morphological data suggesting that the decrease of incisor number during evolution was realized by the integration of the incisor primordia, potentially corresponding to rudiments of ancestral incisors (Peterkova et al., 1993). The progressive gradient of re-individualization of incisor primordia observed in our sprouty gene dosage series (Fig. 6A-E) might correspond to a reversal of how the fusion of these incisor primordia occurred during evolution. The ancestor of Glires possessed more incisors than extant rodents (Fig. 7B), and our results suggest that the loss of incisors during evolution of Glires might have occurred through the merging of dental germs.
Enlarged incisors have been positively selected during evolution and maintained in all Glires, perhaps because possessing enlarged incisors constitutes a functional advantage to cut or gnaw on hard plants. The main groups of Glires arose at the beginning of the Cenozoic, at about 60 Ma (Wang, Y., et al., 2007). It is thus likely that Glires diverged from other placental mammals near the close of the Cretaceous (Asher et al., 2005), meaning that the enlargement of the incisor took place during the Upper Cretaceous. The loss of sprouty gene function in the dosage series reported here leads to incisor phenotypes that resemble those of the earliest Glires, which might have been similar to the phenotype of the Euarchontoglires common ancestor.
Supplementary Material
Acknowledgments
We thank G. R. Martin for providing mice and for insightful observations and N. Strauli, D.-K. Tran and P. Mostowfi for technical assistance. We are grateful to W. v. Koenigswald for intellectual input, and to K. Seidel, C. Li and other members of the Klein laboratory for advice and discussion. This work was funded by a New Faculty Award II (RN2-00933) from the California Institute for Regenerative Medicine to O.D.K. and by the National Institutes of Health through the NIH Director's New Innovator Award Program, 1-DP2-OD007191, to O.D.K. Additional support was provided by the Grant Agency of the Czech Republic (CZ:GA ČR:GA 304/09/1579 and CZ:GA ČR:GA 304/07/0223), MSMT of the Czech Republic (MSM0021620843) and Academy of Science of the Czech Republic (AV0Z50390512). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.069195/-/DC1
References
- Ahn Y., Sanderson B. W., Klein O. D., Krumlauf R. (2010). Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development 137, 3221-3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher R., Meng J., Wible J., McKenna M., Rougier G., Dashzeveg D., Novacek M. (2005). Stem lagomorpha and the antiquity of Glires. Science 307, 1091-1094 [DOI] [PubMed] [Google Scholar]
- Basson M., Akbulut S., Watson-Johnson J., Simon R., Carroll T., Shakya R., Gross I., Martin G., Lufkin T., McMahon A., et al. (2005). Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev. Cell 8, 229-239 [DOI] [PubMed] [Google Scholar]
- Boran T., Peterkova R., Lesot H., Lyons D., Peterka M., Klein O. D. (2009). Temporal analysis of ectopic enamel production in incisors from sprouty mutant mice. J. Exp. Zool. B Mol. Dev. Evol. 312B, 473-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catón J., Luder H. U., Zoupa M., Bradman M., Bluteau G., Tucker A. S., Klein O., Mitsiadis T. (2009). Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev. Biol. 328, 493-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles C., Pantalacci S., Peterkova R., Tafforeau P., Laudet V., Viriot L. (2009a). Effect of eda loss of function on upper jugal tooth morphology. Anat. Rec. 292, 299-308 [DOI] [PubMed] [Google Scholar]
- Charles C., Pantalacci S., Tafforeau P., Headon D., Laudet V., Viriot L. (2009b). Distinct impacts of Eda and Edar loss of function on the mouse dentition. PLoS One 4, e4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobourne M., Sharpe P. (2010). Making up the numbers: The molecular control of mammalian dental formula. Semin. Cell Dev. Biol. 21, 314-324 [DOI] [PubMed] [Google Scholar]
- Cobourne M., Hardcastle Z., Sharpe P. (2001). Sonic hedgehog regulates epithelial proliferation and cell survival in the developing tooth germ. J. Dent. Res. 80, 1974-1979 [DOI] [PubMed] [Google Scholar]
- Danforth C. (1958). The occurence and genetic behavior of duplicate lower incisors in the mouse. Genetics 43, 140-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond I., Owolabi T., Marco M., Lam C., Glick A. (2000). Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J. Invest. Dermatol. 115, 788-794 [DOI] [PubMed] [Google Scholar]
- Fuchs E. (2009). Finding one's niche in the skin. Cell Stem Cell 4, 499-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori S., Novak H., Weissenbock M., Jussila M., Goncalves A., Zeller R., Galloway J., Thesleff I., Hartmann C. (2010). Wnt/beta-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev. Biol. 348, 97-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy G., Wong E., Yusoff P., Chandramouli S., Lo T., Lim J., Fong C. (2003). Sprouty: how does the branch manager work? J. Cell Sci. 116, 3061-3068 [DOI] [PubMed] [Google Scholar]
- Harada H., Kettunen P., Jung H.-S., Mustonen T., Wang Y., Thesleff I. (1999). Localization of putative stem cells in dental epithelium and their association with notch and FGF signaling. J. Cell Biol. 147, 105-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Toyono T., Toyoshima K., Yamasaki M., Itoh N., Kato S., Sekine K., Ohuchi H. (2002). FGF10 maintains stem cell compartment in developing mouse incisors. Development 129, 1533-1541 [DOI] [PubMed] [Google Scholar]
- Hovorakova M., Prochazka J., Lesot H., Smrckova L., Churava S., Boran T., Kozmik Z., Klein O., Peterkova R., Peterka M. (2011). Shh expression in a rudimentary tooth offers new insights into development of the mouse incisor. J. Exp. Zool. B Mol. Dev. Evol. 316B, 347-358 [DOI] [PubMed] [Google Scholar]
- Ji Q., Luo Z., Yuan C., Wible J., Zhang J., Georgi J. (2002). The earliest known eutherian mammal. Nature 416, 816-822 [DOI] [PubMed] [Google Scholar]
- Kaufman M., Chang H., Shaw J. (1995). Craniofacial abnormalities in homozygous Small eye (Sey/Sey) embryos and newborn mice. J. Anat. 186, 607-617 [PMC free article] [PubMed] [Google Scholar]
- Kaufman M., Kaufman D., Brune R., Stark M., Armstrong J., Clarke A. (1997). Analysis of fused maxillary incisor dentition in p53-deficient exencephalic mice. J. Anat. 191, 57-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen P., Laurikkala J., Itäranta P., Vainio S., Itoh N., Thesleff I. (2000). Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 219, 322-332 [DOI] [PubMed] [Google Scholar]
- Kieffer S., Peterkova R., Vonesch J. L., Ruch J. V., Peterka M., Lesot H. (1999). Morphogenesis of the lower incisor in the mouse from the bud to early bell stage. Int. J. Dev. Biol. 43, 531-539 [PubMed] [Google Scholar]
- Kim H. J., Bar-Sagi D. (2004). Modulation of signalling by Sprouty: a developing story. Nat. Rev. Mol. Cell Biol. 5, 441-450 [DOI] [PubMed] [Google Scholar]
- Klein O., Minowada G., Peterková R., Kangas A., Yu B., Lesot H., Peterka M., Jernvall J., Martin G. (2006). Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev. Cell 11, 181-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein O., Lyons D., Balooch G., Marshall G., Basson M., Peterka M., Boran T., Peterkova R., Martin G. (2008). An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135, 377-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopcic B., Maass T., Meyer E., Lehr H. A., Metzger D., Chambon P., Mann A., Blessing M. (2007). TGF-beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur. J. Cell Biol. 86, 781-799 [DOI] [PubMed] [Google Scholar]
- Knudsen P. (1965). Congenital malformations of upper incisors in exencephalic mouse embryos, induced by hypervitaminosis A. I. Types and frequency. Acta Odontol. Scand. 23, 71-89 [DOI] [PubMed] [Google Scholar]
- Kriangkrai R., Chareonvit S., Yahagi K., Fujiwara M., Eto K., Iseki S. (2006). Study of Pax6 mutant rat revealed the association between upper incisor formation and midface formation. Dev. Dyn. 235, 2134-2143 [DOI] [PubMed] [Google Scholar]
- Lesot H., Vonesch J.-L., Peterka M., Turecková J., Peterková R., Ruch J.-V. (1996). Mouse molar morphogenesis revisited by three-dimensionalreconstruction. II. Spatial distribution of mitoses and apoptosis in cap to bell staged first and second upper molar teeth. Int. J. Dev. Biol. 40, 1017-1031 [PubMed] [Google Scholar]
- Luckett W. P. (1985). Superordinal and intraordinal affinities of rodents: developmental evidence from the dentition and placentation. In Evolutionary Relationships Among Rodents (ed. Luckett W. P., Hartenberger J. L.), pp. 227-276 New York: Plenum; [Google Scholar]
- Min H., Danilenko D., Scully S., Bolon B., Ring B., Tarpley J., DeRose M., Simonet W. (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156-3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss-Salentijn L. (1978). Vestigial teeth in the rabbit, rat and mouse; their relationship to the problem of lacteal dentitions. In Development, function and evolution of teeth (ed. Butler P. M., Joysey K. A.), pp. 13-29 London: Academic Press; [Google Scholar]
- Munne P., Tummers M., Jarvinen E., Thesleff I., Jernvall J. (2009). Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development 136, 393-402 [DOI] [PubMed] [Google Scholar]
- Munne P., Felszeghy S., Jussila M., Suomalainen M., Thesleff I., Jernvall J. (2010). Splitting placodes: effects of bone morphogenetic protein and Activin on the patterning and identity of mouse incisors. Evol. Dev. 12, 383-392 [DOI] [PubMed] [Google Scholar]
- Murashima-Suginami A., Takahashi K., Kawabata T., Sakata T., Tsukamoto H., Sugai M., Yanagita M., Shimizu A., Sakurai T., Slavkin H., et al. (2007). Rudiment incisors survive and erupt as supernumerary teeth as a result of USAG-1 abrogation. Biochem. Biophys. Res. Commun. 359, 549-555 [DOI] [PubMed] [Google Scholar]
- Nakatomi M., Wang X., Key D., Lund J., Turbe-Doan A., Kist R., Aw A., Chen Y., Maas R., Peters H. (2010). Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 340, 438-449 [DOI] [PubMed] [Google Scholar]
- Ohazama A., Johnson E., Ota M., Choi H., Choi H., Porntaveetus T., Oommen S., Itoh N., Eto K., Gritli-Linde A., et al. (2008). Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One 3, e4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohazama A., Blackburn J., Porntaveetus T., Ota M. S., Choi H., Johnson E., Myers P., Oommen S., Eto K., Kessler J., et al. (2010). A role for suppressed incisor cuspal morphogenesis in the evolution of mammalian heterodont dentition. Proc. Natl. Acad. Sci. USA 107, 92-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe T. (1980). Embryonary development of the incisors in the rabbit (Oryctolagus-cuniculus L)-dental formula interpretation. Mammalia 44, 259-275 [Google Scholar]
- Parsa S., Kuremoto K., Seidel K., Tabatabai R., Mackenzie B., Yamaza T., Akiyama K., Branch J., Koh C., Al, Alam D., et al. (2010). Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development 137, 3743-3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka M., Lesot H., Peterková R. (2002). Body weight in mouse embryos specifies staging of tooth development. Connect. Tissue Res. 43, 186-190 [DOI] [PubMed] [Google Scholar]
- Peterkova R., Peterka M., Vonesch J.-L., Ruch J.-V. (1993). Multiple developmental origin of the upper incisor in mouse: histological and computer assisted 3-D-reconstruction studies. Int. J. Dev. Biol. 37, 581-588 [PubMed] [Google Scholar]
- Peterkova R., Peterka M., Viriot L., Lesot H. (2002). Development of the vestigial tooth primordia as part of mouse odontogenesis. Connect. Tissue Res. 43, 120-128 [DOI] [PubMed] [Google Scholar]
- Peterkova R., Lesot H., Peterka M. (2006). Phylogenetic memory of developing mammalian dentition. J. Exp. Zool. B Mol. Dev. Evol. 306, 234-250 [DOI] [PubMed] [Google Scholar]
- Peterkova R., Churava S., Lesot H., Rothova M., Prochazka J., Peterka M., Klein O. (2009). Revitalization of a diastemal tooth primordium in Spry2 null mice results from increased proliferation and decreased apoptosis. J. Exp. Zool. B Mol. Dev. Evol. 312B, 292-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J., Thesleff I. (2003). Mechanisms of ectodermal organogenesis. Dev. Biol. 262, 195-205 [DOI] [PubMed] [Google Scholar]
- Prochazka J., Pantalacci S., Churava S., Rothova M., Lambert A., Lesot H., Klein O., Peterka M., Laudet V., Peterkova R. (2010). Patterning by heritage in mouse molar row development. Proc. Natl. Acad. Sci. USA 107, 15497-15502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H., Nüsslein-Volhard C. (2001). Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr. Biol. 11, 503-507 [DOI] [PubMed] [Google Scholar]
- Seidel K., Ahn C., Lyons D., Nee A., Ting K., Brownell I., Cao T., Carano R., Curran T., Schober M., et al. (2010). Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development 137, 3753-3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K., Minowada G., Coling D., Martin G. (2005). Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell 8, 553-564 [DOI] [PubMed] [Google Scholar]
- Sofaer J. (1969). Aspects of the tabby-crinckled-downless syndrome I. The development of Tabby teeth. J. Embryol. Exp. Morphol 22, 181-205 [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W., Rendl M., Fuchs E. (2004). Defining the epithelial stem cell niche in skin. Science 303, 359-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtokari A., Aberg T., Jernvall J., Kerwen S., Thesleff I. (1996). The enamel knot as a signaling center in the developing mouse tooth. Mech. Dev. 54, 39-43 [DOI] [PubMed] [Google Scholar]
- Vaezi A., Bauer C., Vasioukhin V., Fuchs E. (2002). Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell 3, 367-381 [DOI] [PubMed] [Google Scholar]
- Wang X.-P., Suomalainen M., Felszeghy S., Zelarayan L., Alonso M., Plikus M. V., Maas R., Chuong C.-M., Schimmang T., Thesleff I. (2007). An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5, 1324-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Meng J., Ni X., Li C. (2007). Major events of Paleogene mammal radiation in China. Geol. J. 42, 415-430 [Google Scholar]
- Yokohama-Tamaki T., Ohshima H., Fujiwara N., Takada Y., Ichimori Y., Wakisaka S., Ohuchi H., Harada H. (2006). Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development 133, 1359-1366 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Verheyden J. M., Hassell J. A., Sun X. (2009). FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev. Cell 16, 607-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.