Abstract
It has been shown in several organisms that multiple cis-regulatory modules (CRMs) of a gene locus can be active concurrently to support similar spatiotemporal expression. To understand the functional importance of such seemingly redundant CRMs, we examined two CRMs from the Drosophila snail gene locus, which are both active in the ventral region of pre-gastrulation embryos. By performing a deletion series in a ∼25 kb DNA rescue construct using BAC recombineering and site-directed transgenesis, we demonstrate that the two CRMs are not redundant. The distal CRM is absolutely required for viability, whereas the proximal CRM is required only under extreme conditions such as high temperature. Consistent with their distinct requirements, the CRMs support distinct expression patterns: the proximal CRM exhibits an expanded expression domain relative to endogenous snail, whereas the distal CRM exhibits almost complete overlap with snail except at the anterior-most pole. We further show that the distal CRM normally limits the increased expression domain of the proximal CRM and that the proximal CRM serves as a `damper' for the expression levels driven by the distal CRM. Thus, the two CRMs interact in cis in a non-additive fashion and these interactions may be important for fine-tuning the domains and levels of gene expression.
Keywords: Cis-regulatory modules, Gene expression, Drosophila melanogaster, snail, Developmental patterning, Repression, Huckebein
INTRODUCTION
A number of cis regulatory modules (CRMs) have recently been identified that support concurrent expression of individual genes in similar spatiotemporal profiles in early Drosophila embryos, as well as later in development (e.g. Frankel et al., 2010; Hong et al., 2008; Zeitlinger et al., 2007). For the most part, these secondary CRMs were identified as a result of ChIP-chip and ChIP-seq analyses as regions of occupancy located at a distance from genes of interest, up to 10 kb or more (e.g. Li et al., 2008; Ozdemir et al., 2011; Sandmann et al., 2007; Zeitlinger et al., 2007). These newly identified CRMs have been described as being redundant to previously identified promoter-proximal located CRMs and, most recently, it has been proposed that they function to provide robustness to environmental or genetic perturbation (Frankel et al., 2010; Perry et al., 2010). Moreover, in vertebrate genomes it has been shown that many genes have multiple CRMs active concurrently, and that deletion of one cis-regulatory module can have no observable effect on the gene expression pattern (e.g. Ghiasvand et al., 2011; Xiong et al., 2002). Therefore, identifying why multiple CRMs of similar spatiotemporal expression domains are active simultaneously is a problem of general interest.
Here, we focus on analysis of the snail (sna) locus in Drosophila. sna encodes a transcription factor containing Zn-finger DNA-binding domains that predominantly functions to repress the expression of a number of genes from ventral regions of the embryo (e.g. Cowden and Levine, 2002; De Renzis et al., 2006; Ip et al., 1992a). As such, Snail is an important patterning molecule that influences the mesoderm-mesectoderm-neurogenic ectoderm boundary (Kosman et al., 1991; Leptin, 1991). Although a CRM supporting expression similar to sna was isolated almost 20 years ago by standard lacZ reporter gene constructs from a promoter proximal location, even 6.0 kb of upstream sequence failed to completely represent native sna expression, which exhibits very sharp anterior-posterior and lateral boundaries (Ip et al., 1992b). Since then, the predominant view in the field has been that synergy between the Dorsal and Twist transcription factors, which is present in ventral gradients within early embryos, functions to specify the sharp sna dorsal boundary (Ip et al., 1992b; Zinzen et al., 2006), and that the sharp posterior boundary is defined by the repressor Huckebein (Reuter and Leptin, 1994). Yet the promoter proximal CRM of sna does not exhibit either of these sharp borders, despite the fact that it encompasses the region all the way up to the adjacent upstream gene (Ip et al., 1992b).
In general, it is a common assumption in the field that CRMs located in promoter-proximal locations are required to support gene expression. Thus, although it was noticed that the pattern of the promoter-proximal CRM was expanded relative to endogenous sna, the existence of another CRM to serve as a vehicle for repressors was not proposed upon the initial characterization of the reporter gene pattern (Ip et al., 1992b). It is a common finding that CRMs do not always support expression in the exact same domain as the genes they regulate, but in the past this was explained away as a flaw inherent to reporter gene assays. For example, the CRM supporting expression within stripes 3/7 of the even-skipped (eve) gene does not exhibit equivalent effects in knirps mutants as does the endogenous eve gene: the expression of the reporter gene expands into the midsection, whereas stripes 3/7 associated with the endogenous eve gene retain sharp boundaries (Frasch and Levine, 1987; Small et al., 1996).
More recently, however, additional CRMs have been identified sharing similar spatiotemporal profiles to previously characterized CRMs, including one that shares close similarity with the sna expression pattern (Ozdemir et al., 2011; Perry et al., 2010). Another recent study presumably labeled this CRM as a `shadow' enhancer because it is located at a distance from the snail gene, whereas the proximally located CRM was defined as the primary acting enhancer (Perry et al., 2010).
To provide insight into the functions of CRMs associated with the snail locus in the Drosophila early embryo, we undertook a genetic approach towards studying cis-regulatory control using BAC recombineering and site-directed transgenesis to assay the domain and level of expression supported by concurrently functioning CRMs. We focused on the distinction between the proximal and distal snail CRMs, which control early embryonic expression, in particular on the patterns and levels of expression supported by each, as well as their abilities to support Snail function.
MATERIALS AND METHODS
Fly stocks
Adhn7sna1cn1vg1/CyO, and sna18/CyO fly stocks were used (BDSC) after rebalancing with CyO ftz-lacZ marked balancer. The proximal 2.2 kb and 6 kb lacZ reporter lines and F10 line (hsp83-Toll10B-bcd3′UTR) have been published previously (Huang et al., 1997; Ip et al., 1992b).
Cloning and generation of lacZ constructs
Enhancer sequence for the distal enhancer was amplified from genomic DNA using Sna-Dist 2kb-f (5′-AATTGGTACCACAATTAGCTGCCGTTTGCAGC-3′) and Sna-Dist 2kb-r (5′-AATTGGTACCTGTAGCACCCTTGAACTTGTTGTG-3′) and cloned into the KpnI site of the evepromoter-lacZ-attB vector (Liberman and Stathopoulos, 2009). Site-directed transgenesis system was used to create reporter lines (Bischof et al., 2007). The 86Fb fly stock with attP landing site was injected in house with reporter constructs to generate transgenic lines.
Generation of 25 kb sna rescue constructs
The 25 kb sna P[acman] construct was generated using recombineering mediated gap repair performed using SW105 cells as described previously (Venken et al., 2006). The BAC encompassing the sna gene (BACR23I04) was obtained from the BacPac Resource Center and the attB-P[acman]-ApR was modified to contain ∼600 bp homology arms to the region of interest. Insertion of GFP just before the stop codon of sna was performed using a GFP-frt-kan-frt plasmid and the kan cassette was removed after insertion as described previously (Lee et al., 2001).
Deletion, rearrangement and mutation of the enhancer regions was carried out using the galK system (Warming et al., 2005). All final constructs were isolated and electroporated into EPI300 cells (Epicenter) and the copy number was induced using Fosmid Autoinduction Solution (Epicentre) according to the manufacturers instructions. The constructs were isolated using Nucleobond EF plasmid midi prep kits (Clontech). P[acman] constructs were injected into line 23648 (BDSC) at a concentration of 0.5-1 μg/μl in water using standard techniques. All primers used for gap repair and recombineering are listed in Table S1 in the supplementary material.
Rescue experiment
Lines were created that contained sna18/Cyo ftz-lacZ and one of the sna BAC constructs. Males from these lines were crossed to virgin Adhn7sna1cn1vg1/CyO ftz-lacZ. Separate vials were placed at 25°C, 29°C and 18°C. All transgenic flies were counted and the total number of straight wing flies (i.e. sna mutants) was compared with the total number of transgenic flies. The final percentage of straight wing flies for each experiment was then divided by 33%, which would be the expected result were the rescue to be perfect.
We note key distinctions between our construct design and that of another recent study of the snail locus which used a similar approach (Perry et al., 2010): (1) our transgene functions to rescue a sna mutant (i.e. sna1/sna18) to viability, whereas the other group was limited to assaying early gastrulation defects presumably because a large deficiency background was used; (2) our deletions were guided by our own Twist ChIP-seq data (Ozdemir et al., 2011), effectively guiding definition of the distal CRM as a larger region (∼2.0 kb), (3) a spacer sequence (i.e. ampicillin resistance cassette) was not put in place of deletions in our constructs, which allowed us to assay whether native spacing is important; (4) the sna-coding sequence, which may possibly influence cis-regulatory mechanism or stability of transcripts, was left intact within our reporter constructs; and (5) the other group did not assay the gastrulation defects associated with the distal CRM delete large transgene but relied on cDNA rescue data conducted previously (Hemavathy et al., 2004).
In situ hybridization
Embryos were fixed and stained following standard protocols. Antisense RNA probes labeled with digoxigenin, biotin or FITC-UTP were used to detect reporter or in vivo gene expression as described previously (Jiang and Levine, 1993; Kosman et al., 2004). Primary antibodies used were: rabbit anti-Eve (provided by M. Frasch, University of Erlangen-Nürnberg, Germany), guinea pig anti-Twist (provided by M. Levine, UC Berkeley, CA, USA), mouse anti-Dorsal (7A4-s from the Hybridoma Bank) and rabbit anti-Histone H3 (Abcam).
Mean intensity quantification
Images of three embryos from each construct were taken using identical parameters. From each embryo, a square of 345 μm2 was extracted and analyzed for mean intensity using the LSM Image Examiner program (Zeiss). This was repeated three times in each embryo within the snail stripe in consistent locations from embryo to embryo. A negative control square of the same size was also analyzed for each embryo. For each measurement within the snail stripe, the negative measurement from that embryo was subtracted and then the measurements were averaged and a standard deviation was determined from the nine measurements.
RESULTS
Multiple CRMs in proximity to the snail gene support expression in overlapping domains
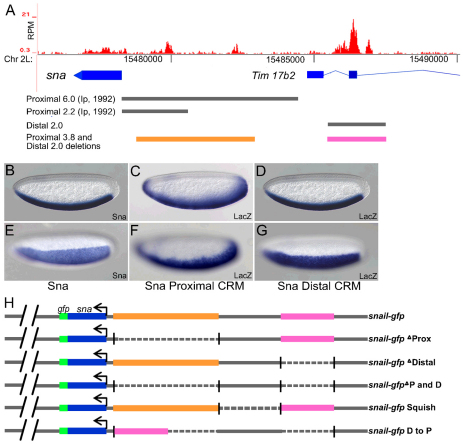
Previously published Twist-ChIP-seq binding data identified multiple peaks of Twist occupancy to DNA in proximity to the snail gene (Fig. 1A) (Ozdemir et al., 2011). By far, the largest peaks were detected ∼7 kb upstream of sna gene within the intron of another gene, Tim17b2. The two proximal Twist occupied regions are covered by the previously studied 2.2 kb and 6 kb enhancer constructs (`proximal CRM') (Ip et al., 1992b). A 2.0 kb DNA fragment from the Tim17b2 intronic sequence, containing several closely positioned peaks of Twist occupancy, was also assayed in a reporter context (`distal CRM') (Ozdemir et al., 2011).
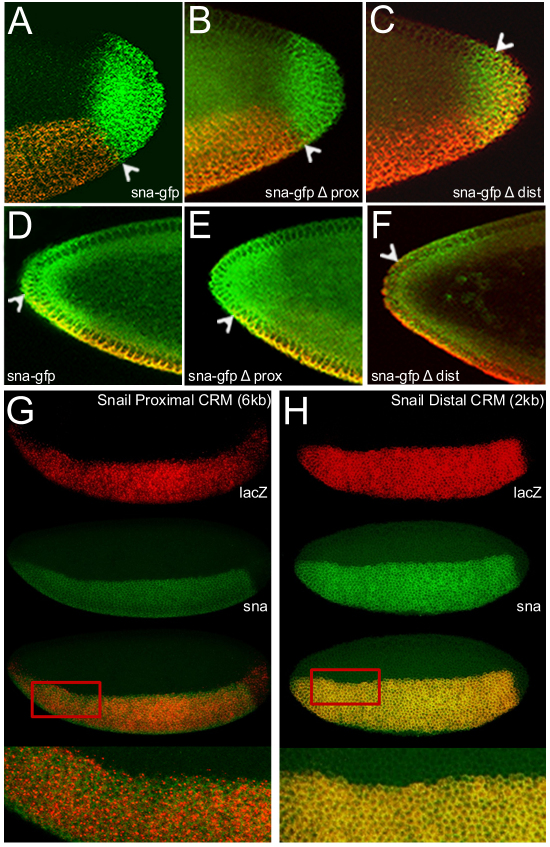
Fig. 1.
Distinct regions in the vicinity of the snail gene regulate expression in ventral regions of early embryos. (A) Twist ChIP-seq defined binding (shown in reads per million, RPM) was identified previously in three domains upstream of snail: –1.6, –3.4 and –7 kb (Ozdemir et al., 2011). We created a lacZ reporter construct of the ∼2 kb distal region in order to encompass the entire region defined by our Twist ChIP-seq analysis, and compared with two lacZ reporter constructs assayed previously: proximal 2.2 kb and 6.0 kb constructs (gray lines) (Ip et al., 1992b), regions deleted in the context of a 25 kb rescue construct are shown in orange (proximal) and pink (distal). (B-G) In situ hybridization data using riboprobes to detect either snail transcript in wild-type embryos (B,E) or lacZ transcript in transgenic embryos containing the snail 2.2 kb promoter proximal reporter (C,F) or the snail distal 2.0 kb reporter (D,G). In this and subsequent figures, embryos are oriented with anterior towards the left. (B-D) Sagittal views; (E-G) ventrolateral surface views. (H) A ∼25 kb snail rescue transgene was modified by insertion of gfp as an in-frame fusion to 3′ end of the snail gene. Various deletions were created as shown.
By analysis of lacZ reporter transgenes, we found that both these CRMs (proximal and distal) supported expression in the ventral region of the early embryo in patterns that are spatiotemporally similar but not identical. In contrast to the broadened expression of the proximal CRM fragment (Fig. 1C,F), the distally located CRM fragment supports high-level expression that is refined, sharp and similar to the endogenous sna expression pattern (Fig. 1D,G, compare with 1B,E). It should be noted that our tested DNA fragment was defined by Twist ChIP-seq analysis and was larger in size than the one recently tested by another group (i.e. 2 kb versus 1.2 kb) (Perry et al., 2010), a study in which no spatial distinctions between the patterns supported by the proximal and distal CRMs was noted.
Assay of CRM function using larger reporter transgenes in which native context is retained or modified
To analyze how expression of the sna gene is controlled in the early embryo, we created a 24.8 kb P[acman] construct encompassing the sna gene, as well as flanking DNA sequences using recombineering methods (Fig. 1H) (Venken et al., 2006). We isolated stable transgenic lines using site-directed methods and determined that this DNA sequence can complement the sna mutant, suggesting that the cis-regulatory information encoded within this ∼25 kb DNA segment is sufficient to support the essential aspects of sna expression. To create the reporter construct, we recombineered the gfp cDNA sequence into the sna locus as an in-frame C-terminal fusion to Snail protein (Fig. 1H, `sna-gfp'), allowing us to monitor transgenic expression of sna-gfp using a gfp riboprobe (see below).
As our goal was to provide insight into cis-regulatory mechanisms regulating snail expression, we created five deletion constructs within the 25 kb sna-gfp construct using our Twist ChIP-seq data as a guide: (1) a sna promoter proximal deletion of 3.8 kb containing two peaks of Twist binding, including most of the 2.2 kb minimal sna enhancer identified by Ip et al. (Ip et al., 1992b), but leaving the 500 bp promoter proximal region and including more upstream sequence that we found was also bound by Twist in the early embryo (`Δ Proximal'); (2) a distal deletion of 2.0 kb, which includes three major peaks of Twist binding, located in the intron of the gene upstream of sna, Tim17b2 (`Δ Distal'); (3) a double-deletion of both the proximal and distal CRMs (`Δ P and D'); (4) a deletion of the intervening sequence, present between the proximal and distal CRMs (`squish'); and (5) a construct in which the distal CRM is moved to the proximal position, in a double-delete background (`D to P') (Fig. 1H). 500 bp directly upstream of the sna-coding sequence was left unmodified in all cases, with the purpose of leaving the promoter intact.
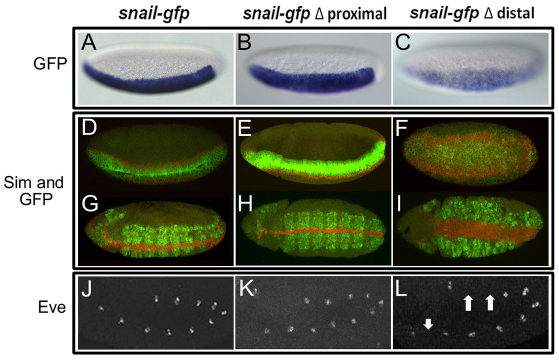
As both the distal and proximal CRMs supported sna expression during early embryogenesis, we investigated whether they function redundantly through analysis of these recombineered reporter transgenes. The proximal CRM deletion (`Δ Proximal') supported gfp expression that was comparable with gfp expression from the full sna-gfp rescue construct (Fig. 2B, compare with 2A). Moreover, gfp expression similar to that supported by sna-gfp was detected in the constructs that moved the distal promoter to a proximal location (`D to P') and the construct that deleted the intervening sequence (`squish') in the early embryo (data not shown). By contrast, deletion of the distal CRM (`Δ Distal') supported weaker expression (Fig. 2C), and the construct that deletes both (`double delete') lacked early expression altogether (data not shown). Based on pattern alone, the distal CRM appeared more faithful to the snail endogenous expression domain.
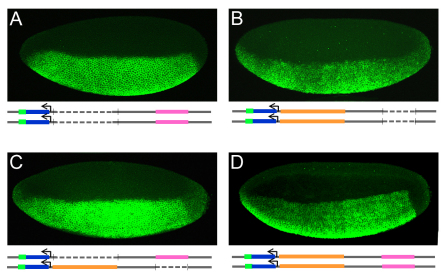
Fig. 2.
The distal CRM is required to rescue gastrulation and Eve cell specification defects. (A-C) In situ hybridization of cellularized wild-type embryos (stage 5) containing sna-gfp construct using a gfp riboprobe and alkaline phosphatase staining procedure. sna-gfp (A) and sna-gfp ΔProximal (B) constructs supported sharp lateral and posterior borders, whereas the sna-gfp ΔDistal (C) construct was weaker and exhibited expanded lateral and posterior boundaries. (D-I) Fluorescent in situ hybridizations of sna1/sna18 mutant embryos using sim (red) and gfp (green) riboprobes to detect sna construct reporter expression and effects on gastrulation through assay of sim. sna mutant embryos containing either the full-length construct sna-gfp (D,G); the proximal delete construct sna-gfp ΔProximal (E,H); or the distal delete construct sna-gfp ΔDistal (F,I) are shown. (J-L) Eve expression in sna mutant germ-band elongated embryos containing sna-gfp (J), sna-gfp ΔProximal (K) or sna-gfp ΔDistal (L). Arrows indicate gaps in eve expression. (See Fig. S1 in the supplementary material for sna-gfp `D to P' and `squish images', also see Fig. S2 in the supplementary material for late Eve expression.)
Genetic assay of CRM function by snail mutant rescue
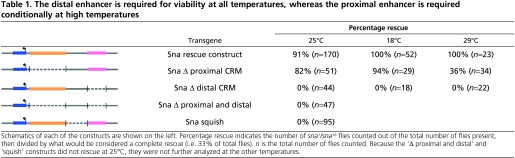
To determine whether snail expression supported by these transgenes was functionally equivalent, we assayed the ability of these transgenes to rescue a sna mutant. The wild-type reporter and five modified versions, were introduced into a sna mutant background (sna1/sna18) and assayed for their ability to support viability. We found that the native sna gene rescued at 91% (Table 1) but there was significant, but only partial, rescue with the sna-gfp fusion constructs (76%) (data not shown). For this reason, we assayed the ability of native sna gene constructs, unmodified with gfp, to support rescue.
Table 1.
The distal enhancer is required for viability at all temperatures, whereas the proximal enhancer is required conditionally at high temperatures
The 25 kb snail transgene and the delete proximal CRM constructs rescued the sna mutant phenotype; 91% and 82% of expected F1 progeny, respectively, were obtained in rescue crosses (Table 1). By contrast, the distal CRM delete construct completely failed to rescue the sna mutant, as did the double delete `Δ P and D' construct. The `squish' construct, which removes sequence between the proximal and distal CRMs, also failed to complement the mutant. These results support the conclusion that the distal CRM is required to support viability. In turn, the fact that more than 80% of the expected flies emerged from the sna rescue cross with proximal CRM delete transgene suggested that the proximal CRM is not required to support viability.
To further study functional differences between CRMs, we examined the ability of our constructs to support viability at various temperatures: 25°C, 29°C and 18°C. The proximal CRM delete construct showed decreased viability at higher temperature, with 36% viability supported at 29°C when compared with 82% at 25°C; yet at 18°C, we found the rescue was also high at 94% (Table 1). However, we found that the distal CRM delete construct did not rescue at any temperature tested: 0% viability at 18°C, 25°C, and 29°C; further evidence that the distal CRM is the primary CRM responsible for supporting sna expression.
Deletion of the distal CRM, specifically, has consequences on gastrulation
Next, we examined whether these CRMs have similar or different roles during gastrulation. The constructs containing the distal CRM rescued the gastrulation defects of sna mutants [i.e. `Δ Proximal' (Fig. 2E,H) and `squish' and `D to P' (see Fig. S1 in the supplementary material), compare with full length sna-gfp (Fig. 2D,G)]. By contrast, constructs without the distal CRM exhibited gastrulation defects (i.e. `Δ Distal', Fig. 2F,I). In the absence of the distal CRM, not only was single-minded (sim) expression aberrant, with expansion into a broad domain compared with the single line of cells found in wild-type embryos, but invagination was non-uniform and presumably contributed to unequal mesoderm spreading (Fig. 2F,I). As sim is directly repressed by the Snail transcription factor in gastrulating embryos (Kasai et al., 1992), these results indicated that the level of snail expression in the sna mutant background supported by sna-gfp Δ Distal is insufficient to fully support function at this stage of development, resulting in an expansion of the sim domain.
As an assay for possible later phenotypes, we examined expression of even-skipped (eve). eve encodes a homeodomain transcription factor necessary for dorsal mesoderm lineage specification (Frasch et al., 1987), and its lateral expression in 11 clusters of cells on either side of the embryo at stage 11 can be used as an indicator for proper mesoderm spreading. In rescue experiments in which the distal CRM was absent, eve expression was aberrant as gaps in expression were detected in all of the embryos examined (Fig. 2L, arrows). By contrast, constructs that removed the proximal CRM, leaving the distal CRM intact, exhibited normal gastrulation (invagination and sim expression, Fig. 2E,H), as well as normal mesoderm spreading and specification even at later stages of embryogenesis (Eve expression; Fig. 2K). Even when the temperature was raised to 29°C, no obvious mesoderm specification defects in the trunk of the embryos were observed in the absence of the proximal CRM (see Fig. S2 in the supplemental material). Our data for rescue of the sna1/sna18 background demonstrated that the distal CRM is required to support gastrulation, but that the proximal CRM is not required or supports a minor role (such as supporting expression at the anterior, see below).
The proximal CRM deletion of 3.8 kb removes multiple tissue-specific enhancers, a minimum of three: one module from 1.2 kb to 2 kb supports expression in ventral regions of the early embryo (e.g. Fig. 1C) and two other modules, one from 0.4-0.9 kb and another from 2.2-2.8 kb, support expression in the peripheral nervous system (PNS) and central nervous system (CNS), respectively, at later stages of embryogenesis (Ip et al., 1994; Ip et al., 1992b). We observed changes in the PNS and CNS expression in constructs that delete the proximal CRM, but no effect on expression in these domains was observed in the constructs that delete the distal CRM (see below).
Multiple CRMs support sna expression in germ-band elongated embryos and are organized on the chromosome in a manner that potentially minimizes dominant effects of repressors
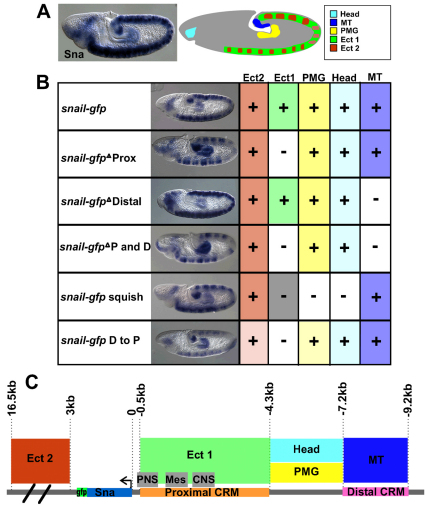
In the course of our sna rescue experiments, we found that a construct removing the intervening sequence between distal and proximal CRMs was not able to complement the mutant (Table 1, `sna-gfp squish'). We hypothesized that either this sequence supports another function required for viability or it influences the ability of the distal enhancer to function. To test the first possibility, we examined expression of sna in slightly older embryos, ones that were undergoing germ-band elongation. Previous studies have documented sna expression at this stage within the ectoderm and in malphigian tubule (MT) precursor cells (Alberga et al., 1991; Ip et al., 1994). From analysis of germ-band elongated embryos (stage 9), we observed that sna was also expressed at this stage in the posterior midgut (PMG) and in the head (possibly marking either anterior midgut and/or head mesoderm) (Fig. 3A) (Alberga et al., 1991; de Velasco et al., 2006).
Fig. 3.
Expression of snail is regulated by at least four CRMs, the spacing of which is important to support patterning. (A) In embryos that have undergone germband elongation (stage 9), snail is expressed in at least five different regions, as detected by in situ hybridization using a snail riboprobe (left) and schematized to demarcate the patterns (right): (1) within the head; (2) in posterior midgut primordium (PMG); (3) in malphigian tubule (MT) precursor cells; and in the ectoderm within two domains – either (4) broadly expressed (Ect1) or (5) in stripes (Ect2). (B) The gain or loss of specific expression patterns in a series of modified large transgenes. Colors in the chart correspond to the region key in the schematic in A, with weaker colors indicating lower expression than wild type and the gray box indicating loss of expression that was potentially due to repression. (C) The genomic region surrounding sna is divided into a minimum of four CRMs and marked with the expression pattern that each CRM supports in the embryo based on the expression data in B. Gray boxes associated with the proximal enhancer denote regions previously described (Ip et al., 1994) to support expression in the peripheral nervous system (PNS), mesoderm (Mes) and central nervous system (CNS).
The patterns of reporter expression supported by each sna-gfp transgene were analyzed (Fig. 3B). When the proximal CRM region was deleted, we found that a subset of expression in the ectoderm was lost (i.e. pattern `Ect1') (Ip et al., 1994). Yet upon loss of the 3.8 kb proximal CRM, expression in the neurogenic ectoderm was retained in stripes within the trunk but was absent in the midsection domain of the embryo (i.e. pattern `Ect2'), suggesting that other sequences also impact ectodermal expression. We deduced that the CRM responsible for supporting expression in the Ect2 pattern is most probably present in the DNA sequence of our rescue construct downstream of sna (∼14 kb), because none of the modified constructs we tested ever affected expression of the reporter in this domain. Next, we found that expression within the MT precursor cells was completely lost when the distal CRM was deleted (Fig. 3B, delete distal: `Δ distal' and double delete: `Δ P and D') and that the pattern was retained as long as the distal CRM was present, even if located in a different location. When the distal CRM was moved to the proximal position (`D to P'), there was an overall diminishment of expression in all domains but the MT precursor cell expression was retained. These results suggested that the 2.0 kb DNA associated with the distal CRM supports expression in the MT precursor cells in addition to its function in supporting early sna expression in ventral regions of the embryo. Consistent with this view, when the distal CRM lacZ construct was examined, expression in MT precursor cells within embryos at stage 9 was also observed (data not shown).
Last, the `squish' construct was the only construct found to cause loss of expression in the head and PMG, suggesting that this intervening sequence contains CRMs that support these sna expression domains. Loss of expression in these domains may be responsible for the inability of this construct to rescue the mutant. The `squish' construct also resulted in a partial to complete loss of expression within the Ect1 region (Fig. 3B, gray box). Although it is possible that deletion of the intervening sequence from –4.3 to –7.2 kb, which was removed by the `squish', could influence neuronal expression; this is unlikely as full sna expression within the CNS and PNS is observed with a transgene that includes only the most proximal 2.8 kb (Ip et al., 1994).
We hypothesized that by moving the two CRMs into closer proximity by deleting the intervening DNA (`squish'), repressors acting within the distal CRM may function to repress expression in the ectoderm normally supported by the proximal CRM. This idea, together with the fact that the distal CRM exhibited spatially refined expression relative to the proximal CRM in the early embryo (e.g. Fig. 1B-G), led us to investigate whether repressor(s) that act to limit snail expression function through the distal CRM.
Repressors predominantly function through the distal CRM to regulate the posterior and dorsal boundaries of the sna expression domain within the early embryo
It has previously been shown that the Huckebein (Hkb) transcription factor, which is expressed at both the anterior and posterior poles, functions as a repressor to define the posterior boundary of sna expression (Goldstein et al., 1999; Reuter and Leptin, 1994). In hkb mutants, posterior sna expression is expanded into the pole and anterior expression is expanded beyond the tip and into the dorsal region of the embryo. Upon examination of the sna-gfp construct in which the proximal CRM was deleted, we found that gfp expression was excluded from the posterior hkb expression domain, similar to endogenous sna expression (Fig. 4B, compare with 4A). This result suggested that Hkb can function to repress the sna posterior boundary, even when the proximal CRM is removed. By contrast, gfp expression was expanded into the posterior end of the embryo upon deletion of the distal CRM (Fig. 4C, compare with 4A).
Fig. 4.
Repressors function predominantly through the distal CRM, whereas expansion toward the anterior pole requires the proximal CRM. (A-F) Fluorescent in situ hybridization of wild-type embryos (stage 5) containing either the sna-gfp (A,D), sna-gfp Δ Proximal (B,E) or sna-gfp Δ Distal (C,F) constructs using riboprobes to detect gfp (red) and hkb (green) transcripts. Magnified images of the poles of stage 5 embryos showing the posterior (A-C) and anterior (D-F) variation in sna-gfp expression (red) with respect to the domain of hkb expression (green). The posterior images are projections, whereas the anterior images represent a single scan. Extent of gfp expression supported at the poles is marked by arrowheads in each case. (G,H) Ventrolateral views of in situ hybridization recognizing lacZ (red), driven by either the proximal CRM (G) or the distal CRM (H), and snail (green). The red rectangle in each indicates the area magnified in the bottom image.
sna and hkb expression domains overlap at anterior regions of the embryo. Upon closer analysis of the sna-gfp proximal delete construct, we found that the gfp expression domain recedes relative to sna, such that the boundary of expression was more ventrally located and sharper relative to wild type (Fig. 4E). A similar effect on sna expression has been observed previously in bicoid mutants (Reuter and Leptin, 1994). However, in comparison with the expression domain supported by the sna-gfp distal CRM delete, we found that the sna expression domain was expanded more dorsally at the anterior of the embryo than normal (Fig. 4F), similar to that seen in hkb mutants (Reuter and Leptin, 1994). Collectively, these results suggest the proximal CRM supports Bicoid-mediated activation at the anterior of the embryo and that the distal CRM supports Hkb-mediated repression at both embryonic poles.
The distal CRM supported a refined sna expression boundary not only at the poles where Hkb is functioning but also in lateral regions of the embryo. Because the snail-gfp reporters are also recognized by a riboprobe to snail (such that endogenous and reporter expression cannot be distinguished by in situ hybridization), we chose to assay the lacZ reporter constructs of the two CRMs in comparison with endogenous snail to visualize the patterns at the dorsal boundary of expression. The distal CRM dorsal border was sharp and comparable with the endogenous snail pattern, whereas the proximal CRM supported a patchy expression that was weak and sporadically extended beyond the snail border (Fig. 4H, compare with 4G). Although the dorsal border of the proximal CRM is mostly encompassed by the endogenous snail border, the extension of the proximal CRM even by a few cells beyond the endogenous snail border indicates that this CRM in isolation has no (or reduced) responsiveness to a putative repressor responsible for supporting the sharp boundary (see below).
Previous studies using intersecting dorsoventral patterning cues had provided evidence that one or more repressors might be required for the establishment of the sna lateral border (Huang et al., 1997). In these experiments, an ectopic gradient of nuclear Dorsal (a transcription factor pivotal for dorsoventral patterning) was specified along the anterior-posterior axis by localizing constitutively active Toll receptor to the anterior end of embryos by a transgene, `F10' (Huang et al., 1997). The result was an embryo containing two dorsal-ventral patterning axes that intersect (see Fig. S3A in the supplementary material), with loss of sna expression at the intersection; the existence of a laterally acting repressor was postulated (Huang et al., 1997). Although the identities of such laterally acting repressors (`repressor X') have remained uncertain, we nevertheless assayed whether the proximal and/or distal CRMs are required to support the ability to repress sna in this manner.
When the distal CRM lacZ reporter construct was introduced into the F10 background, complete overlap with the endogenous sna expression domain was observed (see Fig. S3B in the supplementary material). By contrast, the expression domain was expanded both laterally and posteriorly when the proximal 2.2 kb lacZ construct was introduced into the F10 background (see Fig. S3C in the supplementary material). These results are consistent with the idea that both repressor X and Hkb function predominantly through the distal CRM to refine sna borders in lateral and posterior regions of the embryo, respectively, although we cannot dismiss a more minor role at the proximal CRM.
Non-additive patterns of expression when proximal and distal CRMs are located in trans suggest cis-interactions are necessary to support patterning
Although both CRMs are present in the wild-type locus, we noticed that the snail gene expression pattern was not simply the summed equivalent of the domains of expression supported by the two CRMs. We hypothesized that repressors associated with the distal CRM might also work to define the expression supported by the proximal CRM output. This would explain why the endogenous snail expression domain was absent from the posterior pole and also why its lateral boundary was sharp. However, it was also possible that the level of expression supported by each CRM was so different that when both were present, the pattern supported by the distal CRM effectively masked that supported by the proximal CRM. To distinguish between these possibilities, we examined embryos containing various combinations of the proximal delete and/or the distal delete CRM reporters, in either cis or trans conformation, and analyzed the gene expression outputs supported by each combination in terms of spatial domain (Fig. 5) and level of expression (Fig. 6).
Fig. 5.
The proximal and distal enhancers function in a non-additive manner when organized in cis conformation but not in trans. (A-D) Fluorescent in situ hybridization using a gfp riboprobe of stage 5 embryos expressing one of the following constructs: homozygous `snail-gfp Δ proximal' (A), homozygous `snail-gfp Δ distal' (B), heterozygous `snail-gfp Δ proximal'/`snail-gfp Δ distal' (C) or homozygous `snail-gfp' (D). All images were captured under the same confocal settings but the brightness and/or contrast was modulated to support visual comparison of the domains of expression supported by these four transgenes.
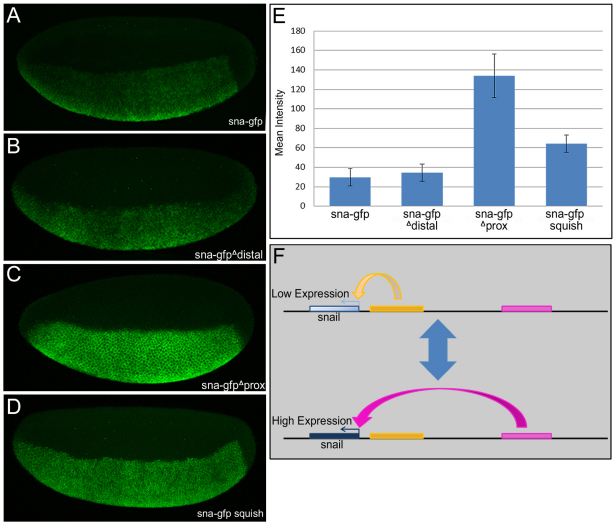
Fig. 6.
The proximal and distal CRMs influence levels of snail expression. (A-D) Fluorescent in situ hybridization using a gfp riboprobe to recognize expression from the gfp reporter constructs. All images were captured under the same settings and there has been no manipulation of the brightness or contrast to allow for visual comparison of the expression levels supported by these four transgenes. (E) The mean intensity of expression in the snail stripe for all four constructs was quantified, showing that deletion of the proximal CRM leads to a greater than fourfold difference in expression levels. Data are mean±s.e.m. (F) Schematic of the interplay between the proximal and distal CRMs and the snail promoter. The proximal CRM drives low level expression, whereas the distal CRM drives high level expression; there is most probably a trade off between the two CRMs, effectively lowering the level of expression that is seen in the full snail construct to a level many times lower than that supported by the distal CRM alone.
At two copies, the proximal CRM delete construct supported refined expression (repressed at the posterior and laterally), whereas the distal CRM delete construct supported expanded expression (extending at the poles and laterally) compared with an unmodified reporter construct (Fig. 5A,B, compare with 5D), similar to expression supported by one copy of the transgenes. However, when reporter expression was assayed in an embryo containing one copy of the proximal CRM delete and one copy of the distal CRM delete transgenes, the pattern supported exhibited an expanded expression domain, most apparent at the posterior pole. This result suggested that the expression supported by the proximal CRM is not simply too weak to be observed in the presence of the expression supported by the distal CRM, but that instead repressors associated with the distal CRM normally function to refine expression at the poles and in lateral regions supported by the proximal CRM. Furthermore, these data demonstrate that repressors associated with the distal CRM cannot function in trans, but instead require a cis conformation relative to position of the proximal CRM in order to have an effect. Our results suggest that the normal pattern is a non-additive reflection of the domains of expression supported by each CRM (see Discussion).
Besides differences in domain of expression, we noticed that these constructs supported differences in levels of expression (Fig. 6). When imaged at a power and gain in which all of the constructs examined were not over-exposed, the mean intensity supported by the sna-gfp and sna-gfp Δ distal constructs were comparable, but in comparison the expression levels supported by the sna-gfp Δ proximal construct were considerably higher (∼3-4 fold). Therefore, in the absence of the proximal CRM, the expression levels increased. At higher gain, however, it was observed that the sna-gfp expression was at least twofold higher than that of the sna-gfp Δ distal (data not shown). Thus, alternately, in the absence of the distal CRM, the expression levels decreased. In addition, the sna-gfp squish construct also supported increased levels of expression relative to the sna-gfp construct (approximately twofold). Collectively, these results suggest that normal levels and patterns of snail gene expression require input from both the proximal and distal CRMs, and that effective regulation of expression levels requires proper organization of these CRMs upon the chromosome.
DISCUSSION
In this study, we provide evidence that early snail expression is regulated by two concurrently acting CRMs that support gene expression patterns that are spatially and functionally different. The distally located CRM is necessary to support gastrulation as well as viability of snail mutants, whereas the proximal CRM is dispensable for viability except at high temperature. Furthermore, our data show these CRMs support distinct expression patterns. Although they probably share many transcription factors, the distal CRM alone is responsive to the repressor Huckebein and the unknown laterally acting `repressor X', whereas the proximal CRM alone responds to an anterior activator.
Our data suggest that the proximal CRM functions as a `damper' to reduce the high levels of expression normally supported by the distal CRM. Multiple CRMs associated with a single gene may support spatiotemporally similar expression patterns, but the mean levels of gene expression supported by each can be very different. In the case of the snail locus, our data show that the distal and proximal CRMs drive high or low levels of expression, respectively, within a similar domain in ventral regions of the embryo. Our results supports a model in which these two CRMs provide dual-control of expression levels, high versus low, to provide flexibility in terms of levels of snail expression (Fig. 6F). The requirement for the proximal CRM at high temperatures could indicate a need to more closely regulate the expression levels of snail in stressful environments. Such flexibility is probably advantageous and may explain why two CRMs that support similar expression patterns may be evolutionarily constrained.
Both the proximal and distal CRMs support expression not only during gastrulation in ventral regions of the embryo but in other domains at later stages of development. The distal CRM also supports expression within malphigian tubule precursors (Fig. 3), and, as was previously shown, the proximal CRM supports expression later within neuroblasts (Ip et al., 1992b). Therefore, these elements can be reused during the course of development, and may be evolutionarily retained for reasons beyond a role in canalization.
CRMs associated with the snail locus function in a non-additive manner to support expression
Our results show that transcription factors associated with the distal CRM can dominantly affect the other proximally located CRM to support expression of sna that is refined and excluded from the posterior pole. Our data support the view that non-autonomous CRM function is responsible for the resulting pattern which is effectively non-additive, i.e. it is not simply the summed equivalent of the domains of expression supported by the two CRMs. Non-autonomous CRM function may be advantageous, providing additional flexibility by allowing individual and combined activities of CRMs based on circumstances, to support canalization. It has been demonstrated that non-additive CRM interactions also play a role defining the expression domain of another Drosophila early patterning gene, sloppy-paired 1 (Prazak et al., 2010). Our data support the view that this is a more common cis-regulatory mechanism than currently appreciated. For example, even in case of the even-skipped gene locus that has received considerably focus, questions remain about why particular CRM behaviors are not equivalent to the behaviors of the eve gene itself. The expansion of a eve stripe 3/7 reporter gene in knirps mutants (Small et al., 1996), but not the eve gene itself (Frasch and Levine, 1987), suggests that another repressor is required to drive proper eve stripe 3/7 expression and that this activity is supported through another DNA fragment. We propose that another CRM associated with the eve locus may aid in definition of eve stripes 3/7 by serving as a vehicle for additional repressors(s), similar in mechanism to regulation of snail gene expression shown here in this study.
CRMs are organized along the DNA to support effective transcription
This study also supports the view that CRMs are organized in the context of the gene locus to support proper patterning and to minimize cross-repressive interactions (see also Cai et al., 1996; Small et al., 1993). We believe that the loss of Ect1 expression that we see in the `squish' construct is the result of dominant repression, owing to the fact that the distal enhancer is moved in proximity to the proximal enhancer (see Fig. 3B). This would suggest that the native context of CRMs within a locus can limit interactions between elements, and may go towards explaining why enhancers in diverged species/animals tend to be found in the same general location (Cande et al., 2009; Hare et al., 2008). Similarly, the dampening of all snail expression patterns we observe in the `D to P' construct may be due to the repressive activity of the distal CRM being moved near the promoter.
Placing binding sites for repressors near the promoter potentially limits the range of activity of a gene. Many genes involved in early development, such as snail, take on different roles later in development and are subject to different molecular inputs during the life of the animal. Like snail, the intermediate neuroblasts defective (ind) gene also has a distally located enhancer and another that is located in the proximal position. Similar to what we see at the snail locus, the distal CRM has documented repression associated with it, whereas the proximally located element functions through positive autoregulatory feedback (Stathopoulos and Levine, 2005; Von Ohlen et al., 2007). We suggest that keeping repressors located at a distance from the promoter supports flexibility in reiterative reactivation of genes throughout the course of development. However, in addition to buffering repressive crosstalk through distance, we propose that linking repression function to the presence of an activator (i.e. between CRMs concurrently active in the same cells) may also serve as an alternate mechanism to moderate non-autonomous CRM interactions; other studies in the past have suggested that repressors may require activators to bind DNA (i.e. `hot chromatin' model) (see Nibu et al., 2001).
Our data show that expression of the Drosophila snail gene in embryos is established through integrated activity of multiple CRMs that function concurrently and, in part, through non-additive interactions. Non-additive activity of CRMs, through sharing of repressors for example, is likely more commonplace than currently appreciated. It is possible that concurrently acting CRMs function coordinately to regulate spatial domain and levels of expression in general, and may provide one explanation why genes in Drosophila and other animals often have multiple CRMs that support similar spatiotemporal patterns of expression.
Supplementary Material
Acknowledgments
We are grateful to Eric Davidson (Caltech) and to the Stathopoulos lab for helpful discussions. In addition, we thank Sagar Damle (Caltech) for advice on BAC recombineering. This work was funded through NIGMS R01 grant GM077668 (A.S.) and corresponding ARRA supplement from the NIH. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.069146/-/DC1
References
- Alberga A., Boulay J. L., Kempe E., Dennefeld C., Haenlin M. (1991). The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development 111, 983-992 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific {varphi}C31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. N., Arnosti D. N., Levine M. (1996). Long-range repression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 93, 9309-9314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande J. D., Chopra V. S., Levine M. (2009). Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136, 3153-3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden J., Levine M. (2002). The Snail repressor positions Notch signaling in the Drosophila embryo. Development 129, 1785-1793 [DOI] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., Wieschaus E. (2006). Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell 10, 257-264 [DOI] [PubMed] [Google Scholar]
- de Velasco B., Mandal L., Mkrtchyan M., Hartenstein V. (2006). Subdivision and developmental fate of the head mesoderm in Drosophila melanogaster. Dev. Genes Evol. 216, 39-51 [DOI] [PubMed] [Google Scholar]
- Frankel N., Davis G. K., Vargas D., Wang S., Payre F., Stern D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Levine M. (1987). Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1, 981-995 [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. (1987). Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6, 749-759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiasvand N. M., Rudolph D. D., Mashayekhi M., Brzezinski J. A. t., Goldman D., Glaser T. (2011). Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. E., Jimenez G., Cook O., Gur D., Paroush Z. (1999). Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126, 3747-3755 [DOI] [PubMed] [Google Scholar]
- Hare E. E., Peterson B. K., Iyer V. N., Meier R., Eisen M. B. (2008). Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 4, e1000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K., Hu X., Ashraf S. I., Small S. J., Ip Y. T. (2004). The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Dev. Biol. 269, 411-420 [DOI] [PubMed] [Google Scholar]
- Hong J.-W., Hendrix D. A., Levine M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. M., Rusch J., Levine M. (1997). An anteroposterior Dorsal gradient in the Drosophila embryo. Genes Dev. 11, 1963-1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Bier E., Levine M. (1992a). The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 6, 1728-1739 [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Yazdanbakhsh K., Levine M. (1992b). dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 6, 1518-1530 [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Levine M., Bier E. (1994). Neurogenic expression of snail is controlled by separable CNS and PNS promoter elements. Development 120, 199-207 [DOI] [PubMed] [Google Scholar]
- Jiang J., Levine M. (1993). Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72, 741-752 [DOI] [PubMed] [Google Scholar]
- Jiang J., Kosman D., Ip Y. T., Levine M. (1991). The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 5, 1881 [DOI] [PubMed] [Google Scholar]
- Kasai Y., Nambu J. R., Lieberman P. M., Crews S. T. (1992). Dorsal-ventral patterning in Drosphila: DNA binding of snail protein to the single-minded gene. Proc. Natl. Acad. Sci. USA 89, 3414-3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Ip Y. T., Levine M., Arora K. (1991). Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254, 118-122 [DOI] [PubMed] [Google Scholar]
- Kosman D., Mizutani C. M., Lemons D., Cox W. G., McGinnis W., Bier E. (2004). Multiplex detection of RNA expression in Drosophila embryos. Science 305, 846 [DOI] [PubMed] [Google Scholar]
- Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., Court D. L., Jenkins N. A., Copeland N. G. (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56-65 [DOI] [PubMed] [Google Scholar]
- Leptin M. (1991). twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 5, 1568-1576 [DOI] [PubMed] [Google Scholar]
- Li X. Y., MacArthur S., Bourgon R., Nix D., Pollard D. A., Iyer V. N., Hechmer A., Simirenko L., Stapleton M., Luengo Hendriks C. L., et al. (2008). Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman L. M., Stathopoulos A. (2009). Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev. Biol. 327, 578-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y., Zhang H., Levine M. (2001). Local action of long-range repressors in the Drosophila embryo. EMBO J. 20, 2246-2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir A., Fisher K., Pepke S., Samanta M., Dunipace L., McCue K., Zeng L., Ogawa N., Wold B., Stathopoulos A. (2011). High resolution mapping of Twist to DNA in Drosophila embryos: efficient functional analysis and evolutionary conservation. Genome Res. 21, 566-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Bothma J. P., Levine M. (2010). Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazak L., Fujioka M., Gergen J. P. (2010). Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev. Biol. 344, 1048-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter R., Leptin M. (1994). Interacting functions of snail, twist and huckebein during the early development of germ layers in Drosophila. Development 120, 1137-1150 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E. E. (2007). A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Arnosti D. N., Levine M. (1993). Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119, 762-772 [PubMed] [Google Scholar]
- Small S., Blair A., Levine M. (1996). Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 175, 314-324 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Levine M. (2005). Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev. Biol. 280, 482-493 [DOI] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747-1751 [DOI] [PubMed] [Google Scholar]
- Von Ohlen T. L., Harvey C., Panda M. (2007). Identification of an upstream regulatory element reveals a novel requirement for Ind activity in maintaining ind expression. Mech. Dev. 124, 230-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong N., Kang C., Raulet D. H. (2002). Redundant and unique roles of two enhancer elements in the TCRgamma locus in gene regulation and gammadelta T cell development. Immunity 16, 453-463 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R. P., Senger K., Levine M., Papatsenko D. (2006). Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 16, 1358-1365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.