Constructing an online database of histones and histone fold-containing proteins has allowed our group to analyze histone sequence variation in some detail (Sullivan et al.1; Sullivan and Landsman2). Here, we describe how we inventoried core histone protein sequences as part of this project. The issues involved in such an undertaking are for the most part not unique to histone sequences. Our methods and observations should be broadly applicable to studies of protein families that are highly represented in public sequence databases.
Considerations
Our initial goal was to collect as many reported histone sequences as we could find. Among the considerations that came into play were the following:
1. Sourcing of sequences
Several excellent public sequence repositories make protein sequences available to researchers. We relied on the protein database maintained by the NCBI, which is updated frequently and has been compiled from worldwide sources, including SwissProt (Boeckmann et al.3), the Protein Information Resource (PIR) (Wu et al.4), the Protein Research Foundation (PRF) (http://www.prf.or.jp/en/), the Protein Data Bank (PDB) (Westbrook et al.5), and translations from annotated coding regions in GenBank (Benson et al.6) and RefSeq (Pruitt et al.7), a curated, non-redundant set of sequences
2. Sequence harvesting tools
Generally a sequence database search is a similarity search of either the actual sequence data or its annotation. We find that both must be targeted in order to maximize the sequence harvest, because sequence-based searches alone can miss small or ambiguous sequence fragments that have been deposited in the public databases, and text-based searches can miss ‘cryptic’ histones, i.e., those with inadequate or incorrect annotation.
For text-based searches of sequence annotation we used the Entrez search engine at the NCBI website http://www.ncbi.nlm.nih.gov/Entrez. For sequence-based searching we used several varieties of the popular BLAST pairwise alignment algorithm. The most commonly used sequence similarity search tools find ‘hits’ based on pairwise alignments of each sequence in the database to either the query sequence alone, e.g., in the case of BLAST, or a query profile derived from a previously aligned set of similar sequences, e.g., in the case of PSI-BLAST or HMMER (Altschul et al.8; Eddy9). The latter tools are better at finding highly divergent members of a protein family but can be expected to return false positives, requiring further filtering of results. PSI-BLAST is actually a hybrid tool that performs one round of standard BLAST using a user-supplied query sequence, then builds a profile from the alignment of the initial BLAST results, which becomes the query for the next round of BLAST. The process is reiterated until ‘convergence’ is reached, that is, until no more new matches are found above the cutoff score. Ideally this should take less than ten iterations, but ‘convergence’ can be elusive when the query sequence matches a diverse and perhaps distantly related set of proteins. This was more difficult to interpret with searches for nonhistone proteins containing the histone fold, than for harvesting core histone sequences. With the latter we found that seven iterations were sufficient to either reach convergence or to reach the point where all of the ‘new’ hits appeared by other criteria to be false positives. PSI-BLAST routinely returned a small number of true positive matches to the query sequences that gapped protein BLAST (BLASTPGP) had missed.
Reasonably fast BLASTPGP and PSI-BLAST servers are available at the NCBI Website http://www.ncbi.nlm.nih.gov/BLAST. One advantage of the NCBI website PSI-BLAST implementation over a command-line version is that the user can edit each set of aligned sequences before it is used to generate a profile. This can redirect a diverging sequence search back towards convergence. Unfortunately, however, it can also happen that a valid match from one iteration falls below the noise cutoff in the next, and in the WWW-based implementation, that match is lost. Therefore we ran PSI-BLAST (and BLASTPGP) from the command line in a UNIX environment, which allowed us to save the results from all of the iterations into one file for subsequent text parsing. It also allows considerable flexibility in setting other BLAST options. Most of default values were adequate for typical BLAST searches, but we commonly increased the number of displayed description lines and alignments (the –b and –v options) to 3000, to ensure retrieval of all of the possible hits for subsequent filtering steps.
3. Query sequences
Histones are ancient proteins, found in all known eukaryote lineages as well as some archaeal microbes. Using a single query sequence, there is the possibility that some valid hits might be missed due to the extreme biodiversity and sequence divergence of the histones, even using a profile-generating protocol. To maximize the identification of eukaryote core histones from the protein databases, we ‘bracketed’ the kingdom evolutionarily by using core histone sequences from human and yeast as search queries. This proved important for the more divergent histones, H2A and H2B, but less so for the more conserved histones H4 or H3. For example, queries with human or yeast H4 or H3 returned almost the same sets of true positive hits. In H3 searches, the most common outliers requiring taxonomic bracketing to capture were sequences fragments from protists, and members of the centromeric H3 subclass (data not shown).
4. Sequence redundancy
Sequence redundancy is the bane of most database searches. In most cases, redundant sequences in a large public sequence repository such as GenBank are often the same sequence from the same organism, automatically harvested from different databases, rather than originating from discrete sequencing projects in different laboratories. Thus, Web-based sequence similarity search tools like PSI-BLAST on the NCBI website, tend to present results in a convenient, non-redundant fashion, with sequence identifiers of identical sequences grouped together with an anchored sequence. To populate the histone database, however, we required every sequence in FASTA format (i.e., each record consisting of only a unique definition line and a sequence), one reason being that homologous histones display remarkable degrees of sequence identity, rather than mere similarity, across species. It is not uncommon that fully ‘redundant’ histone sequences in the public database derive from more than one species. We wanted to start with a set where such identical sequences are properly resolved. Since we are attempting an exhaustive search, the well-intentioned non-redundancy of the public databases was thus for us, a minor obstacle. Our strategy was to extract all of the unique sequence identifiers from the BLAST outputs (in the case of NCBI records, the unique identifier is the ‘gi number’ found at the beginning of the sequence definition line of a FASTA-formatted record), into a file, and use this file to generate a corresponding ‘library’ of FASTA records. NCBI Entrez on the World Wide Web can take a file of gi numbers as input for batch retrieval of records; alternately, we used the SEALS software suite to perform such retrievals in a UNIX environment (Walker and Koonin10). SEALS has a ‘fauniq’ tool for reducing a set of redundant FASTA sequence records to a non-redundant format, based either on definition-line identifiers such as the gi number, or on the sequence itself. This tool proved invaluable for filtering BLAST outputs to remove gi-based redundancies, and for generating non-redundant sequence sets for alignment and variation analysis.
5. Fragmentary, ambiguous, and frameshifted sequences
Some sequences in the public databases are less than full-length; for example, a few records annotated as ‘histone H3’ consist of only two or three amino acid residues. As sequences get shorter their detection becomes more difficult for typical ‘flavors’ of BLAST when querying a large database since they become less distinguishable from chance hits. This problem is compounded if, as is the case with histones, the protein features segments of low sequence complexity, or if the fragment records contain ambiguous (‘X’) residues. To capture sequence fragments, we first divided the full-length query sequence into overlapping segments, with a segment ‘window’ of twenty residues, sampled at intervals of ten residues along the length. This was easily done with the SEALS ‘fenestrate’ command. We then used these segments as queries against the public database, in a modified gapped BLAST search optimized to capture short, nearly exact matches (a search option that is also available on the NCBI website cited earlier). For these searches, low-complexity filters were turned off. The combined results of all the ‘window BLASTS’ for a query sequence were made non-redundant with respect to gi number.
Frameshifted sequences (either authentic or erroneous) can pose a similar problem depending on the size of the frameshifted region. Putative frameshifts are easily identified by visual inspection of multiple alignments of query results, e.g. using the popular Clustal X program (Thompson et al.11), where they manifest as sudden and extensive loss of sequence similarity. To verify a frameshift, assuming access to the genomic DNA or cDNA record for the protein (which are often, but not always, available in public databases), one should translate the DNA in all frames and add those conceptual translations to the alignment; the ‘correct’ frames will be visually evident in a true frameshift. Several tools exist on the web for doing such translations; we commonly use the one at the ExPASy (Expert Protein Analysis) website: http://us.expasy.org/tools/dna.html. A translation tool is also available in the SEALS package.
Comparison of search strategies
There are many available variations on the basic BLAST search protocol. We investigated several parameters for their effects for the identification of histone H3 sequences. Histone H3 is a moderately diverse histone class, with more than half of known full-length sequences displaying >80% identity in their histone fold domains; a figure falling between the more highly conserved H4 and the more diverse H2A and H2B classes (Sullivan and Landsman2). The H3 class comprises two subclasses that are markedly distinct in sequence and in function: replication-dependent H3 (the major H3) and centromeric H3. There is also a third, replication-independent H3.3 class, though its sequence is only marginally divergent from that of the major H3.
We first compiled a redundant reference set of H3 sequences, using a variety of BLAST- and Entrez-based searches, to include as many probable H3 sequence records as we could find in the NCBI protein database. This set was manually reviewed to eliminate false positives, yielding a final set of 1742 good candidate H3 sequences from all three subclasses. We then compared the results of different individual BLAST and Entrez search strategies to the reference set, to determine the efficiency (percentage of hits that are true positives, i.e., that also found in the reference set) and the success (percentage of the reference set found by the search method). The results are shown in Table 1. Entrez searches of eukaryotic sequence record annotation used the terms ‘H3’ or ‘histon’. BLAST parameters that we varied were: query sequence BLAST ‘flavor’ (gapped BLAST vs. gapped PSI-BLAST vs. gapped BLAST for short, nearly exact ‘window’ matches); query sequence (human vs. yeast); database size (all vs. the eukaryotic subset); and SEG low-complexity filtering (off vs. on).
Table 1. Comparison of search strategies for H3 histone sequences.
Entrez queries of the NCBI protein database were conducted from the NCBI website www.ncbi.nlm.nih.gov/Entrez. BLAST searches using human or yeast histone H3 sequences were performed from the command line in a Unix environment:
| reference H3 set | uniquegi |
H3 |
success | efficiency |
|---|---|---|---|---|
| 1742 | 1742 | |||
| ENTREZ “eukaryota[ORGN]” | 1143461 | 1742 | 100.0% | 0.2% |
| ENTREZ “H3” | 3303 | 1452 | 83.4% | 44.0% |
| ENTREZ “histon” | 9297 | 1653 | 94.9% | 17.8% |
| ENTREZ “eukaryota[ORGN] and H3” | 2703 | 1452 | 83.4% | 53.7% |
| ENTREZ “eukaryota[ORGN] and histon” | 7453 | 1653 | 94.9% | 22.2% |
| BLASTPGP H3human | 1747 | 1719 | 98.7% | 98.4% |
| BLASTPGP H3human+seg | 1747 | 1719 | 98.7% | 98.4% |
| BLASTPGP H3human+eukgi | 1754 | 1722 | 98.9% | 98.2% |
| BLASTPGP H3human+eukgi+seg | 1754 | 1722 | 98.9% | 98.2% |
| BLASTPGP H3yeast | 1777 | 1718 | 98.6% | 96.7% |
| BLASTPGP H3yeast+seg | 1777 | 1718 | 98.6% | 96.7% |
| BLASTPGP H3yeast+eukgi | 1780 | 1718 | 98.6% | 96.5% |
| BLASTPGP H3yeast+eukgi+seg | 1780 | 1718 | 98.6% | 96.5% |
| PSIBLASTPGP H3human | 1897 | 1726 | 99.1% | 91.0% |
| PSIBLASTPGP H3human+seg | 1897 | 1726 | 99.1% | 91.0% |
| PSIBLASTPGP H3human+eukgi | 1949 | 1727 | 99.1% | 88.6% |
| PSIBLASTPGP H3human+eukgi+seg | 1949 | 1727 | 99.1% | 88.6% |
| PSIBLASTPGP H3yeast | 2011 | 1726 | 99.1% | 85.8% |
| PSIBLASTPGP H3yeast+seg | 2011 | 1726 | 99.1% | 85.8% |
| PSIBLASTPGP H3yeast+eukgi | 2077 | 1727 | 99.1% | 83.1% |
| PSIBLASTPGP H3yeast+eukgi+seg | 2077 | 1727 | 99.1% | 83.1% |
| WINBLASTPGP H3human | 69678 | 1730 | 99.3% | 2.5% |
| WINBLASTPGP H3human+eukgi | 60821 | 1732 | 99.4% | 2.8% |
| WINBLASTPGP H3human+eukgi+seg | 1697 | 1646 | 94.5% | 97.0% |
| WINBLASTPGP H3yeast | 70864 | 1730 | 99.3% | 2.4% |
| WINBLASTPGP H3yeast+eukgi | 63949 | 1730 | 99.3% | 2.7% |
| WINBLASTPGP H3yeast+eukgi+seg | 1788 | 1646 | 94.5% | 92.1% |
BLASTPGP = gapped protein blast; PSIBLASTPGP = interated gapped protein blast using profiles; WINBLASTPGP = gapped protein BLAST for short, nearly exact matches, using sequence windows as queries; eukgi = search restricted to sequences from eukaryotes; seg = SEG filtering of low-complexity regions enabled. All results were compared to a curated reference_H3_set of sequences. Column headers: uniq gi = number of unique sequence records retrieved; H3 = number of retrieved unique gis shared with the reference set; efficiency = percent H3/uniq gi; success = percent H3/reference set.
The Entrez results indicate that almost 20% of H3 sequences in the public database are ‘cryptic', lacking specific annotation as H3 histones. The search results for ‘histon’ as a query term recovered 95% of the reference sequences, with a trade-off of many more false positives, as one would expect. The ‘histon’ query also captured all of the true positive ‘H3’ query results (data not shown).
Any of the BLAST-based strategies was sufficient to capture at least 94% of the reference set from the public databases. The best combination of efficiency and success was achieved using gapped BLAST. The effects of differences in query sequence, database size, and filtering were minor compared to the difference between using BLAST, PSI-BLAST or ‘windowed’ BLAST, because the latter two BLAST implementations return far more false positives while increasing the success rate only marginally. Low entropy sequence filtering appeared to make no difference whatsoever except in the case of ‘windowed’ searches, in which the query sequence was divided into overlapping segments 20 residues each in length, with several gapped BLAST parameters altered to facilitate finding short, nearly exact matches to the query segments. Using the low-complexity filter here vastly increased efficiency by greatly reducing false positives, though success suffered in comparison to non-filtered strategies, reflecting the presence of short, often basic low-complexity regions that are a hallmark of core histone sequences.
Unfortunately, as these results show, no single method captures all of the relevant sequence records. A combination of strategies is the only way to achieve 100% success. However, the results of our comparison suggest a rational way to mine the maximum number of histone sequence records of a class from a database. The first step is to perform a single-round gapped BLAST search, making sure that the options for ‘number of descriptions’ and ‘number of alignments’ returned are set high (e.g. several thousand each). This should return most of the true positives with high efficiency. This set should be inspected carefully, using a variety of tools including text-search of the definition lines, multiple alignment, and further BLAST searches with a different query sequence, to remove false positives. The resulting ‘validated’ set becomes most useful in subsequent searches employing other strategies, such as PSI-BLAST or text-based searches. The validated set can be used to ‘subtract’ known positives from subsequent search results, using difference-finding tools such as the SEALS ‘fanot’ command, which finds the logical exclusion of two sets of FASTA records or definition lines. This leaves a much shorter list of candidates from the new search results to be examined for new true positives. As these are identified they are added to the validated set, increasing its usefulness as a filter. This search strategy has also served us well in harvesting histone H4, H2A, and H2B sequences, and should work for any well-conserved class of protein sequences.
Histone Sequence Variants
Histone variants have been divided into ‘homomorphous’ and ‘heteromorphous’ categories (West and Bonner12; reviewed in Ausio et al.13), also sometimes referred to as ‘minor’ and ‘major’ variants. Homomorphous variants have relatively minor sequence differences and require high-resolution separation methods to distinguish them biochemically (reviewed in von Holt et al.14). They are found in all four core histone classes, and are presumed to be functionally identical. Heteromorphous variants are readily distinguished by conventional biochemical separation methods and tend to be distinct from other histones in their class with respect to function and/or spatiotemporal localization, as well as sequence. The distinction between the two categories of variants is not rigid – e.g., the ostensibly ‘homomorphous’ H3.3 appears to be functionally distinct from the major H3 – and may become less so as the functions of more variants are experimentally tested. In clustering trees made from multiple sequence alignments of each histone class, heteromorphous variants tend to form biodiverse clades distinct from the major form, indicating early branching off from major histones, while homomorphous variants tend to co-mingle with the major form in clades that are more strongly delineated by phylogeny than by any other factor, suggesting the variants arose after the founding speciation event (data not shown; see also Thatcher and Gorovsky15). For all core histone classes, sequence alignments show clear distinctions between metazoan, plant, fungal, and various basal eukaryote subclasses. Distinct subclasses within the metazoan sequences are also common (e.g., insect or echinoderm sequences). Nomenclature is only occasionally helpful in classifying histone variants. It is not standardized, and thus ‘H3.2’ in one species may not be similar to ‘H3.2’ in another. The only other constant among aligned histone sequences apparent in Figures 1-4, is that there tends to be less variation in the α-helical regions of the histone fold, than in the inter-helical loops and the N- and C-terminal regions flanking the histone fold. This pattern of variation is common in other α-helix-containing protein families.
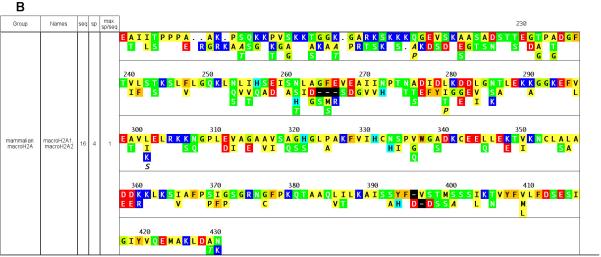
Figure 1. Summary of H2A subclasses and variants.
A. A consensus sequence of all aligned H2A sequences is shown at top. Dots in the sequences below indicate identity to the consensus. Groups are named based on clustering patterns observed in neighbor-joining trees of aligned H2A sequences (not shown); Names = a selection of sequence descriptors found in the definition lines of the sequence records; seq = number of unique sequences in the group; sp = number of species in the group; max sp/seq = the greatest number of species having the same sequence in the group. For each group the first line is the consensus sequence for that group. Variations from the group consensus are indicated below it. Italic = ‘singleton’, i.e., the residue was found in only one sequence from one species in the group. * = singleton identity or gap. Background color key: white = identity to the anchored consensus; black = gap; orange= aromatic; yellow=aliphatic/hydrophobic; light green = glycine; green = hydrophilic; light blue = histidine; blue = basic; red = acidic B. C-terminal section of macroH2A.
Figure 4. Summary of H4 subclasses and variants.
See legend for Figure 1.
H2A
The H2A class is the more diverse of the four core histone classes both functionally and in terms of sequence, comprising four subclasses of known or putative functional variants in addition to typical phylogeny-based subclasses (Figures 1A and 1B). H2A.X is found in species spanning the eukaryotic spectrum and features a conserved serine four residues from the carboxyl terminus (part of an SQ motif, positions 208-9 in Figure 1A) that is phosphorylated in response to double-stranded DNA breaks, perhaps marking the site for repair (reviewed in Redon et al.16). Interestingly, the fungal H2A subclass clusters near the H2A.X subclass, and also features a conserved SQ motif at its C-terminus. H2A.F/Z sequences constitute another pan-eukaryotic subclass, and are necessary but not sufficient for H2A function in organisms tested. Characteristic H2A.F/Z residues in a C-terminal, H3-binding portion of the protein (positions 145-193 in Figure 1A) have been suggested to impart a specific, though as yet unknown, function, as have the lysine residues in the amino-terminal portion (reviewed in Redon, Pilch16). Of these lysine residues, two (at positions 11 and 42 in Figure 1A) appear to be specific to H2A.F/Z and not the major metazoan H2A. MacroH2A is a large bipartite histone divided into a recognizable H2A portion with many subclass-characteristic substitutions, and a long C-terminal extension found in no other histone subclass (residues 227-430 in Fig 1C). MacroH2A has been found only in vertebrates and is concentrated in the inactive female X chromosome (reviewed in Brown17). H2A-Bbd is a highly divergent subclass, so far found only in mammals, which displays a complementary localization to macroH2A, i.e., it is excluded from inactive chromosomes (Chadwick and Willard18).
H2B
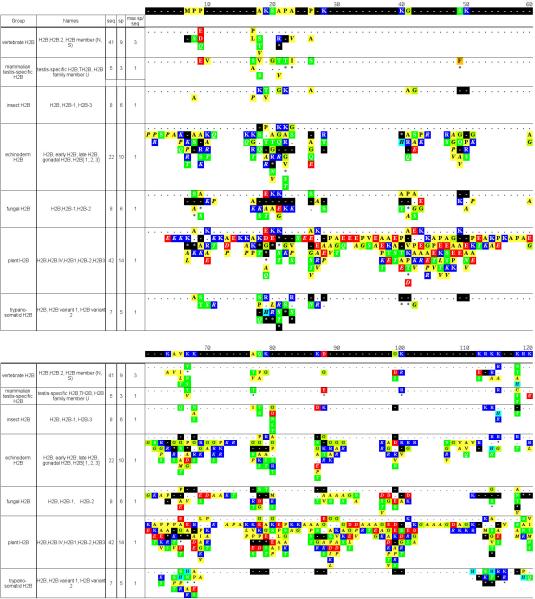
Functional subclasses of H2B sequences have not been positively identified, though at least one tissue-specific form has been identified in mammalian testis (Figure 2). An echinoderm sperm variant featuring a repeating pentapeptide has also been described (reviewed in Von Holt et al.19), indicating that the echinoderm group in Figure 2 probably could be subdivided further. The N-terminal diversity seen within the plant subclass in Figure 2 suggests that it, too, could be further subdivided.
Figure 2. Summary of H2B subclasses and variants.
See legend for Figure 1.
H3
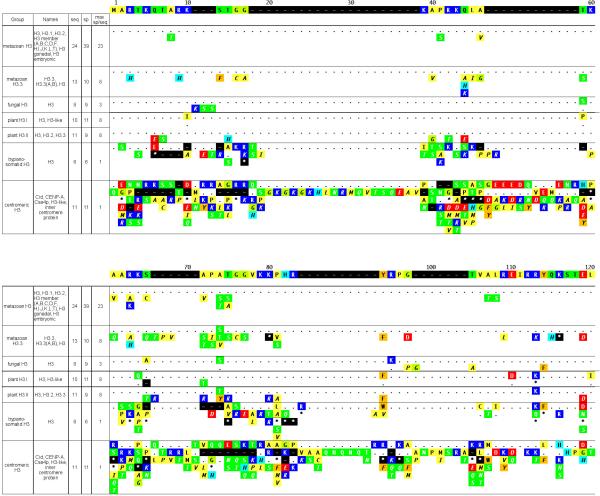
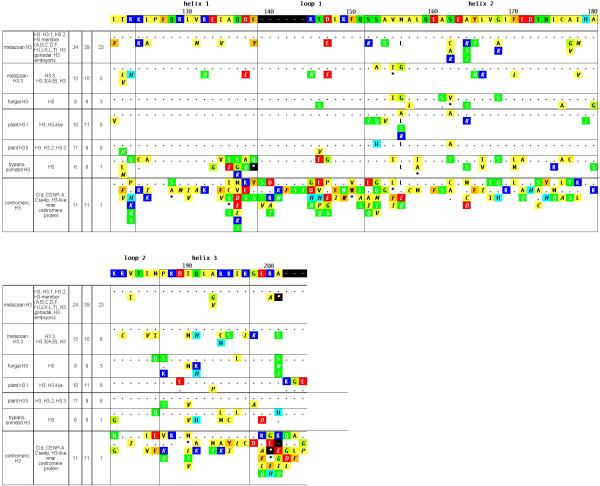
The H3 class notably contains two subclasses of replication-independent variants that are differentially localized within the cell. Histone H3.3 is an ostensibly ‘homomorphous’ metazoan subclass that varies significantly from the predominant H3 in only four positions (positions 73, 153, 155, and 156 of Figure 3). H3.3 can be deposited in nucleosomes of replicating DNA like the major H3, but can also be deposited in non-replicating DNA, preferentially in actively transcribed regions (Ahmad and Henikoff20). The replication-independence of H3.3 may be mediated by any of the three H3.3-specific residues in positions 153-156 (Ahmad and Henikoff21). Centromere-specific H3 is found in species ranging from yeast to human, and its deposition has been shown to be replication-independent (reviewed in Smith22). It is thought to help specify centromere regional identity within the chromosome. Centromeric H3 displays somewhat more subclass specificity (and considerably more diversity) within the histone fold than other H3 subclasses (Figure 3), which may reflect a role in forming specialized nucleosomes.
Figure 3. Summary of H3 subclasses and variants.
See legend for Figure 1.
H4
The H4 class is the most conserved of the four core histones. No functional, localization, or expression variants are known, and thus the clustering of its sequences falls entirely along phylogenetic lines. (Figure 4)
References
- 1.Sullivan S, Sink DW, Trout KL, Makalowska I, Taylor PM, Baxevanis AD, Landsman D. Nucleic Acids Res. 2002;30:341. doi: 10.1093/nar/30.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan SA, Landsman D. Proteins. 2003 doi: 10.1002/prot.10441. in press. [DOI] [PubMed] [Google Scholar]
- 3.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. Nucleic Acids Res. 2003;31:365. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. Nucleic Acids Res. 2003;31:345. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westbrook J, Feng Z, Chen L, Yang H, Berman HM. Nucleic Acids Res. 2003;31:489. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. Nucleic Acids Res. 2003;31:23. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2003;31:34. doi: 10.1093/nar/gkg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy SR. Bioinformatics. 1998;14:755. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 10.Walker DR, Koonin EV. Proc Int Conf Intell Syst Mol Biol. 1997;5:333. [PubMed] [Google Scholar]
- 11.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West MH, Bonner WM. Biochemistry. 1980;19:3238. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- 13.Ausio J, Abbott DW, Wang X, Moore SC. Biochem Cell Biol. 2001;79:693. [PubMed] [Google Scholar]
- 14.von Holt C, Brandt WF, Greyling HJ, Lindsey GG, Retief JD, Rodrigues JD, Schwager S, Sewell BT. Methods Enzymol. 1989;170:431. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 15.Thatcher TH, Gorovsky MA. Nucleic Acids Res. 1994;22:174. doi: 10.1093/nar/22.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Curr Opin Genet Dev. 2002;12:162. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 17.Brown DT. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-7-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chadwick BP, Willard HF. J Cell Biol. 2001;152:375. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Holt C, Strickland WN, Brandt WF, Strickland MS. FEBS Lett. 1979;100:201. doi: 10.1016/0014-5793(79)80337-3. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad K, Henikoff S. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16477. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad K, Henikoff S. Mol Cell. 2002;9:1191. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 22.Smith MM. Curr Opin Cell Biol. 2002;14:279. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]