Abstract
THO/TREX is a conserved nuclear complex that functions in mRNP biogenesis and prevents transcription-associated recombination. Whether or not it has a ubiquitous role in the genome is unknown. Chromatin immunoprecipitation (ChIP)-chip studies reveal that the Hpr1 component of THO and the Sub2 RNA-dependent ATPase have genome-wide distributions at active ORFs in yeast. In contrast to RNA polymerase II, evenly distributed from promoter to termination regions, THO and Sub2 are absent at promoters and distributed in a gradual 5′ → 3′ gradient. This is accompanied by a genome-wide impact of THO–Sub2 deletions on expression of highly expressed, long and high G+C-content genes. Importantly, ChIP-chips reveal an over-recruitment of Rrm3 in active genes in THO mutants that is reduced by RNaseH1 overexpression. Our work establishes a genome-wide function for THO–Sub2 in transcription elongation and mRNP biogenesis that function to prevent the accumulation of transcription-mediated replication obstacles, including R-loops.
Keywords: replication impairment, R-loops, Rrm3, THO/TREX, transcription
Introduction
THO/TREX is a conserved nuclear complex that functions in mRNP biogenesis and has a role in preventing transcription-associated genome instability (Luna et al, 2008). First identified in the yeast Saccharomyces cerevisiae as consisting of four tightly bound proteins (Tho2, Hpr1, Mft1 and Thp2; Chavez et al, 2000), THO interacts with other proteins involved in mRNA export, such as the Sub2 RNA-dependent ATPase and the RNA-binding protein Yra1 to form TREX together with Tex1 (Jimeno et al, 2002; Strasser et al, 2002). The integrity of yeast THO requires the four THO proteins but not Sub2 (Huertas et al, 2006). Chromatin immunoprecipitation (ChIP) experiments demonstrated that Hpr1, Sub2 and Yra1 are recruited to specific genes such as PMA1, ACT1, DBP2, or GAL1 under active transcription and that Hpr1 travels with the RNA polymerase II (RNAPII) and it is required for the efficient recruitment of Sub2 and Yra1 (Lei et al, 2001; Strasser et al, 2002; Zenklusen et al, 2002). Thus, the current view is that THO associates with the transcription machinery and recruits Sub2 and Yra1 to the nascent transcript (Rondon et al, 2010). The association with the transcription machinery seems to be lost downstream of the polyadenylation site (Kim et al, 2004).
THO null mutants are viable but show impaired transcription elongation, hyper-recombination and RNA export defects (Chavez et al, 2000; Strasser et al, 2002; Rondon et al, 2003b; Mason and Struhl, 2005). THO was proposed to be required for efficient transcription elongation of long and G+C-rich genes (Chavez et al, 2001) and of genes with internal repeats (Voynov et al, 2006). Transcription of short and low G+C-content genes is also affected in THO mutants with the strongest phenotypes such as hpr1Δ and tho2Δ (Garcia-Rubio et al, 2008). In this respect, sub2Δ mutants show phenotypes as hpr1Δ and tho2Δ. Notably, however, the abundance of THO subunits has been estimated in 500–5000 molecules/cell whereas this value is about 50 000 for Sub2 (Ghaemmaghami et al, 2003). It is therefore unclear whether THO function extends to all transcribed genes or is specific to a subset of genes and whether THO recruitment is restricted to RNAPII genes or is extended to other DNA regions.
One of the most intriguing features of yeast THO mutants is their high transcription-associated recombination (TAR) levels. In the absence of proteins functioning at the interface between transcription and mRNA export such as THO, mRNP biogenesis does not occur properly and the nascent mRNA can form stretches of DNA:RNA hybrids (R-loops; Huertas and Aguilera, 2003). These observations make THO mutants a paradigmatic example to evaluate the importance of R-loop formation in transcription and recombination (Yu et al, 2003; Li and Manley, 2005). Consistent with the role of recombination as a major DNA repair pathway for lesions occurring during replication and the ability of transcription to become an obstacle for replication and to compromise genome integrity (see Aguilera and Gómez-González, 2008 and Branzei and Foiani, 2010), several reports suggest that replication fork progression is impaired in specific plasmid and chromosomal DNA regions in THO mutants (Huertas and Aguilera, 2003; Wellinger et al, 2006; Gómez-González et al, 2009).
Here, we analysed genome-wide expression profiles of THO–Sub2 mutants and recruitment of Hpr1 and Sub2 to chromatin by ChIP-chip experiments in asynchronously and synchronously growing yeast cultures. The impact of the mutations in gene expression is general and not restricted to a specific functional class of genes, but long, high G+C-content and highly expressed genes are those preferentially down-regulated in THO–Sub2 mutants. Interestingly, THO–Sub2 binds to virtually all RNAPII actively transcribed genes. Hpr1 and Sub2 are not observed at promoters, and their recruitment into ORFs follows an increasing gradient in the 5′ → 3′ direction. The pattern of replication fork pausing, as determined by ChIP-chip analysis of the Rrm3 helicase required for progression of replication through obstacles (Ivessa et al, 2000, 2003), reveals a genome-wide impairment of replication progression in transcribed genes in the wild type (WT) that is clearly increased in the hpr1Δ mutant. Notably, the higher accumulation of Rrm3 in THO mutants is reduced by RNaseH overexpression, consistent with the idea that R-loops contribute to replication fork impairment. Consequently, our work establishes a genome-wide function for THO and Sub2 in transcription and in preventing RNAPII-mediated replication obstacles.
Results
Genome-wide down-regulation of long, G+C-rich and highly expressed genes in THO–Sub2 null mutants
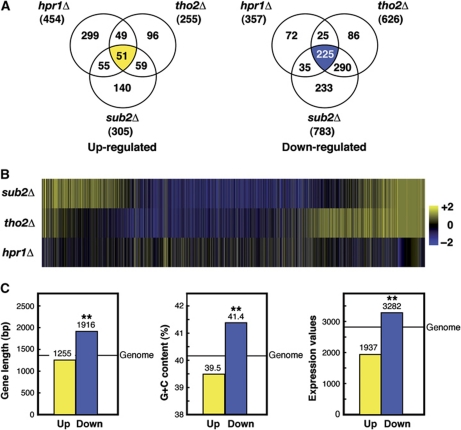
To determine whether the consequences of THO–Sub2 depletion extend to all transcribed genes of the genome or is specific to only a subset of them, we performed genome-wide microarray analyses in hpr1Δ, tho2Δ and sub2Δ strains. mRNA levels that were at least 1.5-fold above or below the WT values were found for 811 genes in hpr1Δ, 881 genes in tho2Δ and 1088 genes in sub2Δ (Supplementary Dataset I). Consistent with the similarity of the phenotypes described for these mutations (Garcia-Rubio et al, 2008), overlap analyses between the three sets of genes revealed a high correlation (Figure 1A and B). Therefore, the genome-wide expression data are consistent with the idea that these proteins form a functional and structural unit.
Figure 1.
Comparative analysis of genes affected by the different THO–Sub2 mutations. (A) Venn diagrams representing the overlap between genes whose expression is changed in hpr1Δ, tho2Δ and sub2Δ mutants. See also Supplementary Dataset I. (B) Heatmap of genes affected by THO/TREX null mutants. The heatmap of the 50% genes with major intensities is shown. (C) Statistical analyses of length, G+C content and model-based expression values of genes whose expression levels are coincidently affected in hpr1Δ, tho2Δ and sub2Δ mutants as compared with the genome average. **P<0.0001 (Mann–Whitney U-test). See also Supplementary Figure S1.
Next, we assessed by statistical analyses whether genes whose expression was dependent on THO and Sub2 could be catalogued into a subset of genes with specific structural or functional features. We selected the 51 genes up-regulated and 225 genes down-regulated that we found to overlap in all sets of genes affected by hpr1Δ, tho2Δ or sub2Δ (see Figure 1A). First, we performed basic statistical analyses of the average length, G+C content, and expression levels of these genes. The set of down-regulated genes (Figure 1C; Supplementary Figure S1) were longer, G+C richer and with higher expression in the WT than the genome average (P<0.0001, Mann–Whitney U-test), where long genes tend to be G+C-poor and lowly expressed (Marin et al, 2003). However, the values of these variables in the set of up-regulated genes were more similar to the average yeast gene. We conclude that the effects of constitutive THO–Sub2 depletion are stronger in long, G+C-rich and highly expressed genes of the genome.
Gene ontology analysis indicated that there is no relevant functional class of genes specifically affected by any of the mutations tested (Supplementary Dataset I). The impact of THO/TREX deletions is clearly visible in long, G+C-rich and highly expressed genes consistent with results with artificial reporters (Chavez et al, 2001; Garcia-Rubio et al, 2003). These results not only indicate a global functional relevance of THO/TREX, but a functional equivalence between their different subunits. Thus, all subsequent studies were performed with Hpr1 and Sub2.
Genome-wide recruitment of THO and Sub2 to RNAPII-transcribed genes
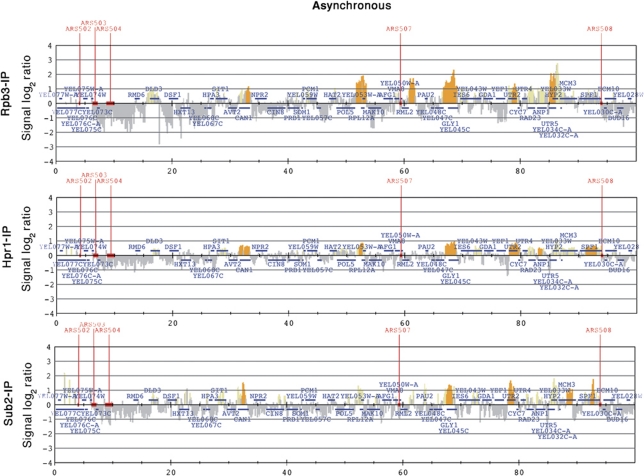
We performed ChIP-chips with FLAG-tagged Hpr1 and Sub2 proteins and compared the results with those of RNAPII recruitment as determined with a PK-tagged Rpb3 subunit. Asynchronous log-phase cultures of Hpr1-FLAG, Sub2-FLAG and Rpb3-PK-tagged strains were cross-linked with formaldehyde in vivo. Samples were then collected and processed for ChIP with anti-FLAG and anti-PK-specific antibodies, respectively, and hybridized to a S. cerevisiae tiling array (ChIP-chip method; Katou et al, 2003). No-tag strains (No-FLAG and No-PK) were used as controls and ChIP-chip data of these strains showed no recruitment at all, as expected (Supplementary Figure S2). Data were subjected to computational analysis to obtain a genomic map distribution (Figure 2; Supplementary Dataset II). Statistical analyses revealed that Rpb3, Hpr1 and Sub2 were significantly enriched in 2380, 2057 and 2629 genes, respectively, most of which overlapped (Supplementary Figure S3). Linear regression between the three data sets yielded correlation coefficients consistent with a linear relationship between each pair (R2=0.71, 0.60 and 0.65; Supplementary Figure S3). We also found a significant correlation between the Rpb3, Hpr1 and Sub2 clusters (Supplementary Table I), in which the significance of the correlations is computed as the actual distribution with respect to randomly simulated positions (see Materials and methods), indicating that the coincidence of Rpb3, Hpr1 and Sub2 is statistically significant. Importantly, 94% of Hpr1, 92% of Sub2 and 81% of Rpb3 ChIP hits peak inside ORFs. Similar results were obtained for Tho2 (data not shown). Thus, THO is mostly recruited to the same genes as Rpb3; that is, to RNAPII-transcribed genes. Sub2 is recruited to RNAPII-transcribed genes as THO, consistent with the physical association of Sub2 with THO.
Figure 2.
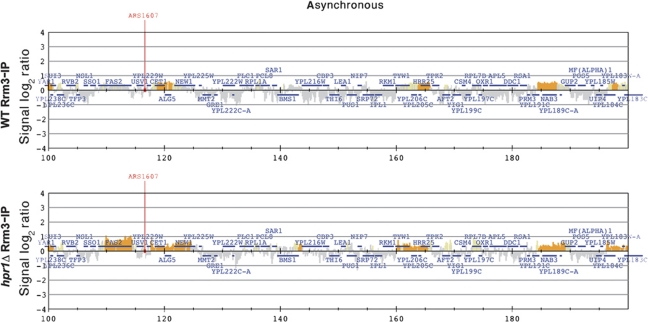
Hpr1 and Sub2 recruitment correlates with RNAPII binding and transcription levels. Rpb3-IP (top), Hpr1-IP (middle) and Sub2-IP (bottom) histogram bars in the y axis show the average signal ratio of loci significantly enriched in the immunoprecipitated fraction along the indicated regions in log2 scale (grey), and whether they fulfil one of the P-value criteria (yellow) or both (orange) (see Materials and methods). The x axis shows the chromosomal coordinates. Positions of ARS elements are indicated. The horizontal bars mark the positions of the indicated ORFs. See also Supplementary Dataset II and Supplementary Figures S2 and S3.
Basic statistical analysis of the average length, G+C content, and mRNA expression levels showed that the genes recruiting Rpb3, Hpr1 and Sub2 are highly expressed as compared with the genome average (Supplementary Figure S4), indicating that the higher the transcription levels of genes the better the detection capacity of transcription proteins by ChIP. Altogether the data imply that the THO complex is recruited to most RNAPII-transcribed genes regardless of their function, length, G+C content and expression level.
Hpr1, Tho2 and Sub2 are absent at promoters and increasingly recruited towards the 3′ end regions
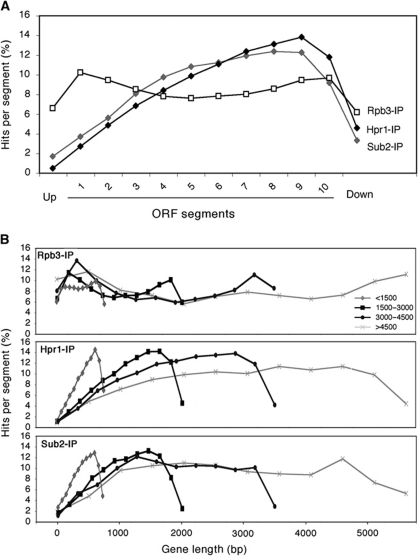
To analyse the distribution of THO and Sub2 along the length of all ORFs, these were subdivided into 10 segments independent of ORF size. In addition, we included two flanking segments with the same size upstream (5′) and downstream (3′) of each ORF. 5′ and 3′ ends of genes were defined by the positions of the start and stop codons. The occupancy for Rpb3, Hpr1 and Sub2 was determined by the number of hits (see Materials and methods) mapping on each segment across a given ORFs. The binding profile of Rpb3 was similar along the length of the ORFs (Figure 3A), although it showed a slight over-accumulation at the start site and termination regions. A similar pattern has just been reported in a genome-wide analysis of Rpb3 recruitment, even though in this case the analysis was performed only on the genes with no close neighbours and did not show up the peak at the 3′ end (Mayer et al, 2010). Notably, the binding profiles for Hpr1 and Sub2 were different from that of Rpb3, showing an increase from the 5′ towards the 3′ end of the genes. Thus, THO and Sub2 recruitment to ORFs occurs in a gradient manner, being absent at promoter regions and highly abundant towards the 3′ end of genes. The low recruitment at promoter with respect to termination regions can also be observed when analysing genes individually (Supplementary Figure S5) or when analysing the intergenic spacers (Supplementary Figure S6).
Figure 3.
THO–Sub2 recruitment increases towards the end of ORFs and at terminator regions. (A) Composite profile of Rpb3, Hpr1 and Sub2 occupancy (detected by ChIP-chip) across the average ORF plotted as Rpb3, Hpr1 and Sub2 percentage of ChIP hits per segment (see Materials and methods). (B) Composite profile of Rpb3, Hpr1 and Sub2 occupancy across the average ORF for different intervals of gene length. Other details as in (A). See also Supplementary Figures S4–S6.
This result implies that chromatin recruitment of THO and Sub2 does not depend on RNAPII or transcription activation itself, but on some feature or signal that is modified along the length of any ORF. One possible feature could be the length of the nascent mRNA. If THO sensed the nascent mRNA, we would expect that the longer the ORF, the higher the levels of THO would accumulate towards the 3′ ends. However, the percentage of hits per segment for different intervals of gene length in all genes revealed that the increase in Hpr1 and Sub2 recruitment is independent of gene length (Figure 3B). The overall levels of recruitment towards the 3′ ends is the same regardless of whether the nascent RNA is <1.5 kb long or longer than 4.5 kb. We can therefore conclude that THO–Sub2 recruitment does not rely, at least not only, on the length of synthesized RNA.
Recruitment to RNAPII-transcribed genes is independent of cell cycle
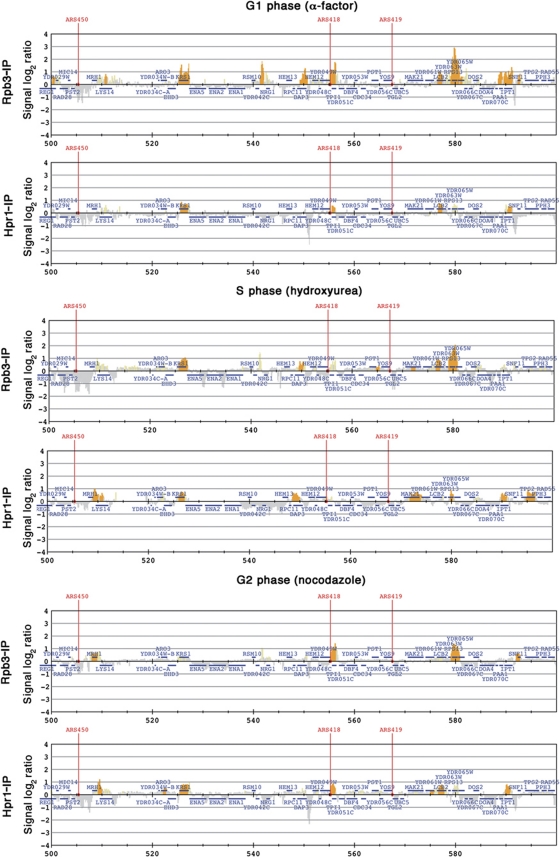
Next, given the known impact of THO mutations on genome instability, we assayed whether THO recruitment to transcribed genes changed, depending on the cell-cycle phase. We analysed Hpr1-FLAG and Rpb3-PK ChIPs in cells synchronized in G1, S and G2 cell-cycle phases using α-factor, hydroxyurea or nocodazol, respectively (Supplementary Dataset III; Supplementary Figure S7A). As shown in Figure 4, among the three cases, profiles were highly similar to those obtained in asynchronous cultures. The few differences observed are mainly explained by the different expression pattern of genes throughout the cell cycle. Therefore, recruitment of Hpr1 to RNAPII-transcribed genes is independent of the cell-cycle phase, consistent with a global and general role of THO along the genome, regardless of gene function or any structural feature.
Figure 4.
Hpr1 binding correlates with RNAPII binding in every cell-cycle phase. Histogram bars of Rpb3 (top) and Hpr1 (bottom) ChIP-chip data in different cell-cycles phases: α-factor arrested for G1, hydroxyurea arrested for S and nocodazol arrested for G2. Other details as in Figure 2. See also Supplementary Dataset III and Supplementary Figure S7.
High genome-wide accumulation of Rrm3 at transcribed genes in hpr1Δ cells
In order to study the global effects of THO depletion in replication progression, we performed Rrm3-FLAG ChIP-chip analysis (Supplementary Dataset IV). Rrm3 is a helicase required for the progression of the replication fork through obstacles in the DNA and its accumulation at specific DNA sites has been used to identify replication fork pauses or stalls (Ivessa et al, 2000, 2003). ChIP-chips were done in asynchronous cultures according to a previous study showing that replication fork pausing is captured in asynchronous cells (Azvolinsky et al, 2009).
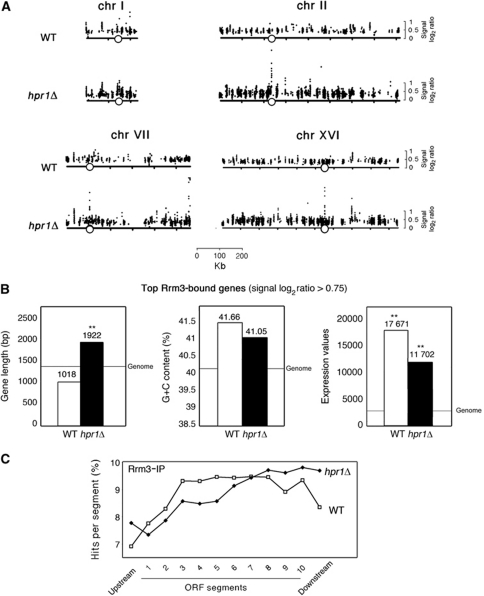
As shown in Figure 5, there is a strong overlap between the clusters of Rrm3 recruitment in WT and hpr1Δ cells, for which there were 78 and 75% of the hits that mapped in ORFs for each strain, respectively, and there was a high coincidence between the ORFs recruiting Rrm3 in WT and hpr1Δ cells. Among these ORFs, we observed a 45% overlap with the genes recruiting Rpb3 in WT cells (Supplementary Figure S7B), similar to previous results (Azvolinsky et al, 2009). Importantly, 43% of the genes recruiting Hpr1 in WT cells overlapped with genes recruiting Rrm3 in hpr1Δ cells. Therefore, Rrm3 recruitment appears mostly at highly transcribed regions in both WT and hpr1Δ cells. These are indeed the regions at which THO is more frequently accumulated. Importantly, the hits of Rrm3 recruitment were clearly enhanced in hpr1Δ (Figures 5 and 6A; Supplementary Figure S8), implying that natural replication fork pause sites are stronger or occur more frequently in hpr1Δ cells. Additional pausing regions can also be seen recruiting Rrm3 in hpr1Δ with respect to the WT. Interestingly, among these additional replication fork pause sites are most centromeres and telomeres, suggesting an additional impact of THO function on replication through these heterochromatic regions (Figure 6A; Supplementary Figure S8). However, this is less clear at the HMR or HML regions of chromosome III (data not shown). Further analyses would be required to define the physiological relevance of this observation.
Figure 5.
Genomic view of Rrm3 recruitment in WT and hpr1Δ mutant. Rrm3 ChIP-chip data of a region of chromosome XVI in WT (top) and hpr1Δ (bottom). Other details as in Figure 2. See also Supplementary Dataset IV and Supplementary Figure S7.
Figure 6.
Rrm3 recruitment in highly transcribed DNA regions in WT and hpr1Δ cells. (A) Genome-wide recruitment of Rrm3 to different chromosomes in WT and hpr1Δ mutants. A representation of chromosomes I, II, VII and XVI with the signal log2 ratio values for the significant hits is plotted. The x axis shows chromosomal coordinates in kb. Positions of centromeres are indicated as open circles. See also Supplementary Figure S8. (B) Statistical analysis of length, G+C content and model-based expression values of genes showing Rrm3 recruitment with signal log2 ratios above 0.75 in WT (52 genes) or hpr1Δ cells (137 genes). Grey lines indicate the genome average value. **P<0.0001 (Mann–Whitney U-test). (C) Composite profile of Rrm3 occupancy (detected by ChIP-chip) across the average ORF plotted as percentage of hits per segment in WT or hpr1Δ cells. Other details as in Figure 3.
To determine the functional and structural features of the genes to which Rrm3 preferentially binds, we selected the genes in which the signal log2 ratio of hits was above 0.75 (52 in WT and 137 in hpr1Δ). Statistical analysis revealed that on average these are the most highly transcribed genes in both WT and hpr1Δ, indicating that highly transcribed RNAPII genes are more prone to affect replication fork progression (Figure 6B). Among them, there was the same proportion of genes with co-directional versus converging transcription and replication (see Materials and methods). Gene ontology analysis revealed that genes involved in translation and expression were significantly over-represented (Supplementary Dataset I), which may in part be due to the fact that genes involved in these processes are highly transcribed. Furthermore, the average length of this top class of Rrm3-immunoprecipitated genes is higher in hpr1Δ than in the WT (P<0.0001, Mann–Whitney U-test), suggesting that replication through long and highly transcribed genes is the most dependent on the THO complex. The average length was also higher when we considered the genes that specifically showed Rrm3 binding in hpr1Δ and not in the WT (data not shown).
While the final mechanisms for the formation of R-loops remains to be understood, in vitro experiments have shown that R-loops only form when transcription occurs in the direction that results in a G-rich transcript (Yu and Lieber, 2003; Roy and Lieber, 2009). This is consistent with a positive (0.028) (G−C)/(G+C) ratio for the average genome, which means a sequence bias towards G in the non-transcribed strand for the whole genome. Lower ratios were obtained for the 52 and 137 top Rrm3-bound genes in WT and hpr1Δ (a 0.001 and a −0.014 ratio, respectively), indicating that a higher G proportion in the non-transcribed strand is not required for R-loop formation and subsequent replication fork stalling and Rrm3 accumulation in hpr1Δ cells. This is consistent with our conclusions that, in hpr1Δ cells, it is the suboptimal mRNP formation what stimulates genome instability regardless of which DNA strand is the G-rich strand, as analysed in class switch S regions inserted in the yeast genome (Ruiz et al, 2011).
Finally, it is worth noting that even though Rrm3 is recruited all over the length of genes in both WT and hpr1Δ cells, the binding profile shows a slight tendency to increase towards the 3′ ends in hpr1Δ (Figure 6C). This is coincident with the higher presence of Hpr1 observed in ChIP-chips in these regions.
R-loop-dependent Rrm3 accumulation at transcribed ORFs in hpr1Δ cells
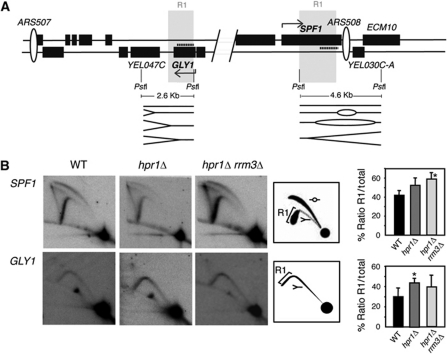
Accumulation of Rrm3 at specific sites may be the result of a replication fork pause or delay (Ivessa et al, 2003). In order to test replication fork progression in more detail, we performed 2D-gel electrophoresis through two highly transcribed genes such as SPF1 and GLY1, which are close to replication origins in chromosome V. As shown in Figure 7, we observed no retention of replication forks at discrete sites. This is expected from the fact that Rrm3 binding in hpr1Δ cells covered the whole ORFs. However, densitometry analysis of the arc signals revealed a weak but consistent delay of replication fork progression along the ORFs. While ChIP-chips have shown that Rrm3 is accumulated in highly transcribed genes, 2D-gel replication patterns are not affected by deleting RRM3 in the WT (Azvolinsky et al, 2009). This implies that the DNA regions in which replication fork finds obstacles (as recorded by Rrm3 accumulation) do not necessarily accumulate specific replication fork pauses or stalls. In any case, a slight but statistically significant replication delay could be observed in the GLY1 or SPF1 genes analysed in hpr1Δ or hpr1Δ rrm3Δ, respectively, although it was not evident in the other cases (Figure 7B). These differences are not sufficient as to be detected in asynchronous cultures (data not shown).
Figure 7.
Replication fork analysis by 2D-gel electrophoresis. (A) Scheme of the DNA region analysed. The indicated 2.6 and 4.6 kb PstI restriction fragments were analysed for GLY1 and SPF1, respectively. Probes are indicated as dash lines. Direction of transcription is indicated with an arrow. The R1 region is shown. (B) Analysis of replication fork progression through the highly expressed GLY1 and SPF1 genes in WT, hpr1Δ and hpr1Δ rrm3Δ cells. The ratio of the signal in the descending Y arc (R1) versus the total replicating molecules (Y plus bubble arcs) is plotted on the right as a way to determine the relative delay of replication fork progression in mutant versus WT strains. One representative gel is shown for each experiment. *P<0.05 as calculated by the Student's t-test.
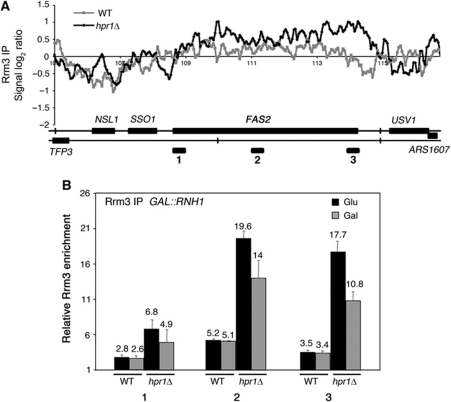
Finally, as one hallmark feature of hpr1Δ mutants is the accumulation of transcription-dependent R-loops, we wondered whether Rrm3 accumulation was related to R-loop formation. For this, we first performed specific Rrm3-FLAG ChIP analyses on selected genes, such as FAS2 on chromosome XVI, which is highly expressed and proximal to a replication origin located downstream of it. As can be seen in Figure 8B, there was a consistent and highly significant enhancement of Rrm3 recruitment to this gene that was up to five-fold higher in hpr1Δ than in WT cells, validating the ChIP-chip results. A similar result was obtained with the GLY1 gene (Supplementary Figure S9). We then repeated the analyses in cells overexpressing RNaseH, which removes the RNA counterpart from the RNA–DNA hybrids of R-loops. Rrm3-FLAG ChIP analysis shows that RNH1 overexpression reduces the levels of Rrm3 detected at the transcribed FAS2 gene (Figure 8B). This result suggests that the genome-wide effect of THO–Sub2 in preventing transcription-dependent replication impairment is linked to the ability of THO to prevent R-loop formation.
Figure 8.
Rrm3 recruitment at the FAS2 gene in WT and hpr1Δ cells and effect of RNAse H1 overexpression. (A) Detailed analysis of ChIP-chip data of Rrm3-FLAG at the FAS2 region. (B) Specific Rrm3-FLAG ChIP analyses using RT–qPCR of three regions (depicted as black rectangles on A) of FAS2 in WT and hpr1Δ cells with normal and overexpressed levels of RNH1. Overexpression was achieved with plasmid pRS315-GALRNH1 under galactose conditions. See also Supplementary Figure S9.
Discussion
We demonstrate in this study a genome-wide function for THO–Sub2 in transcription of all RNAPII-dependent genes regardless of their function and in preventing the accumulation of transcription-mediated replication obstacles all over the genome. First, microarray analyses revealed a major down-regulation on highly expressed, long and G+C-rich genes all over the genome in mutants of THO–Sub2 subunits, but with no significant gene ontology. Second, Hpr1 and Sub2 have genome-wide distributions on transcriptionally active ORFs independent of cell-cycle stage. In contrast to RNAPII, THO and Sub2 are not detected at promoters but are distributed according to an increasing 5′ → 3′ gradient in all active genes. Importantly, the presence of the Rrm3 helicase at active genes in THO mutants was enhanced, which suggests a global impairment of replication fork progression, as supported by 2D-gel analyses of replication fork progression of specific DNA regions. The higher accumulation of Rrm3 in THO mutants is reduced by RNaseH overexpression, consistent with the idea that R-loops are a major contributor to replication fork impairment. We therefore provide evidence for the role of specific eukaryotic mRNP functions in preventing genome-wide replication–transcription collisions that compromise global genome integrity.
A genome-wide impact of THO–Sub2 deletion mutation that is enhanced in long, high G+C and highly expressed genes regardless of their function
Microarray analyses of deletions of either of the THO subunits analysed reveal that they have a significant overlap of genes that are either down-regulated or up-regulated. This overlap is high between sub2 and the most affected mutants of THO, hpr1 and tho2, confirming the structural and functional relationship of Sub2 and THO and despite the much higher (above 10-fold) cell abundance of Sub2 (Ghaemmaghami et al, 2003). THO–Sub2 impact on gene expression is global and independent of gene function. Instead, there is a clear over-representation of down-regulated genes that are longer than average, with a high G+C content and a high expression level, which extends our previous findings using artificial reporters under the GAL1 or tet promoters (Chavez et al, 2001; Garcia-Rubio et al, 2008).
Although the profile of gene expression of deletion mutants does not provide clues about which gene could be a primary target for a putative role of a protein in the control of gene expression, in a study performed in Drosophila, genes involved in DNA metabolism increased their levels of expression in THO mutants (Rehwinkel et al, 2004). This is not evident in our study and further research would be required to assess if there is a correlation between overexpression of specific DNA metabolism genes and the genome instability phenotypes associated with THO–Sub2 mutants.
A global genome-wide recruitment of THO and Sub2 to all RNAPII-transcribed genes
Despite their different cell abundance, THO and Sub2 are present at all transcribed genes all over the genome, regardless of gene function, length, G+C content or levels of expression or cell-cycle stage. THO and Sub2 are mostly found inside the ORF but not at the promoter and only poorly after termination. Therefore, loading of these proteins at transcribed chromatin does not occur through the initiating RNAPII, as is the case of many transcription factors (see Sims et al, 2004).
Importantly, accumulation of THO and Sub2 increases in a continuous manner towards the 3′ end of the genes. It is likely that the signal that facilitates THO and Sub2 recruitment could also increase in a cooperative manner. It would be interesting to know whether some RNAPII subunits or specific transcription factors are modified during transcription elongation to allow THO and Sub2 loading. Lack of THO at initiation sites, its increasing recruitment towards the 3′ end, the reported activity of human UAP56 as an RNA-dependent ATPase (Shi et al, 2004) and the relevance of THO in preventing transcription-dependent R-loop formation (Huertas and Aguilera, 2003) would be consistent with the idea that these factors sense and bind directly the nascent RNA or a hnRNP bound to the RNA. Indeed, it has been shown that yeast-purified THO binds RNA in vitro (Rondon et al, 2003a) and that for two endogenous genes tested, Sub2 recruitment is partially dependent on THO and RNA, whereas this is not the case for THO (Zenklusen et al, 2002; Abruzzi et al, 2004). The observation that the levels of THO and Sub2 accumulated towards the 3′ end of genes are similar regardless of the length of a particular gene and consequently, the length of the nascent RNA, indicates that these proteins do not only sense RNA length. We believe that THO and/or Sub2 may sense some other signal that marks the stage of the transcriptional cycle of RNAPII. The recent observation that RNAPII CTD Ser2 phosphorylation follows the same increasing pattern towards the 3′ end of ORFs in genome-wide ChIP-chip analyses (Kim et al, 2010; Mayer et al, 2010) opens the possibility that THO–Sub2 recruitment and CTD Ser2 phosphorylation are linked phenomena.
The observation that more Hpr1 hits accumulate in intergenic regions for genes with convergent transcription than for divergent transcription (Supplementary Figure S6), could suggest a possible role for Hpr1 in relieving the torsional stress created when two transcription bubbles converge. However, further research would be required to ascertain this possibility. Interestingly, hpr1Δ and tho2Δ cells show synthetic growth defect with top1 and top2 mutations (Aguilera and Klein, 1990).
The low abundance of intracellular THO complex (Ghaemmaghami et al, 2003) may not easily fit with a stable presence in all transcribed genes with a gradient increase towards the 3′ ends. This may be explained by a transient and dynamic recruitment of THO/TREX to transcribed genes, in contrast to a stable and permanent presence of RNAPII. As has been proposed previously, THO may be required to load Sub2 (Zenklusen et al, 2002), which is highly abundant (∼50 000 molecules/cell) and therefore can be stably recruited to chromatin. The fact that THO is recruited in a gradient manner but is not present in promoters or in early part of the transcript—where Rpb3 is present—may explain part of the difference. Our data suggest that the loading of THO is transient. THO is likely more stably loaded at the 3′ end, where it may also be important for polyA+ processing (Rougemaille et al, 2008). Consequently, the engagement of THO in transcription may be highly dynamic as to be able to span the whole transcriptome. The major abundance towards the 3′ end of genes may be related to the fact that the longer the ORF, the higher the number of molecules recruited. Therefore, the impact of THO mutations on gene expression would be stronger for long ORFs, which might require a more stable association of the complex. It is also likely that a high G+C content makes the RNA prone to adopt suboptimal mRNP structure in the absence of THO/TREX.
Rrm3-dependent replication obstacles in RNAPII-transcribed genes in THO mutants
Rrm3 is a DNA helicase with an important role in facilitating the progression of replication fork through nucleo-protein obstacles in the DNA (Ivessa et al, 2003; Azvolinsky et al, 2006). A recent genome-wide ChIP-chip analysis revealed that Rrm3 can be used as a marker to identify DNA regions in which replication has a higher probability of suffering pause or stalling (Azvolinsky et al, 2009). Our ChIP-chips show an over-representation of Rrm3 hits in highly transcribed genes, confirming that transcription is a major source of genome instability by interfering with the progression of the replication fork (Prado and Aguilera, 2005; Azvolinsky et al, 2009; Bermejo et al, 2009). Importantly, the abundance of Rrm3 at transcribed ORFs is clearly enhanced in hpr1Δ mutants, suggesting that DNA regions prone to replication pauses in WT cells are highly sensitive to the absence of THO–Sub2. These results were confirmed by a fine-tuned study of two genes, in which the enhancement of the Rrm3 signal by ChIP is evident. Therefore, the data support a general function of THO in preventing the formation of RNAPII-mediated obstacles to the progression of replication forks.
It has been shown that highly transcribed genes that recruit high levels of Rrm3 were similarly distributed among genes in which transcription and replication are co-directional or convergent, thus implying that the direction of transcription is not relevant to determine replication fork impairment in a WT (Azvolinsky et al, 2009). Consistently, and given the fact that we observe a high overlap between our Rrm3-binding data in both WT and hpr1Δ cells and the data previously described in WT (Azvolinsky et al, 2009), we observed no difference in the proportion of genes that are transcribed co-directionally or head-on with respect to replication. Therefore, our results suggest an orientation-independent ability of R-loops to stall replication fork progression (measured as Rrm3 accumulation in hpr1Δ). This is consistent with the fact that instability of direct repeats in THO mutants is independent of the orientation in which the repeats are transcribed with respect to the replication fork (our unpublished results). It is worth mentioning that in contrast to the rDNA region or highly expressed tRNA and mRNA plasmid constructs in yeast and bacteria (Takeuchi et al, 2003; Prado and Aguilera, 2005; Mirkin and Mirkin, 2005; de la Loza et al, 2009; Boubakri et al, 2010), no site-specific replication fork pause is observed in hyper-recombinant hpr1Δ mutants (Wellinger et al, 2006; Gómez-González et al, 2009; Figure 7B). This strengthens our conclusion that the type of obstacle generated in THO mutants does not localize to specific sites along the genome, making it hard to probe them via 2D-gel analysis as has also been shown by the Zakian's laboratory for this type of obstacles (Azvolinsky et al, 2009).
In contrast to the sharp pause or stalling sites that are enhanced by rrm3Δ in different reports (Ivessa et al, 2003), replication impairment in THO mutants is observed along DNA regions rather than being localized at particular and discrete sites, as it has also been reported recently in WT cells (Azvolinsky et al, 2009). Our study at the SPF1 and GLY1 ORFs, in which Rrm3 is over-represented, confirms that the delay in replication progression is not detectable as pausing or stalling sites, as THO is needed to prevent stalling along the entire ORF. Therefore, the function of THO during RNAPII transcription elongation contributes to the prevention of transcription–replication collisions, avoiding chromosome fragility and hyper-recombination.
A number of specific factors functioning in mRNP biogenesis, including THO/TREX and THSC/TREX-2, which functions in the proximity of the nuclear pore complex, link genome stability to RNA functions (Luna et al, 2005; Gonzalez-Aguilera et al, 2008; Faza et al, 2009). Notably, it has been shown that R-loops are formed in these mutants and are linked to the transcription-dependent hyper-recombination (Huertas and Aguilera, 2003; Gómez-González and Aguilera, 2007; Gonzalez-Aguilera et al, 2008). Evidence for the importance of R-loops in the origin of genetic variation and instability exists for class switch recombination of immunoglobulin genes in B cells and in HeLa and chicken DT40 cells depleted of the SF2/ASF splicing factor (Yu et al, 2003; Li and Manley, 2005). Interestingly, a recent genome-wide search of siRNAs causing accumulation of phosphorylated H2AX in human cell lines permitted the identification of a relevant group of RNA metabolism proteins whose depletion may also lead to R-loop formation (Paulsen et al, 2009). It is likely that the global impact of Hpr1 depletion along the genome is linked to the formation of transcription-dependent R-loops. Consistently, we have shown that over-expression of RNH1 strongly reduces the accumulation of Rrm3 at FAS2, which supports the previous observations that RNH1 overexpression minimizes the transcription-dependent hyper-recombination and pausing of the replication fork at a lacZ reporter (Huertas and Aguilera, 2003; Wellinger et al, 2006) and the HU sensitivity of hpr1Δ (Gómez-González et al, 2009).
The mechanism of R-loop formation is unclear. However, it should involve RNA re-invasion of the DNA duplex to form a DNA–RNA hybrid with the concomitant displacement of the non-transcribed strand (Aguilera, 2005). This re-invasion process could be favoured by an unusually high stability of the RNA–DNA hybrid, that is local G-richness of the nascent RNA as it has been shown in vitro (Roy and Lieber, 2009; Belotserkovskii et al, 2010). Alternatively, the re-invasion process must be favoured by the negative supercoiling behind the transcribing RNAPII as we have previously proposed (Huertas and Aguilera, 2003; Aguilera, 2005; Gómez-González and Aguilera, 2007). The known synthetic growth defect of THO mutants with top1 and top2 mutations would support this view (Aguilera and Klein, 1990; data not shown). Interestingly, recent data suggest that the anchoring of the nascent RNA to its DNA template could also generate negative supercoiling between the anchoring point and the transcribing RNA polymerase, which would also promote R-loop stability (Belotserkovskii and Hanawalt, 2011). It is also possible that the suboptimal formation of the mRNP particle could make the RNA more prone to react with the template DNA or could help to stabilize longer stretches of DNA–RNA hybrids as we have previously proposed (Gómez-González and Aguilera, 2007). In this sense, we cannot discard that a G-rich RNA molecule would be more dependent on the action of THO/TREX for its proper assembly than a ribonucleoparticle as we have previously proposed for the mammalian S regions (Gómez-González and Aguilera, 2007). In any case, the influence of other elements such as the chromatin structure remains to be tested.
Whether the molecular nature of transcription–replication collisions leading to genome instability in WT cells versus THO/TREX and other RNA processing/export mutants occurs by the same mechanism is yet to be seen. However, different genetic evidence suggests that TAR has a lower dependence on the RNA molecule in WT cells than in THO mutants (Garcia-Rubio et al, 2003; Gómez-González and Aguilera, 2007). Previous observations indicating that the S-phase checkpoint is activated and is required to respond to replication fork obstacles linked to the accumulation of R-loops in hpr1Δ cells (Gómez-González et al, 2009) supports the relevance of mRNP biogenesis factors and of R-loops in preventing replication fork progression impairment that could compromise genome integrity. It is worth remarking that although our study has been performed with the THO complex, similar results would be expected for mutations in the THSC/TREX-2 complex and other mRNP biogenesis factors that have been previously shown to control transcription-associated genome instability in a RNA-dependent manner (Gonzalez-Aguilera et al, 2008).
In summary, we demonstrate a genome-wide function and recruitment of THO and Sub2 towards the 3′ end of RNAPII genes in a transcription-dependent manner, which is consistent with a global role of THO in elongation and in the formation of the mRNP particle. THO–Sub2 contributes to reduce the collisions between elongating RNA polymerases and the replication fork. Our study, therefore, demonstrates the genome-wide physiological relevance of mRNP functions in the maintenance of genome integrity and opens new perspectives to understand the molecular nature of this phenomenon.
Materials and methods
Strains and plasmids
Yeast strains used are shown in Supplementary Table II. Plasmid pRS315-GALRNH1 was constructed by sub-cloning the SalI–SpeI GAL∷RNH1 fragment from pRS416GALRNH1 (Huertas and Aguilera, 2003) into SalI–SpeI digested pRS315. FLAG-tagged strains were constructed by PCR amplification of the FLAG sequence from pU6H10F (De Antoni and Gallwitz, 2000) with the following oligos:
HPR1up 5′-TTCGAACATTTCTAATGGTTCATCTACCCAAGATATGAAATCCCACCACCATCATCATCAC-3′ and HPR1low 5′-TTTCTTATCAGTTTAAAATTTCTATTAAGAGGATAATTTAACTATAGGGAGACCGGCAGATC-3′, SUB2up 5′-CCCAGAAGAAGGCATTGATCCGTCCACTTATTTGAATAATTCCCACCACCATCATCATCAC-3 and SUB2low 5′-ATATAATCTATATAAAAACGTATCTTTTTTCCTTTATTAACTATAGGGAGACCGGCAGATC-3′, RRM3up 5′-AAGAGTAAAAGATTTCTATAAACGTTTAGAAACTTTGAAATCCCACCACCATCATCATCAC-3′ and RRM3low 5′-GAAAACTTCAACTAGAGTATATGCATTTATTCGTTGCAAGACTATAGGGAGACCGGCAGATC-3′, and integration into the SY2201 strain to yield SYHPR1, SYSUB2 and SYRRM3. PCR products from genomic DNA of the U678-4C strain (Piruat and Aguilera, 1998) obtained with oligos hpr1A 5′-GAAAGGAATAGGGAGC-3′ and hpr1B 5′-GAAAAGCGTGGTGATAA-3′ were used to replace HPR1 in SYRRM3 by hpr1Δ∷HIS3 thus giving rise to strain SYHRRM3. All strains were verified by PCR and Southern. Correct expression of FLAG-tagged proteins was confirmed by western. Strains were grown to a final concentration of 1 × 107 cells/ml. Growth of all epitope-tagged strains was similar to the corresponding untagged strains (Supplementary Figure S10).
Microarray gene expression analysis
Microarray determination of total RNA was performed using the Affymetrix platform (see Supplementary Dataset I). For each experimental microarray determination, total RNA was isolated from each of the three deletion strains used and compared with RNA isolated from its isogenic WT strain in the same conditions. The relative RNA levels for all yeast genes were determined using an Affymetrix microarray scanner. For each strain, microarray analysis was conducted in triplicate. All values presented represent the average of these three determinations. The expression data for each mutant can be accessed at Gene Expression Omnibus (GSE24802).
ChIP-chip experiments
S. cerevisiae oligonucleotide tiling microarrays were provided by Affymetrix. The high-density oligonucleotide arrays used are able to analyse yeast chromosomes at a 300-bp resolution, each of the 300-bp region being covered by at least 60 probes. Cells were arrested in G1 with 3 μg/ml of α-factor during 2 h (G1 phase) and then released in new media with the corresponding treatment: 200 mM HU for 1 h (S phase) or 15 μg/ml Nocodazole for 3 h (G2 phase). ChIP-chip was carried out as described (Katou et al, 2003, 2006; Bermejo et al, 2007, 2009). Briefly, we disrupted 1.5 × 108 cells by multi-beads shocker (MB400U, Yasui Kikai, Japan), which was able to keep cells precisely at lower than 6°C during disruption by glass beads. Anti-FLAG monoclonal antibody M2 (Sigma-Aldrich) or anti-PK SV5-Pk1 antibody (AbD Serotec) were used for ChIP. ChIP DNA was purified and amplified by random priming using a WGA2 kit (Sigma-Aldrich) and following the manufacturer's procedure. A total of 4 μg of amplified DNA was digested with DNaseI to a mean size of 100 bp, purified, and the fragments were end labelled with biotin-N6-ddATP23. Genomic profiles of all proteins studied are provided in Supplementary Datasets II, III and IV. The entire set of data can be accessed at Gene Expression Omnibus (GSE24960).
Statistical analysis
Microarray data analysis of gene expression was performed using the dChip application software (http://www.dchip.org; Li and Wong, 2001). All arrays were normalized with PM probes (invariant set normalization and model-based expression) and the outliers treated as missing data in the subsequent analyses. For each strain, the expression profile was compared with its isogenic WT strain and the genes showing at least a 1.5-fold expression change considered as altered (parameters: absolute difference between signal in mutant versus WT strain >100; difference P-value <0.05). Heatmaps were done with the Multiexperiment Viewer application software.
ChIP-chip analysis was carried out as described (Katou et al, 2003, 2006; Bermejo et al, 2007, 2009). Data were analysed using the GCOS software from Affymetrix with a custom Chip Description File that summarizes the probes within 50 bp of genomic position into a single probe set. GCOS produces signal, detection P-value and change P-value for each probe set as described. The extent of protein binding across the genome was estimated by the number of loci that showed P-values <0.025 for both ‘detection’ and ‘change’ and that were flanked by loci fulfilling the two above criteria. Therefore, a hit showing a positive signal for the binding involves three contiguous loci filling the two critical P-values. A gene was considered to have positive binding when there was at least one hit that mapped within its ORF.
Data were also analysed using a modified version of the Tiling Array Suite software (TAS) from Affymetrix. TAS produces per each probe position the signal and the change P-value taking into account the probes localized within a given bandwidth around the inspected probe. Protein chromosomal distribution was then analysed by detecting binding clusters, which where defined as ranges within the chromosome respecting the following conditions: estimated signal (IP/SUP-binding ratio) positive in the whole range; change P-value of the Wilcoxon signed rank test <0.2 in the whole range, except for segments within the range shorter than 600 bp; size of the region at least 600 bp. The significance of such protein cluster distribution was evaluated by confrontation against a null hypothesis model generated with a Montecarlo-like simulation as described (Bermejo et al, 2009). Briefly, for each pair of data sets (binding clusters of two specific proteins), 1000 randomizations of the positions of the genomic ranges were produced. For each randomized set, the number of peaks falling within the ranges was counted and taken as a random score of the model. The random scores’ distribution was validated to be approximately normal (∣Skew∣ <0.25 and ∣Kurtosis excess∣ <0.25) and then the average and standard deviation for the random model was taken as null hypothesis. The increase or decrease ratio for the scores of the actual positions with respect to the expected value for the null hypothesis (the average score of random attempts) was then calculated, and the P-value for the drift was estimated as standard normal CDF of actual—mean deviation (for further details see Bermejo et al, 2009). See Supplementary Table I for the detailed correlations and statistical significances.
To visualize the distribution of binding sites along genes (Figures 3 and 6), ORF longer than 500 bp were divided into 10 equivalent segments from the start and end coordinates. Two additional segments of the same size were considered upstream (5′) and downstream (3′) each ORF. The number of hits mapping on each of the 12 segments was counted (for every gene). The sum of hits for each segment across genes was plotted as a percentage of the total number of hits. The distribution of hits per segment remains the same when considering only hits with a signal log ratio >0.5, >0.75 or even >1 and P<0.01 instead of P<0.025 for both change and detection (see Supplementary Figure S11), thus bolstering the reliability of the binding profiles.
To be confident on the orientation of replication versus transcription (co-directional (C) or head-on (H)), we considered only those genes that are within 10 kb of an early firing, efficient origin (defined as replication timing <25.6 min in Yabuki et al, 2002) and obtained 5C versus 9H in the WT and 16C versus 23H in hpr1Δ. Similar proportions (26C versus 26H and 73C versus 63H in WT and hpr1Δ, respectively) were obtained when considering all genes (not only those within 10 kb of an efficient, early origin) or when considering all identified origins in Yabuki et al, 2002 (22C versus 28H and 59C versus 71H in WT and hpr1Δ, respectively).
ChIP analysis
Recruitment of Rrm3 to chromatin was determined by ChIP analyses on the endogenous FAS2 gene as previously described (Hecht et al, 1999). The GFX purification system (Amersham) was used for the last DNA purification step. We used the PCR of the intergenic region at positions 9716–9863 of chromosome V as a negative control. Real-time quantitative PCR and calculations of the relative abundance of each DNA fragment was performed as described (Huertas et al, 2006). For each experiment, the DNA ratios in the different regions were calculated from the DNA amount of these regions relative to the intergenic region. Median and s.d. of three independent experiments are shown.
2D-gel electrophoresis
WT, hpr1Δ and hpr1Δrrm3Δ cells were arrested with α-factor and released into minimal medium containing 40 mM hydroxyurea for 30 min before DNA extraction. DNA extraction was performed with the cetyltrimethylammonium bromide method, and neutral–neutral 2D-gel electrophoresis was performed as described previously (Liberi et al, 2006). Probes for 2D-gel analyses were purified PCR fragments from genomic DNA with oligos 5′-TCAAACTTTAATGTGCGGTG-3′ and 5′-GCAAGTGCCAAACCTGTCAC-3′ for SPF1 and oligos 5′-TGATGGAGGCCGCTTTAGAG-3′ and 5′-CGGAGATGGCCAATTTGACT-3′ for GLY1.
Supplementary Material
Acknowledgments
We thank C Tous, E Andújar and M Pérez for technical assistance in microarray analysis and D Haun for style supervision. This work was funded by grants from the Spanish Ministry of Science and Innovation (BFU2006-05260 and Consolider Ingenio 2010 CSD2007-015), Junta de Andalucía (BIO102 and CVI4567) and the European Union (FEDER) to AA, Telethon and AIRC to MF, Spanish Ministry of Science (BFU2007-60791) and Junta de Andalucía (BIO162) to AM, and supported by a UICC Yamagiwa-Yoshida Memorial International Cancer Study Grant.
Author contributions: BG-G, MG-R and RB performed the experiments. BG-G, MG-R, RB, HG, KS and AM analysed the data. BG-G, MG-R, RB, HG, AM, MF and AA discussed and prepared the results, and revised and contributed to the final version of the manuscript. BG-G and AA wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abruzzi KC, Lacadie S, Rosbash M (2004) Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A (2005) mRNA processing and genomic instability. Nat Struct Mol Biol 12: 737–738 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gómez-González B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Klein HL (1990) HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol 10: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA (2009) Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Hanawalt PC (2011) Anchoring nascent RNA to the DNA template could interfere with transcription. Biophys J 100: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC (2010) Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA 107: 12816–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, Foiani M (2009) Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138: 870–884 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M (2007) Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev 21: 1921–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B (2010) The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Garcia-Rubio M, Prado F, Aguilera A (2001) Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 21: 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Gallwitz D (2000) A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246: 179–185 [DOI] [PubMed] [Google Scholar]
- de la Loza MC, Wellinger RE, Aguilera A (2009) Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair 8: 620–626 [DOI] [PubMed] [Google Scholar]
- Faza MB, Kemmler S, Jimeno S, Gonzalez-Aguilera C, Aguilera A, Hurt E, Panse VG (2009) Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol 184: 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio M, Chavez S, Huertas P, Tous C, Jimeno S, Luna R, Aguilera A (2008) Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics 279: 123–132 [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A (2003) Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics 165: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gómez-González B, Aguilera A (2007) Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci USA 104: 8409–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B, Felipe-Abrio I, Aguilera A (2009) The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Gómez-González B, Huertas P, Luna R, Aguilera A (2008) The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1999) Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol Biol (Clifton, NJ) 119: 469–479 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A (2006) An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol Cell Biol 26: 7451–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kaneshiro K, Aburatani H, Shirahige K (2006) Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol 409: 389–410 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL (2005) Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Liberi G, Cotta-Ramusino C, Lopes M, Sogo J, Conti C, Bensimon A, Foiani M (2006) Methods to study replication fork collapse in budding yeast. Methods Enzymol 409: 442–462 [DOI] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A (2008) Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma 117: 319–331 [DOI] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A (2005) Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 18: 711–722 [DOI] [PubMed] [Google Scholar]
- Marin A, Gallardo M, Kato Y, Shirahige K, Gutierrez G, Ohta K, Aguilera A (2003) Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast 20: 703–711 [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM (2005) Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol 25: 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A (1998) A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (2005) Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E (2004) Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A (2003a) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J 22: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Aguilera A (2010) The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A (2003b) Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278: 39037–39043 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, Libri D (2008) THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321 [DOI] [PubMed] [Google Scholar]
- Roy D, Lieber MR (2009) G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 29: 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Gómez-González B, Aguilera A (2011) AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet 7: e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Cordin O, Minder CM, Linder P, Xu RM (2004) Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci USA 101: 17628–17633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voynov V, Verstrepen KJ, Jansen A, Runner VM, Buratowski S, Fink GR (2006) Genes with internal repeats require the THO complex for transcription. Proc Natl Acad Sci USA 103: 14423–14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A (2006) Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cell 8: 781–789 [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451 [DOI] [PubMed] [Google Scholar]
- Yu K, Lieber MR (2003) Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair (Amst) 2: 1163–1174 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F (2002) Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.