Abstract
The cytosolic chaperonin CCT is a 1-MDa protein-folding machine essential for eukaryotic life. The CCT interactome shows involvement in folding and assembly of a small range of proteins linked to essential cellular processes such as cytoskeleton assembly and cell-cycle regulation. CCT has a classic chaperonin architecture, with two heterogeneous 8-membered rings stacked back-to-back, enclosing a folding cavity. However, the mechanism by which CCT assists folding is distinct from other chaperonins, with no hydrophobic wall lining a potential Anfinsen cage, and a sequential rather than concerted ATP hydrolysis mechanism. We have solved the crystal structure of yeast CCT in complex with actin at 3.8 Å resolution, revealing the subunit organisation and the location of discrete patches of co-evolving ‘signature residues’ that mediate specific interactions between CCT and its substrates. The intrinsic asymmetry is revealed by the structural individuality of the CCT subunits, which display unique configurations, substrate binding properties, ATP-binding heterogeneity and subunit–subunit interactions. The location of the evolutionarily conserved N-terminus of Cct5 on the outside of the barrel, confirmed by mutational studies, is unique to eukaryotic cytosolic chaperonins.
Keywords: actin, CCT, chaperonin, crystallography, protein folding
Introduction
The cytosolic chaperonin containing TCP1 (CCT) is a 1-MDa protein complex that has a critical function in cellular protein folding in all eukaryotes (Valpuesta et al, 2002). Its interactome shows involvement in the folding or assembly of a range of proteins linked to central and essential cellular processes such as cytoskeleton assembly, cell-cycle regulation and chromatin remodelling (Dekker et al, 2008; Yam et al, 2008). Electron microscopy (EM) studies of CCT revealed a classic chaperonin architecture, with two rings each containing eight different subunits that form a double-toroid with a central cavity (Llorca et al, 1999a, 1999b). Like all chaperonins, CCT acts upon binding and hydrolysis of ATP to assist in the folding of non-native proteins or in the assembly of substrates with their respective partners. However, the mechanism by which CCT assists folding is distinct from other chaperonins (Horovitz and Willison, 2005), with no hydrophobic wall lining a potential Anfinsen cage (Pappenberger et al, 2002; Horwich et al, 2007), and with the eight subunits within one ring hydrolysing ATP in a sequential, rather than a concerted, manner (Llorca et al, 2001b; Rivenzon-Segal et al, 2005). We show the crystal structure of CCT from yeast at 3.8 Å resolution, which was crystallised in complex with rabbit α-actin and the yeast cofactor protein Plp2p (PLP2). The CCT used for crystallisation contains mutation G345D in the apical domain of the actin-binding Cct4 subunit, which was shown to cause a two-fold reduction in the rate of actin folding and perturb the allosteric behaviour of the intrinsic ATPase activity of CCT (Shimon et al, 2008). Residual electron density locates bound actin within the folding cavity proximal to the Cct4 subunit consistent with previous cryo-EM studies (Llorca et al, 1999a) and reveals the location of a patch of co-evolving ‘signature residues’ that mediate specific interaction with substrates such as actin and tubulin.
We test the location of the evolutionarily conserved NH2-terminus of the Cct5 subunit on the outside of the chaperonin barrel using antibody labelling and EM. We perform crosslinking experiments with succinimidyl-ester diazirine-labelled yeast actin (ACT1) and rabbit α-actin (ACTA) and subdomain 4 of yeast actin (ACT1sub4) and PLP2 to validate the identity of the residual electron density in one of the CCT cavities as unfolded actin. This study and previous mutational analysis (Shimon et al, 2008; Amit et al, 2010) clearly show that CCT is a highly asymmetrical assembly both structurally and functionally. The asymmetrical properties of CCT are essential for generating a sequential allosteric mechanism that facilitates the specific and asymmetric initial association of CCT with the multi-domain actin and tubulin proteins, which are subsequently folded by CCT to their respective native states. The crystal structure of CCT polypeptide will allow further analysis of our mechanistic model of the productive yeast CCT–ACT1–PLP2 folding complex (McCormack et al, 2009, Stuart et al, 2011) and provide a secure framework on which to base further mutational studies aimed at understanding the sequential allostery of CCT and the substrate specificity determining features of eukaryotic chaperonins.
Results
Structure determination and overall structure
Yeast CCT, containing an internal tag, CCT–3CBP (Pappenberger et al, 2006), and harbouring mutation CCT4ANC2 (Shimon et al, 2008) was complexed with rabbit α-actin, that is bound but not folded by yeast CCT (Altschuler et al, 2009). The CCT–α-actin complex was established in the presence of PLP2 that acts as a cofactor and stabilises CCT–actin interaction (Stirling et al, 2007; McCormack et al, 2009; Supplementary Figure S1). The ternary complex of CCT–Actin–PLP2 was crystallised in the presence of ATP and beryllium fluoride. A 3.8-Å data set was collected at the Swiss Light Source; data statistics are shown in Supplementary Table I. A molecular replacement solution was obtained using the structure of the hexadecameric assembly of Thermococcus KS1 α-CPN (Shomura et al, 2004) (PDB entry 1Q3Q) as a search model, and the individual yeast CCT chains built by exhaustive manual examination of difference Fourier maps, and automated refinement. The final model refined to R/Rfree=30/34 consists of two independent copies of CCT in the asymmetric unit with a total molecular mass of ∼2 MDa. Subunit identification (Figure 1A–C) was determined by careful examination of electron density (Figure 1E and F; Supplementary Figure S1F–H); the order within each CCT ring was consistent with the order defined for mouse testis CCT, via biochemical (Liou and Willison, 1997) as well as 14 various independent antibody labelling EM experiments (reviewed in Valpuesta et al, 2005). The positioning of the two rings relative to each other (Figure 1C) differs, by one subunit counter-clockwise, from our previous model of bovine testis CCT based on antibody labelling EM (Martin-Benito et al, 2007) and can be attributed to the pronounced right-handed twist in each subunit relative to the ring-perpendicular, which was not evident in the earlier low-resolution EM reconstructions.
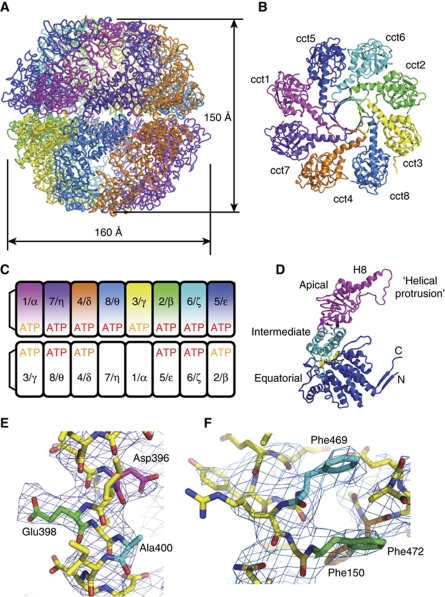
Figure 1.
(A) Overall structure of CCT with dimensions as shown. The colour scheme identifying the subunits is the same for all figures in this paper and reads Cct1=magenta, Cct2=green, Cct3=yellow, Cct4=orange, Cct5=blue, Cct6=cyan, Cct7=purple, Cct8=marine. (B) Top-down view of the apical domains of one ring, showing the non-symmetric iris-like closure of the helical protrusions, including helix H8, at the top of the complex. The views in (A) and (B) reveal the right-handed twist in each subunit. (C) Subunit organisation, showing both the nomenclature for yeast (numbers) and mammalian CCT (greek symbols) and the nucleotide occupancy in the crystal structure; nucleotide pockets that were fully occupied in both molecules in the asymmetric unit have ATP printed in red, nucleotide pockets that had nucleotide in one but not in the other molecule are printed in orange. Where no ATP was bound, an SO4 ion was modelled in the nucleotide pocket. (D) Domain structure of Group II chaperonin fold, with equatorial domain in blue, intermediate domain in cyan and apical domain in magenta; shown in yellow is a bound nucleotide. (E) The individual CCT chains were identified by careful inspection of density for unique stretches of sequence. The (2Fo-Fc) density is plotted at 1.0σ level. The absence of side-chain density for Ala400 (blue) and the presence of Glu398 (green) and Asp396 (magenta) indicate a sequence motif that is unique for Cct8. (F) Similarly, Phe150 (orange) occurs in both Cct5 and Cct6 but the combination with Phe469 (cyan) and Phe472 (green) makes this stretch unique to Cct5.
Both rings in the yeast CCT structure are in a closed conformation (Figure 1A and B). Each ring encloses a cavity volume of ∼145 000 Å3, with dimensions comparable to the cavity of the archaeal thermosome (Ditzel et al, 1998) (PDB entry 1A6D). The heterogeneous nature of the eukaryotic chaperonin complex is reflected in its ring structure, which is asymmetric (Figure 1B) and each individual subunit within one ring is at a different distance and angle with respect to each of the others. Each CCT subunit has a typical Group II chaperonin fold (Figure 1D), with an equatorial or ATP-binding domain, an intermediate domain and an apical or substrate binding domain that contains the helical protrusion that forms the lid in the closed state. A top-down view of the apical domains (Figure 1B) shows the variability in conformation of the helical protrusions; the curvature in Helix 8 in Cct4 may indicate a wide range of motion.
Subunit orientation
The orientation of each of the two rings with respect to the other, revealed by the crystal structure (Figure 1), involves six heteromeric-subunit interactions and two homomeric-subunit interactions across the ring–ring interface. The crystallographic implications of such a subunit organisation are that, of a total of eight possible pseudo two-fold symmetry axes per molecule relating the homologous subunits, one axis is a proper two-fold axis relating pairs of identical subunits. This is indeed the case for our P1 data, as shown by the self-rotation function in Supplementary Figure S2A and B. Moreover, the angle between the two proper two-fold axes (Supplementary Figure S2C) is consistent with the angle between the axes bisecting the cct4–cct6 plane in each molecule, and hence provides additional support for the model shown in Figure 1. At the current resolution, it is not appropriate to comment on the conformations of individual side chains; however, residues that are likely to be involved in subunit interactions across the ring–ring interface can be identified, showing that this interface is primarily polar and ionic.
Unique position of the Cct5 N-terminus
The N- and C-termini of all subunits, except one, come together on the inside of the ring, in a network of β-sheets at the ring–ring interface, which forms the ‘cavity floor’. Neighbouring subunits form their pairwise interactions using both the N- and C-termini of one subunit (the anti-clockwise subunit in the top view in Figure 1B) to donate two strands to a 4-stranded β-sheet that also comprises two strands from the equatorial domain of its neighbouring subunit; this is illustrated for the Cct1/Cct5 pair in Figure 2A and B. The exception to this rule is subunit Cct5 that interacts with its other neighbour Cct6 via a 3-stranded β-sheet, donating only its C-terminus (Figure 2C and D). The N-terminus of subunit Cct5 does not run from the inside of the cavity, like the N-termini of all other subunits, but threads in from the outside of the CCT complex through a channel formed by four interacting subunits (Figure 2E). Although only visible from residue 30 in the crystal structure, mass-spectrometry analysis shows the N-terminus is present in the CCT preparation, and is presumably disordered on the surface of the complex. Sequence alignments of paralogues (Supplementary Figure S3A) show that the N-terminus of Cct5 is much longer (∼25 amino-acid residues) than any other yeast Cct subunit, whereas an alignment of orthologues (Supplementary Figure S3B) reveals that this extended N-terminus has been highly conserved during eukaryotic evolution, with a potential Asp-Glu (DE) electrostatic motif near the extreme N-terminus.
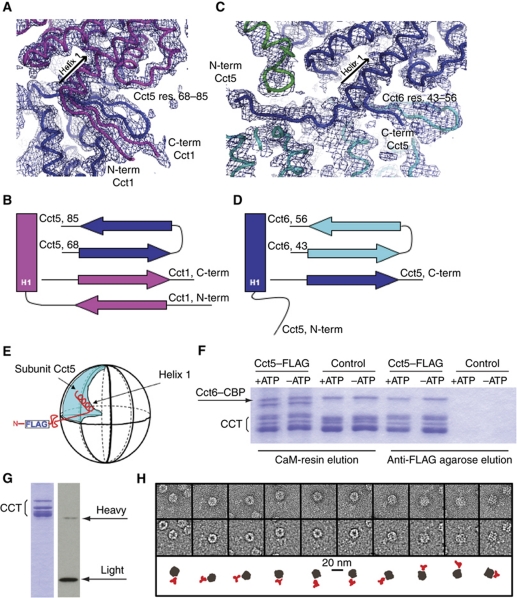
Figure 2.
All N- and C-termini are involved in subunit–subunit interaction (A, B) except for Cct5 (C, D) where the N-terminus is at the periphery of the complex and threads through a channel formed by four subunits. In panels (A–D), Cct5 is coloured blue, Cct1 is magenta, Cct6 is cyan and Cct2 is green, with (2Fo-Fc) density at 1.2σ. (E) Schematic representation of the expected location of the extreme N-terminus of Cct5 at the periphery of the complex, indicating the position of the FLAG-tag after residue number 9. (F) SDS–PAGE showing CCT pull-down using either CaM-resin or anti-FLAG agarose with or without ATP. The control is CCT complex harbouring the CBP-tag but not the FLAG-tag. (G) Coomassie-stained SDS–PAGE showing CCT in complex with anti-FLAG antibody in substoichiometric amounts. The heavy chain is just visible by Coomassie stain, as confirmed by western blot (right-hand panel). (H) Negative stain EM images of representative particles of CCT–FLAG in complex with anti-FLAG MAb. Top row: example of MAb complexes extracted from the raw micrographs; middle row: same molecular views low pass filtered to 30 Å to enhance the contrast; bottom row: cartoon representation of the complexes with CCT depicted in grey and the MAb in red.
To test biochemically that the location of the Cct5 N-terminus is on the outside of the complex at the periphery near the ring–ring interface, we successfully inserted an N-terminal FLAG-tag in the Cct5 subunit (Figure 2E). This FLAG-tagged Cct5 subunit was incorporated into a CCT complex, in which the Cct6 subunit harboured the internal CBP-tag. Consequently, we were able to purify CCT using either anti-FLAG agarose or calmodulin-resin (CaM-resin), whereas the control CCT complex containing only the CBP-tag in the Cct6 subunit does not bind to the anti-FLAG agarose (Figure 2F). We also used this antibody affinity column to investigate the effects of the nucleotide state of CCT towards access of the antibody; the Cct5 N-terminal FLAG-tag is accessible both in the presence and absence of ATP (Figure 2F). As both the CCT cavities are known to close in the presence of excess ATP (Llorca et al, 2001b), the N-terminal tag would not have been accessible to the large, 160 kDa, IgG1 anti-FLAG-antibody molecule were it located on the inside of the complex.
The presence of a FLAG-tag in the Cct5 subunit also facilitated the use of anti-FLAG antibodies for single particle EM analysis. We coupled substoichiometric amounts of anti-FLAG antibody with Cct-5–FLAG (molar ratio CCT:Ab=1:0.5 and mass ratio CCT:Ab=12.5:1) in the presence of 10 mM ATP which closes the chaperonin lids (McCormack et al, 2009). The antibody complex was subsequently purified from free antibody using a sucrose gradient. Analysis on SDS–PAGE (Figure 2G) reveals the CCT complex and antibody co-migrate in the same fraction, with the heavy chain in substoichiometric amounts just visible using Coomassie stain, and confirmed by western blotting for both heavy and light chains (Figure 2G). The specific binding of anti-FLAG antibody to the Cct5 N-terminus allowed visualisation by negative stain EM. The purified CCT–antibody complexes were deposited on EM grids in the presence of 2 mM ATPγS which blocks the ATP cycle (Pappenberger et al, 2006). Figure 2H shows six selected top views and four side views, revealing the antibody bound to the CCT complex on the outside and in a range of conformations compatible with a flexible conformation of both the FLAG-tag and the Cct5 N-terminus to which it is attached.
Complex dissociation experiments
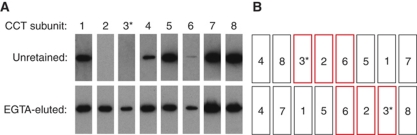
The CCT subunit order within a ring was established previously based on our biochemical dissociation experiments with mouse testis CCT (Liou and Willison, 1997). The yeast CCT complex when isolated and purified remains as a complex only when kept in 20% glycerol (Pappenberger et al, 2006). This dependency of the yeast CCT complex stability upon glycerol provides an alternative way of identifying stable segments of the ring system by making use of the purification tag in the Cct3 subunit located on the outside of the CCT complex. CCT–3CBP was bound to the affinity resin and left overnight in low-glycerol buffer. Then, unretained subunits were washed off the resin and identified by western blot using eight anti-CCT subunit antibodies (McCormack et al, 2009; Figure 3A, top row). The remaining CCT subunits, bound to the resin via the CBP-tag in Cct3, were eluted specifically using EGTA-containing buffer and identified by western blotting (Figure 3A, bottom row). Signals from all eight subunits were detected in the EGTA elution step because not all the CCT–3CBP was disassembled under the chosen conditions. The absence of subunits Cct2, Cct3 and Cct6 in the unretained-subunit-wash demonstrates that these subunits were still associated with the resin. Since the CBP-tag is only present in subunit Cct3, this indicates that Cct2 and Cct6 are located in a stable, trimeric, segment associated with Cct3. The crystal structure shows that Cct3 and Cct6 are located on either side of Cct2 within one ring (Figure 3B). We propose that the strength of the neighbouring subunit interactions between Cct5 and Cct6 is weakened within the ring, due to the 3-stranded β-sheet rather than 4-stranded β-sheet interaction (Figure 2D). This helps to explain why CCT dissociates more easily at the Cct5/Cct6 interface. Cct3 is also flanked by Cct8. Previous work on the dissociation of mouse CCT always showed Cct8 as a monomeric entity and never as part of microcomplexes (Liou and Willison, 1997). We, therefore, postulated a weak subunit–subunit interface between Cct8 and its neighbours, which is corroborated by our current results. Also in our previous study, CCTβ/2–CCTζ/6 complexes were found in mouse testis CCT and CCTβ/2–CCTγ/3 complexes in human 293-T cells (Liou and Willison, 1997), which is consistent with this neighbour analysis of yeast CCT. Altogether, these experiments demonstrate that in CCT, both yeast and mammalian, the relative strengths of interaction between subunits within a ring vary considerably.
Figure 3.
(A) Western blotting analysis, with the eight anti-Cct subunit antibodies (McCormack et al, 2009), of CCT–3CBP complexes bound to CaM agarose after overnight incubation in low-glycerol buffer (5%) which destabilises yeast CCT. Unretained (top panels) is the supernatant sample containing subunits, which became unbound during the incubation. EGTA-eluted (bottom panels) is the material eluted from the column with 2 mM EGTA. (B) Subunit arrangement within and between the CCT rings highlighting, in red boxes, the Cct3, Cct2 and Cct6 neighbours that are retained on the column. The CBP-tagged Cct3 subunit is denoted by an asterisk.
Nucleotide status
Difference Fourier maps showed clear density for bound nucleotide in a number of ATP-binding pockets, but not all of them. Typical nucleotide density (Supplementary Figure S4A) was fitted with a model of ADP–BeF. In those nucleotide pockets that showed insufficient density to accommodate the base and sugar moieties of a nucleotide, the residual density could be satisfactorily interpreted as sulphate ions, also present in the crystallisation condition (Supplementary Figure S4B).
CCT–actin interaction
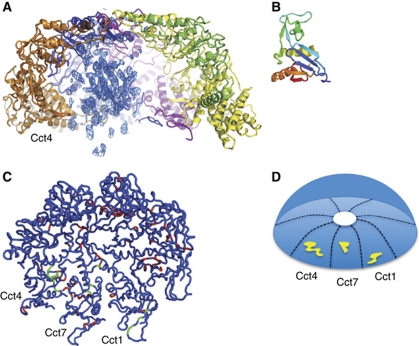
The protein used for crystallisation was a complex of CCT, α-actin and PLP2. It was not possible to locate the PLP2 cofactor protein, which constitutes <3% of the complex and is expected to bind on the outside and across the top of CCT (Martin-Benito et al, 2004) via two binding sites either side of the thioredoxin (TRX) fold (McCormack et al, 2009) and may not occupy a fixed position within the crystal packing. A further complication was the bias of MR-based searches towards the TRX-like fold present in the apical domains of CCT (Dekker et al, 2011), which are in 16-fold excess compared with the TRX-fold of PLP2. Based on EM studies of the protein complex before crystallisation (Figure 2 in Altschuler et al, 2009), α-actin is able to bind inside the yeast CCT cavity in a folding intermediate state with its ‘small’ and ‘large’ domains in an extended conformation, as previously shown for mouse testis CCT–α-actin complexes by EM (Llorca et al, 1999a). Residual electron density, not accounted for by the CCT model, was clearly visible inside the cavity of one ring in both crystallographically independent CCT complexes in the crystal (Supplementary Figure S5). Enhancement of the density inside the cavity using averaging techniques reveals continuous proteinaceous density with β-strand features (Figure 4A) of a volume comparable to actin's small domain (Figure 4B). This density, which we attribute to bound actin, is located near the apical domains of CCT subunits Cct1, Cct7 and Cct4 (Figure 4C and D).
Figure 4.
(A) Residual density on the inside of the cavity is attributed to actin bound to CCT. One CCT ring with a slice of density to reveal the asymmetric distribution of the residual density with respect to the cavity is shown. The (2Fo-Fc) density calculated at 5 Å reveals β-strand features, showing the ‘actin’ density which is in the proximity of Cct1, Cct7 and Cct4. The Cct4 subunit is depicted in orange on the left-hand side of this slice. (B) Actin's ‘small’ domain (residues 1–137 from PDB entry 1ATN) drawn to the same scale as panel (A). (C) Signature residues (red) for CCT apical domains mapped onto the structure of the closed conformation. The view is from the inside of the cavity looking towards the ‘ceiling’ of the dome formed by the helical protrusions. Signature residues located near the ‘actin density’ are shown in green. (D) Schematic representation of view shown in (C); yellow denotes the patch of signature residues in the vicinity of the ‘actin density’.
Actin, actin subdomain 4 and PLP2 crosslinking experiments
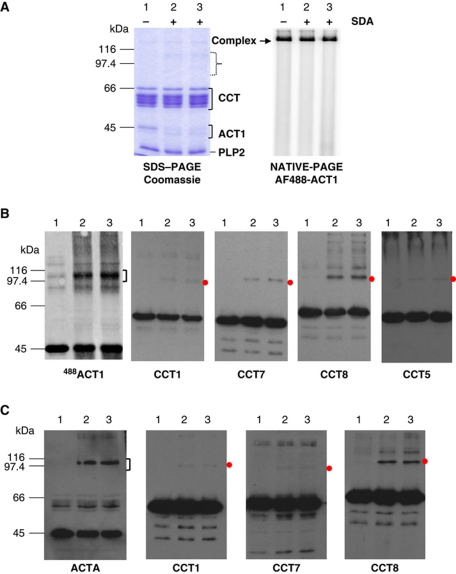
To confirm the location of actin binding with respect to the CCT ring, native yeast actin (ACT1), labelled with AlexaFluor488 on cysteine 374, was derivatised with a bifunctional crosslinker, succinimidyl-ester diazirine (sulfo-SDA), and CCT–ACT1–PLP2 complexes were assembled by EDTA-mediated unfolding of actin in the presence of CCT and PLP2 (McCormack et al, 2009) and crosslinked using UV irradiation (Figure 5A). Samples were probed with all eight anti-CCT subunit antibodies as previously described (McCormack et al, 2009). Crosslinked species compatible with a crosslink formed between one chain of the respective CCT subunit and one yeast actin polypeptide were observed for Cct1, Cct5, Cct7 and Cct8 (Figure 5B). Since the ternary complex of crystallised CCT–Actin–PLP2 was formed from rabbit α-actin, we repeated the crosslinking analysis with this complex (Figure 5C) and obtained similar results: crosslinks of α-actin to Cct1, Cct7 and Cct8. This crosslinking confirms that the rabbit actin binds CCT in a similar manner to yeast actin, namely near the apical domains of Cct1 and Cct7, which is compatible with much of the density we attribute to actin.
Figure 5.
(A) Crosslinking of sulfo-SDA-labelled yeast and rabbit actins to yeast CCT. Analysis of CCT–ACT1–PLP2 complexes by Coomassie-stained 8% SDS–PAGE (left-hand panel) or 6% native-PAGE and Typhoon imaging of the AlexaFluor-488-labelled yeast actin signal (right-hand panel). AlexaFluor-488-labelled yeast G-actin (ACT1) was coupled with sulfo-SDA and assembled into yeast CCT and PLP2 complexes as described in McCormack et al (2009). Lane 1 shows control complexes with sulfo-SDA-unlabelled actin. Lanes 2 and 3 show CCT-sulfo-SDA-labelled 488ACT1–PLP2 complexes assembled in ATPγS (lane 2) or in the absence of nucleotide (lane 3). All three samples were exposed to UV light for 30 min to induce crosslinking via the diazirine group. The ACT1 signal at 45 kDa is more diffuse in the Coomassie-stained SDS–PAGE analysis due to altered electrophoretic mobilities of the sulfo-SDA adducts of ACT1 (lanes 2 and 3; bracket) and higher molecular weight adducts are also visible (lanes 2 and 3; dotted bracket). However, all the 488ACT1 signals are equivalent in native-PAGE demonstrating that the sulfo-SDA crosslinking is compatible with complex assembly and stability. (B) Eight percent SDS–PAGE western blotting analysis of CCT–sulfo-SDA–labelled 488ACT1–PLP2 complexes. Replicate blots were probed with antibodies for all eight yeast CCT subunits or MAb-1f to PLP2 (McCormack et al, 2009). Four anti-CCT subunit antibody blots (CCT1, CCT7, CCT8 and CCT5) reveal species migrating between the 97.4 and 116 kDa markers (red dots) compatible with a crosslink formed between one chain of the respective CCT subunit and one ACT1 polypeptide. After collecting ECL exposures, these blots were washed and Typhoon scanned for the 488ACT1 signal 24 h later. The 488ACT1 signal was identical on all the blots as expected (only one shown here; labelled 488ACT1) and the main cluster of crosslinked products between 97.4 and 116 kDa is indicated with a bracket. No well-defined crosslinks were observed for PLP2. (C) Eight percent SDS–PAGE western blotting analysis of CCT SDA-rabbit α-actin PLP2 complexes; assembled as described for yeast actin. Replicate blots were probed with antibodies to all eight yeast CCT subunits or an mAb-C4 (Chemicon) to mammalian actins (McCormack et al, 2009). The anti-rabbit actin antibody blot (labelled ACTA) revealed a main cluster of crosslinked products between 97.4 and 116 kDa as indicated with a bracket. Three anti-CCT subunit antibody blots (CCT1, CCT7 and CCT8) reveal species migrating between the 97.4 and 116 kDa markers (red dots) compatible with a crosslink formed between one CCT subunit and one rabbit actin polypeptide. No well-defined crosslinks were observed for PLP2.
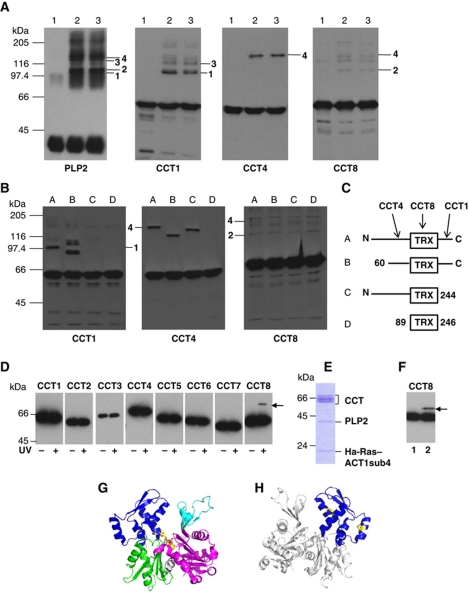
We performed further crosslinking experiments using sulfo-SDA PLP2 (Figure 6A) and three PLP2 deletion mutants (Figure 6B) which mapped interactions between PLP2 and Cct1, Cct4 and Cct8 (Figure 6C), identifying also a potential Cct4–Cct8–PLP2 complex (species 4). The PLP2 interaction with Cct1 and Cct8, across the ring equator, is consistent with previous EM models of CCT–phosducin-like protein interactions (Martin-Benito et al, 2004). Finally, when crosslinking sulfo-SDA ACT1sub4 with CCT and PLP2 we find that ACT1sub4 crosslinks to Cct8 (Figure 6D–H).
Figure 6.
(A) Analysis of CCT–ACT1–sulfo-SDA–PLP2 complexes via 8% SDS–PAGE western blotting. PLP2 was coupled with sulfo-SDA and assembled into complexes by EDTA unfolding of ACT1 in the presence of yeast CCT (McCormack et al, 2009). Lane 1 shows control complexes with unlabelled PLP2. Lanes 2 and 3 show CCT–ACT1–SDA–PLP2 complexes with ATPγS added post assembly (lane 2) or in the absence of nucleotide (lane 3). All three samples were exposed to UV light for 30 min to induce crosslinking via the diazirine group. Replicate blots were probed with antibodies to all eight yeast CCT subunits or an mAb-1f to PLP2 (McCormack et al, 2009). Several higher molecular weight anti-PLP2 antibody species are detected and four are highlighted (RHS of PLP2 panel: 1, 2, 3 and 4). In the anti-CCT subunit blots species 1 and 3 overlay with anti-CCT1 antibody signals, species 2 overlays with an anti-CCT8 signal and species 4 overlays with both an anti-CCT4 signal and an anti-CCT8 signal (RHS of panels). (B) Eight percent SDS–PAGE western blotting analysis of CCT–ACT1–PLP2-deletion mutant complexes. PLP2 deletion mutants were previously described (McCormack et al, 2009) and are shown schematically in panel (C). Complexes with the mutant PLP2 proteins (lanes B–D) were prepared as described above for wild-type PLP2 (lane A). Replicate blots were probed with antibodies to the C-termini of all eight yeast CCT subunits or anti-PLP2 mAb-6i (McCormack et al, 2009). This analysis clearly maps the PLP2 interaction sites of the three CCT subunits (C shows mapping results arrowed). The ternary complex of PLP2–CCT4–CCT8 (species 4) with an apparent Mr 170 kDa is indicated in both the anti-CCT4 and anti-CCT8 blots. (C) The three PLP2 deletion mutants used in the analysis: wild-type PLP2 (A), N-terminal truncation; 60-C-terminus (B), C-terminal truncation; N-terminus-244 (C) and thioredoxin fold only; 89–246 (D) as previously described (McCormack et al, 2009). (D, E) Analysis of Ha-Ras–ACT1sub4 in complex with CCT and PLP2. The L-photo-methionine-labelled Ha-Ras–ACT1sub4 fusion protein was purified and refolded as previously described (McCormack et al, 2009). CCT–Ha-Ras–ACT1sub4–PLP2 complex was assembled and purified using 10–40% sucrose gradient sedimentation (McCormack et al, 2009). An aliquot of the peak fraction, electrophoresed on 12.5% SDS–PAGE and Coomassie stained is shown in panel (E) 200 μl of peak sucrose gradient fraction was irradiated with UV for 30 min on ice to initiate crosslinking. Samples were analysed on 10% SDS–PAGE western blots. Eight duplicate blots (plus and minus UV treatment step) were probed with antibodies to the C-termini of all eight yeast CCT subunits. A crosslinked species corresponding to CCT8–Ha-Ras–ACT1sub4 is clearly observed (D, arrow). (F) Analysis of ACT1sub4 in complex with CCT and PLP2. The ACT1sub4 protein was labelled in vivo in E. coli with L-photo-methionine as described in the legend to panel (D). The L-photo-methionine-labelled ACT1sub4 protein was purified and refolded as previously described (McCormack et al, 2009). CCT–ACT1sub4–PLP2 complex was assembled and purified by 10–40% sucrose gradient. 200 μl of peak sucrose gradient fraction with ATPγS added to a final concentration of 3.5 mM and was irradiated with UV for 30 min on ice to initiate crosslinking. Samples were analysed on 8% SDS–PAGE western blots and eight duplicate blots were probed with antibodies to all eight yeast CCT subunits; only the anti-CCT8 blot is shown; lane 1 minus L-photo-methionine crosslinker, lane 2 plus L-photo-methionine crosslinker (F). A single crosslinked species corresponding to CCT8–ACT1sub4 is observed (arrow) which migrates faster than the CCT8–Ha-Ras–ACT1sub4 species because it lacks the 90 amino-acid residues from Ha-Ras. Ha-Ras–ACT1sub4 is a fusion protein linking residues M67–L168 of Ha-Ras to residues I178–F262 of S. cerevisiae actin. Apart from the initiation methionine there are only two methionine residues in the ACT1sub4 protein (blue domain shown in panels G and H); M190 and M227 (coloured yellow in panel H) both located on the surface-exposed sides of helices on either side of the CCT-binding site II (McCormack et al, 2001a). Ha-Ras–ACT1sub4 fusion protein contains two additional methionines from the Ha-Ras domain, M72 and M111. (G) Schematic representation of the domain structure of actin, based on the actin chain in PDB entry 2BTF where domains 1 and 2 together form the ‘small domain’, and domains 3 and 4 combined the ‘large domain’. (H) View 180 degrees rotated relative to (G), showing the location of Cys374 at the C-terminus within domain 1, and the two methionines (in yellow ball-and-stick model) targeted by crosslinkers in domain 4.
Discussion
Implications of structure for sequential allosteric mechanism
The 3D cryo-EM structures of mouse testis CCT–actin and CCT–tubulin complexes in the open (Llorca et al, 1999a, 2000) and closed states (Llorca et al, 2001b) led to the model that, upon nucleotide-induced lid-closure, these two substrates remain bound to the inner wall of the cavity and progress towards more compact, native-like states (Llorca et al, 2001b). The X-ray structure of CCT presented here is compatible with this model, since residual electron density is detected inside one of the closed cavities of CCT, which we attribute to the bound actin; crosslinking experiments support this assignment. The density is located near the apical domains of Cct1, Cct7 and Cct4 (Figure 4A), and Cct4 is known to bind actin in the open state as shown by antibody decoration of the Cct4 subunit in the CCT–actin open complex (Llorca et al, 1999a). However, because of the absence of antibody labelling data on closed CCT complexes, it is not known formally if actin's so-called subdomain 2 disengages from Cct4 in the closed state as implied in the early model of the closed state of the CCT–actin complex (Llorca et al, 2001b). We now consider that more complex reconfigurations of the partially folded actin molecule on the apical domain surfaces can occur upon lid-closure, based upon protease-protection mapping experiments with yeast CCT–ACT1sub4–PLP2 complexes (McCormack et al, 2009) and spectroscopic analysis of CCT–ACT1–PLP2 folding kinetics (Stuart et al, 2011). The X-ray structure shown here with its novel interactions between actin and Cct1 and Cct7 supports the model of more complex reconfigurations occurring during the two-step folding cycle (Stuart et al, 2011). The PLP2 interaction with Cct1 and Cct8, across the ring equator of the CCT–ACT1–PLP2 ternary complex, is consistent with previous EM models of CCT–phosducin-like protein interactions (Martin-Benito et al, 2004). Our interpretation of all the combined data is that the actin density in the crystal structure corresponds to actin's ‘small’ domain (Figure 4B) and that the ‘large’ actin domain, containing the CCT-binding sites IIi and IIii in ACT1sub4, rotates about the gly146/gly150 hinge region (McCormack et al, 2001a, 2001b; Stuart et al, 2011) and resides in the vicinity of Cct8 since ACT1sub4 crosslinks to Cct8.
To obtain the crystal structure described here, it was imperative to use an allosterically impaired mutant of CCT (Shimon et al, 2008) in combination with rabbit α-actin, whose folding cycle stagnates on yeast CCT (Altschuler et al, 2009). Hence, the detailed characterisation of the intermediate states in one complete actin folding cycle may depend upon future high resolution 3D cryo-EM structures of wild-type yeast CCT in complex with yeast actin and Plp2 in different nucleotide states.
EM imaging of bovine testis CCT in the presence of different ATP concentrations, after single particle alignment using anti-CCT1/α and CCT4/δ antibody decoration led to the conclusion that ATP-induced conformational changes spread around the ring in a sequential manner in accordance with the KNF allosteric model (Rivenzon-Segal et al, 2005). In our yeast CCT structure, the nucleotide is coordinated by highly conserved residues within the equatorial domain of each of the eight subunits which are all homologous to the structure of the ATP-binding sites in the α/β thermosome structure (Ditzel et al, 1998). In the final model, only subunits Cct5, Cct6 and Cct8 are fully occupied with nucleotide in all four copies in the asymmetric unit. The remaining subunits have differential occupancies in different rings (Figure 1): an observation that strongly supports a sequential, weakly cooperative intra-ring mechanism for CCT.
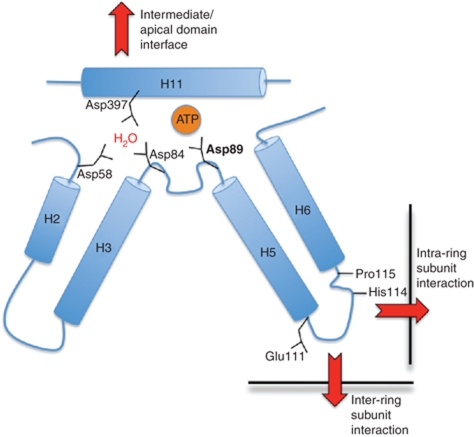
Interestingly, Cct8 lacks one of the three aspartates coordinating the catalytic water, shown diagrammatically for Cct6 (Figure 7), and has instead a lysine at position 93, illustrating ATP-binding site heterogeneity between the eight CCT subunits. In a recent study, one of these catalytic aspartates, the conserved aspartic acid residue in the GDGTT motif (shown diagrammatically for Cct6-Asp89 in Figure 7 and structurally for Cct2 and Cct4 in Supplementary Figure S4), was systematically replaced with glutamic acid in each of the eight subunits, leading to dramatically different phenotypes in vivo in yeast. Importantly, whereas this virtual identical mutation resulted in lethality in the case of Cct4, it only showed mild growth and benomyl sensitivity defects in Cct8 (Amit et al, 2010). Taken together with our observation of the unusual Cct8 catalytic water coordination, this may suggest that the ATP hydrolysis potential of the Cct8 subunit has limited importance for the completion of an entire CCT folding cycle. Another unexpected observation in the aforementioned study was the extreme sensitivity of the Cct7 mutant to the actin inhibitor latrunculin (Amit et al, 2010), which can now be reconciled by the fact that the actin density in the crystal structure colocalises near the Cct7 apical domain and actin can be crosslinked to Cct7 (Figure 4C and D).
Figure 7.
Schematic representation of key allosteric elements involved in nucleotide binding and subunit–subunit interactions. The residue numbering corresponding to Cct6 and Asp89 is in bold as this is the catalytic Asp in the GDGTT motif which is strictly conserved in all chaperonins and was mutated to Glu in each of the eight Cct subunits in a study by Amit et al (2010). Cct8 is the only subunit not to contain aspartate at the residue equivalent to Cct6, Asp84; instead, the residue at position 93 is lysine. Red arrows indicate the direction in which nucleotide-induced conformational changes can be transduced throughout the structure.
The sequential mechanism could be mediated by a combination of ATP-binding site variation and heterogeneity in intra-ring and inter-ring subunit interactions. Our structure reinforces this model, since we can define the connections between the ATP-binding sites in each subunit and the intra-ring subunit–subunit interaction interfaces (Figure 7) as previously shown for the thermosome. Our structure also reveals a heteromeric ring–ring interaction between Cct7 and Cct8, the only two subunits that lack the otherwise strictly conserved His-Pro motif around residues 114–115 (Figure 7). This His-Pro motif is in all other subunits directly involved in intra-ring subunit–subunit interactions, while its location close to the ring–ring interface suggests it could also modulate crosstalk between the rings.
Comparison of yeast X-ray structure with previous cryo-EM-derived models
The crystal structure of CCT reveals a subunit organisation within each ring that is consistent with all our original biochemical and antibody labelling experiments conducted with mouse testis CCT (reviewed in Valpuesta et al, 2005), whereas the relative orientation of the two rings exposed unexpected homomeric interactions and a proper two-fold axis which was not evident in the low-resolution EM reconstructions of bovine CCT (Martin-Benito et al, 2007). Our subunit organisation also differs from the 4.0-Å single particle cryo-EM-derived structure of bovine testis CCT (Cong et al, 2010), both within and between the rings. We performed a comparison of the yeast X-ray structure model with the 4.0-Å single particle model in the region adjacent to the GDGTT ATP-site motif which was used by Cong et al (2010) to define the Cct5 subunit in their model (Figure 8). Since the subunit order within one ring is predicted to be different in each of the two models, a simple yeast Cct5-bovine Cct5 comparison has no meaning. Instead, we have taken the two-fold symmetry axis as point of reference, assuming that this symmetry axis is the same in both models. If this assumption holds true, then yeast Cct5 is located at 45° with respect to the two-fold axis and bovine Cct5 is located at 90° with respect to the two-fold axis (Figure 8A). Therefore, the data sets need to reflect the distinction between a 45° and 90° location, for example, between Cct5 and Cct4 in the bovine structure, corresponding to Cct1 and Cct5, respectively, in the yeast model (relevant amino-acid sequences are aligned in Figure 8B). Figure 8C and D shows the fits for yeast Cct1 and Cct5 subunits revealing clear helical density and associated side-chain density. Since Cong et al (2010) have only deposited a poly-alanine Cα trace for their structure, we fitted a Cα trace to yeast Cct5 (Figure 8E) and compared the fit with a Cα trace of their primary 4.7 Å data (Figure 8F) and their symmetrised 4.0 Å model (Figure 8G). Figure 8G and H shows the density for bovine Cct5 and Cct4, respectively. In our view, it is impossible to assign subunits conclusively or distinguish between bovine Cct5 and Cct4 based on the available EM data provided by Cong et al (2010). Critically, for the unambiguous identification of the yeast subunits in the crystal structure determination, we did not rely on this particular stretch of sequence due to the conserved nature of this region (Figure 8B). Instead, we relied on the combination of multiple sequences as described in Figure 1E and F and Supplementary Figure S1F–H.
Figure 8.
Side by side comparison of yeast CCT X-ray model and bovine CCT EM model. Shown is a characteristic stretch of sequence in the equatorial domain that according to Cong et al (2010) displays a unique fit that allowed subunit identification (see Cong et al, 2010 and Supplementary Figure S4). Since the subunit order within one ring is predicted to be different in the two models, a simple yeast Cct5-bovine Cct5 comparison has no meaning. Instead, the two-fold symmetry axis was taken as point of reference, assuming that this symmetry axis is the same in both models. If this assumption holds true, then yeast Cct5 is located at 45° with respect to the two-fold axis and bovine Cct5 is located at 90° with respect to the two-fold axis as shown schematically in the ring diagrams in panel (A). Therefore, the data sets need to reflect the distinction between a 45° and 90° location, for example, between Cct5 and Cct4 in the bovine structure, corresponding to Cct1 and Cct5, respectively, in the yeast model. (A) Ring diagrams of top views of the yeast X-ray structure (this study) and the bovine electron microscopy model of Cong et al (2010). The Cct5 subunit is indicated in red in each model and the two-fold axis is indicated by the blue line. (B) Alignment of the yeast Cct1–8 and bovine Cct4 and Cct5 amino-acid sequences N-terminal to the GDGTT motif region located in the equatorial domain of all CCT subunits. The 3.8-Å CCT X-ray model is shown in panels (C–E) in the 2Fo-Fc density map, plotted at 1.1σ. (C) Density and X-ray model for Cct1 residues 95–130 with His 121 and Pro 122 in green. (D) Density and X-ray model for Cct5 residues 116–151 with His 142 and Pro 143 in green and Ile 144 and Lys 145 in cyan corresponding to the unique bovine Ile 109 and Arg 110 as marked out by Cong et al (2010) and highlighted in red in the bovine Cct5 sequence in the alignment above in (B). (E) The same as (D) but showing only Cα tracing to allow comparison with EM model. (F) Density at 4.7 Å (emd_5145) and EM model for bovine Cct5 for which only Cα-coordinates are available in the PDB (entry 3IYG). (G) Density at 4.0 Å (emd_5148) and EM model for bovine Cct5. (H) Density at 4.0 Å and EM model for bovine Cct4. Density figures were created using the author's recommended contour levels as indicated in the EMDB entry.
Importance of Cct5 N-terminus localisation
The N-terminus of Cct5 is much longer than any of the other subunits, and this feature is highly conserved among orthologues (Supplementary Figure S3B), suggesting that it has an important role in CCT function, the nature of which remains to be defined. It is likely that the strength of the neighbouring subunit interactions between Cct5 and Cct6 is weakened within the ring, due to the 3-stranded β-sheet rather than 4-stranded β-sheet interaction (Figure 2D), and across the ring interface since Cct5 is flanking the Cct6-Cct6 homomeric interface (Figure 1C). Cct6 is the only subunit of CCT that has coexisting isoforms in mammalian testis and perhaps the arrangement of this region of the ring favoured the evolution of the testis-specific Cct6 subunits in vertebrates 200 million years ago (Kubota et al, 1997) by enabling a subunit exchange mechanism that does not require complete disassembly of the complex. It is also possible that the N-terminus of Cct5 acts as an electrostatic binding site, which binds to sites on CCT or other protein partners of CCT.
Asymmetric distribution of discrete patches of signature residues
We have previously suggested that sites of interaction of substrate proteins on CCT could be identified by the presence of ‘signature residues’ (Pappenberger et al, 2002), which are strongly conserved among orthologues, without any level of conservation among paralogues (Supplementary Table II). The signature residues of all apical domains, mapped onto the crystal structure (Figure 4C; Supplementary Figure S6), highlight potential sites for substrate interaction. Here, we focus on the distribution of signature residues in a global sense. In Supplementary Figure S6C, it is clear that residues that are part of the consensus sequence, comprising residues conserved among all eight subunits in one species, mainly localise to the equatorial and intermediate domains, in particular around the nucleotide pocket, and are more likely to be involved in the ATP hydrolysis mechanism or stability of the complex. Out of 28 identical residues in the consensus sequence, nine are glycines, highlighting the importance of flexibility for the mechanism of this dynamic folding machine. Signature residues (Supplementary Figure S6D–K) localise predominantly to the apical domain and are more likely involved in specific interactions between ligands and a particular CCT subunit. Within the apical domains, the signature residues are distributed differently in the eight different subunits, with some facing the inside of the cavity and others facing the outside of the complex (Supplementary Figure S6A and B). Also of interest is the number of signature residues within the apical domain when comparing the eight subunits; Cct2, Cct4, Cct6 and Cct7 that have the highest number of signature residues, respectively 16, 17, 14 and 17, have all been implicated in the binding of CCT's main substrates actin and tubulin. Cct1, Cct3 and Cct5 have a lower number of signature residues, respectively 11, 7 and 9, whereas Cct8 has only four signature residues in its apical domain. Signature residues within the apical domains are expected to be involved with substrate interaction, although some are buried and might therefore have another role. Signature residues in the proximity of the ‘actin density’ (Figure 4C and D) reveal a defined substrate interaction patch. It is likely that different CCT substrates will utilise different interaction patches given the fact that we already know that actins and tubulins have both shared and unique contacts with CCT subunits (Llorca et al, 2000, 2001a).
Concluding remarks
The crystal structure of CCT completes the set of representative chaperonin structures for bacteria, archeae and eukaryae and will open up ways to study the evolutionary origin of this unique class of protein folding machines. CCT is a highly dynamic complex, as demonstrated by variability in EM class averages induced by nucleotide status (Rivenzon-Segal et al, 2005). This is reflected in the crystal structure by different conformations of the individual subunits, most notably the helical protrusion regions, and the mixed nucleotide occupancies, which strongly support the sequential ATP hydrolysis mechanism, that is unique to eukaryotic chaperonins. The structure reveals the subunit organisation of CCT as a complex and reveals that two subunits, Cct4 and Cct6, form homomeric interactions across the ring–ring interface. Both of these subunits are known to be prominent in the folding of CCT's main substrates, actin and tubulin. The location of the evolutionary conserved signature residues in Cct1, Cct7 and Cct4 and their direct interactions with actin strongly supports the hypothesis of co-evolution of CCT and substrates, in this case actin, during the earliest phases of eukaryotic cell evolution. The presence of a substrate interaction patch spanning at least three subunits illustrates how CCT achieves high selectivity towards its different substrates via multiple contact sites.
Materials and methods
Crystallisation of CCT
Yeast cells contain ∼2000 copies per cell of the chaperonin CCT, and using our tagging strategy (Pappenberger et al, 2006) we were able to obtain crystallisation-quality protein. Variability in ATP status can have major effects on the conformational flexibility of the apical domains, as shown previously in EM studies (Rivenzon-Segal et al, 2005). To overcome the intrinsic heterogeneity stemming from the sequential ATP hydrolysis mechanism, we chose to work with CCT mutant CCT-ANC2 that has reduced ATP kinetics as well as a loss of allosteric signalling (Shimon et al, 2008). Also α-actin was introduced as a substrate. Rabbit α-actin binds to yeast CCT effectively but the folding cycle stagnates, preventing α-actin from proceeding beyond the open conformation (Altschuler et al, 2009). Hence, the species incompatibility provides us with a homogeneous complex of CCT and α-actin in a defined actin folding intermediate state.
The CCT–α-actin complex was established in the presence of Plp2 that acts as a cofactor to CCT (Stirling et al, 2007; McCormack et al, 2009) resulting in >90% occupancy, based on analysis by SDS–PAGE (Supplementary Figure S1). To favour a close packing of the molecules in the crystal, beryllium fluoride (BeF) was added, as ADP–BeF is known to induce the closed state in chaperonins (Melki et al, 1997). The addition of sulphate, or for that matter selenate (SeO4), also induces the closed state of the chaperonin (Ditzel et al, 1998). The complex of CCT–Actin–PLP2 was purified as described (Altschuler et al, 2009) and crystallised in hanging drop in the presence of ATP and BeF, which was added as BeSO4 and KF. Equilibration buffer contained 100 mM Hepes pH 7.6, 50 mM MgCl2, 300 mM Na2SeO4, 6% PEG8k, 1.0 mM TCEP and 20% glycerol. Nucleation typically took 10–14 days after which streak seeding was done in pre-equilibrated drops to yield single crystals.
Data collection and structure solution
A 3.8-Å data set was collected at the Swiss Light Source (Villigen, Switzerland) and data statistics are shown in Supplementary Table I. Phases were obtained from placing a molecular replacement model derived from PDB entry 1Q3Q (α-chaperonin from Thermococcus strain KS-1 complexed with AMP-PNP) (Shomura et al, 2004) in the P1 cell based on self-rotation function and self Patterson data. This was subsequently refined in PHENIX (Adams et al, 2002) using the rigid body refinement option only. Creating OMIT maps by systematically leaving out two neighbouring subunits at a time resulted in improved difference Fourier maps that allowed subunit identification within one ring, guided by careful inspection of density for distinct and subunit-unique side-chain constellations (Figure 1). This was an important step, since the homology between the CCT subunits is low, and sequence identity varies from 23 to 35%. This unbiased method revealed the subunit order within a ring to be as predicted previously. The same method was then extended to identify subunits in the opposite ring, which revealed a ring–ring interacting orientation depicted in Figure 1. After successfully placing all 32 chains in the density, limited NCS averaging was used but only between identical chains in order to ensure that, at no stage in the process, was eight-fold averaging within one ring used. Nucleotide pockets with strong difference density were fitted with a model of ADP–BeF or with SO4, depending upon the density. None of the nucleotide pockets was completely void of residual density.
Enhancing density inside the cavity
As stated in the main text, locating models of either Plp2 or actin by molecular replacement methods failed, most likely due to the unfavourable mass ratio (<5% of total mass in the unit cell). Actin is expected to bind inside the cavity, based on EM imaging of the same protein preparation as used for crystallisation (Altschuler et al, 2009). To enhance density of potentially ordered actin residues inside the cavity, we performed the following routine. First, the cavity was filled with water molecules with low occupancy to avoid solvent flattening in that region. Second, the resolution was cut back to 5 Å, and 32-fold NCS averaging was applied while refining in PHENIX. The resulting density showed continuous stretches of β-strand-like density, allowing partial Cα tracing but not side-chain identification.
Data submission
The original PDB entry code deposited in June 2009 (3I8Q) has been withdrawn because we were asked to submit the two molecules separately. The coordinates and structure factors entry codes are now 3P9D and 3P9E.
EM imaging of CCT–antibody complexes
The CCT anti-FLAG antibody complex at a final concentration of 10 ng ml−1 in 20 mM HEPES pH 8, 50 mM KCl, 10 mM MgCl2, 1 mM TCEP buffer containing 2 mM ATPγS was applied to glow-discharged quantifoil grids coated with thin carbon and stained directly in 2% uranyl acetate. Images were collected on a FEI Tecnai F20 electron microscope using a 4 × 4k TVIPS CCD detector at a nominal magnification of 50 k. A data set of 206 Mab complex views from 45 micrographs was chosen manually using Boxer, part of the EMAN software package (Tang et al, 2007). Subsequent processing was carried out using Imagic (van Heel et al, 1996). A typical field would usually show about 10 free IgG particles on a total of 100 CCT-bound particles.
UV crosslinking of CCT–actin complexes
Diazirine-based photo-crosslinkers are activateable by long wave UV light (330–370 nm). In all, 200 μl of assembled complex was dispensed in the bottom of a 24-well tissue culture placed on ice. A 60 W, 365-nm lamp (LF-215.L lamp, Uvitec) was placed 2 cm above the sample and exposure was for 30 min. In vitro labelling with sulfo-NHS-Diazirine (Pierce) was performed with 20–50 molar excess of crosslinker over protein concentration. In vivo labelling in E. coli of the Ha-Ras–ACT1sub4 and ACT1sub4 fusion proteins was performed with the diazarine-based methionine analogue L-2-amino-5,5′-azi-hexanoic acid (Pierce; L-photo-methionine). L-photo-methionine was added at 125 μg ml−1 to the Novagen overnight express Autoinduction system 2. Yeast and rabbit actins were purified and labelled with AlexaFluor 488 (McCormack et al, 2009). PLP2 purification and mutants are described by McCormack et al (2009). Ternary complexes of actin or actin subdomains with PLP2 and yeast CCT were assembled and purified according to McCormack et al (2009).
Signature residues
The definition of signature residues (Pappenberger et al, 2002) remained unchanged, but the protocol was modified in order to exclude potential sequence errors in CCT subunits from poorly annotated organisms in the UniProt database. In short, sequences from yeast (Saccharomyces cerevisiae), slime mold (Dictyostelium discoideum), worm (Caenorhabditis elegans), cress (Arabidopsis), fly (Drosophila), mouse and man were aligned using T-Coffee (Notredame et al, 2000) per subunit and as a global alignment. The global alignment gave a consensus sequence as shown in Supplementary Table II. The alignment per subunit indicated residues that were completely conserved among these seven species (orthologues). If a residue was marked as fully conserved among orthologues but had no similarity index in the global alignment and was not part of the consensus sequence, then this residue was identified as a signature residue. Previously, the signature residues were mapped onto the apical domain of the 2.2 Å mouse CCTγ structure (Pappenberger et al, 2002), whereby an additional restriction was imposed, that the residue should be surface exposed and facing the inside of the chaperonin cavity.
Supplementary Material
Acknowledgments
We thank Dr Edward Morris and Dr Paula da Fonseca for advice with electron microscopy experiments and analysis of models. We thank the Swiss Light Source, Paul Scherrer Institut, for access to the X06SA beamline. This work was funded by Cancer Research UK, the Institute of Cancer Research and Human Frontiers Science Program (RGP63/2004).
Author contributions: KRW and CD conceived project. CD produced crystals, collected diffraction data, solved structure and built the model. SMR collected diffraction data and helped with analysis and model building along with LHP. EAM performed crosslinking experiments with KRW. EAM and FB performed EM analysis of Cct5–MAb complexes with KRW. CD and KRW wrote paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Altschuler GM, Dekker C, McCormack A, Morris EP, Klug DR, Willison KR (2009) A single amino acid residue is responsible for species-specific incompatibility between CCT and alpha-actin. FEBS Lett 583: 782–786 [DOI] [PubMed] [Google Scholar]
- Amit M, Weisberg SJ, Nadler-Holly M, McCormack EA, Feldmesser E, Kaganovich D, Willison KR, Horovitz A (2010) Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J Mol Biol 401: 532–543 [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W (2010) 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci USA 107: 4967–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR (2008) The interaction network of the chaperonin CCT. EMBO J 27: 1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Willison KR, Taylor WR (2011) On the evolutionary origin of the chaperonins. Proteins 79: 1172–1192 [DOI] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S (1998) Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93: 125–138 [DOI] [PubMed] [Google Scholar]
- Horovitz A, Willison KR (2005) Allosteric regulation of chaperonins. Curr Opin Struct Biol 15: 1–6 [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW (2007) Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol 23: 115–145 [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes GM, Kerr SM, Willison KR (1997) Tissue-specific subunit of the mouse cytosolic chaperonin-containing TCP-1. FEBS Lett 402: 53–56 [DOI] [PubMed] [Google Scholar]
- Liou AK, Willison KR (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J 16: 4311–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Gomez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM (2001a) Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J Struct Biol 135: 205–218 [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Grantham J, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM (2001b) The ‘sequential allosteric ring’ mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. EMBO J 20: 4065–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J 19: 5971–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM (1999a) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402: 693–696 [DOI] [PubMed] [Google Scholar]
- Llorca O, Smyth MG, Carrascosa JL, Willison KR, Radermacher M, Steinbacher S, Valpuesta JM (1999b) 3D reconstruction of the ATP-bound form of CCT reveals the asymmetric folding conformation of a type II chaperonin. Nat Struct Biol 6: 639–642 [DOI] [PubMed] [Google Scholar]
- Martin-Benito J, Bertrand S, Hu T, Ludtke PJ, McLaughlin JN, Willardson BM, Carrascosa JL, Valpuesta JM (2004) Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc Natl Acad Sci USA 101: 17410–17415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J, Grantham J, Boskovic J, Brackley KI, Carrascosa JL, Willison KR, Valpuesta JM (2007) The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep 8: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack EA, Altschuler GM, Dekker C, Filmore H, Willison KR (2009) Yeast phosducin-like protein 2 acts as a stimulatory co-factor for the folding of actin by the chaperonin CCT via a ternary complex. J Mol Biol 391: 192–206 [DOI] [PubMed] [Google Scholar]
- McCormack EA, Llorca O, Carrascosa JL, Valpuesta JM, Willison KR (2001b) Point mutations in a hinge linking the small and large domains of beta-actin result in trapped folding intermediates bound to cytosolic chaperonin CCT. J Struct Biol 135: 198–204 [DOI] [PubMed] [Google Scholar]
- McCormack EA, Rohman MJ, Willison KR (2001a) Mutational screen identifies critical amino acid residues of beta-actin mediating interaction between its folding intermediates and eukaryotic cytosolic chaperonin CCT. J Struct Biol 135: 185–197 [DOI] [PubMed] [Google Scholar]
- Melki R, Batelier G, Soulie S, Williams RC (1997) Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry 36: 5817–5826 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Pappenberger G, McCormack EA, Willison KR (2006) Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J Mol Biol 360: 484–496 [DOI] [PubMed] [Google Scholar]
- Pappenberger G, Wilsher JA, Roe SM, Counsell DJ, Willison KR, Pearl LH (2002) Crystal structure of the CCT gamma apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J Mol Biol 318: 1367–1379 [DOI] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A (2005) Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol 12: 233–237 [DOI] [PubMed] [Google Scholar]
- Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A (2008) ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J Mol Biol 377: 469–477 [DOI] [PubMed] [Google Scholar]
- Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K (2004) Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J Mol Biol 335: 1265–1278 [DOI] [PubMed] [Google Scholar]
- Stirling PC, Srayko M, Takhar KS, Pozniakovsky A, Hyman AA, Leroux MR (2007) Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol Biol Cell 18: 2336–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart SF, Leatherbarrow RF, Willison KR (2011) A two-step mechanism for the folding of actin by the yeast cytosolic chaperonin. J Biol Chem 286: 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Carrascosa JL, Willison KR (2005) Structure and function of the cytosolic chaperonin CCT. In Protein Folding Handbook, Buchner J, Kiefhaber T (eds) Vol. II, pp 725–755. Weinheim: Wiley-VCH [Google Scholar]
- Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR (2002) Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett 529: 11–16 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HTJ, Burlingame A, Gerstein M, Frydman J (2008) Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15: 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.