Abstract
Cell-selective glucocorticoid receptor (GR) binding to distal regulatory elements is associated with cell type-specific regions of locally accessible chromatin. These regions can either pre-exist in chromatin (pre-programmed) or be induced by the receptor (de novo). Mechanisms that create and maintain these sites are not well understood. We observe a global enrichment of CpG density for pre-programmed elements, and implicate their demethylated state in the maintenance of open chromatin in a tissue-specific manner. In contrast, sites that are actively opened by GR (de novo) are characterized by low CpG density, and form a unique class of enhancers devoid of suppressive effect of agglomerated methyl-cytosines. Furthermore, treatment with glucocorticoids induces rapid changes in methylation levels at selected CpGs within de novo sites. Finally, we identify GR-binding elements with CpGs at critical positions, and show that methylation can affect GR–DNA interactions in vitro. The findings present a unique link between tissue-specific chromatin accessibility, DNA methylation and transcription factor binding and show that DNA methylation can be an integral component of gene regulation by nuclear receptors.
Keywords: DNA methylation, DNaseI hypersensitivity, enhancer, glucocorticoid receptor, tissue specificity
Introduction
The glucocorticoid receptor (GR) is a member of the steroid receptor superfamily of transcription factors (TFs). GR is ubiquitously expressed and controls a wide spectrum of physiological processes including metabolism, differentiation and immune responses. However, glucocorticoid-mediated gene expression profiles observed in various cell types overlap minimally. Several mechanisms have been implicated in cell-selective regulation by GR, including tissue-specific expression of co-regulatory proteins (Xu et al, 1999; O’Malley, 2007), and the organization of local chromatin structures (Zaret and Yamamoto, 1984; Wang et al, 2004; John et al, 2008).
Chromatin access imposes an important constraint for TF interactions with the template (Wiench et al, 2011). Regulatory elements can become available by localized remodelling of nucleosome structures, and these perturbations can be detected by an increased susceptibility to chemical or enzymatic (e.g., DNaseI) digestion. DNaseI hypersensitive (DHS) sites thus serve as hallmarks of regulatory protein interactions at the template as well as sites of chromatin remodelling (John et al, 2008). Mapping these sites provides an accurate method for identifying the position of functional elements including promoters, enhancers and locus control regions (Xi et al, 2007; Boyle et al, 2008; Auerbach et al, 2009; Hesselberth et al, 2009).
We characterized GR interactions in the context of chromatin accessibility in two murine cell lines: mammary epithelial (3134) and pituitary corticotroph (AtT-20) (John et al, 2008). Regions of remodelled chromatin were mapped as sites of DHS, and GR-binding profiles were determined by chromatin immunoprecipitation (ChIP); both were found to be highly cell type specific. These findings have recently been extended to the genome scale, using Digital DNaseI sequencing (DNaseI-seq) and ChIP-seq, respectively (John et al, 2011). GR binds mostly to distal regulatory elements (enhancers) and almost exclusively within regions of accessible chromatin, suggesting that nucleosome reorganization is a universal requirement for GR–chromatin interactions. Two types of interactions are observed: ∼88% of GR binding takes place at constitutively accessible chromatin regions (pre-programmed DHSs), while ∼10% of GR binding occurs at hormone-dependent sites (i.e., regions that are remodelled only after hormone activation); these are designated de novo or inducible DHSs (Figure 1A). Thus, we conclude that tissue-specific transcriptional profiles are critically defined by GR occupancy at cell-specific glucocorticoid response elements (GREs), and propose that availability of these elements for binding is regulated by remodelling of local chromatin structure. The mechanisms that create and maintain these accessible chromatin regions are not understood but are clearly central to the regulation of tissue selective receptor function. They are likely determined by combinatorial binding and interactions between different chromatin regulators with DNA methylation possibly being one of them.
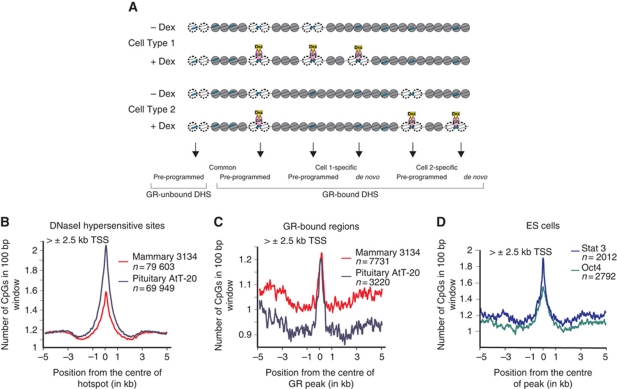
Figure 1.
DNaseI hypersensitive regions and GR-binding sites are characterized by an increased density of CpG dinucleotides. (A) A schematic overview describing cell type specificity of GR-binding sites (GREs, blue boxes) and DNaseI hypersensitive regions. Dex, dexamethasone. (B) All distal (>±2.5 kb) DHS sites identified by DNaseI-seq were scanned from 5 kb upstream to 5 kb downstream and the average number of CpG dinucleotides was determined for 3134 and AtT-20 cell lines. The average density of CpG dinucleotides for all DHSs as well as for a subset of DHSs exclusive of CpG islands is shown in Supplementary Figure S1 (n, number of sites analysed in each group). (C) The same analysis was performed for GR-bound regions identified by GR ChIP-seq and located 2.5 kb away from the TSS. All GR-binding sites and GR-binding sites outside of CpG islands were also analysed and showed similar distribution of CpGs (Supplementary Figure S2). (D) Oct4 and Stat3 binding sites are also enriched in CpG dinucleotides. Binding data were from data sets previously published (Chen et al, 2008) and available in GEO Database ID no. GSE11431.
In differentiated mammalian cells, cytosine methylation (5mC) is established exclusively in a CpG context by a family of DNA methyltransferases (Dnmts) (Klose and Bird, 2006; Clouaire and Stancheva, 2008; Lister et al, 2009). The vast majority (98%) of CpG dinucleotides is located within CpG-poor regions, and is mostly methylated. The remaining 2% is densely grouped as CpG islands located at the 5′ end of the genes (Saxonov et al, 2006; Suzuki and Bird, 2008). In normal differentiated cells, CpG islands stay mostly unmethylated (Shen et al, 2007; Weber et al, 2007; Illingworth et al, 2008). Thus, the unmethylated state of CpG islands is not a good indicator of the transcriptional activity of associated promoters.
DNA methylation has been shown to be subject to changes during differentiation at sequences outside of core promoters and CpG islands (Weber et al, 2007; Meissner et al, 2008; Yagi et al, 2008; Ball et al, 2009; Brunner et al, 2009; Maunakea et al, 2010), where most GR binding occurs. Furthermore, selective demethylation has been suggested to be associated with the formation of DHS chromatin regions (Thomassin et al, 2001; Kim et al, 2007; Santangelo et al, 2009) while methylated regions are relatively refractory to DNaseI (Groudine and Weintraub, 1981) or MspI (Antequera et al, 1989) digestion. Although the formation of accessible chromatin within distal enhancers is highly tissue-specific (Xi et al, 2007; Heintzman et al, 2009) DNA methylation at these elements has not been systematically studied.
We have utilized the hormone-inducible GR as a model system to examine DNA methylation at tissue-specific enhancer regions. We find that distal regulatory elements are enriched in CpG dinucleotides when compared with the surrounding genomic regions. CpG methylation at GR-associated DHS sites is a cell type-specific event, with hypomethylation correlating with chromatin accessibility and GR binding. We further observe that this feature is characteristic for the pre-programmed sites while de novo DHSs are different both in CpG content and methylation pattern. They specifically occur at low CpG density sequences and are thus devoid of the strong suppressive effect of methylated cytosines. Furthermore, tissue-specific methylation of de novo DHSs is restricted to a few CpG dinucleotides and reflects a state established before ligand-triggered activation. When a CpG is located within the core GRE motif, the methylation can directly destabilize GR–DNA interactions in vitro. We also provide evidence that the restricted CpGs serve as sites of dynamic, hormone-induced changes in methylation. Global changes in DNA methylation alone, in Dnmt1-depleted cells, moderately increase chromatin accessibility but are not sufficient to enhance GR binding, suggesting that complex chromatin structures like DHSs are maintained by the combined action of multiple contributing factors.
Results
CpG content of GR-bound DHS regions
CpG dinucleotides are underrepresented in most of the genome due to the increased mutability of methylated cytosine (Shen et al, 1994). The exception is unmethylated CpGs within CpG islands, which have been maintained under selective pressure at proximal promoters. To determine the CpG content of DHS regions, we assessed the number of CpGs within ±5 kb of the centre of all DHSs. We observe a major enrichment in CpG density (3–3.5-fold increase) localized within 2 kb of the centre of the DHS elements in both 3134 and AtT-20 cells (Supplementary Figure S1A). A large component of this enrichment is due to the global overlap between CpG islands and promoter-proximal DHS sites (Supplementary Figure S1B and C), as previously reported (Auerbach et al, 2009; Hesselberth et al, 2009). GR binding is rare within CpG islands (∼1%) and regions within 2.5 kb from transcription start site (TSS) (∼7%) (Supplementary Figure S2A) and occurs predominantly at distal DHS sites. When only distal DHS sites are analysed they also show an increase in the occurrence of CpG dinucleotides when compared with surrounding sequences (Figure 1B; Supplementary Figures S1D, E and S2B). CpG enrichment is further observed when only GR ChIP-seq peaks are considered, both in 3134 and AtT-20 cells, despite markedly different genomic locations of GR binding (Figure 1C; Supplementary Figure S2C–E). These distal CpG occurrences are not, however, clustered in typical ‘island’ structures. They are smaller, and do not satisfy the criteria typically applied to CpG islands (GC content ⩾50% and observed/predicted CpGs ratio ⩾0.6). Moreover, we show that enrichment of CpG dinucleotides at enhancers is not unique for GR-bound regions, but is detected also for Oct4 and Stat3 binding in mouse embryonic stem cells (Chen et al, 2008). As observed for GR, these factors bind mostly distal elements (∼80%) (Supplementary Figure S2A), and these elements show a local increase in CpG density, even when promoter-proximal binding sites are excluded from the analysis (Figure 1D).
We postulate different mechanisms of GR binding within pre-programmed and de novo DHSs, as the latter require hormone-induced nucleosome rearrangement to increase template accessibility after stimulation (Figure 1A). We, therefore, examined the complete set of GR-bound DHSs and compared the subsets of pre-programmed and de novo sites (Figure 2A and B). This analysis reveals that the observed increase in CpG content within GR-bound DHSs is due to CpG enrichment at pre-programmed sites only. These elements are even more enriched in CpG elements when shared between the 3134 and AtT-20 cell lines (Figure 2C; Supplementary Figure S2F). Further analysis shows that pre-programmed sites are always characterized by CpG density higher than surrounding sequences and this feature is independent of the CpG content of sequences they lie within (high versus medium versus low CpG density) (Figure 2D). In contrast, de novo sites show a preference for CpG content <1.4 CpG per 100 bp. Thus, the CpG content does not differ from the surrounding sequences if the sites are located within genomic regions of low (∼0.8/100 bp) or medium (∼1/100 bp) CpG density. The localized decrease in CpG density within de novo sites is observed only when they are located within regions with high (∼1.4/100 bp) average CpG content (Figure 2E). Based on the observed contrast in CpG content between pre-programmed and de novo DHSs, we suggest that DNA methylation might be one of the features that contribute to the formation of these different chromatin entities and that DHS sites inducible by GR represent a distinct class of enhancer elements.
Figure 2.
Pre-programmed and de novo DNaseI hypersensitive sites have different CpG contents. (A) Chromatin organization around the GR-activated gene (Arrdc2) in 3134 cell line. DHSs were identified by DNaseI-seq and GR binding by ChIP-seq. The 5′ end of the gene overlaps with a CpG island (black box) with an extensive open chromatin structure present before (upper track, light blue) and after (middle track, dark blue) Dex stimulation. Two enhancer elements located downstream from the transcribed region are marked by GR binding (bottom track, red) and exemplify two different patterns of CpG distribution (black bars): widespread enrichment for the pre-programmed DHSs (Arrdc2-R1) and scarce CpGs at the de novo sites (Arrdc2-R2). (B) CpG density was analysed separately for the subsets of pre-programmed and de novo DHSs that overlap with GR binding in 3134 mammary epithelial cells (n, number of sites analysed in each group). (C) Comparison of CpG enrichment between GR-bound pre-programmed DHSs unique to 3134 and shared with AtT-20. To balance the difference in number of observations bootstrapping was applied (s.d. shown by dotted line). (D) Distribution of CpGs ±5 kb from the centre of GR peak in three subgroups with low, medium and high CpG density of pre-programmed sites or (E) de novo sites.
CpG demethylation at GR-binding sites correlates with cell type-specific chromatin accessibility
We next analysed the methylation status of tissue-specific GR enhancers associated with GR-regulated genes. We compared DNA methylation levels at 17 selected sites in untreated 3134 and AtT-20 cells using the bisulphite sequencing method (Supplementary Table S1). Seven of these sites are localized within pre-programmed DHSs, eight are found at de novo DHS regions, and two are localized at regions described as hormone dependent in 3134 and hormone independent in AtT-20. None of the sites overlap with a CpG island. The CpG density within a peak of GR binding varies between 0.7 and 4. Between 3 and 12 CpGs were analysed for methylation at each site.
The pre-programmed DHS sites shared between 3134 and AtT-20 are fully demethylated in both cell types (Per1-R1 and Arrdc2-R1) (Figure 3A; Supplementary Figure S3A). However, tissue-specific DHSs are demethylated in the cell type where they are accessible, but hypermethylated in the cell where they are inaccessible and lack GR binding (3134 specific: Dusp1, Fkbp5 and AtT-20 specific: Arhgef3, Ttr) (Figure 3B and C; Supplementary Figure S3B and C). As was reported for CpG islands (Meissner et al, 2008; Brunner et al, 2009), the methylation pattern at pre-programmed DHSs is mostly bimodal and prevalent across the whole hypersensitive region (bimodal equals methylation >80% or <20%). Moreover, cell type-specific pre-programmed DHSs that are independent of GR binding also show differential methylation patterns, possibly associated with other TFs (Supplementary Figure S4). Thus, the correlation between demethylation status and accessibility appears to be a general feature of pre-programmed chromatin at enhancer elements.
Figure 3.
Cytosine demethylation marks the regions of chromatin transition and correlates with GR binding in a cell type-specific manner. DNA was extracted from untreated 3134 and AtT-20 cells and analysed by bisulphite sequencing. The following pre-programmed DHSs are shown: (A) Per1-R1, shared by 3134 and AtT-20; (B) Dusp1, 3134-specific; (C) Arhegf3, AtT-20-specific and (D) de novo, 3134-specific DHS site (Tsc22d3). Additional examples are shown in Supplementary Figures S3 and S5. The GR-binding profile (red track) is shown with reference to the gene location and position of CpG island (black boxes). Black frames indicate the GR-binding site analysed further by bisulphite sequencing. The distribution of CpGs (vertical lines), location of a GRE (green oval) and region covered by bisulphite sequencing (restricted by arrows) are shown below the GR-ChIP track. The CpG density is indicated (# of CpGs/100 bp) . The bisulphite sequencing results are described as follows: each line represents one clone with CpGs shown as open (unmethylated) or filled (methylated) dots and CpG number indicated on the bottom; the green bars show the position of GREs; the left side of each graph shows the DHS-seq and GR ChIP-seq tracks for the regions being queried. (D) CpG methylation pattern of Sgk regulatory elements reflects their tissue-specific utilization. The three upper tracks represent the treated (Dex, 1 h) and untreated DHS profiles and GR ChIP-seq profile detected in 3134 cells; the three lower tracks are specific for AtT-20 cells. The results of bisulphite sequencing of the three GR-binding regions (Sgk-R1: 3134 specific, de novo; Sgk-R2: AtT-20 specific, pre-programmed; Sgk-R3: common, de novo in 3134 and pre-programmed in AtT-20) are compared between 3134 and AtT-20 cells and presented as above. In (D, E), the differentially methylated CpGs, with at least 35% difference in methylation levels between the two cell lines are marked (*).
In contrast to pre-programmed DHS sites, de novo sites are characterized by low CpG content, especially when located within genomic regions of high CpG density (Figure 2B and E). Moreover, the CpGs present in these elements lack a clear methylation signature. They are characterized by a non-homogenous pattern of DNA methylation, unlike the bimodal distribution and with tissue-specific demethylation restricted to a subset of CpGs (Figure 3D; Supplementary Figure S5). We refer to them as ‘differentially methylated CpGs’ if the difference in methylation levels between cell types is at least 35%. This pattern is consistently observed in a set of enhancer elements located near many GR-activated genes, including Tsc22d3, Snf1lk, Chrna5, Sgk (elements R1 and R3), Per1 (element R2), Ptprg, Suox, Mef2b and Arrdc2 (element R2) (Figure 3D and E; Supplementary Figure S5). The location of differentially methylated CpGs relative to the core GRE sequence varies, and factor(s) active at these sites remain to be specified. However, the observed pattern suggests that this differential methylation is a common and prerequisite feature of de novo DHSs, marking these sites as closed chromatin that is poised for remodelling.
The correlation between DNA methylation, chromatin accessibility and GR binding is further exemplified by the Sgk gene (Serum and glucocorticoid-regulated kinase), a major GR target that is activated by corticosteroids in many cell types (Wang et al, 2004; John et al, 2009). Despite its ubiquitous expression, Sgk is regulated by different sets of GR-binding elements (Figure 3E). A comparison of 3134 and AtT-20 revealed the presence of cell type-specific GR-binding sites with DNA methylation strongly reflecting chromatin accessibility in a given cell type. The Sgk-R1 element is a 3134-specific de novo site within which cell type-specific demethylation occurs at CpG #1 in 3134 cells. The Sgk-R2 element is an AtT-20-specific pre-programmed (accessible) site within which all CpGs are demethylated in AtT-20 cells. In contrast, this element is constitutively methylated and inaccessible in 3134 cells. In particular, the Sgk-R3 site provides an example of an element that is commonly utilized in both cell types, but hormone dependent in 3134 cells and hormone independent in AtT-20 cells. Within this element, demethylation of CpG #3 is cell type specific (4% in AtT-20 versus 57% in 3134).
Next, we attempted to answer the question whether demethylation of DNA alone is sufficient to affect chromatin accessibility of inactive enhancers and facilitate GR binding to these elements. Using siRNAs to Dnmt1, we decreased the levels of DNA methylation in 3134 cells (Supplementary Figure S6) and observed a 2–3-fold increase in chromatin accessibility both at silent enhancers (AtT-20 specific: Sgk-R2, Arhgef3 and Ttr) and at de novo sites (Suox and Ptprg) (Figure 4A). Chromatin accessibility at control sites characterized by a complete lack of methylation was not affected by the Dnmt1 knockdown (Sgk-CpG island, Per1-R1). However, the observed increase in accessibility was well below the degree of enrichment (5–30-fold change) present at AtT-20 cells where these elements are established as pre-programmed DHS sites (Figure 4A, left panel; Supplementary Figure S7). Additionally, GR binding (GR-ChIP) was not enhanced after Dnmt1 depletion (Figure 4B). Thus, we conclude that the change in DNA methylation alone is not sufficient to promote the conversion of silent chromatin into active chromatin despite the partial restoration of chromatin accessibility.
Figure 4.
Dnmt1 depletion increases the accessibility of previously methylated regions without impacting GR binding. (A) DNA demethylation was accomplished in 3134 cells by transfection with siRNA to Dnmt1. Increase in chromatin accessibility was monitored by FAIRE at AtT-20-specific DHS sites and compared with the level of accessibility observed in AtT-20 cells (left panel). Additionally, changes in partially methylated 3134-specific de novo elements are shown (middle panel). The unmethylated, pre-programmed regions are used as control sites (right panel). (B) ChIP–qPCR using GR antibody at AtT-20-specific (left panel) and 3134-specific, de novo (right panel) regulatory elements. The results show the normalized enrichment above untreated samples (NH) from three independent experiments (or two independent experiments in AtT-20 cells shown at left panel, Figure 4A). Error bars indicate s.e.m.
DNA methylation at the core GRE can interfere with GR binding in vitro
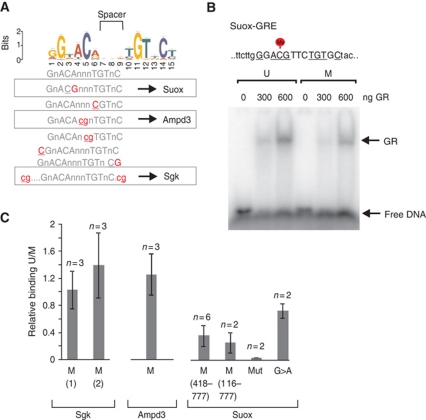
Besides the indirect effect that methylated cytosines have on generating repressive chromatin environment, they can also directly interfere with the ability of TFs to bind regulatory elements and many TFs have been shown to include the CG dinucleotide within their recognition motif (Klose and Bird, 2006). The GRE consensus site derived from the global GR-binding data is a 15-mer sequence with palindromic hexamer ACA/TGT, separated by a 3-bp spacer with highly degenerate nucleotides (John et al, 2011). Each half of the hexamer provides contact sites for interactions with GR (Luisi et al, 1991; So et al, 2007; Meijsing et al, 2009). However, the GR consensus sequence (Figure 5A) does not include CpG dinucleotides, and we consider that GR binding is influenced primarily by methylation in an indirect manner. Nevertheless, nucleotide substitutions at GREs even within non-degenerate positions exist in vivo. Among 8236 GR-binding sites characterized in 3134 cells (John et al, 2011), 576 GREs were identified with a substitution creating a CpG site either at the first (ACG) or second (CGT) half of the GRE hexamer (Figure 5A). When located within an ACG sequence, the methylated cytosine is in close proximity to the GR arginine 466 which is one of the main contact sites for GR–DNA interaction (Supplementary Figure S8A; Meijsing et al, 2009). In addition, CpGs were also observed at less critical nucleotides within the 3-bp spacer, or at regions flanking the GRE. Using electrophoretic mobility shift assay (EMSA), we evaluated the potential effect of methylation on GR binding in vitro to sequences where a CpG is located at the core hexamer, spacer or flanking region. The reaction was performed with purified GR protein fragments containing both ligand- and DNA-binding domains and with methylated or unmethylated probes based on the sequences from three genomic loci: Suox-GRE (ACG), Ampd3-GRE (spacer) and Sgk-GRE (CpGs located +1 and −5 bp from GRE) (Figure 5B and C; Supplementary Figure S8B–E). We observed that GR binding was compromised by 5mC when present at a critical GRE base (Suox-GRE). In contrast, the interaction was unaffected when cytosine was located within a spacer or outside of the core 15-mer (Ampd3-GRE and Sgk-GRE) (Figure 5C; Supplementary Figure S8E).
Figure 5.
GR binding can be directly affected by CpG methylation. (A) The position weight matrix for GREs derived from the global GR-binding data. The locations of possible substitutions (highlighted) leading to an occurrence of CpG site are shown. (B) EMSA gel shift assays were carried out with titrated amounts of purified GR fragment (418–777aa) and 32P-labelled probe for Suox-GRE, either unmethylated or methylated at the indicated position. (C) The quantified results of independent gel shift experiments comparing three naturally occurring GREs (Sgk, Ampd3, Suox; representative gels are shown in Supplementary Figure S8E) (mean values±s.d., n=2–6); M, methylated. For Suox-GRE, similar results were obtained by repeating the experiment with bigger GR fragment (116–777aa). The oligo probes with mutations disrupting Suox-GRE motif (mut) or mutation disrupting CpG element within Suox-GRE and reversing to a perfect motif (G>A) were used as controls.
The results of the in vitro assay demonstrate that a subset of GREs can be potentially affected, and thus potentially regulated by the presence of a methylated cytosine if it is located at critical nucleotide positions within the GRE sequence.
Hormone-dependent demethylation at GR-binding sites
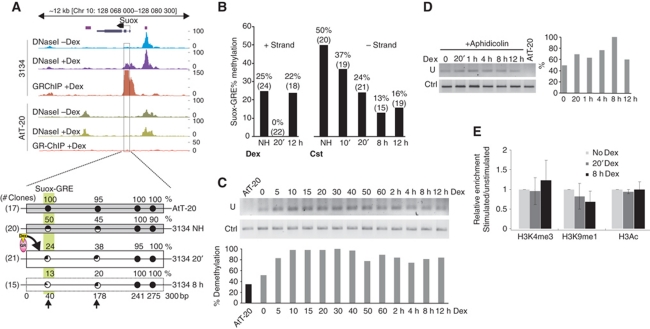
In 3134 cells, the location of the hormone-induced GR-binding site encompassing Suox-GRE coincides with induced (de novo) hypersensitivity while in AtT-20 this chromatin region remains inaccessible and methylated (Figure 6A). The regulatory CpG at Suox-GRE shows partial, 50% methylation in 3134 cells before induction. Further analysis of this site after hormone stimulation revealed a rapid demethylation process initiated 5 min after activation (Figure 6B and C). Demethylation of CpGs at the Suox-GRE was observed after stimulation with both synthetic (dexamethasone, Dex) and natural (corticosterone, Cst) ligands, and affected the regulatory CpGs on both DNA strands (Figure 6B) while more distant CpGs remained unchanged after hormone treatment and were methylated independent of cell type (Figure 6A; Supplementary Figure S9). It is unlikely that this fast response is caused by passive, replication-dependent demethylation. Additionally, the kinetics of demethylation was similar after blocking the cell cycle with aphidicolin (Figure 6D). Thus, this observation supports rather a mechanism of active demethylation, as described recently for promoter regions of other nuclear receptor-regulated systems (Kangaspeska et al, 2008; Metivier et al, 2008; Kim et al, 2009). Prolonged hormone treatment (12 h) resulted in partial remethylation. The timing of remethylation coincided with a decrease in the transcriptional activity of the Suox gene (John et al, 2009).
Figure 6.
GR-binding site is subject to rapid hormone-dependent demethylation. (A) Localization of GR-binding site within the first exon of Suox gene shows an overlap with 3134-specific de novo DHS. Below: Schematic representation of DNA methylation status across the Suox GR-binding site as detected on the negative DNA strand. DNA encompassing 300 bp and four CpGs were PCR amplified and sequenced. Each circle represents a CpG dinucleotide in relation to GRE location (vertical box), with indicated percent of methylation and number of analysed clones shown to the left. Grey narrow rectangles, closed chromatin; white wide rectangles, open chromatin; dotted lines, unknown chromatin accessibility. (B) In vivo, bisulphite sequencing analysis of Suox-GRE shows tissue-specific and hormone-triggered demethylation of a CpG located within core GRE. DNA methylation changes within Suox-GRE were detected on both positive and negative DNA strands after treatment with with 100 nM Dex (left graph) or 1 μM corticosterone, Cst (right graph) for the indicated time. The number of analysed clones is shown above each bar. (C) Extended time course analysis of DNA methylation by MS-PCR on bisulphite-treated genomic DNA. PCR was performed with primers for negative DNA strand recognizing converted unmethylated cytosine (U) or with methylation-insensitive primers (Ctrl). Lower panel shows quantified and normalized data. DNA from AtT-20 cells served as a methylated control. (D) DNA methylation changes observed at the presence of cell-cycle block with aphidicolin (1 μM). (E) ChIP–qPCR analysis of untreated and Dex-treated (20 min and 8 h) 3134 cells with the antibodies as indicated and primers encompassing Suox GR-binding site (mean values±s.d., n=3).
Regulatory elements of actively transcribed genes are marked by a pattern of active modifications including acetylation of H3, trimethylation of H3K4 and monomethylation of H3K9 (Barski et al, 2007; Wang et al, 2008; Heintzman et al, 2009). ChIP with antibodies against H3 modifications at the Suox-GRE locus showed no significant changes in histone modifications following hormone treatment (Figure 6E). The example of the Suox-GRE demonstrates that the CpG element that affects GR–DNA interactions in in vitro binding also shows dynamic tissue-dependent demethylation and methylation in vivo. Importantly, the process of chromatin remodelling and GR binding within this region is associated with changes in methylation, but not histone modifications.
To evaluate whether hormone-triggered demethylation is a common feature of a GR regulatory network, we analysed the status of DNA methylation during dexamethasone treatment at two additional de novo sites: Mef2b and Ptprg (Supplementary Figures S10 and S11; DHS and GR-binding profiles are shown in Supplementary Figure S5). Both sites encompass a GRE sequence with an ACA->ACG substitution (Supplementary Table S1) and similar to the Suox example, partial demethylation of a single CpG within Mef2b and Ptprg enhancer regions was observed shortly after stimulation. Prolonged dexamethasone treatment restored the basal methylation level (indicated by arrows in Supplementary Figure S10). However, despite similar time courses for DNA methylation changes, the three examples discussed here display differences in the type of CpG involved: differentially methylated and within a GRE (Suox), non-tissue specific and within a GRE (Mef2b), and differentially methylated but outside of a GRE (Ptprg). The common feature appears to be the presence of partial (40–80%) methylation observed in naive cells. We propose that the methylation states measured at a given site in cell populations are composite averages that reflect dynamic enzymatic processes of methylation and demethylation. Activation of the receptor could then impact the 5mC equilibrium state at a given site.
Discussion
In the present study, we highlight the role of DNA methylation as an integral component of transcriptional regulation mediated by nuclear receptors. An unexpected finding was a highly localized enrichment of CpG dinucleotides at distal enhancers, identified by genome-wide GR-binding and global DHS profiles. Spontaneous deamination of 5mC to thymidine is a frequent event and a major cause of global depletion of CpG dinucleotides across the mammalian genome (Shen et al, 1994). However, activity-related demethylation of proximal promoters has led to the conservation of CpGs within these regions in the form of CpG islands (Saxonov et al, 2006). Furthermore, the correlation between the presence of unmethylated CpG islands and hypersensitive chromatin has been previously reported (Xi et al, 2007; Boyle et al, 2008; Auerbach et al, 2009). Due to the observed CpG enrichment for pre-programmed DHSs, we propose that DNA methylation may also have a role in the formation of open chromatin regions and maintenance of enhancer elements. We hypothesize that the utilization of distal regulatory elements is accompanied by cytosine demethylation, and this link to enhancer activity has also maintained the increased density of CpG dinucleotides. In fact, we observe that shared DHSs are more enriched in CpGs than tissue-specific elements. Our study supports the observation that pioneer factor interactions in embryonic stem cells might leave unmethylated windows at enhancers as a mark of permissive chromatin for subsequent activation of tissue-specific genes (Xu et al, 2007, 2009). Thus, chromatin structures can be established early in development by one set of factors and then maintained during differentiation by others. Whether DNA methylation is causal or reflective of processes regulating the occurrence of enhancer elements remains an unanswered question. Our results suggest that once chromatin states have been established in a given cell line the global decrease in DNA methylation (Dnmt1 knockdown) only partially increases chromatin accessibility and is not sufficient to induce GR binding. Therefore, the demethylated state is a feature of hypersensitive sites and the hypersensitive site, as a structure, is a prerequisite for GR binding. Similarly, increase in histone acetylation alone does not lead to the increase of chromatin accessibility at the Tat gene (Flavin et al, 2004).
Much of our understanding of DNA methylation and gene regulation comes from the study of CpG islands. There is agreement that high density CpG promoters are rarely methylated (3–4%) and are associated with ubiquitous housekeeping genes or highly regulated developmental genes (Shen et al, 2007; Weber et al, 2007; Illingworth et al, 2008), while low CpG density promoters are generally methylated and linked to tissue-specific genes (Meissner et al, 2008). However, in both cases no correlation has been found between gene expression and DNA methylation: global demethylation of CpG islands does not ensure transcriptional activity, while low density CpG promoters are frequently methylated in inactive as well as in active states (Weber et al, 2007). In addition, a considerable number of methylated CpG islands have been recently found in the vicinity of actively expressed genes in CD4+ T cells (Hughes et al, 2010). We demonstrate here that, in contrast to CpG islands, the methylation pattern of enhancer elements is a common tissue-specific feature that correlates well with both chromatin accessibility and TF binding. Recently, two groups reported a general decrease in methylation at active enhancers, suggesting that sustaining these elements in an unmethylated state might be important for their availability for protein–DNA interactions (Lister et al, 2009; Schmidl et al, 2009). These reports did not, however, examine local chromatin structure as a critical feature.
A persistent difficulty in this type of study is the assignment of enhancer function to transcriptional activity of a given gene. This complexity is exemplified by activity of the Sgk gene. Although this locus is active in both cell types utilized in this study, unique GR-binding elements are employed in each cell. Thus, to evaluate the role of methylation, we have focused specifically on GR-binding efficiency at a given GRE. As clearly shown in the several examples presented, demethylation of CpG dinucleotides is uniformly associated with tissue-specific GR binding.
Many regulatory elements in mammals, especially those that are tissue specific, as well as ‘covered’ promoters of inducible genes in yeast, show high nucleosome occupancy in vivo that might effectively compete with TF binding (Cairns, 2009; Tillo et al, 2010). Restricted access to regulatory information is a key element of inducible or cell type-specific control of gene expression. In the current report, we discriminate between pre-programmed and de novo DHSs as distinct chromatin structures that are characterized by different CpG content and CpG methylation patterns. Pre-programmed DHSs are similar to CpG islands in their bimodal and widespread methylation pattern, with 5mC acting as a strong suppressive mark, supporting closed chromatin conformations with demethylation shifting the balance towards hypersensitive chromatin. However, agglomeration of CpG dinucleotides at pre-programmed DHSs is much lower in comparison with CpG islands and this feature may render these sites easier targets for de novo methylation. The CpG density of not >1.4 CpG per 100 bp seems to be required for the majority of de novo DHSs. This feature could protect de novo DHSs from a strong inhibitory effect of methylated cytosines which, if densely grouped, could draw in repressory complexes and prevent dynamic changes during remodelling of the site. Thus, the methylation state of scarce CpG dinucleotides at de novo elements is not necessarily an obstacle for chromatin transitions and rapid response to stimuli. We show here that demethylation of limited CpGs accompanies the chromatin transition of de novo sites after hormone treatment. The change in methylation status could reflect the occurrence of unstable nucleosome or follow changes in nucleosome positioning (Chodavarapu et al, 2010). Alternatively, a methylation shift could lead to an exchange between TFs with opposite affinities to modified cytosines within their responsive elements, similarly to what has been reported for CREB and C/EBPα (Rishi et al, 2010). Furthermore, single CpGs are likely the most efficient targets for active demethylation.
We demonstrate that ligand-triggered demethylation of single CpGs at de novo sites is rapid and replication independent, suggesting an active enzymatic mechanism. Once controversial, evidence is now accumulating to support the presence of active demethylation processes in vertebrates, involving a deamination-glycosylation-base excision repair pathway (Kangaspeska et al, 2008; Metivier et al, 2008; Ooi et al, 2008; Rai et al, 2008; Gehring et al, 2009; Kim et al, 2009). This is consistent with the observation of DNA breaks accompanying DNA demethylation at the Tat-GRU (Kress et al, 2006). Although this example was reported to require several hours, other rapidly occurring events have been previously published and have argued that demethylation was carried out by an active mechanism which affected several CpGs within promoter regions (Kangaspeska et al, 2008; Metivier et al, 2008; Kim et al, 2009). We propose that such events might be frequent at enhancer elements, especially at de novo DHSs, which are characterized by a very limited number of CpG dinucleotides. Demethylation affecting a single CpG has been recently reported at the RET gene enhancer in response to retinoic acid treatment (Angrisano et al, 2010). Consistently, examples of active demethylation discussed above as well as one of the first cases of activity-dependent demethylation of BDNF gene in neurons (Martinowich et al, 2003) describe small changes of 20–40%, similar to our observations.
Cytosine methylation can affect the transcriptional response through multiple mechanisms (Klose and Bird, 2006). This modification can directly interfere with the ability of a TF to bind to regulatory elements, or it can affect local chromatin structure. A primary methylation signal is mediated through the action of methyl binding domain proteins and their interactions with co-repressor complexes (Klose and Bird, 2006; Clouaire and Stancheva, 2008). Alternatively, the unmethylated state can be maintained by proteins (i.e., Cfp1) that bind specifically to unmethylated sequences (Thomson et al, 2010). Many differentially methylated CpGs described in this paper are within sequences flanking GR-binding elements, suggesting involvement of GR-associated factors. However, we have identified a subset of GREs where substitutions at a non-degenerate base create CpGs within the GRE core element. We further demonstrate that in these cases GR binding in vitro can be affected by the presence of 5mC. We also show that in the case of the Suox-GRE, DNA methylation rather than histone modifications have a leading dynamic role during the hormone response.
These findings underscore the importance of DNA methylation analysis at base pair resolution (Weber et al, 2007; Hodges et al, 2009; Lister et al, 2009; Zhang et al, 2009), within regulatory elements and distinct from CpG islands. Further investigation of dynamic DNA methylation at these elements, particularly using genome-wide and high-throughput methods (Ball et al, 2009; Hodges et al, 2009; Lister et al, 2009) should considerably advance our understanding of the modulation of the epigenetic landscape, and the contribution of these mechanisms to the regulation of transcriptional activity.
Materials and methods
Cell lines and culture
The 3134 cell line is a subclone of 904.13 (Fragoso et al, 1998) derived originally from a mammary tumour of the RIII mouse (Lowy et al, 1978). The AtT-20 cell line is an anterior pituitary corticotroph of murine origin (ATCC). Both cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, and 2 mM L-glutamine and 5 mg/ml of penicillin–streptomycin. Cells were kept in 37°C incubator with 5% CO2. Before hormone treatment, growth medium was replaced with DMEM supplemented with 10% charcoal-dextran-treated serum (Invitrogen) for 24 h. Hormone stimulation was performed with either 100 nM dexamethasone (Dex) or 1 μM corticosterone (Cst) for indicated time. For cell-cycle arrest, 3134 cells were cultured with DMEM supplemented with 10% charcoal-dextran-treated serum and 1 μM aphidicolin for 24 h before dexamethasone stimulation. The aphidicolin effect on cell cycle was confirmed by propidium iodide staining and flow cytometry.
Genomic DNA preparation, bisulphite conversion and methylation analysis
Cells were harvested at the designated time points and genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen). One microgram of DNA was bisulphite converted and then purified using the EpiTect Bisulfite Kit (Qiagen). DNA was eluted with nuclease-free water and used for PCR. The primers for amplification of converted DNA were designed using MethPrimer software (Li and Dahiya, 2002). The primers sequences are listed in Supplementary Table S2. PCR products were gel extracted (Qiagen) and ligated into PCR2.1 TOPO vector (Invitrogen). The colonies were used directly in the PCR. PCR products were screened for positive clones, purified using ExoSAP-IT (USB Corporation) and sequenced (Macrogen USA, Rockville, MD). For each condition, 15–25 separate clones were analysed using MethTools 2.0 software (Grunau et al, 2000).
Primers for methyl-specific PCR were designed to be complementary to the methylated cytosine (‘C’ in bisulphite-converted DNA) on the 3′ end. The primers’ sequences and PCR conditions using AmpliTaq Gold 2 × PCR Mix (Applied Biosystems) are shown in Supplementary Table S3. PCR products were subsequently analysed in 2% agarose gels and band intensity was quantified using TotalLab Quant software. The results were normalized to the control (Ctrl) reactions with non-discriminative primer sets for each fragment analysed (listed previously in Supplementary Table S2).
Methylation-specific restriction enzyme analysis of bisulfite-treated DNA was used to detect methylation changes after Dnmt1 depletion. Each analysed fragment was PCR amplified using primers showed in Supplementary Table S2 and then subjected to restriction analysis with BstBI, EcoRV, TaqαI, HpyCH4III and RsaI restriction endonucleases (New England Biolabs). The details are presented in Supplementary Figure S6C.
Dnmt1 siRNA transfection
We transfected 3134 cells with Dnmt1 siRNA ON-TARGET smart pool (Dharmacon, L-056796-01-0010), siCONTROL Non-Targeting siRNA (D-001810-10-20) or empty control cuvette and grown 2 days. The transfections were then repeated with the same siRNA and controls and grown additional 2 days before harvest. Briefly, five million washed 3134 cells were mixed with 14 μg of siRNA or control and subjected to three 10-microsecond pulses using a BTX Electro Square Porator T820. Surviving cells were immediately re-plated in growth media in 100 or 150 mm dishes for formaldehyde-assisted isolation of regulatory element (FAIRE) and ChIP analyses, respectively. In addition, the efficiency of Dnmt1 knockdown was monitored by western blot, RT–qPCR and methylation-specific restriction enzyme analysis (Supplementary Figure S6). Cell extracts for western blots were made in RIPA buffer with protease inhibitors and quantified by Bradford assay. In all, 40 μg of total cell extract was used for the western blots with 1:2000 dilution of anti-Dnmt1 antibody (Active Motif 39204). For Dnmt1 and GR expression analysis, total RNA was extracted using the RNeasy Mini Kit (Qiagen), including a DNaseI digestion step (RNase free DNase Set, Qiagen). One microgram DNA was reverse transcribed (iScript cDNA Synthesis Kit, BioRad) in 20 μl reaction volume, and 1 μl was used per qPCR using SYBR Green and a BioRad IQ system (BioRad) with the following primers: Dnmt1-For 5′-TCAACGAGGCAGACATCAAG-3′, Dnmt1-Rev 5′-GTTCACCACAGCTTCCTCGT-3′ and GR-For 5′-AGGCCGCTCAGTGTTTTCTA-3′, GR-Rev 5′-TACAGCTTCCACACGTCAGC-3′.
Chromatin immunoprecipitation
We seeded 3134 cells in DMEM supplemented with 10% charcoal-dextran-treated serum in 150 mm tissue culture plates and collected the next day for ChIP experiments after indicated time of 100 nM dexamethasone treatment. ChIP was performed using standard protocols (Upstate Biotechnology) with crosslinking step (1% formaldehyde, 37°C, 10 min) followed by a 10-min quenching step with 150 mM glycine. Chromatin was sonicated using Bioruptor sonicator (Diagenode) with 30 s ‘on’ and 30 s ‘off’ for 15 min. In all, 400 μg of sonicated chromatin was immunoprecipitated with antibodies against H3K4me3 (5 μg of ab8580, Abcam), H3K9me1 (15 μg of ab8896, Abcam), H3ac (5 μg of 06-599, Upstate/Millipore) and GR (cocktail of antibodies 7.5 μg of PA1-511A, ABR; 15 μg of MA1-510, ABR; and 3 μg of sc-1004, Santa Cruz). DNA isolated from immunoprecipitates as well as input DNA was used as a template for qPCR (primer pairs are listed in Supplementary Table S4). Standard curves were created by 10-fold serial dilution of an input template. The data present mean values from three independent experiments (error bars s.e.m.).
Formaldehyde-assisted isolation of regulatory elements-qPCR
Cells were seeded on 100 mm tissue culture plates, collected and sonicated according to the ChIP protocol. After sonication, samples were subjected to repeated phenol/chloroform extraction as previously described (Giresi et al, 2007). Next, the FAIRE samples and input samples were incubated overnight at 65°C in order to reverse the crosslinks, subsequently followed by Proteinase K and RNase treatment, phenol/chloroform purification and ethanol precipitation. The dried pellets were resuspended in 50 μl of water and the FAIRE enrichment was analysed by qPCR using primers listed in Supplementary Table S4. The method was first validated in order to confirm whether tissue-specific and hormone-dependent chromatin accessibility described by DNaseI-seq (John et al, 2011) could be detected. Two independent sets of samples were collected and compared between AtT-20 cells and 3134 cells, either naive or treated with dexamethasone for 1 h. The FAIRE enrichment was then compared between untransfected, Dnmt1 siRNA-transfected and control siRNA-transfected 3134 cells. The results were obtained in three independent experiments, normalized to input samples and displayed as mean values with error bars (s.e.m.). Two-tailed Student's t-test was applied to calculate the statistical significance between Dnmt1 siRNA-transfected samples and the remaining conditions.
Electrophoretic mobility shift assay
EMSA was performed to determine whether sequence-specific DNA–GR interactions within the Suox, Ampd3 and Sgk GREs are altered by the presence of methylated CpG. Oligonucleotides and methylated oligonucleotides were purchased from Operon Technologies, Inc. Oligonucleotides encompass 32 bases of each enhancer element with 15 bases defined as a consensus GRE site centrally located (Supplementary Table S5). The following controls for Suox-GRE were used: Mut Suox-GRE (four substitutions disrupting Suox-GRE motif) and G>A Suox-GRE (G>A substitution disrupting CpG element and reversing GRE to the perfect motif).
The sense oligonucleotides were annealed to the corresponding antiparallel partners to create EMSA probes. Purified hGR proteins (aa418–777 and aa116–777) were used in EMSAs. In vitro activities were determined using BIAcore DNA-binding, gel shift and fluorescence polarization assays. Indicated amounts of protein in 10 μl reaction (10 mM Tris–HCl, pH 7.0, 5% glycerol, 50 mM NaCl, 5 mM MgCl2, 1 mM EDTA, pH 8.0, 1 mM freshly added dithiothreitol and 100 nM Dex) were incubated with or without 40 ng of double-stranded probes for 30 min on ice. DNA–protein complexes were resolved on 6% non-denaturing polyacrylamide gel electrophoresis (2 min 400 V followed by 15 min 200 V) and stained with SYBR Green for 45 min.
In order to increase the detection sensitivity of shifted and free DNA, the results for Suox-GRE were repeated using EMSA probes phosphorylated with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P] ATP. The binding reaction with GR aa418–777 was performed as above using 25 000 c.p.m. of a probe and 100 ng of poly(dI·dC).
DNaseI-seq, ChIP-seq data analysis and CpG distribution analysis
DNAseI-seq and ChIP-seq experiments were described previously (John et al, 2011). High-throughput sequencing output was processed for both DNaseI and ChIP data by detecting regions of significant tag enrichment as ‘hotspots’ using algorithm described in John et al (2011).
For CpG distribution analysis, we counted the number of CpG dinucleotides contained in 100-bp window centred at each genomic base pair within 5 kb upstream and downstream from the centre of given regions. Then, we calculated the average counts over the DHS hotspots or GR ChIP peaks as specified in the respective figures. The bootstrapping analysis was applied to mediate the difference in the number of observations between unique and shared pre-programmed sites (Figure 2C). In all, 140 hotspots were chosen randomly from unique DHSs and average CpG counts were calculated repeatedly 100 times. The mouse genome assembly mm8 (UCSC Genome Browser) was used as a reference genome. The genomic coordinates of TSSs and CpG islands were downloaded from the UCSC genome browser (http://hgdownload.cse.ucsc.edu/goldenPath/mm8/database/refGene.txt.gz and http://hgdownload.cse.ucsc.edu/goldenPath/mm8/database/cpgIslandExt.txt.gz). The ChIP-seq data sets for Oct4 and Stat3 binding were obtained from GEO database ID number GSE11431 (Chen et al, 2008). CGI-associated regions were excluded from the analysis.
Supplementary Material
Acknowledgments
We thank Yamini Dalal, Tina Miranda, Tom Misteli, Ty Voss and Sam Clokie for critical discussions and reading of the manuscript. We acknowledge Anindya Indrawan for technical support and Katherine McKinnon and the NCI Vaccine Branch FACS Core facility for the assistance with the cell-cycle analysis. This research was supported by Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author contributions: MW and GLH designed research, with important contributions from SJ. MW, TE, SJ, TAJ, SCB and PJS performed experiments. MW, SB, M-HS, RET and JAS analysed data. KHP and CAS contributed purified GR fragments. MW, SJ and GLH wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Angrisano T, Sacchetti S, Natale F, Cerrato A, Pero R, Keller S, Peluso S, Perillo B, Avvedimento VE, Fusco A, Bruni CB, Lembo F, Santoro M, Chiariotti L (2010) Chromatin and DNA methylation dynamics during retinoic acid-induced RET gene transcriptional activation in neuroblastoma cells. Nucleic Acids Res 38: 1993–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Macleod D, Bird AP (1989) Specific protection of methylated CpGs in mammalian nuclei. Cell 58: 509–517 [DOI] [PubMed] [Google Scholar]
- Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrancois P, Struhl K, Gerstein M, Snyder M (2009) Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci USA 106: 14926–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, Leproust EM, Park IH, Xie B, Daley GQ, Church GM (2009) Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM (2009) Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res 19: 1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Stancheva I (2008) Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci 65: 1509–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin M, Cappabianca L, Kress C, Thomassin H, Grange T (2004) Nature of the accessible chromatin at a glucocorticoid-responsive enhancer. Mol Cell Biol 24: 7891–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G, Pennie WD, John S, Hager GL (1998) The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol 18: 3633–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S (2009) DNA demethylation by DNA repair. Trends Genet 25: 82–90 [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD (2007) FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M, Weintraub H (1981) Activation of globin genes during chicken development. Cell 24: 393–401 [DOI] [PubMed] [Google Scholar]
- Grunau C, Schattevoy R, Mache N, Rosenthal A (2000) MethTools--a toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res 28: 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Zhang Z, Sabo PJ, Chen X, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, McCombie WR, Wigler M, Hannon GJ, Hicks JB (2009) High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res 19: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH (2010) DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun 11: 554–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Koch-Paiz CA, Davis SR, Walker R, Meltzer P, Hager GL (2009) Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150: 1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL (2008) Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G (2008) Transient cyclical methylation of promoter DNA. Nature 452: 112–115 [DOI] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S (2009) DNA demethylation in hormone-induced transcriptional derepression. Nature 461: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Kim ST, Fields PE, Flavell RA (2007) Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci USA 104: 17052–17057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kress C, Thomassin H, Grange T (2006) Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA 103: 11112–11117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy DR, Rands E, Scolnick EM (1978) Helper-independent transformation by unintegrated Harvey sarcoma virus DNA. J Virol 26: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352: 497–505 [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–893 [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M et al. (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- O’Malley BW (2007) Coregulators: from whence came these ‘master genes’. Mol Endocrinol 21: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH, Shen JC, Rideout WM III, Jones PA (2008) The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C (2010) CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA 107: 20311–20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo S, Cousins DJ, Winkelmann N, Triantaphyllopoulos K, Staynov DZ (2009) Chromatin structure and DNA methylation of the IL-4 gene in human T(H)2 cells. Chromosome Res 17: 485–496 [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA 103: 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, Rehli M (2009) Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res 19: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Rideout WM III, Jones PA (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res 22: 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP (2007) Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR (2007) Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinas ML, Grange T (2001) Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 20: 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR (2010) High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5: e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR (2004) Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA 101: 15603–15608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Wiench M, Miranda TB, Hager GL (2011) Control of nuclear receptor function by local chromatin structure. FEBS J 278: 2211–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST (2007) Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci USA 104: 12377–12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST (2009) Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev 23: 2824–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev 9: 140–147 [DOI] [PubMed] [Google Scholar]
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Hattori N, Ohgane J, Tanaka S, Liu XS, Shiota K (2008) DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res 18: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Yamamoto KR (1984) Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell 38: 29–38 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, Jeltsch A (2009) DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 5: e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.