Abstract
Like spliceosomal introns, the ribozyme-containing group II introns are excised as branched, lariat structures: a 2′–5′ bond is created between the first nucleotide of the intron and an adenosine in domain VI, a component which is missing from available crystal structures of the ribozyme. Comparative sequence analysis, modelling and nucleotide substitutions point to the existence, and probable location, of a specific RNA receptor for the section of domain VI that lies just distal to the branchpoint adenosine. By designing oligonucleotides that tether domain VI to this novel binding site, we have been able to specifically activate lariat formation in an engineered, defective group II ribozyme. The location of the newly identified receptor implies that prior to exon ligation, the distal part of domain VI undergoes a major translocation, which can now be brought under control by the system of anchoring oligonucleotides we have developed. Interestingly, these oligonucleotides, which link the branchpoint helix and the binding site for intron nucleotides 3–4, may be viewed as counterparts of U2–U6 helix III in the spliceosome.
Keywords: allosteric ribozyme, group II intron, lariat branchpoint, self-splicing, spliceosome
Introduction
Group II introns, when fully functional, are retrotransposons composed of a large ribozyme and the coding sequence of a reverse transcriptase. The ribozyme catalyses splicing of the intron-containing precursor transcript and reverse splicing of the excised intron into DNA targets, while the intron-encoded protein is essential to copy the inserted intron RNA into DNA (Lambowitz and Zimmerly, 2004). Both the ribozyme components of group II introns and the eukaryotic spliceosome excise introns as branched, lariat structures. Lariats result from a 2′–5′ phosphodiester bond being formed between an adenosine internal to the intron and the first intron nucleotide. In group II introns, the adenosine whose 2′OH group will attack the 5′ splice site during the first step of splicing bulges out of ribozyme domain VI, on its 3′ side (Figure 1A). After the branching reaction, the newly formed 2′–5′ dinucleotide is removed from the (apparently) single ribozyme catalytic centre and replaced by the 3′ splice site in order for exon ligation to take place (Chanfreau and Jacquier, 1994, 1996).
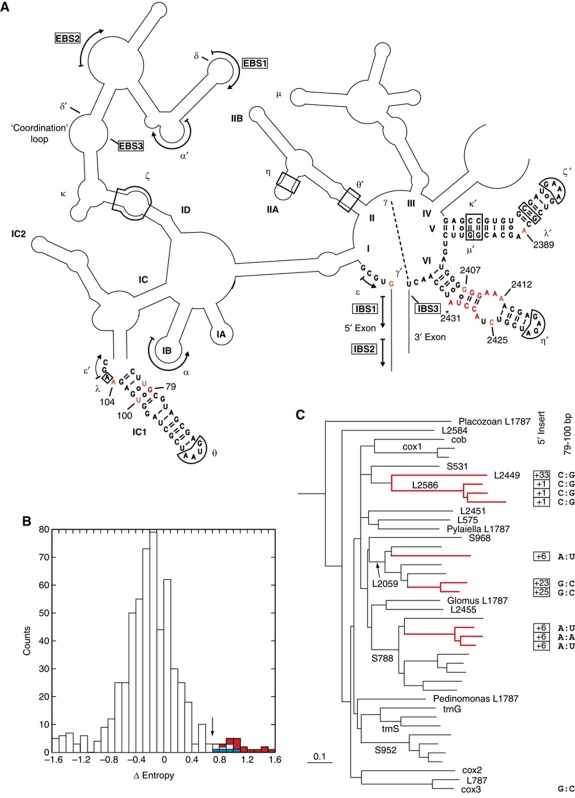
Figure 1.
Identification of a candidate site for binding the branchpoint-carrying domain of a group II intron. (A) Schematic secondary structure of the Pl.L1787 (Pl.LSU/2) ribozyme, a representative mitochondrial member of subgroup IIB1. Only the sequences of domains V and VI and the distal part of subdomain IC1 are shown, the asterisk next to domain VI indicates the branchpoint. Greek letters and arrows correspond to prominent tertiary interactions, which are generally conserved in group II introns (Michel et al, 2009). Sites in red and orange are those at which the difference in sequence entropy between the set of introns with and without a 5′ terminal insert exceeds 1.0 or is included in the 0.70–1.0 range, respectively (see (B)). (B) Statistical distribution over aligned ribozyme sites of the difference in sequence entropy between sets of introns with and without a 5′ terminal insert. Ordinates: number of sites; abscissa: difference in sequence entropy at homologous sites between the two intron sets, calculated as in Materials and methods (numbers are positives when site entropy is larger for the set of introns with a 5′ insert). The arrow points to the 0.70 differential entropy threshold (for sites highlighted in (A); red and blue rectangles correspond to sites in domains VI and IC1, respectively). (C) Phylogenetic relationships of mitochondrial subgroup IIB1 introns based on an alignment of their ribozyme sequences (the tree is redrawn from Li et al, 2011). Introns and intron clades are designated by their host gene (Li et al, 2011). Thick red lines correspond to lineages of introns that possess a 5′ terminal insert, the length of which is indicated at right (boxed numbers). When not G and U, the nucleotides at positions 79 and 100 (of the Pl.L1787 ribozyme) are indicated at the far right.
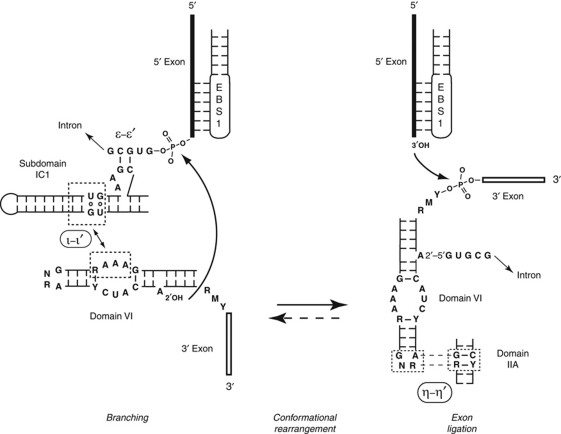
Except for its branchpoint adenosine, the rather small domain VI is poorly conserved between subgroups of group II ribozymes (Michel et al, 2009) and its sequence and secondary structure may vary even within sets of closely related introns. Nevertheless, an RNA tertiary contact involving domain VI and domain II (η–η′ in Figure 1A), which had been identified by Chanfreau and Jacquier (1996) in a screen for interactions specific to the exon ligation step, was subsequently shown to be present in both major subdivisions, IIA and IIB, of the group II intron family. In addition to dramatically reducing the rate of exon ligation, disruption of η–η′ promotes branching: it increases the rate of first step transesterifications (branching and its reverse reaction, debranching) and, in a subgroup IIA intron (Costa et al, 1997a), it was shown to favour branching over hydrolysis at the 5′ splice site. The latter is usually a minor reaction that only prevails when the branchsite is missing or mutated (Van der Veen et al, 1987), when the 5′ splice site is separated from the rest of the intron (Jacquier and Jacquesson-Breuleux, 1991) or else, in the presence of potassium ions (Jarrell et al, 1988). These data were rationalized by postulating that group II ribozymes exist in two conformations, one in which η–η′ contributes to the specific positioning of the 3′ splice site for exon ligation and another one in which domain VI and the branch site are somehow poised for branching.
By contrast to the identification of η–η′, the search for interactions that, by being specific to the branching step, could contribute to our understanding of the mechanism by which formation of the lariat bond is activated, proved particularly frustrating. Only in 2006 was a candidate receptor for the domain VI branchpoint finally proposed by Hamill and Pyle, based on crosslinking experiments. This receptor consists of a subdomain ID internal loop that had previously been shown to contain the binding site for the 3′ exon of subgroup IIB introns and to be indirectly involved as well in the binding of the 5′ exon (Costa et al, 2000); it was accordingly dubbed the ‘coordination loop’ by Hamill and Pyle (Figure 1A). However, no counterpart for the subgroup IIB coordination loop can be discerned in secondary structure models of subgroup IIA ribozymes (see Michel et al, 2009), which is surprising, given the nearly universal conservation of the branchpoint adenosine and bulge. Also, some nucleotide substitutions in the coordination loop do reduce dramatically the rate at which precursor molecules react (Hamill and Pyle, 2006), but they have not been shown to affect branching specifically (i.e. with respect to hydrolysis).
The first atomic-resolution structure of a group II ribozyme, by Toor et al (2008a), lacked both the coordination loop and domain VI. Subsequent refinements of this structure have made it possible to visualize the coordination loop and its predicted interactions with both the 3′ exon and the EBS1 loop, which binds the 5′ exon (Toor et al, 2010; Wang, 2010), but domain VI remains invisible, possibly because its flexibility leads to its degradation (see Discussion in Pyle, 2010).
This situation, and our recent finding that the ability to initiate splicing by branching was recurrently lost during the evolution of a subclass of natural group II introns (Li et al, 2011) prompted us to reexamine the sequences of group II ribozymes in search for a potential receptor site that would bind the nucleotides that surround the branchpoint, in the middle part of domain VI. We now show that there exists such a candidate site, located in subdomain IC1 (Figure 1), at which nucleotide substitutions specifically affect branching, rather than hydrolysis. In a second stage, by taking advantage of the currently available group II ribozyme structure, we were able to model the possible interaction of domain VI with this receptor and from there, to create an allosteric ribozyme (Tang and Breaker, 1997), whose ability to form the lariat bond depends on oligonucleotides that anchor domain VI to its binding site.
Results
Comparison of introns with and without branchsites points to a potential first-step receptor for domain VI
It has long been known that some rare group II introns in organelles lack a bulging A on the 3′ side of domain VI (Michel et al, 1989; Li-Pook-Than and Bonen, 2006) and at least one of these introns, in the tRNAVal (UAC) gene of plant chloroplasts, is excised indeed as a linear molecule, rather than a lariat (Vogel and Börner, 2002). Such cases used to be regarded as oddities but recently, an evolutionary process that recurrently created intron lineages with additional nucleotides at the intron 5′ extremity, and no apparent branchpoint, was shown to be at play in mitochondria (Li et al, 2011; one member of this subset was confirmed to be unable to generate other than linear excised intron molecules in vitro). In these lineages, not only is the branchpoint adenosine missing, but the middle part of domain VI next to it, which normally consists, in the IIB1 intron subclass from which these lineages originated, of a 3-bp helix and a well-conserved 6-nt internal loop (Figure 1A), is highly variable, in contrast to the basal and distal sections of the same domain VI (Li et al, 2011). This suggests that not merely the branchpoint and its two flanking G:U base pairs (Figure 1; Chu et al, 1998), but the entire middle part of domain VI could be involved in branching, presumably by binding to one or several specific receptor sites. We sought to identify candidate sites for such receptors by taking advantage of the fact that their sequences and structures may no longer be constrained in molecules that have lost the ability to carry out branching.
Only 10 sequences of introns with a 5′ terminal insert are currently known, but these sequences belong to four to five independent lineages (Li et al, 2011; Figure 1C), which should ensure some measure of statistical significance in comparisons. In fact, when those 10 sequences are aligned with 32 sequences of mitochondrial introns that belong to the same intron subclass, but lack a 5′ terminal insert (and possess a potential branchpoint), and the sequence entropy in each subset is systematically compared site by site (Figure 1B; see Materials and methods), a small number of intron positions at which the difference in sequence entropy (ΔE) lies well beyond the main distribution stand out from the rest. In simple terms, these sites are very well conserved as long as the branchpoint is present, but very poorly so otherwise.
Among the 20 sites with the highest ΔE scores, two were discarded because their nucleotide composition was too variable (entropy above 0.3) in the no-5′-insert subset. Out of the remaining 18 sites (Figure 1), 12 are concentrated in the middle part of domain VI, which, as already emphasized, is quite variable in the 5′-insert subset; one corresponds to the first intron nucleotide, that no longer forms a 2′–5′ bond in that subset; another one is at position 2389 (generally an A), which, in the crystal-derived atomic-resolution model of the Oceanobacillus ribozyme (Toor et al, 2008a), lies next to the 5′ splice site and binds two metal ions that have been proposed to be critical for catalysis; and yet another one, at position 104, is also known to be part of the catalytic core. Remarkably, however, the remaining three sites (positions 78, 79, 100) belong to two consecutive G:U pairs in the IC1 distal helix, a component whose terminal loop (θ) is known to have an important structural role by contacting domain II, but which lies rather far away from the reaction centre and had not yet been proposed to be implicated in catalysis.
The distribution of bases at positions 79 and 100 is especially striking. These nucleotides form a G:U pair in all but one of the 32 intron sequences with a recognizable branchpoint, whereas 9 out of the 10 sequences with a 5′ terminal insert have a Watson–Crick pair instead and one has an A:A mismatch (Figure 1C). Such a nearly perfect correlation suggests that presence of a G:U pair at positions 79:100 is particularly important for the initiation of splicing by branching, whereas in the absence of a functional branchpoint, the type of base pairing at that site affects only the overall stability and precise geometry of the IC1 stem.
Nucleotide substitutions in domain VI and its IC1 candidate receptor site
In constructs that lack domain VI or have an altered branchpoint, hydrolysis at the 5′ splice site substitutes for branching and the intron is excised in linear, rather than lariat form. By contrast, introns with fully functional, well-folded ribozymes are expected to initiate splicing almost exclusively by transesterification. As seen in Table I, that is the case for the Pylaiella L1787 intron (Pl.LSU/2; Costa et al, 1997b), which we have been using as a model subgroup IIB1 molecule: about 90% of excised intron products are lariats when the in vitro self-splicing reaction takes place in the presence of ammonium and magnesium counterions.
Table 1. Kinetic parameters of dVI and IC1 mutants.
| Construct | Fraction of products branched | kbranching (min−1) | khydrolysis (min−1) | kbr/khy |
|---|---|---|---|---|
| Ammonium | ||||
| wta | 0.90±0.07 | 0.136±0.019 | 0.024±0.010 | 5.5 |
| 0.88±0.11 | 0.166±0.032 | 0.023±0.008 | 7.2 | |
| dVI-7 bp | 0.89±0.04 | 0.092±0.006 | 0.019±0.002 | 5.0 |
| dVI-4 bp | 0.84±0.06 | 0.058±0.006 | 0.014±0.002 | 4.2 |
| dVI-2 bp | 0.02b | <0.008±0.002c | 0.013±0.002 | <0.62 |
| IC1 Δθ | ND | ND | ND | ND |
| IC1 UA:UA | 0.89±0.09 | 0.028±0.003 | 0.024±0.004 | 1.3 |
| IC1 Δθ/UA:UA | ND | ND | ND | ND |
| IC1-2 bp | 0.90±0.11 | 0.016±0.004 | 0.024±0.004 | 0.69 |
| Potassium | ||||
| wta | 0.76±0.08 | 0.160±0.030 | 0.064±0.023 | 2.5 |
| 0.77±0.06 | 0.149±0.020 | 0.065±0.009 | 2.3 | |
| dVI-7 bp | 0.41±0.04 | 0.132±0.021 | 0.057±0.012 | 2.3 |
| dVI-4 bp | 0.15±0.01 | 0.045±0.006 | 0.072±0.008 | 0.63 |
| dVI-2 bp | 0 | 0 | [0.135±0.011]d | 0 |
| IC1 Δθ | 0.69±0.05 | 0.097±0.006 | 0.019±0.004 | 5.1 |
| IC1 UA:UA | 0.10±0.007 | 0.028±0.002 | 0.042±0.005 | 0.67 |
| IC1 Δθ/UA:UA | 0.067±0.005 | 0.025±0.003 | 0.029±0.002 | 0.84 |
| IC1-2 bp | 0.063±0.025 | 0.026±0.013 | 0.031±0.011 | 0.85 |
| ND, not determined. | ||||
| aDeterminations from different RNA preparations. | ||||
| bObserved value at 180 min. | ||||
| cEstimated from the fraction branched at 180 min. | ||||
| dDetermined at 50 mM Mg. | ||||
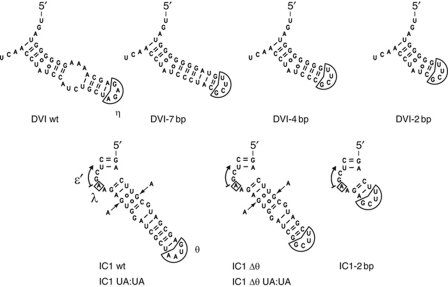
Assuming, as suggested by sequence analyses, that both the middle section of domain VI and the 79:100 and 78:101 G:U pairs are specifically involved in the branching reaction, nucleotide substitutions at these sites should shift the balance between transesterification and hydrolysis towards the latter process. However, replacement of the internal loop of domain VI by canonical base pairs and trimming of the resulting, extended helix down to 4 bp (Figure 2) have barely detectable effects on the fraction of products branched when reactions are carried out in the presence of 1 M ammonium chloride (Table I). The observed rate constant for branching (kbranching) does decrease (by less than three-fold), but so does that for hydrolysis, so that their ratio is barely affected. Only by bringing the length of the helix distal to the branchpoint down to 2 bp (mutant dVI-2 bp) do consequences suddenly become dramatic, with splicing proceeding almost exclusively by hydrolysis (Table I, line 4).
Figure 2.
Ribozyme constructs with altered dVI and IC1 structures.
One possibility was that under optimal in vitro self-splicing conditions, processes other than the positioning of domain VI are rate limiting for transesterification at the 5′ splice site of precursor molecules. Among monovalent counterions, potassium has long been known to favour hydrolysis relative to branch formation (Jarrell et al, 1988). Compared indeed to the situation in ammonium, the observed rate constant for hydrolysis is increased by almost three-fold for the wild-type Pl.LSU/2 ribozyme, and there is also a significant decrease in the fraction of lariats among intron excision products (Table I). Interestingly, all of the mutant domain VI (dVI) constructs in Figure 2 are further affected in their ability to react when assayed in the presence of potassium. Removal of the dVI internal loop significantly decreases the fraction of molecules that initiate splicing by branching compared with the wild type, while truncation of the resulting helix to 4 bp not only reduces this fraction further, but specifically affects the observed rate constant for branching, by about four-fold relative to the wild type.
Since any nucleotide substitution in the IC1 stem might affect the relative position in three-dimensional space of the ε′ and θ loops, we deemed it preferable to try and delete the latter component before assessing our IC1 constructs in potassium. Fortunately, removal of θ turned out to be without severe consequences on the ability of precursor molecules to react under the experimental conditions we had chosen; in fact, transesterification is even less affected than hydrolysis, resulting in an elevated kbranching/khydrolysis ratio (Table I). By contrast, when the two G:U pairs at positions 79:100 and 78:101, which 16 out of 32 mitochondrial IIB1 introns with a recognizable branchpoint share, are simultaneously substituted by A:U pairs, the observed rate constant of branching and the fraction of intron molecules excised in lariat form are both markedly affected and this, whether in a wild-type or Δθ context (Table I; in ammonium, kbranching is specifically affected as well, but the fraction of molecules that react by branching is left unchanged; see also Supplementary Figures S1 and S2). Interestingly also, trimming of the IC1 helix down to only 2 bp is without further effects on kinetic parameters. Thus, these experiments are consistent with the conclusions of comparative sequence analyses, which pointed to the tandem G:U pairs in IC1 as major potential contributors to the ability to perform branching.
Modelling of the interaction between domain VI and its proposed IC1 receptor
The G79:U100 pair is highly conserved in a majority of group II intron subclasses (Dai et al, 2003), including the somewhat divergent subgroup IIC, to which the Oceanobacillus intron belongs. We have explored the possibility that this pair constitutes part of the first-step receptor site for domain VI by attempting to model the missing domain VI (Figure 3A) into the latest atomic-resolution models (Toor et al, 2010; Wang, 2010) of the Oceanobacillus group II ribozyme.
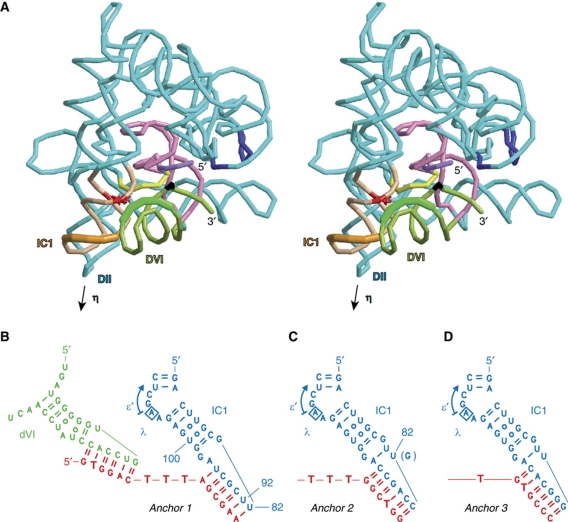
Figure 3.
Three-dimensional model of the interaction between ribozyme domains VI and IC1 and optimization of oligonucleotides anchoring dVI to IC1. (A) Stereo views were generated from the coordinate set of Toor et al (2010) for the Oceanobacillus iheyensis subgroup IIC ribozyme (PDB accession number 3IGI). Only domain VI, the 3-nt dV–dVI linker and intron residues 1–2 were modelled de novo; the last three nucleotides of the intron and the 3′ exon are missing (see Results and Materials and methods). Colour scheme: black, branchpoint adenosine; green, domain VI; pink, domain V; violet, 5′ exon; yellow, intron nt 1–5; tan, subdomain IC1; red, bp 79:100 (81:101 in the Oceanobacillus ribozyme); deep blue, ‘coordination loop’. Thickened sections of dVI (G2409–A2413) and IC1 (A83–A87) correspond to the base-paired segments (‘handles’) of our anchoring oligonucleotides (see panels B–D). The arrow points to the location in the Pl.LSU/2 ribozyme of the η receptor (see Figure 1A); assuming stems II and IIA are stacked, the latter should be situated about one helical turn beyond the tip of what was left of domain II in the molecule crystallized by Toor et al (2008a). (B) Scheme for anchoring dVI to IC1, showing IC1 anchor 1 with a 3-T tether. (C) Anchor 2, with a 3-T tether; at position 82, G was introduced before switching back to U. (D) Anchor 3, with a 1-T tether.
Currently available structures of the Oceanobacillus ribozyme reflect the final stage of splicing, after exon ligation. They lack domain VI and the last three intron residues as well as the first intron nucleotide (G1). The latter must move away from the catalytic centre after the first step of splicing in order to make way for the 3′ splice site and the segment at the intron 5′ extremity that gets relocated may include also U2 (although not G3, for the ε–ε′ interaction—Jacquier and Michel, 1990—is believed to persist throughout splicing). As first pointed out by Steitz and Steitz (1993) (see also Michel and Ferat, 1995; Jacquier, 1996), the best way to reconcile data on the inhibition of individual splicing steps by phosphorothioate stereoisomers of the reactive phosphate group with the generally accepted existence of a single catalytic site is to postulate that the O3′-P-O5′ dihedral angle at the 5′ splice site undergoes a 120° rotation away from the helical geometry that prevails at the splice junction of the intron-bound ligated exons (Costa et al, 2000; Toor et al, 2008b). In the predicted structure of the Oceanobacillus precursor RNA, such a sharp bend is required anyway in order to ensure connectivity within the segment that extends between the last nucleotide of the 5′ exon, which is expected to remain bound to EBS1 throughout the splicing process, and G3 (see Wang, 2010). Modelling of the phosphodiester bond at the 5′ splice site then makes it possible to position precisely the attacking 2′OH group of the branchpoint adenosine, which sets in turn the stage for placing the basal and distal helices of domain VI.
We found that in order for the 5′ strand of the basal dVI helix to bridge the distance between the branchpoint and domain V, the first two base pairings at the base of the latter in Figure 1 of Toor et al (2008a) need to be disrupted: these pairings, the existence of which is not supported by comparative sequence analysis (note their absence in Figure 1A), may owe their presence in the Oceanobacillus ribozyme structure to the absence of domain VI. As for the section of domain VI that lies distal to the branchpoint, we chose to model it as a continuous helix despite the presence of a very well-conserved internal loop (Figure 1A; Li et al, 2011) in mitochondrial subgroup IIB1 introns. The reasons for this are (i) most bacterial members of this subclass lack an internal loop in their distal dVI stem, even though they share tandem IC1 G:U pairs with their mitochondrial counterparts and (ii) substitution of canonical base pairing for the internal loops of introns Sc.cox1/5γ (Chu et al, 1998) and Pl.LSU/2 (Figure 2; Table I) has limited effects on their ability to carry out branching.
As shown in Figure 3A, it is possible indeed to position a continuous dVI distal helix in such a way that its base is connected to, and stacked on, the proximal section of the domain (consisting of the basal dVI helix and branchpoint adenosine), while its 5′ backbone fits neatly into the shallow (‘minor’) groove of the IC1 stem. This model is consistent with our comparative sequence analysis and nucleotide substitution experiments, since the section of IC1 that is specifically contacted by domain VI encompasses the G79:U100 base pair (G81:U101 in the Oceanobacillus intron). For the sake of consistency with η–η′, we propose to name ι–ι′ (iota–iota′) this novel interaction between the IC1 shallow groove at, and immediately distal to, positions 79 and 100 (ι) and the middle part of the dVI distal stem (ι′).
Activation of lariat formation by oligonucleotides that anchor domain VI to its binding site
As apparent from Figure 3A, optimal positioning of the dVI distal helix into the shallow groove of helix IC1 results in placing IC1 nucleotides A83–A87 (Pl.LSU/2 numbering) in near continuity of A2413 in the 5′ strand of domain VI. This peculiar arrangement suggested to us that it might be possible to replace part of the 5′ strands of the dVI and IC1 helices by an oligonucleotide that would at the same time restore the dVI helical structure and anchor it to its proposed receptor. The complete setup, consisting of such an ‘anchoring’ DNA oligonucleotide with segments (‘handles’) that are complementary to the terminal loops of the truncated dVI and IC1 stems and are connected with one another by a tether made out of deoxythymidines, is shown in Figure 3B.
As expected from the data in Table I, the construct in Figure 3B, in which the dVI distal helix has been truncated down to 2 bp, with a 7-nt terminal loop, has only residual branching activity (Figure 4A, intercept with the y axis). However, the same precursor transcript, when incubated in the presence of increasing concentrations of an oligonucleotide capable of restoring base pairing in both the dVI and IC1 stems (Figure 3B), gradually recovered the ability to initiate splicing by transesterification, with up to ca 58% of reaction products consisting of the lariat intron at 200 μM oligonucleotide (not shown). A plot of the fractional rate of branching (observed rate of branching relative to total rate of conversion of precursor into products) as a function of the concentration of oligonucleotide can be fitted indeed to a saturation curve (see Materials and methods) with an estimated Km equal to 58±20 μM (Figure 4A).
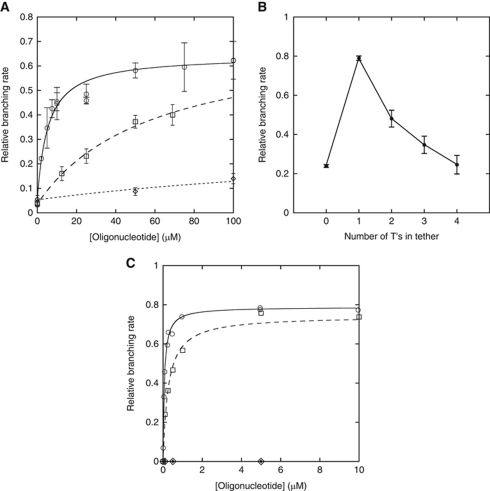
Figure 4.
Branching by dVI–IC1 constructs as a function of the concentration of anchoring oligonucleotides, their tether length and their complementarity to the terminal loop of truncated IC1 stems. See Materials and methods for calculation of relative branching rates and standard errors. (A) Optimization of IC1 anchors. Relative branching rate as a function of oligonucleotide concentration for individual construct–oligonucleotide combinations (Figure 3B and C) was fitted to equation (3) of Materials and methods. Empty squares and dashed curve, construct in Figure 3B with matched oligonucleotide 5′-GTGGAC-TTT-AGCGAA, Km=58±20 μM, Pearson's R=0.9969; empty circles and solid curve, construct in Figure 3C with matched oligonucleotide 5′-GTGGAC-TTT-GGCTGG, Km=5.4±1.0 μM (Kd=7.5 μM), R=0.9825; lozenges and dotted curve, construct in Figure 3C with 5′-GTGGACT (no anchor). (B) Relative branching rate of construct in Figure 3C as a function of the number of T's in the tether of oligonucleotide 5′-GTGGAC[T]nGGCTGG. The concentration of oligonucleotide was set at 5.0 μM, close to the observed Km for a 3-T tether (see (A)). (C) Abscissa and ordinates as in (A). Empty circles and solid curve, construct in Figure 3C with matched oligonucleotide 5′-GTGGAC-T-GGCTGG, Km=0.073±0.009 μM (Kd=0.17 μM), R=0.9946; empty squares and dashed curve, construct in Figure 3D with matched oligonucleotide 5′-GTGGAC-T-GTGCCC, Km=0.27±0.05 μM (Kd=0.55 μM), R=0.9938; filled lozenges, construct in Figure 3C with mismatched oligonucleotide 5′-GTGGAC-T-GTGCCC; empty lozenges, construct in Figure 3D with mismatched oligonucleotide 5′-GTGGAC-T-GGCTGG.
Subsequent experiments showed that this Km could be decreased by playing with both the geometry of the IC1 terminal loop and its sequence. Among the combinations we tried, the one shown in Figure 3C turned out to be optimal, with a Km of 5.4±1.0 μM (Figure 4A; a G which had been introduced at position 82 so as to leave unspecified the junction between the IC1 and anchoring helices proved suboptimal). As a control, reactions in the presence of increasing concentrations of a 7-mer, no-anchor oligonucleotide that merely restored the dVI helix resulted in only minimal recovery of branching activity (Figure 4A). Additional controls (Table II) performed in the presence of oligonucleotide concentrations (100 μM) well above the observed Km for the combination in Figure 3C demonstrate that: (i) whether the structure of IC1 is wild type (setup 1), truncated (setup 3) or (presumably) restored by a complementary 7-mer oligonucleotide (setup 5), only residual branching activity is observed as long as the terminal loop of the truncated dVI stem is left unpaired; (ii) restoration of the dVI stem by a complementary 7-mer, whether in a wild-type (setup 2) or IC1 mutant context (setup 4; Figure 4A) only slightly improves branching activity; and (iii) simultaneous restoration of base pairing in both the dVI and IC1 stems by two 7-mers (setup 6) is not sufficient: branching activity remains very modest unless anchoring is achieved by creating a covalent link between these oligonucleotides (setup 7).
Table 2. Rate of branching relative to total reaction rate in the presence of a 15-mer anchoring oligonucleotide and 7-mer controls.
| Setup | IC1a | Oligonucleotide(s) (100 μM) | Anti-dVI handle | Anti-IC1 handle | Relative rate of branching |
|---|---|---|---|---|---|
| 1 | wt | No | 0.040±0.009 | ||
| 2 | wt | 7-mer | GTGGACT | 0.126±0.012 | |
| 3 | Figure 3C | No | 0.040±0.009 | ||
| 4 | Figure 3C | 7-mer | GTGGACT | 0.145±0.021 | |
| 5 | Figure 3C | 7-mer | TGGCTGG | 0.068±0.017 | |
| 6 | Figure 3C | 7-mer + 7-mer | GTGGACT | TGGCTGG | 0.150±0.037 |
| 7 | Figure 3C | 15-mer | GTGGACT-T-TGGCTGG | 0.530±0.045 | |
| aDomain VI of all constructs was truncated as in Figure 3B. | |||||
The next step in optimizing this system consisted in keeping the sequence of the anchor in Figure 3C constant and varying the length of the tether from zero to four T's (Figure 4B) at an oligonucleotide concentration (5 μM) about equal to the Km determined for a 3-T tether (Figure 4A, full curve). A sharp optimum was observed for a tether consisting of just one T, with a relative rate of branching equal to 0.790±0.011. The latter value should be close to saturation, as was verified indeed by determining the corresponding Km (0.073±0.009 μM; Figure 4C).
Final proof that complementarity between the IC1 terminal loop and an anchoring oligonucleotide is both necessary and sufficient to activate branching was obtained by nucleotide substitutions (Figures 3D and 4C): whereas mismatched combinations devoid of potential for base pairing exhibit no detectable branching activity, restoration of complementarity by substitution of both the oligonucleotide anchor and the IC1 terminal loop was found to result in almost complete recovery of the ability to initiate splicing by branching (relative rate of branching at saturation, 0.744±0.022; Km equal to 0.270±0.047 μM). Finally, it should be noted that for the setup of Figure 3C, we verified the oligonucleotide-induced branching reaction to be an authentic one, in the sense that the same branchpoint is used as in a wild-type molecule and the resulting ligated exons have the same sequence (see Materials and methods and also the analytical gel in Supplementary Figure S3).
Discussion
A first-step-specific receptor for the branchpoint-carrying domain VI
We have shown that by using oligonucleotides that bring together domain VI and what we propose to be a first-step RNA receptor for this domain, in subdomain IC1 (Figure 5), it is possible to specifically activate the branching reaction in a defective precursor molecule that is otherwise essentially unable to initiate self-splicing, except by 5′ splice site hydrolysis. The dVI and IC1 helices must truly come in contact in the active first-step complex, for we found the optimal connecting segment between the dVI and IC1 handles of the anchoring oligonucleotide to consist of just one thymidine residue (Figure 4B). The use of longer tethers leads to a gradual decrease in the efficiency of branching, as would be predicted by a random-coil model (Jacobson and Stockmayer, 1950), whereas, conversely, when the single connecting nucleotide is removed, restoration of branching is much less efficient, presumably because the anchoring oligonucleotide and its targets must give up one or several base pairs in order to release the resulting strain.
Figure 5.
Conformational rearrangements and tertiary interactions involving domain VI. Tentative delimitation of the ι and ι′ motifs is based on our modelling of the interaction in Figure 3A. During the splicing process, domain VI is successively bound by ribozyme subdomain IC1 (ι–ι′ interaction—this work—which positions domain VI for the branching step) and subdomain IIA (η–η′ interaction—Chanfreau and Jacquier, 1996—which positions domain VI for exon ligation; a 90° rotation was chosen for convenience of drawing, the actual value must be less, see Figure 3A). In reverse splicing into a DNA or (possibly) RNA target, formation of ι–ι′ should follow that of η–η′ (dashed arrow). Bases shown are consensus ones for mitochondrial subgroup IIB1 introns (Li et al, 2011). Curved arrows symbolize reactions.
While compatible with all available data, our modelling of the interaction between domain VI and the IC1 distal helix was dictated by our identification of the G79:U100 base pair as a likely receptor for domain VI. Current ignorance of the exact configuration of the branchpoint adenosine, which has alternatively been proposed to be extrahelical (Schlatterer et al, 2006), to be stacked between 2 bp (Erat et al, 2007) or to be part of a two-nucleotide bulge (Zhang and Doudna, 2002), is such that in fitting the middle part of domain VI optimally into the shallow groove of IC1, we opted to care primarily about the need to retain connectivity to the dVI proximal helix: the two dVI helices are actually stacked on top of one another in Figure 3 and in connecting the branchpoint ribose to its immediate neighbours, we chose to bulge it out from the helical stem, without taking stands on its exact geometry.
In this context, our finding that the optimal dVI–IC1 tether consists of only one nucleotide is important and clearly pleads in favour of our own working model of the ribozyme first-step configuration (Figure 3A), when compared with another recently proposed arrangement of domain VI (Wang, 2010), which attempted to meet previous claims that the coordination loop serves as receptor for the branchpoint (Hamill and Pyle, 2006). In the latter model (Figure 10 of Wang, 2010), which includes a hypothetical ‘mispair’ between the universal branchpoint adenine and A393 (Oceanobacillus ribozyme numbering), a nucleotide that is poorly conserved by evolution, domain VI is oriented right towards the coordination loop, away from IC1. In yet another recently published sketch of a possible first-step conformation (Figure 13 of Pyle, 2010), the location of domain VI, which is represented only as a cylinder, is somewhat intermediate between ours and Wang's since it is placed in between IC1 and the coordination loop, though in a position that would still not allow it to contact our proposed IC1 receptor. It is also important to note that even though they clearly differ, Wang's, Pyle's and our own modelling of the ribozyme first-step conformation all imply a major rotation of domain VI after the branching step in order for its tip to dock into its domain II, second-step receptor (inasmuch as the position of the latter can be modelled precisely, see Figure 3A and its legend).
When interpreting crosslinks between the dVI branchsite and the coordination loop as evidence that the latter constitutes the binding site for the former, Hamill and Pyle (2006) implicitly assumed that domain VI should be stably docked in its first-step receptor before the branching reaction. However, it seems more likely that domain VI keeps toggling between different states, as initially proposed by Costa et al (1997a), based on kinetic analysis of mutant ribozymes, and, more recently, by Toor et al (2010) to account for the absence of that domain in the crystal structure of the Oceanobacillus intron. In fact, when Hamill and Pyle's sites of crosslinking are mapped on the atomic-resolution model of ribozyme domains I–V (Toor et al, 2008a, 2008b), it becomes apparent that essentially every nucleotide that would have been accessible to the branch site and its two flanking nucleotides in a dVI molecule that could freely rotate around the dV–dVI junction did give rise to a crosslink (only residues in the proximal helix of domain V are missing, since crosslinks at those sites are internal to the D56 piece and, therefore, could not be recovered in the experiment). When domain VI is bound to its IC1 receptor, photoactivable bases at the branchpoint and its two neighbours are predicted from our model to crosslink neither to the coordination loop, nor to stem IC1, but to the first two nucleotides of the intron and the last nucleotide of the 5′ exon: these three positions were indeed among those recovered by Hamill and Pyle. Moreover, among the latter crosslinks, those to the G1 nucleotide (and perhaps also to the second residue of the intron) are liable to be compatible with splicing, which provides a ready explanation for the reactivity of part of the XL1 material of Hamill and Pyle (2006).
Interestingly, some published pieces of data in the literature already hinted at the possible involvement of the IC1 distal helix in the branching process. Stabell et al (2009) noted that in a paraphyletic subset of group II introns that share additional secondary structures 3′ of domain VI, the section of the IC1 stem that lies immediately distal to the ε′ loop is unexpectedly conserved. Several nucleotide substitutions were introduced, among which was the replacement of the (counterpart of the) 79:100 G:U pair by A:U. That mutation was found to markedly decrease the rate of reaction of precursor molecules, but in the absence of 5′ splice site hydrolysis, branching could not be shown to be specifically affected.
Much earlier, Boudvillain and Pyle (1998) had published a map of domains I–III of the subgroup IIB1 Sc.a5γ ribozyme (a close relative of Pl.LSU/2) that showed, based on NAIM (Nucleotide Analogue Interference Mapping; see Strobel, 1999), which nucleotides were important for a branching reaction with domains V and VI (unfortunately, the authors’ setup did not make it possible to discriminate between nucleotides required specifically for branching and those involved in catalysis in general or in binding of domain V by domains I–III). Removal of the NH2 at position 2 of G79 (Pl.LSU/2 numbering) and also of the 2′ OH groups of U78 and U100 was reported to interfere with activity, thus pointing to the importance of the shallow groove in this section of the IC1 distal helix; remarkably, these three residues are none other than the ones that generate a statistical signal when molecules with and without a recognizable branchpoint are compared (Figure 1; it is also worth noting that no hit was found in the coordination loop proper, whether by NAIM or our comparative sequence analyses, despite its claimed function as a receptor for domain VI—Hamill and Pyle, 2006). In fact, our phylogenetic approach may rightly be regarded as related to NAIS (Nucleotide Analogue Interference Suppression, also called ‘chemogenetics’; Strobel, 1999), a method in which nucleotide interference maps (rather than sequence conservation maps) are compared for the wild type and a molecule that includes a specific defect.
Towards atomic resolution
It is now generally agreed that group II ribozymes exist in at least two major states (Figure 5), one in which domain VI is prepositioned for the branching reaction and another one in which it interacts with domain II (whether the latter interaction helps positioning the 3′ splice site for exon ligation is still a matter of debate—see Pyle, 2010—despite the fact that disruption of η–η′ was found to impair specifically the second step of splicing—Chanfreau and Jacquier, 1996). The identification of a second-step-specific receptor for domain VI (Chanfreau and Jacquier (1996) was a breakthrough, if only since it made it possible, by playing with the strength of the interaction between diverse loops of the GNRA family and their RNA receptors (Costa and Michel, 1997), to place introns into a well-defined configuration that could be probed by biochemical and biophysical methods. Our use of anchoring oligonucleotides that force domain VI and its IC1 first-step receptor to interact should similarly open the way to trapping the ribozyme into its branching-ready configuration, something which could presumably be achieved by replacing our current DNA ‘handles’ by higher-affinity, RNA or perhaps LNA (Locked Nucleic Acid; Petersen et al, 2002) counterparts (the affinity of even our best anchoring oligonucleotides for their targets—see legend to Figure 4 and Materials and methods for estimated Kd values—is still too low to prevent ‘breathing’ of helices, which also explains why we did not observe accumulation of the lariat-3′ exon reaction intermediate—not shown). This approach might even make it possible to obtain crystals and visualize at last the ribozyme branchpoint and its molecular context at atomic resolution.
One possible objection to the use of anchoring oligonucleotides for biochemical and biophysical probing is that despite the fact that the authentic branchpoint is being used (see Materials and methods) the resulting arrangement in space of domain VI and subdomain IC1 might be an unnatural one. However, because the segment of IC1 that was engineered to interact with the oligonucleotide anchor is located distal to the section that we believe to constitute the natural receptor for domain VI (Figures 3 and 5), that receptor is likely to remain structurally intact in the complex (our initial choice of a 3-nucleotide tether reflected our concern that shorter connecting segments might distort proximally located contacts). It may prove possible also to reconstruct an authentic middle dVI section by replacing our current DNA handle by an RNA counterpart with the appropriate sequence to generate the characteristic internal loop of mitochondrial subgroup IIB1 introns (Figure 1A). This would open the way to the substitution of individual chemical groups in the 5′ strand of that loop, which we propose to be the site of contact with the IC1 receptor (in this respect, it is interesting to note that besides the branchpoint adenosine, the only other sites in domain VI to give rise to interference signals in the NAIM experiments of Boudvillain and Pyle (1998) were positions 2411–2413 (Pl.LSU/2 numbering), which are precisely the ones that should contact the IC1 shallow groove according to the model in Figure 3A). Up to now, the introduction of atomic substitutions and, therefore, the use of NAIS to explore interactions in this section of the ribozyme was made difficult (though not impossible) by the fact that domain VI cannot be supplied alone in a two-piece intron system, but needs to be covalently connected to domain V in order to be bound by the rest of the ribozyme (Jarrell et al, 1988).
Conclusion
Now that a tertiary contact between the branchpoint-carrying component of group II introns and the rest of the group II ribozyme has been found and shown to be essential for the efficiency of lariat formation, the stage is set at last to explore the atomic surroundings of the branchpoint itself. In the meantime, pending a high-resolution structure of an entire intron, our newly acquired ability to control at will the conformation of the ribozyme through the use of oligonucleotides should prove particularly useful for detailed mechanistic investigations of individual steps in the splicing and transposition processes carried out by the sophisticated molecular machinery that we call a group II intron. Finally, it did not escape our notice that in tinkering with the architecture of the group II ribozyme, we may have been preceded by nature: U2–U6 helix III (Sun and Manley, 1995) which, in the spliceosome, links together the branchpoint helix and the segment of U6 that, like ε′, binds the first intron nucleotides, may be regarded as a counterpart of our dVI-anchoring oligonucleotides.
Materials and methods
Sequence analyses
The set of 42 subgroup IIB1 mitochondrial intron sequences collected and aligned by Li et al (2011) (the aligned set is accessible at http://rnajournal.cshlp.org/content/suppl/2011/05/05/rna.2655911.DC1.html) was divided into a subset of 32 intron sequences in which the 5′ splice site is followed by the GUGCG consensus at the intron 5′ end and a subset of 10 intron sequences with a 5′ terminal insert. Entropy (as defined in BioEdit; Hall, 1999: H(l)=−Σf(b,l)ln(f(b,l)), where f(b,l) is the frequency of base b at position l) was calculated for each subset at each of the 577 positions of the alignment and values for the no-insert subset were subtracted from those for the insert-carrying subset in order to generate a ‘Δ Entropy’ measure, the distribution of which is plotted in Figure 1C. In the phylogenetic tree of Figure 1A, host genes were abbreviated as follows: L and S designate the large and small subunit rRNA genes, respectively, and the following number corresponds to the site of insertion, according to Escherichia coli numbering—see Johansen and Haugen, 2001; cob: cytochrome b; cox1, 2, 3: subunits 1, 2, 3 of cytochrome c oxidase.
Modelling
Modelling and refinement were carried out with Rastop 2.2 and the Assemble 1.0 software (Jossinet et al, 2010).
DNA constructs and precursor transcripts
Wild-type precursor transcripts were generated from plasmid pPl.LSU2 (Costa et al, 1997b), a pBluescript II KS (−) (Stratagene) derivative. All mutant constructs in Figures 2 and 4 were verified by sequencing the entire length of the insert. Transcription and RNA purification were carried out as in Costa et al (1997b).
Kinetic analyses
Monomolecular reactions of the wild-type and mutant constructs listed in Table I were initiated by addition of 2 × -concentrated splicing buffer (final concentrations: 40 mM Tris–HCl pH 7.5 at 25°C, 1 M NH4Cl or KCl, 10 mM MgCl2, 0.02% sodium dodecyl sulphate) to an equal volume of a water solution of 32P-labelled precursor RNA molecules (final molar concentration 20–40 nM), which was preequilibrated at the reaction temperature (45°C) after having been denatured for 2 min at 90°C. Reactions were stopped by addition of an equal volume of formamide loading buffer containing Na2EDTA (final concentration 20 mM; each time point—from 0.5 to 180 min—was generated from a separate initial mix). Samples were run on denaturing polyacrylamide gels (50% urea w:v, 4% total acrylamide, with 1:20 bis-acrylamide), and bands associated with the precursor and reaction products were quantitated with a PhosphorImager (Molecular Dynamics).
Accumulation of branched and linear intron products was fitted (with Kaleidagraph 3.6) to simple exponentials,
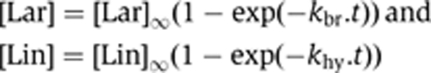 |
where [Lar] and [Lin] are the molar fractions of branched and linear molecules at time t, [Lar]∞ and [Lin]∞, the corresponding, estimated final values, and kbr and khy, the observed rate constants for branching and hydrolysis. As already noted by others (Chu et al, 1998), values obtained for kbr and khy typically differ (Table I), which means that refolded precursor molecules do not form a single population, but rather exist in multiple conformations that do not readily interconvert during the time course of experiments. In ammonium buffer, about 90% of molecules remain committed to forming lariats, even in mutants with a 10-fold reduced rate constant for branching (the only exception is the dVI-2 bp mutant). In potassium buffer, however, changes in rate constants for branching and hydrolysis tend to be reflected in correspondingly altered proportions of branched and linear molecules among reaction products (bottom part of Table I). Importantly, measurements were found to be highly reproducible, whether for the wild-type (Table I) or mutant constructs.
For reactions in the presence of an oligonucleotide (Sigma-Aldrich), the latter was added to concentrated splicing buffer (final concentrations: 40 mM Tris–HCl pH 7.5 at 25°C, 1 M NH4Cl, 10 mM MgCl2, 0.02% sodium dodecyl sulphate) before mixing with the solution of purified precursor molecules (final molar concentration 20 nM) at reaction temperature (37°C). Reaction time courses were modelled according to the following scheme, in which pre:oligo is the unreacted complex between a precursor and an oligonucleotide molecule (whereas hydrolysis at the 5′ splice site is irreversible, transesterification is expected to be reversible; however, the intron 3′ exon lariat intermediate was either absent or barely detectable, even at short reaction times, for all construct and oligonucleotide combinations we tested, so that in this experimental system, branching may be regarded as irreversible for all practical purposes).
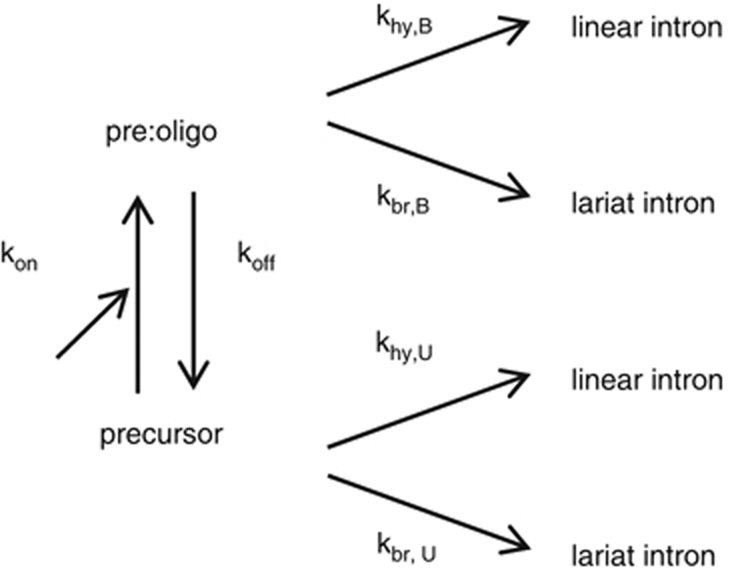
Provided koff and kon are much larger than the rate constants for reactions, the rates of formation of lariat and linear intron products become:
where [Pre] is the molar fraction of unbound precursor molecules at time t; kbr,U, khy,U, kbr,B and khy,B are rate constants for branching (br) and hydrolysis (hy) in the absence (Unbound) and presence of a bound (B) oligonucleotide, respectively; Kd=koff/kon; and [OLI] is the molar concentration of oligonucleotide. Let f be the fractional (relative) rate of formation of lariat intron (f0 and fmax are initial and final values of f):
with
In practice, (i) the accumulation of lariat and linear intron forms for a given oligonucleotide concentration was fitted to a simple exponential or, exceptionally, when reaction was both slow and limited, to a linear function; (ii) initial rates at t=0 and their standard errors were obtained from these fits, f was calculated and plotted as a function of oligonucleotide concentration (the relative error of f was estimated by adding the relative errors of branching and total reaction rates, which were calculated from standard errors associated with initial rates); (iii) the resulting plot was fitted with equation (3) to determine f0, fmax and Km; and (iv) Kd was extracted from equation (6) after khy,U and kbr,B had been obtained from initial reaction rates in the absence and at saturating concentrations of the oligonucleotide, respectively.
Verification of splice junctions and the branchpoint
The identity of splice junctions and the branchpoint were verified for the construct–oligonucleotide combination shown in Figure 3C by purifying the ligated exons and intron lariat from a denaturing polyacrylamide gel before reverse transcription, as described in Costa et al, 1997b (an analytical version of that gel is shown in Supplementary Figure S3). After reverse transcription of the ligated exons with primer 5′-GAGGTCGACGGTATCGATAA (which matches positions 70–89 of the 3′ exon), PCR amplification was carried out with the same primer and 5′-AGCTTTTATCTTTGACACAAAATCGGGGGTG (positions −19 to −49 of the 5′ exon) and products cloned with the pGEM-T vector system (Promega): all clones examined had the expected sequence for the ligated exons. After reverse transcription with primer 5′-GCAGGTACATTGTCTCCAGA (complementary to intron positions 58–77) and PCR amplification with the same primer and 5′-GAAAGGCTGCAGACTTATTA (corresponding to part of ribozyme domain III), five clones were sequenced and found to contain the intron sequence preceding the branchpoint followed by the beginning of the intron, as expected. However, in three clones, an A rather than a T had been incorporated by the reverse transcriptase at the position facing the adenine of the branchpoint, one clone lacked both the branchpoint A and the preceding T and the fifth one lacked that T: these are typical of the errors made by the Superscript II reverse transcriptase when trying to bypass a 2′–5′ branched structure (Vogel and Börner, 2002).
Supplementary Material
Acknowledgments
We are grateful to Eric Westhof for helpful comments on our manuscript. This work was made possible by recurrent funding from the Centre National de la Recherche Scientifique and the generosity of our colleagues at the Centre de Génétique Moléculaire. C-FL was supported by a Joseph Fourier fellowship from the French Government and the National Science Council of Taiwan.
Author contributions: C-FL and MC designed, carried out and analysed the experiments. FM carried out comparative sequence analysis and structural modelling, conceived some experiments and contributed to their interpretation. All authors contributed to drafting the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Boudvillain M, Pyle AM (1998) Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J 17: 7091–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A (1994) Catalytic site components common to both splicing steps of a group II intron. Science 266: 1383–1387 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A (1996) An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J 15: 3466–3476 [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Liu Q, Podar M, Perlman PS, Pyle AM (1998) More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA 4: 1186–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Deme E, Jacquier A, Michel F (1997a) Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol 267: 520–536 [DOI] [PubMed] [Google Scholar]
- Costa M, Fontaine JM, Loiseaux-de Goër S, Michel F (1997b) A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J Mol Biol 274: 353–364 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F (1997) Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J 16: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E (2000) A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19: 5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S (2003) Database for mobile group II introns. Nucleic Acids Res 31: 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erat MC, Zerbe O, Fox T, Sigel RK (2007) Solution structure of domain 6 from a self-splicing group II Intron ribozyme: a Mg(2+) binding site is located close to the stacked branch adenosine. Chembiochem 8: 306–314 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98 [Google Scholar]
- Hamill S, Pyle AM (2006) The Receptor for branch-site docking within a group II intron active site. Mol Cell 23: 831–840 [DOI] [PubMed] [Google Scholar]
- Jacobson H, Stockmayer WH (1950) Intramolecular reaction in polycondensations. i. The theory of linear systems. J Chem Phys 18: 1600–1606 [Google Scholar]
- Jacquier A (1996) Group II introns: elaborate ribozymes. Biochimie 78: 474–487 [DOI] [PubMed] [Google Scholar]
- Jacquier A, Jacquesson-Breuleux N (1991) Splice site selection and role of the lariat in a group II intron. J Mol Biol 219: 415–428 [DOI] [PubMed] [Google Scholar]
- Jacquier A, Michel F (1990) Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II self-splicing introns. J Mol Biol 213: 437–447 [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS (1988) Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439 [PubMed] [Google Scholar]
- Johansen S, Haugen P (2001) A new nomenclature of group I introns in ribosomal DNA. RNA 7: 935–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F, Ludwig TE, Westhof E (2010) Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 26: 2057–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S (2004) Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Li C-F, Costa M, Bassi G, Lai Y-K, Michel F (2011) Recurrent insertion of 5′-terminal nucleotides and loss of the branchpoint motif in lineages of group II introns inserted in mitochondrial preribosomal RNAs. RNA 17: 1321–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L (2006) Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res 34: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Costa M, Westhof E (2009) The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci 34: 189–199 [DOI] [PubMed] [Google Scholar]
- Michel F, Ferat JL (1995) Structure and activities of group II introns. Annu Rev Biochem 64: 435–461 [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Petersen M, Bondensgaard K, Jacobsen JP (2002) Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J Am Chem Soc 124: 5974–5982 [DOI] [PubMed] [Google Scholar]
- Pyle AM (2010) The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol 45: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatterer JC, Crayton SH, Greenbaum NL (2006) Conformation of the Group II intron branch site in solution. J Am Chem Soc 128: 3866–3867 [DOI] [PubMed] [Google Scholar]
- Stabell FB, Tourasse NJ, Kolstø AB (2009) A conserved 3′ extension in unusual group II introns is important for efficient second-step splicing. Nucleic Acids Res 37: 3202–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel SA (1999) A chemogenetic approach to RNA function/structure analysis. Curr Opin Struct Biol 9: 346–352 [DOI] [PubMed] [Google Scholar]
- Sun JS, Manley JL (1995) A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Tang J, Breaker RR (1997) Rational design of allosteric ribozymes. Chem Biol 4: 453–459 [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM (2010) Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM (2008a) Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM (2008b) Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15: 1221–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Kwakman JH, Grivell LA (1987) Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J 6: 3827–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T (2002) Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J 21: 3794–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J (2010) Inclusion of weak high-resolution X-ray data for improvement of a group II intron structure. Acta Crystallogr D Biol Crystallogr 66: 988–1000 [DOI] [PubMed] [Google Scholar]
- Zhang L, Doudna JA (2002) Structural insights into group II intron catalysis and branch-site selection. Science 295: 2084–2088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.