Abstract
Breast cancer is a heterogeneous disease and several distinct subtypes exist based on differential gene expression patterns. Molecular apocrine tumours were recently identified as an additional subgroup, characterised as oestrogen receptor negative and androgen receptor positive (ER− AR+), but with an expression profile resembling ER+ luminal breast cancer. One possible explanation for the apparent incongruity is that ER gene expression programmes could be recapitulated by AR. Using a cell line model of ER− AR+ molecular apocrine tumours (termed MDA-MB-453 cells), we map global AR binding events and find a binding profile that is similar to ER binding in breast cancer cells. We find that AR binding is a near-perfect subset of FoxA1 binding regions, a level of concordance never previously seen with a nuclear receptor. AR functionality is dependent on FoxA1, since silencing of FoxA1 inhibits AR binding, expression of the majority of the molecular apocrine gene signature and growth cell growth. These findings show that AR binds and regulates ER cis-regulatory elements in molecular apocrine tumours, resulting in a transcriptional programme reminiscent of ER-mediated transcription in luminal breast cancers.
Keywords: androgen receptor, breast cancer, chromatin, FoxA1, molecular apocrine
Introduction
The sex steroid hormones oestrogen and androgen are critical for the normal homeostasis of breast and prostate tissue. Deregulation of these hormones leads to the development of tumours and a number of hormone therapies have been developed to successfully target these nuclear receptors. Approximately, two thirds of breast cancers are dependent upon oestrogen receptor-α (ER) while the androgen receptor (AR) is classically thought of as the driver of prostate cancer development and progression (Cunha et al, 1987; Wilding, 1992). However, the majority of breast cancers, both ER positive and negative, also express AR (Isola, 1993; Peters et al, 2009; Hu et al, 2011). Furthermore, modulation of AR by specific ligands can stimulate or inhibit breast cancer cell growth depending on the cell line model used (Birrell et al, 1995, 1998).
In ER-positive breast cancer cells, AR has been shown to function in a growth inhibitory manner, by associating with oestrogen responsive elements (EREs) and impeding ER-mediated transcription (Peters et al, 2009). This would suggest that AR can have an inhibitory effect on ER transcription by occupying similar cis-regulatory elements within the genome. This hypothesis is supported by data showing that ER-positive breast tumours with high AR possess a better clinical prognosis (Peters et al, 2009; Park et al, 2010).
The role of AR in ER-negative breast cancer is less well understood. Recently, a new subtype of breast cancer, termed Molecular Apocrine, has been characterised (Farmer et al, 2005; Doane et al, 2006; Teschendorff et al, 2007). Molecular apocrine tumours are ER negative, but AR positive and in many cases they express genes that are normally expressed in ER-positive luminal tumours, including XBP-1, SCUBE2, SPDEF and FOXA1 (Doane et al, 2006). Molecular apocrine tumours constitute 8–12% of breast cancers assessed by expression profiling in two separate studies (Farmer et al, 2005; Doane et al, 2006). Interestingly, a cell line model of molecular apocrine breast cancer, termed MDA-MB-453, exists (Doane et al, 2006), that are AR positive (Hall et al, 1994) and have a gene expression profile most similar to molecular apocrine tumours when compared with a range of ER-negative breast cancer cell lines (Doane et al, 2006). There is speculation that in these ER− tumours, AR may be able to reside on common cis-regulatory domains, driving transcription of genes that influence cell growth in an ER-independent manner.
We aimed to clarify the role that AR plays in breast cancer cells that are ER−. Using the molecular apocrine cell line model, we mapped AR binding events by chromatin immunoprecipitation (ChIP) sequencing and determined the mechanisms underlying AR-mediated chromatin interactions and transcriptional potential.
Results
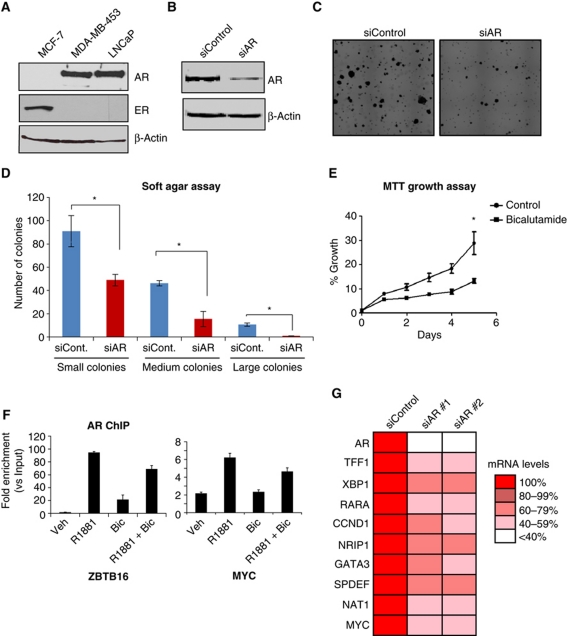
We aimed to define the transcription factor properties that govern molecular apocrine breast cancer cell transcription and growth. Previous work has shown that molecular apocrine tumours are AR+ ER− (Farmer et al, 2005) and the breast cancer cell line, MDA-MB-453, has been shown to have an expression profile most similar to that of molecular apocrine tumours (Doane et al, 2006). We utilised MDA-MB-453 breast cancer cells, MCF-7 breast cancer cells and LNCaP cell, a model of prostate cancer, and assessed for expression of AR and ER by western blot. Figure 1A shows that MDA-MB-453 cells express comparable AR levels to LNCaP cells, but do not express ER. The western blots suggested that LNCaP cells do not express ER. Since AR or ER ligands can, in some cases, be promiscuous, we directly assessed whether MDA-MB-453 cells are dependent on the presence of the AR protein for soft agar colony formation. We transfected MDA-MB-453 cells with siControl or siRNA to AR and confirmed effective silencing of AR (Figure 1B). In MDA-MB-453 cells transfected with siRNA to AR, soft agar colony formation was significantly decreased (Figure 1D), confirming that molecular apocrine breast cancer cells are dependent on AR for their transformed phenotype. This experiment was confirmed with an independent siRNA targeted to AR (Supplementary Figure S1). To confirm a requirement of AR for proliferation of MDA-MB-453 cells, we treated cells growing in the presence of 10% fetal bovine serum (FBS) with 1 μM of the anti-androgen bicalutamide or vehicle control, for 5 days and assessed growth using an MTT assay (Figure 1E). Cells cultured in the presence of bicalutamide had significantly less growth than control cells (P-value <0.001) confirming previous results that anti-androgens can partially inhibit growth of MDA-MB-453 cells (Doane et al, 2006).
Figure 1.
Molecular apocrine breast cancer cells are dependent on AR for growth and transcription. (A) Western blot for AR and ER in MCF-7 breast cancer cells, MDA-MB-453 molecular apocrine breast cancer cells and LNCaP prostate cancer cells. (B) MDA-MB-453 cells were transfected with siControl or siAR. Western blot showing AR silencing. Following AR silencing, MDA-MB-453 colony formation was assessed in a soft agar assay. An example image is shown in (C) and quantification of number and size of colonies from three replicates is shown in (D). *Denotes P-value <0.05. (E) MTT growth assay with MDA-MB-453 cells treated with 1 μM bicalutamide or ethanol control, average of three independent replicates, * denotes P-value <001. (F) AR ChIP of MDA-MB-453 cells after 4 h treatment with vehicle, R1881, bicalutamide or R1881 plus bicalutamide followed by real-time PCR. (G) Transcript levels of a number of genes expressed in luminal tumours were assessed in cells transfected with siControl or two different siRNA to AR, average of three individual replicates.
As AR can exhibit both genomic and non-genomic effects (Heinlein and Chang, 2002), we aimed to determine whether AR was acting via direct interaction with the chromatin. MDA-MB-453 cells were hormone deprived and treated with vehicle, the synthetic androgen R1881 or bicalutamide for 4 h and AR ChIP was performed, followed by quantitative PCR of two genomic regions shown previously to be AR binding regions in LNCaPs (Massie et al, 2011) and ER binding regions in MCF7 cells (Ross-Innes et al, 2010), respectively (Figure 1F). These data suggest that AR mediates its effects by binding to chromatin in a ligand-dependent manner, which can be perturbed by the addition of the anti-androgen, bicalutamide. To understand the AR transcriptional response, MDA-MB-453 cells were transfected with two independent siRNA pools to AR, total RNA collected and assessed for transcript levels of genes previously shown to be classic ER targets in luminal breast tumours (Sorlie et al, 2003). Nine of these ER target genes were significantly decreased when AR was specifically silenced (Figure 1G) or treated with bicalutamide (Supplementary Figure S1). This suggests AR mediates transcription of these typical ER targets, many of which are also signature genes in molecular apocrine breast cancers (Sanga et al, 2009).
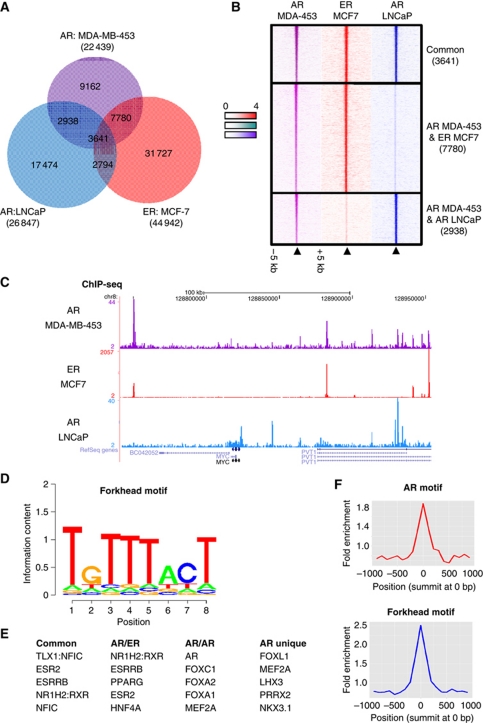
We mapped AR binding events using ChIP followed by high-throughput sequencing (ChIP-seq) in asynchronous MDA-MB-453 breast cancer cells and LNCaP prostate cancer cells. We also mapped ER in MCF7 breast cancer cells. Binding events were called using MACS and 22 439 AR binding events in MDA-MB-453 cells, 26 847 AR binding events in LNCaP cells and 44 942 ER binding events in MCF7 cells could be identified (Figure 2A). Unexpectedly, the AR binding profile in MDA-MB-453 cells was more similar to ER binding in breast cancer cells (50.9% overlapping regions) and substantially distinct from AR binding in prostate cancer cells (29.3% overlapping regions). We define an overlapping region as one where the binding events shared at least one base pair. The relationship between the three cell lines is also observed at more lenient thresholds, where any ER or AR binding event in MCF7 or LNCaP cells are compared with the reproducible AR binding events in MDA-MB-453 cells (Supplementary Figure S2), illustrating that we are not missing shared binding events due to the stringency of our peak calling method. The strongest AR binding events in MDA-MB-453 cells are shared with either ER in MCF7 cells or AR in LNCaP cells (Figure 2B). Furthermore, binding events that we claim to be cell type specific are unique to that ChIP (Supplementary Figure S2). An example of binding events in all three cell lines is shown in Figure 2C. Our findings suggest that AR–chromatin interactions in molecular apocrine breast cancer cells occur in a manner that is more similar to ER in the breast than AR in the prostate.
Figure 2.
AR binding in molecular apocrine breast cancer is more similar to ER binding in the breast than AR binding in the prostate. (A) AR binding was mapped in asynchronous MDA-MB-453 breast cancer cells and asynchronous LNCaP prostate cancer cells, and ER binding mapped in asynchronous MCF-7 breast cancer cells. (B) Heatmap showing AR and ER binding signal intensity for overlapping regions in a window of ±5 kb. The heatmap represents binding events ranked from strongest to weakest AR binding in MDA-MB-453 cells, and the adjacent columns represent signal from the corresponding ChIP-sequencing experiment in the MCF7 and LNCaP cells. (C) Example of binding events which are unique or shared by AR in MDA-MB-453 cells. (D) Motifs enriched in AR binding events in MDA-MB-453 cells. (E) A comparison of differentially enriched motifs between the different categories of binding events: Common represents regions bound by AR in MDA-MB-453 and LNCaP cells and by ER in MCF-7 cells; AR/ER is regions shared by AR in MDA-MB-453 and ER in MCF-7 cells; AR/AR represents regions shared by AR in MDA-MB-453 and LNCaP cells; AR unique represents the AR binding sites unique to MDA-MB-453 cells only. (F) Enrichment for AR and Forkhead motif in the centre of the AR MDA-MB-453 binding events.
To elucidate the mechanism dictating AR behaviour in MDA-MB-453 cells, we conducted a de novo motif search for transcription factors using the discovery tool Weeder to generate a position weight matrix (PWM) for all AR binding sites. This resulting PWM was similar to that of Forkhead proteins; FoxD1, P-value=3.64e−6, HNF3α, P-value=1.03e−5 and FoxA1 P-value=1.23e−5 (Figure 2D). Forkhead motifs have been identified in almost all previous ER mapping experiments in MCF-7 cells (Carroll et al, 2005, 2006; Laganiere et al, 2005; Lupien et al, 2008). The shared and unique MDA-MB-453 AR binding events were also mined for overrepresented DNA sequence motifs. Genomic regions with a shared AR binding region in both MDA-MB-453 and LNCaP cells were called AR/AR, shared AR in MDA-MB-453 and ER in MCF-7 cells were termed AR/ER, a region found in all three contexts (AR in both MDA-MB-453 and LNCaP cells and ER in MCF-7 cells) were called common regions. We found the AR/ER regions contained motifs with similar properties to EREs including ESR2 and PPARG. The AR/AR shared regions were enriched for AR motif and the common binding regions were enriched for ER motifs (Figure 2E). It appears that traditional mechanisms are utilised by ER and AR to mediate binding in MCF7 and LNCaP cells, however, in the MDA-MB-453 cells AR may be able to utilise ERE-like motifs to bind to DNA. This was supported by data showing that the enriched motifs were in the centre of the binding regions, suggesting that AR–chromatin interactions are the direct mechanisms for AR binding to the genome, rather than a tethering mechanism (Figure 2F). Interestingly, the Forkhead motif was also enriched at the centre of the AR binding regions, implying potential cooperativity between AR and Forkhead proteins in mediating AR binding.
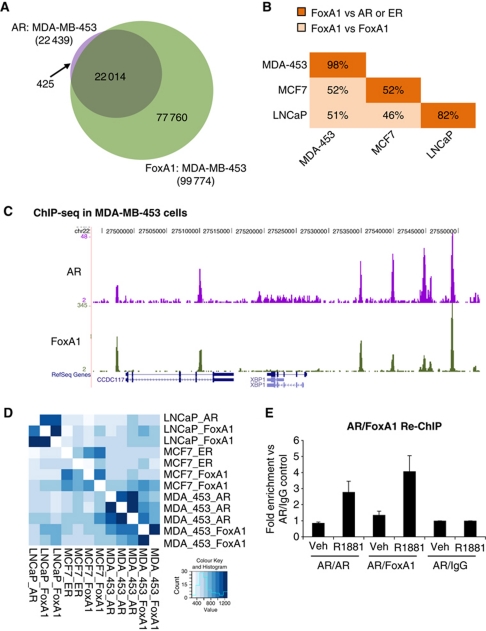
FoxA1 has recently been identified and characterised in mediating AR–chromatin interactions and ER–chromatin interactions and function in prostate and breast cancer cells (Carroll et al, 2005; Eeckhoute et al, 2006; Lupien et al, 2008; Wang et al, 2009; Bernardo et al, 2010; Hurtado et al, 2011). Since Forkhead motifs were enriched within AR binding regions in molecular apocrine MDA-MB-453 breast cancer cells, we hypothesised that the Forkhead protein FoxA1 may be having a similar role with AR in the breast. This hypothesis is supported by the observation that FoxA1 is a signature gene of molecular apocrine tumours (Doane et al, 2006). We subsequently mapped FoxA1 binding events by ChIP-seq in MDA-MB-453 breast cancer cells, combining two biological replicates. This resulted in a total of 99 774 FoxA1 binding regions in MDA-MB-453 cells. Surprisingly, we found AR binding events to be a near complete subset of the FoxA1 binding regions, with 98.1% of all AR binding events overlapping with a FoxA1 binding region (Figure 3A; an example region shown Figure 3C). This overlap is substantially higher than the ∼50% overlap between ER and FoxA1 (Lupien et al, 2008; Hurtado et al, 2011), suggesting that all AR binding events in molecular apocrine breast cancer cells may be mediated by FoxA1. The overlap at a more relaxed threshold is shown in Supplementary Figure S3. As an independent validation, we used the Genome Structure Correction (GSC) statistic (Birney et al, 2007) to assess the extent to which co-binding between replicates and across conditions is likely to have occurred by chance (Figure 3D). Unsupervised clustering of the z scores shows high correlation not only between replicates but also between AR and FoxA1 in MDA-MB-453 cells.
Figure 3.
Almost all AR binding events occur at FoxA1 binding regions. (A) FoxA1 binding was determined by ChIP-seq in MDA-MB-453 breast cancer cells and overlap with AR is shown. (B) The proportion of overlapping AR/FoxA1 binding events is more similar than either the ER/FoxA1 overlap in MCF7 or FoxA1/FoxA1 across the three cell lines. Table shows FoxA1 binding overlap between cell lines in pale orange. FoxA1 and nuclear receptor overlap within the same cell line is in dark orange, that is, AR/FoxA1 MDA-MB-453, AR/FoxA1 LNCaP or ER/FoxA1 MCF7. (C) Example of a genomic region showing AR and FoxA1 binding in MDA-MB-453 cells. (D) Unsupervised clustering GSC Heatmap showing z scores for the comparisons between each ChIP-seq experiment. (E) Re-ChIP showing co-occupancy of AR and FoxA1 on the chromatin in a ligand-dependent manner.
In order to compare FoxA1 occupancy across the three cell lines, we also mapped FoxA1 binding events by ChIP-seq in LNCaP prostate cell lines (65 371 binding events) and compared both data sets to published MCF7 FoxA1 binding data (Hurtado et al, 2011). The FoxA1 binding regions across each cell line were distinct from each other, ∼50% overlapping regions between the two breast cancer cell lines (Figure 3B). Interestingly, FoxA1 and AR in the LNCaP prostate cell line also have a high level of concordance (82%) while FoxA1 and ER in MCF7 breast cancer cell only overlap by 52% (Figure 3B). To confirm that AR and FoxA1 form complexes on the chromatin, Re-ChIP experiments were performed. MDA-MB-453 cells were hormone deprived and treated with vehicle or androgen. AR/FoxA1 Re-ChIP was performed, followed by real-time PCR of a region found by ChIP-seq to be co-bound by AR and FoxA1. As a control, AR/IgG Re-ChIP was performed. The data confirm that AR and FoxA1 could co-occupy the chromatin, but only in a ligand-dependent manner (Figure 3E).
To explore the possibility that FoxA1 may be dictating AR binding in molecular apocrine cells, we transfected MDA-MB-453 cells with siControl or siFoxA1 (two independent siRNAs) and assessed for effective silencing of FoxA1 (Figure 4A; Supplementary Figure S4). After silencing of FoxA1, AR binding was assessed by ChIP–qPCR at a number of the regions identified by AR ChIP-seq. The data confirmed that FoxA1 is required for AR to bind chromatin (Figure 4B). Critically, the silencing of FoxA1 did not alter total AR protein levels (Figure 4A). We extended this experiment and enriched for the chromatin fraction following silencing of FoxA1, followed by western blotting for AR. Total AR–chromatin recruitment was substantially inhibited when FoxA1 was silenced (Figure 4C; Supplementary Figure S4), confirming a strong dependency on FoxA1 for AR–chromatin interactions.
Figure 4.
FoxA1 is required for AR binding and transcription of target genes. (A) Western blot showing silencing of FoxA1 in MDA-MB-453 cells. (B) MDA-MB-453 cells were transfected with siControl or siFoxA1. AR ChIP was performed followed by real-time PCR of AR binding regions. (C) MDA-MB-453 cells were transfected with siControl or siFoxA1, total chromatin fraction was collected and western blotted. Total chromatin-associated AR is decreased when FoxA1 is silenced. (D) Microarray analysis of gene expression changes following transfection of siControl or siFoxA1. Only genes with an FDR <0.01 were considered which resulted in 730 FoxA1-dependent genes. (E) The 730 FoxA1-dependent genes were analysed for enriched biological pathways. (F) Soft agar assay following transfection of MDA-MB-453 cells with siControl or siFoxA1. (G) Heatmap showing log2 fold change in gene expression of the 100 gene molecular apocrine signature induced by two different siRNAs targeted to FoxA1. Black box indicates the presence of AR and FoxA1 binding event ±10 kb from TSS of the gene.
To explore the role that FoxA1 plays in transcription in MDA-MB-453 cells, we conducted a microarray analysis of differentially expressed genes after silencing of FoxA1. Eight biological replicates (composed of two independent siRNAs to FoxA1) were performed and 730 FoxA1 regulated genes were identified, which included a number of classic ER target genes such as TFF1, XBP1 and RARA (Figure 4D; Supplementary Figure S5). Pathway analysis using GeneGo pathway maps shows enrichment for regulatory networks involved in cell cycle, development and cytoskeleton remodelling (Figure 4E). Following this, we investigated FoxA1's role in the growth of MDA-MB-453 cells in 3D culture. Cells were transfected with siRNA to FoxA1, plated in soft agar and after 12 days, colonies were counted. Silencing of FoxA1 resulted in fewer, smaller colonies (Figure 4F) implying a requirement of FoxA1 for growth.
To assess the significance of FoxA1-mediated transcription in a clinical setting, we compared the siFoxA1 gene expression values to the 100 gene molecular apocrine signature derived from meta-analysis of two independent patient data sets (Sanga et al, 2009). The list of genes is provided in Supplementary Table 2. The majority (91%) of the genes in the molecular apocrine gene signature were significantly affected when FoxA1 was silenced in MDA-MB-453 cells (Figure 4G). This is a highly significant enrichment compared with all genes on the microarray of which 26% were affected by FoxA1 silencing (P-value <1e−10). Interestingly, 47% of these genes also have an AR and FoxA1 binding event within 10 kb of their transcription start sites. This is substantially higher than the expected value of 25% (P-value=4.2e−9). As such, FoxA1 is required for AR binding to chromatin and transcriptional activity of the genes that define AR+ ER− molecular apocrine breast cancers.
Discussion
Based on various clinical parameters, different subtypes of breast cancer are a well-known phenomenon. Gene expression microarrays allow for refinement of the breast cancer subtypes, based on the global expression profiles (Perou et al, 2000; Sorlie et al, 2001, 2003). Luminal breast cancers are characterised as ER positive, but a spectrum of ER-negative tumours exist. Recently, a novel subtype of breast cancer was reported, termed molecular apocrine, that is ER− and AR+ (Farmer et al, 2005; Doane et al, 2006). Interestingly, many of the genes expressed in molecular apocrine tumours are ER target genes, traditionally expressed in ER+ luminal breast cancer. It is estimated that ∼8–12% of breast tumours possess features of molecular apocrine breast cancer.
Most ER-positive breast cancers are also AR+ (Park et al, 2010; Hu et al, 2011). In an ER+ context, activation of AR appears to have an anti-proliferative effect on breast cancer cell proliferation and AR positivity predicts a beneficial clinical outcome in ER+ breast cancers (Peters et al, 2009; Park et al, 2010). It has been suggested that AR can bind to similar cis-regulatory regions as ER, and given the correct stoichiometric ratio, AR can physically impede ER transcriptional activity (Peters et al, 2009). Our data suggest that in a cell line model of molecular apocrine breast cancer, MDA-MB-453 cells (Doane et al, 2006), that more than half of AR binding to the genome occurs in a similar pattern to ER binding in breast cancer cells. Therefore, in the absence of ER, it appears that AR is capable of mimicking ER in its ability to bind to DNA, at specific cis-regulatory elements in a transcriptionally active manner.
Interestingly, AR binding in molecular apocrine breast cancer cells, utilise the same mechanisms as ER in luminal breast cancer cells, namely the requirement for FoxA1 to mediate its interaction with chromatin. Almost all AR binding events occurred precisely at a FoxA1 binding event and it appears that essentially all AR binding is dependent on the presence of FoxA1 (Figure 4C). Since FoxA1 is a molecular apocrine signature gene marker (as is AR) (Doane et al, 2006; Sanga et al, 2009), development of a molecular apocrine tumour may depend on the presence of both the nuclear receptor (AR) and the pioneer factor (FoxA1). FoxA1 and AR binding in the LNCaP prostate cell line are also highly concordant while only half of ER and FoxA1 binding events overlap. As such, AR binding may be more dependent on the pioneering qualities of FoxA1, unlike ER where some binding to nucleosome-depleted euchromatic regions are less dependent on FoxA1 for direct chromatin associations (Hurtado et al, 2011). The overlap in FoxA1 binding across the three cell lines was similar to what has been reported across three ER-positive breast cancer cell lines, MCF7, ZR75-1 and T-47D (Hurtado et al, 2011).
Breast cancer and prostate cancer share many similar biological features and common components (Risbridger et al, 2010). In the ER− context, AR is likely to utilise similar mechanisms and factors as ER, to regulate target gene transcription. These gene targets are pro-proliferative and growth of the cells is dependent on the presence of AR. Given the observation that most ER+ luminal breast cancers are also positive for AR (Peters et al, 2009), the loss of ER expression may result in a shift from ER- to AR-mediated transcription. The switch from ER to AR transcriptional regulation would result in similar genes being expressed from common cis-regulatory domains, but growth would occur in a manner that is refractory to traditional breast cancer therapies. The lack of clinical benefit from ER antagonists, such as tamoxifen and aromatase inhibitors, is reflected in the poor clinical outcome of molecular apocrine tumours, when compared with luminal breast cancers (Farmer et al, 2005). There has been speculation about the use of anti-androgens for the treatment of apocrine breast cancers (Farmer et al, 2005), a hypothesis that warrants clinical investigation in light of our findings.
Materials and methods
Cell culture
MDA-MB-453, MCF7 and LNCaP human cell lines were obtained from ATCC and grown in DMEM or RPMI (LNCaP) supplemented with 10% FBS and standard antibiotics. In all experiments, R1881 was added at a final concentration of 100 nM and bicalutamide (Caslodex) was added at a final concentration of 1 μM.
Small interfering RNA (siRNA)
Cells were transfected with siRNA using Lipofectamine2000 (Invitrogen). AR was silenced using two different siRNA pools SMARTpool, siAR #1, and siGenome, siAR #2, both purchased from Dharmacon (Catalogue numbers L-003400-00 and M-003400-02, respectively). Two different siRNAs targeting the sequence of FoxA1 were used, a single siRNA, siFoxA1#1, (GAGAGAAAAAAUCAACAGC) and siGenome pool, siFoxA1#2 (Dharmacon, Cat). AllStars Negative Control siRNA (Qiagen) was used as a negative control.
Anchorage-independent growth
MDA-MB-453 cells were transfected with siRNA, trypinised 24 h later and reseeded in soft agar as described in Fiebig et al (2004). After 12 days, colonies were stained with tetrazolium chloride (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride (1 mg/ml)).
Cell growth assay
MDA-MB-453 cells were plated at equal confluence and treated with vehicle or 1 μM bicalutamide for 5 days. Number of live cells was analysed daily using MTT assay (Millipore).
Gene expression by RT–PCR
MDA-MB-453 cells were transfected with AR or FoxA1 siRNA. Total RNA was collected 48 h later and RT–PCR was performed as described (Carroll et al, 2006). Primers used in qRT–PCR are listed in Supplementary Table 1.
Western blots
Western blots were processed as previously described in Ross-Innes et al (2010). Antibodies used were anti-FoxA1 (ab5089), anti-histone H3 (ab1791) and anti-β-actin (ab6276) from Abcam, anti-ER (6F11) from Novocastra and anti-AR (sc-816) from Santa Cruz Biotechnologies.
Chromatin immunoprecipitation
ChIP experiments were performed as described previously (Schmidt et al, 2009). Antibodies used were anti-AR (sc-816) and anti-ER (sc-543) from Santa Cruz Biotechnologies, and anti-FoxA1 (ab5089) from Abcam. Primer sequences used for real-time PCR are provided in Supplementary Table 1.
ChIP-sequencing experiments
The ChIP DNA was verified by real-PCR before being processed for Illumina sequencing as previously described (Schmidt et al, 2009). Single end 36-bp ChIP-seq data were generated by the Illumina analysis pipeline version 1.6.1, and reads were aligned to the Human Reference Genome (assembly hg18, NCBI Build 36.6, March 2008 using BWA version 0.5.5; Li and Durbin, 2010). Reads were filtered by an alignment quality score removing all reads with a score <15. Peaks were called using MACS, version 1.3.7.1 (Zhang et al, 2008). For AR ChIP-seq in MDA-MB-453, three biological replicates were performed and only binding events that occurred in two out of three replicates were considered. For all other ChIP-seq experiments, two biological replicates were performed binding events that occurred in both replicates were considered, except AR in LNCaPs, were only one replicate was conducted and high confidence binding events are defined as regions called by two peak callers, MACS and SWEMBL.
Motif analysis
De novo motif analysis was conducted using Weeder to create a PWM for all AR binding events in MDA-MB-453 (Pavesi et al, 2006). The resulting PWM was compared with JASPAR, Transfac and UNIPROBE database by MEME's TOMTOM application (Gupta et al, 2007). Overrepresented motifs in the AR binding event overlapping regions were determined using patser (version 3e) (Hertz and Stormo, 1999). Enrichment of all PWMs from JASPAR (Vertebrate subset) was compared with intergenic sequences, using a P-value cutoff of 0.01 to determine significant matches.
Enrichment of motifs across peaks was carried out using the MOtif Occurrence Detection Suite (MOODS) via its BioPerl API to scan for matches to PWMs (Korhonen et al, 2009). In all, 1 kb flanking either side of the centre of the consensus peaks was scanned in 100 bp non-overlapping windows using a P-value cutoff of 0.0001. Enrichment was calculated as the ratio between the number of significant matches to the PWM and the mean number matches from 1000 randomly permuted versions of the PWM, in each window. Randomly permuted PWMs were generated by shuffling the columns of the matrix, removing any sequence specificity but maintaining base composition. PWMs for AR (MA0007.1) and Forkhead (MF0005.1) were obtained from the JASPAR database (Sandelin et al, 2004).
Re-ChIP
Cells were grown in phenol red-free DMEM supplemented with 5% charcoal dextran-treated media for 3 days and treated with either R1881 for 4 h. The Re-ChIP was performed as described in Ross-Innes et al (2010). Antibodies used were same as ChIP and control anti-goat IgG (sc-34665).
Microarray analysis
Cells were transfected with siControl or siFoxA1 for 48 h. RNA was collected from eight biological replicates (five replicates for second siRNA for verification purposes). The Illumina BeadChIP (HumanWG-12 version 4) bead-level data were preprocessed, log2-transformed and quantile normalised using the bead array package (Dunning et al, 2007; Cairns et al, 2008) in Bioconductor (Gentleman et al, 2004). Differential expression analysis was performed using limma eBayes (Smyth, 2004) with a Benjamini and Hochberg multiple test correction procedure (Benjamini and Hochberg, 1995) to identify statistically significant differentially expressed genes (FDR 0.01).
GO pathway analysis
GO pathway enrichment was determined using GeneGo MetaCore.
Statistical analysis
All statistical analyses were performed using the two-tailed Student's t-tests or the hypergeometric distribution. Only values with a P-value <0.05 were considered statistical. Error bars represent standard deviations.
Supplementary Material
Acknowledgments
We would like to acknowledge the Cancer Research UK genomics and bioinformatics core. We thank the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. Jason S Carroll is supported by an ERC starting grant.
Footnotes
The authors declare that they have no conflict of interest.
References
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57: 289–300 [Google Scholar]
- Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, Abdul-Karim FW, Montano MM, Keri RA (2010) FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 137: 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell SN, Bentel JM, Hickey TE, Ricciardelli C, Weger MA, Horsfall DJ, Tilley WD (1995) Androgens induce divergent proliferative responses in human breast cancer cell lines. J Steroid Biochem Mol Biol 52: 459–467 [DOI] [PubMed] [Google Scholar]
- Birrell SN, Hall RE, Tilley WD (1998) Role of the androgen receptor in human breast cancer. J Mammary Gland Biol Neoplasia 3: 95–103 [DOI] [PubMed] [Google Scholar]
- Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG (2008) BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics (Oxford, England) 24: 2921–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y (1987) The endocrinology and developmental biology of the prostate. Endocr Rev 8: 338–362 [DOI] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL (2006) An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25: 3994–4008 [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics (Oxford, England) 23: 2183–2184 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M (2006) A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz AL, Brisken C, Fiche M, Delorenzi M, Iggo R (2005) Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 24: 4660–4671 [DOI] [PubMed] [Google Scholar]
- Fiebig HH, Maier A, Burger AM (2004) Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur J Cancer 40: 802–820 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RE, Birrell SN, Tilley WD, Sutherland RL (1994) MDA-MB-453, an androgen-responsive human breast carcinoma cell line with high level androgen receptor expression. Eur J Cancer 30A: 484–490 [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C (2002) Androgen receptor (AR) coregulators: an overview. Endocr Rev 23: 175–200 [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics (Oxford, England) 15: 563–577 [DOI] [PubMed] [Google Scholar]
- Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM (2011) Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res 17: 1867–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isola JJ (1993) Immunohistochemical demonstration of androgen receptor in breast cancer and its relationship to other prognostic factors. J Pathol 170: 31–35 [DOI] [PubMed] [Google Scholar]
- Korhonen J, Martinmaki P, Pizzi C, Rastas P, Ukkonen E (2009) MOODS: fast search for position weight matrix matches in DNA sequences. Bioinformatics (Oxford, England) 25: 3181–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V (2005) Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA 102: 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, Bon H, Zecchini V, Smith D-M, DeNicola GM, Mathews N, Osborne M, Hadfield J, MacArthur S, Adryan B, Lyons SK et al. (2011) The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J (advance online publication, 20 May 2011; doi:10.1038/emboj.2011.158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH, Park BW, Lee KS (2010) Expression of androgen receptors in primary breast cancer. Ann Oncol 21: 488–492 [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Zambelli F, Stefani M, Mauri G, Pesole G (2006) MoD Tools: regulatory motif discovery in nucleotide sequences from co-regulated or homologous genes. Nucleic Acids Res 34 (Web Server issue): W566–W570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, Jindal S, Segara D, Jia L, Moore NL, Henshall SM, Birrell SN, Coetzee GA, Sutherland RL, Butler LM, Tilley WD (2009) Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res 69: 6131–6140 [DOI] [PubMed] [Google Scholar]
- Risbridger GP, Davis ID, Birrell SN, Tilley WD (2010) Breast and prostate cancer: more similar than different. Nat Rev 10: 205–212 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS (2010) Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev 24: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32 (Database issue): D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanga S, Broom BM, Cristini V, Edgerton ME (2009) Gene expression meta-analysis supports existence of molecular apocrine breast cancer with a role for androgen receptor and implies interactions with ErbB family. BMC Med Genomics 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT (2009) ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C (2007) An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 8: R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM et al. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding G (1992) The importance of steroid hormones in prostate cancer. Cancer Surv 14: 113–130 [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.