Abstract
Regions of amino-acid sequence that are compatible with multiple folds may facilitate evolutionary transitions in protein structure. In a previous study, we described a heuristically designed chameleon sequence (SASF1, structurally ambivalent sequence fragment 1) that could adopt either of two naturally occurring conformations (α-helical or β-sheet) when incorporated as part of the C-terminal dimerization subdomain of two structurally divergent transcription factors, P22 Cro and λ Cro. Here we describe longer chameleon designs (SASF2 and SASF3) that in the case of SASF3 correspond to the full C-terminal half of the ordered region of a P22 Cro/λ Cro sequence alignment (residues 34–57). P22-SASF2 and λWDD-SASF2 show moderate thermal stability in denaturation curves monitored by circular dichroism (Tm values of 46 and 55°C, respectively), while P22-SASF3 and λWDD-SASF3 have somewhat reduced stability (Tm values of 33 and 49°C, respectively). 13C and 1H NMR secondary chemical shift analysis confirms two C-terminal α-helices for P22-SASF2 (residues 36–45 and 54–57) and two C-terminal β-strands for λWDD-SASF2 (residues 40–45 and 50–52), corresponding to secondary structure locations in the two parent sequences. Backbone relaxation data show that both chameleon sequences have a relatively well-ordered structure. Comparisons of 15N-1H correlation spectra for SASF2 and SASF3-containing proteins strongly suggest that SASF3 retains the chameleonism of SASF2. Both Cro C-terminal conformations can be encoded in a single sequence, showing the plausibility of linking different Cro folds by smooth evolutionary transitions. The N-terminal subdomain, though largely conserved in structure, also exerts an important contextual influence on the structure of the C-terminal region.
Keywords: chameleon sequence, protein design, NMR spectroscopy
Introduction
Evolutionary transitions in protein folds may be mediated by ‘metamorphic’ protein sequences that can adopt multiple conformations (Meier and Ozbek, 2007; Murzin, 2008; Yadid et al., 2010). Recent studies on a variety of domain families in which natural structural evolution has occurred point to the plausibility or actuality of intermediate sequences that are structurally heterogeneous. These may act as bridges in sequence space between different folded forms (Van Dorn et al., 2006; Meier et al., 2007; Yadid et al., 2010). Design, mutagenesis and directed evolution studies have shown that even pairs of protein folds unrelated by natural evolution can be encoded by overlapping or intersecting regions of sequence space (Cordes et al., 2000; Ambroggio and Kuhlman, 2006a,b; Alexander et al., 2009). Theoretical investigations have also suggested that continuous pathways of mutation can lead to switches in protein folds (Meyerguz et al., 2007). Finally, the plausibility of evolutionary bridges to new protein structures is supported by the observation that some naturally occurring sequences switch between different folds (Luo et al., 2004; Tuinstra et al., 2008).
The Cro proteins from lambdoid bacteriophages, a family of single-domain, sequence-specific DNA-binding proteins, are useful natural model systems for studying sequence changes that can lead to protein fold evolution. Some family members, such as bacteriophage P22 Cro, have an all-α helical fold (Newlove et al., 2004), while others, such as bacteriophage λ Cro, have a mixed α+β fold (Fig. 1) (Albright and Matthews, 1998; Ohlendorf et al., 1998). The N-terminal Cro subdomain, which includes the helix-turn-helix DNA recognition motif, is conserved as mostly α-helix across all Cro proteins, while the C-terminal subdomain, primarily responsible for mediating functionally critical dimerization, adopts a β-sheet conformation in some Cro proteins and α-helix in others (Ohlendorf et al., 1998; Newlove et al., 2004; Dubrava et al., 2008; Roessler et al., 2008). Sequence and structure analyses strongly suggest that the all α-helical Cro fold is ancestral, and that sequences encoding α-helical C-terminal Cro subdomains are homologous to those encoding C-terminal β-sheet (Newlove et al., 2004; Roessler et al., 2008). Evolutionary replacement of C-terminal secondary structure thus occurred through α-to-β conformational switching induced by accumulation of substitution mutations and/or small insertion/deletion events, rather than through wholesale replacement of large C-terminal sequence fragments, brought about by heterologous recombination or frameshifting.
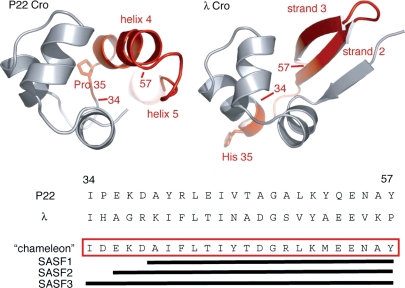
Fig. 1.
Ribbon diagrams of P22 Cro (left) and λ Cro (right) illustrating the conserved N-terminal DNA-recognition subdomain (residues 1–33; gray) and the highly divergent C-terminal dimerization subdomain (residues 34-end; red). At bottom are sequences of the C-terminal region for both proteins, as well as the sequence of a ‘chameleon’ designed to be capable of adopting either the β-sheet C-terminal conformation of λ Cro or the α-helical C-terminal conformation of P22 Cro. The chameleons SASF1, SASF2 and SASF3 have different lengths as shown, and all contain a subset of the full-length design incorporating residues 34–57. Structures were rendered using PyMol (DeLano Scientific, San Carlos, CA).
For fold evolution in the Cro family to occur smoothly by such continuous cumulative sequence change, without major disruption of stability, would require either a series of stable structural intermediates encodable by a continuous range of sequences, or the ability to encode both the limiting α-helical and β-sheet conformations in a single sequence. The latter possibility requires the existence of sequences encoding the C-terminal subdomain that have the chameleon-like ability to form both α-helix and β-sheet folds. It also requires the existence of N-terminal subdomain sequences that can interact interchangeably with both C-terminal conformations. While both issues are important, most of our efforts to date have focused on the possibility of chameleonism in the C-terminal subdomain; in addition, to simplify considerations we have intentionally not considered the additional evolutionary requirement that the protein maintain competence to dimerize and bind to DNA.
In a previous study, we conducted extensive scanning mutagenesis of P22 Cro and λ Cro to assess the differential stability determinants of the C-terminal regions in the two Cro folds (Van Dorn et al., 2006). We then used this knowledge to design and characterize a ∼19-residue chameleon sequence (named SASF1, where SASF stands for structurally ambivalent sequence fragment) that folded stably as either α-helical or β-sheet structure when used to replace part of the C-terminal subdomain of a Cro protein (residues 39–57 of P22 Cro or λ Cro; see Fig. 1). The chameleon represented ∼80% of the structurally diverse C-terminal half of a sequence alignment (Fig. 1) between P22 Cro and λ Cro, including strands 2 and 3 of λ Cro's β-sheet and most of helix 4 and helix 5 from P22 Cro. Hα chemical shift analysis showed SASF1 to adopt a helical structure when incorporated into P22 Cro, but a β-sheet structure when incorporated into λ Cro. Thus, a single primary sequence can encode both the ancestral and descendant forms of the bulk of the Cro C-terminal structure, suggesting the plausibility of smooth evolutionary transitions in which β-sheet secondary structure replaced α-helix while conserving folding stability along the mutational pathway.
One major remaining question is whether the adoption of different structures by SASF1 is governed largely by the influence of the N-terminal subdomain, or by key determinants at the very beginning of the C-terminal region. To address this issue, we sought to extend the chameleon designs to encompass the entire C-terminal ‘half' of the P22 Cro/λ Cro alignment, including residues 34–57 of each protein. In our previous study we reported limited characterization of a ∼22-residue chameleon (SASF2; residues 36–57). This sequence folded with moderate thermal stability in both the P22 and λ backgrounds, but could not be thoroughly characterized by NMR spectroscopy due to protein precipitation for λ-SASF2 during long experiments at 20°C. At that time, we did not attempt a ∼24-residue design to include the full span of residues 34–57, mostly because position 35 loomed as a major obstacle. In P22 Cro, this residue is a proline immediately preceding the N-terminus of helix 4; in λ Cro, it is a histidine preceding the C-terminus of helix 3, part of the helix-turn-helix motif (see Fig. 1). Introduction of a proline residue at position 35 of λ Cro was found to be highly destabilizing, presumably due to disruptive kinking of helix 3, while introduction of histidine (or a number of other polar residues) at position 35 of P22 Cro was also quite destabilizing, possibly because replacement of the proline generates an amide hydrogen that faces the interior of the protein.
Here we describe (i) a detailed characterization of the backbone structure and dynamics of SASF2, made possible by use of a stabilized λ Cro sequence background, (ii) a full 24-residue C-terminal chameleon design (SASF3; residues 34–57), made possible by the discovery that an aspartate residue at position 35 can coexist with either of the Cro folds. The level of characterization of SASF2 far exceeds that reported previously for SASF1, and includes a full analysis of carbon, nitrogen and proton backbone chemical shifts, J couplings and nitrogen relaxation/NOE experiments. For SASF3, we report only a comparison with SASF2 based on overlays of 15N-1H HSQC spectra. This limited analysis, however, strongly supports retention of chameleonism in the longer design. In addition to their relevance to natural fold evolution, SASF2 and SASF3 are among the longest-designed chameleon sequences.
Materials and methods
Cloning, mutagenesis, protein expression and purification.
Plasmids for the expression of P22-SASF2 and λWDQ-SASF2 with C-terminal-LEHHHHHH sequence tags were constructed in a previous study (Van Dorn et al., 2006). λWDD-SASF2 and λWDD-SASF3 were constructed by using QuikChange mutagenesis (Stratagene) to introduce Q26D and H35D mutations into λWDQ-SASF2; P22-SASF3 was similarly constructed by introduction of a P35D mutation into P22-SASF2. Unlabelled and uniform 15N-labelled proteins were expressed and purified essentially as described previously (Van Dorn et al., 2006). Uniform 13C, 15N-labelled proteins were produced as described for uniform 15N-labelled proteins, except that 2.5 g/L 13C6-glucose was substituted for unlabelled glucose as the sole carbon source.
Circular dichroism spectroscopy
Far-ultraviolet CD spectra and melts of Cro variants were obtained on an Olis DSM-20 CD spectrometer. Wavelength scans were obtained using 25 μM protein in SB250 buffer (50 mM Tris [pH 7.5], 250 mM KCl and 0.2 mM EDTA) in 1 mm pathlength cylindrical cells at 10°C. Reported scans represent an average of three replicate scans from 260 to 205 nm at 1 nm intervals with an integration time of 10 s, with a buffer baseline spectrum subtracted. Thermal denaturation curves were obtained on 25 μM protein samples in SB250 buffer in 2 mm pathlength cylindrical cells. Samples were heated from 10 to 80°C in 2°C intervals with a 2 min equilibration time per point and a 55 s signal integration time. A baseline curve of SB250 buffer was subtracted from each denaturation profile. Tm values were obtained by fitting to the following relationship (Becktel and Schellman, 1987):
wherein ΔGu was related directly to the fraction of molecules in the unfolded state, and this fraction in turn was related to the position of the measured ellipticity value between the unfolded and folded baselines. The heat capacity of unfolding (ΔCp) was fixed at 840 cal mol−1 K−1 based on an estimate of 14 cal mol−1 K−1 per residue for a 60-residue-folded region (Myers et al., 1995). All other parameters, including unfolded and folded baseline slopes and intercepts, ΔHu,Tm and Tm, were allowed to vary during fitting.
NMR spectroscopy
Unless otherwise indicated, NMR spectra were recorded at 293 K on a Varian Inova 600 MHz equipped with a cryogenic triple-resonance probe. All samples contained 50 mM sodium phosphate (pH 6.3), 150 mM KCl, 10% 2H2O, 0.01% sodium azide and 1 mM 3-(trimethylsilyl)-propionate (TSP). Comparisons of SASF2 and SASF3 15N-1H correlation spectra were conducted at 288 K and included addition of 0.5 M trimethylamine oxide (TMAO) as a stabilizing osmolyte in the SASF3 samples. Cα, Cβ, C′, N and HN resonance assignments of P22-SASF2 and λWDD-SASF2 were obtained from analysis of 3D HNCO and HNCACB spectra obtained on uniform 13C,15N-labelled samples, both at protein concentrations of 1.1 mM. Hα resonance assignments and 3JHNHα coupling constants were obtained from analysis of HNHA spectra (Vuister and Bax, 1993) obtained on uniform 15N-labelled samples at protein concentrations of 0.7 mM for P22-SASF2 and 1.2 mM for λWDD-SASF2. 15N T1, 15N T2 and 15N-{1H} NOE experiments were conducted on the same set of 15N-labelled samples, essentially as described previously for λWDQ. Reported errors were based on the standard deviation of the noise in the analyzed spectra, and represent 95% confidence intervals. CSI analysis to obtain secondary structure assignments was performed by submitting Cα, Cβ, C′ and Hα chemical shifts to the RCI Server (http://wishart.biology.ualberta.ca/rci) (Berjanskii and Wishart, 2007) using default random coil values (Schwarzinger et al., 2000). 1H chemical shifts used in this analysis were referenced to a water carrier signal at 4.835 ppm, based on referencing of TSP in a P22-SASF2 sample at a chemical shift of -0.015 ppm (Wishart et al., 1995). This value is close to the expected chemical shift of HDO at 20°C at 150 mM salt (4.811 ppm). 15N and 13C chemical shifts were referenced indirectly from the 0 ppm 1H frequency, using frequency ratios listed in Wishart et al.(Wishart et al., 1995).
Results
New designs and background sequence modifications
The designed chameleon sequence SASF2 (Fig. 1) was previously introduced into both the wild-type P22 Cro background and a monomeric version (Newlove et al., 2006) of λ Cro containing the three point mutations A33W/F58D/Y26Q (abbreviated as WDQ). The A33W and F58D mutations monomerize λ Cro by ablating a hydrophobic dimerization interface (LeFevre and Cordes, 2003), while Y26Q is a stabilizing but structurally nonperturbing substitution in the N-terminal subdomain. P22-SASF2 and λWDQ-SASF2 exhibit comparable stabilities by thermal denaturation (Tm ∼ 45°C); λWDQ-SASF2, however, did not yield to detailed NMR analysis due to aggregation of samples during long experiments.
Reasoning that sample precipitation might occur through a weakly populated unfolded or partially folded state, we bolstered the stability of λWDQ-SASF2 using a Q26D mutation. Based on previous mutational studies, we expected replacement of glutamine at position 26 with aspartate to thermally stabilize the protein by an additional ∼7°C (Pakula and Sauer, 1990). Indeed, the modified version, named λWDD-SASF2, had a Tm of 55°C (Fig. 2b). Moreover, NMR samples of 13C,15N-labelled λWDD-SASF2 in 50 mM phosphate (pH 6.3), 150 mM sodium chloride buffer showed negligible precipitation at near millimolar protein concentrations over weeks of experiments at a probe temperature of 20°C. The increased sample stability permitted a detailed NMR comparison between P22-SASF2 and λWDD-SASF2 (see below).
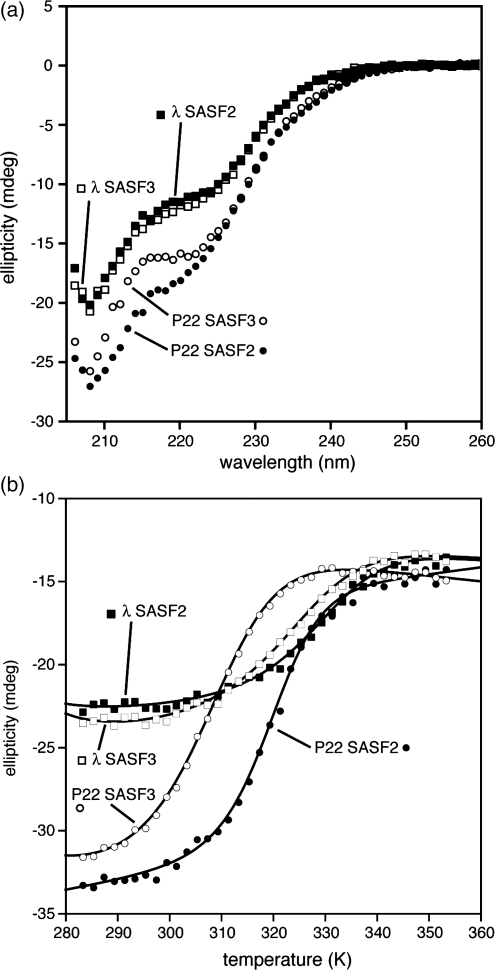
Fig. 2.
Circular dichroism studies of chameleon-containing Cro protein hosts: (a) wavelength scans of P22-SASF2, λWDD-SASF2, P22-SASF3, λWDD-SASF3 at 10°C, 25 μM protein, 1 mm pathlength, (b) thermal denaturation curves from 10–80°C, 25 μM protein, 2 mm pathlength with best fits superimposed (see Materials and Methods).
We also extended the chameleon design itself to include the entire C-terminal subdomain, residues 34–57. Since residue 34 is already isoleucine in both P22 Cro and λ Cro, this extension required only inclusion of residue 35, corresponding to the N-cap residue of helix 4 in P22 Cro and to the penultimate residue in helix 3 of λ Cro. Through trial and error mutational studies (results not shown) aspartate emerged as the best among a variety of plausible N-cap residues to replace proline 35 in P22-SASF2. Incorporation of aspartate into the full C-terminal subdomain design (giving the longer design named SASF3) caused some destabilization of P22-SASF3 relative to P22-SASF2, but the protein remained mostly folded (Fig. 2b), with a Tm of 33°C (Fig. 2b). More critically, aspartate, unlike proline, does not disrupt helix 3 in λ Cro, and λWDD-SASF3 showed thermal stability (Tm = 49°C) reasonably close to that of λWDD-SASF2 (Fig. 2b). Both SASF3 constructs also maintained the distinct far ultraviolet CD wavelength scans seen in the SASF2 versions, with the P22 host contexts giving greater helicity than the λ contexts (Fig. 2a).
NMR of SASF2 and SASF3
Standard triple-resonance techniques permitted complete backbone resonance assignment for P22-SASF2 and λWDD-SASF2, including Hα, Cα, Cβ, C′, N and HN chemical shifts. CSI analysis of chemical shifts, combined with 3JHNHα couplings from HNHA experiments, then yielded secondary structure assignments for each variant (Fig. 3). Secondary structures of P22-SASF2 and λWDD-SASF2 are clearly different in the C-terminal subdomain, despite identical primary sequences from 36–57. Moreover, the secondary structure assignments for P22-SASF2 resemble those in wild-type P22, and those for λWDD-SASF2 correspond approximately to those in parent monomeric λWDQ. These results confirm that SASF2 is a chameleon sequence.
Fig. 3.
NMR-based secondary structure analysis of hosts containing SASF2 chameleons. Sequences of P22-SASF2 and λWDD-SASF2 at bottom are colored with CSI analysis based on four backbone chemical shifts: green indicates a secondary shift favoring helix, blue indicates strand, and orange indicates ambiguity. Blank spaces in the sequences indicate absence of a resonance assignment. Below the sequences are measured values for backbone three-bond coupling constants, again colored by which secondary structure they favor. At the top, shown in green and blue bars, are consensus secondary structure assignments based on the four-shift CSI analysis.
Interestingly, strand 3 of the β-sheet in λWDD-SASF2 appears shorter than that in λWDQ, with some ‘fraying' possible in the region of residues 53–56. In addition, λWDD-SASF2 shows some evidence for helical structure in the region corresponding to the end of helix 5 of P22 Cro (around residue 56 and even beyond). Although the overall structure of the SASF2 chameleon is clearly quite different in the two backgrounds, there may be more similarity than expected near the C-terminal boundary of the domain, and in a very tentative sense λWDD-SASF2 might be regarded as having a structure intermediate between the two Cro folds (further discussion below).
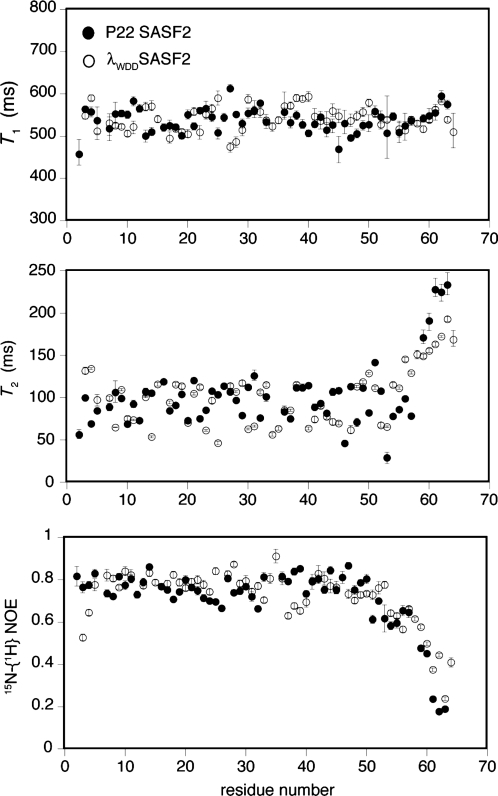
We used 15N relaxation experiments to probe and compare backbone motions in P22-SASF2 and λWDD-SASF2 (Fig. 4). The behavior of the two proteins are similar, with a rise in T2 values observed for residues 58 and higher, as expected based on the folded domain boundaries for the parent proteins (∼56 in the case of monomeric λWDQ and 57–58 in the case of P22 Cro). Similarly, reduced 15N-{1H} NOE values are seen for both proteins around residue 55 and beyond. For both proteins, scattered low T2 values (30–75 ms) occur throughout the folded region of the domain, against a background of longer values in the neighborhood of 110 ms. T2 values near 110 ms are similar to the average of 108 ms previously reported for structured regions of the λWDQ monomer (Newlove et al., 2006) and likely represent global tumbling of the domains; the scattered lower T2 values may reflect significant milli- to microsecond fluctuations. T1 and NOE values for the folded region of the domain are rather uniform (around 0.8 for the NOE and 550 ms for T1), and resemble previously reported averages of 0.77 and 514 ms, for NOE and T1 values of the structured region of the λWDQ monomer (Newlove et al., 2006). Residues 37–40 of λWDD-SASF2 show both longer T1 values and lower NOEs, potentially indicative of altered dynamic behavior in the region connecting helix 3 and strand 2. Notably, the beginning of strand 2 interacts with the end of strand 3, which as noted above appears frayed in λWDD-SASF2.
Fig. 4.
Comparative backbone dynamics of P22-SASF2 and λWDD-SASF2, including 15N T1 values, 15N T2 values and 15N-{1H} NOE values.
Moving beyond the end of the folded domain (near residues 56–57) toward the C-terminus, λWDD-SASF2 shows slightly greater persistence of higher NOE and lower T2 values than does P22-SASF2, hinting at attenuated dynamic behavior and residual structure in this region. Notably, CSI and J coupling analyses show slightly stronger evidence of helical structure for λWDD-SASF2 in the region of 58–61, though the differences are not large. We suggest that λWDD-SASF2 has partial, fluctuating helical structure in this region.
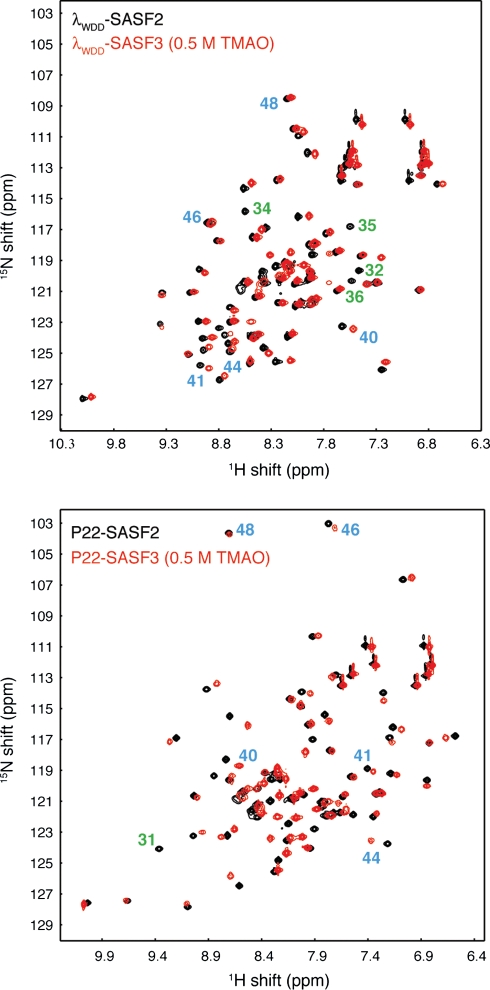
Finally, we conducted an NMR comparison of SASF2 and SASF3 (Fig. 5). The lowered stability of P22-SASF3 limited spectral quality at 293 K (not shown), presumably due to dynamic exchange between the folded state and significantly populated unfolded states. Spectra of P22-SASF3 could be improved, however, by lowering the sample temperature to 288K, and could be further improved by adding the osmolyte TMAO to a concentration of 0.5 M, likely owing to stabilization of the folded state (Wang and Bolen, 1997). Comparison with SASF2 HSQC spectra (Fig. 5) clearly shows high similarity between P22-SASF2 and P22-SASF3, and between λWDD-SASF2 and λWDD-SASF3. Unlike P22 SASF3, λWDD-SASF3 required neither lower temperature nor TMAO for reasonable spectral quality, though we used these conditions for purposes of comparison in Fig. 5. We conclude that the chameleon behavior seen in SASF2 is also present in the full-length C-terminal subdomain chameleon SASF3. This result agrees with conclusions based on the circular dichroism wavelength scan data (Fig. 2a).
Fig. 5.
Comparison of 15N-1H correlation spectra for SASF2 (black) and SASF3 (red) chameleon sequences in λWDD host background (top) and P22 host background (bottom). SASF3 samples include 0.5 M TMAO to enhance protein stability, and all spectra were obtained at 288 K. Blue numbers denote key residues in the structurally divergent C-terminal region containing the chameleon sequence: the small movements in chemical shift indicate retention of a common C-terminal structure between SASF2 and SASF3. Green numbers indicate selected residues assigned in SASF2 constructs that are at or near the site of specific sequence differences between SASF2 and SASF3. Such residues would be expected to show large chemical shift differences even when structure is retained, and indeed the resonances for these amide groups lack obvious corresponding resonances in SASF3.
Discussion
Single C-terminal Cro subdomain sequences designed in this study can adopt both α-helix and β-sheet secondary structure in the context of a full-length Cro protein. Which fold is adopted depends on whether the sequence is fused to the N-terminal subdomain of an α-helical Cro protein or to that of an α+β Cro protein. The chameleon sequence adopts the secondary structure present in the parent, showing that (i) the designed C-terminus is structurally ambivalent and (ii) the N-terminal subdomain acts as a biasing template to determine the C-terminal structure. We thus expand and extend our previous findings (Van Dorn et al., 2006), in which the characterized chameleons did not encompass the first several residues in the C-terminal half of the P22-λ alignment.
The structurally ambivalent sequences designed here largely preserve the folding of the parent sequence, but also exhibit reduced thermal stabilities. P22-SASF2 and P22-SASF3 have melting temperatures of 46 and 33°C, respectively, compared with ∼55–56°C for wild-type P22 Cro (Newlove et al., 2004; Van Dorn et al., 2006). λWDD-SASF2 and λWDD-SASF3 have melting temperatures of 55 and 49°C, respectively, compared with an estimate of ∼70°C for λWDD, based on the known value of 61°C for λWDQ (Newlove et al., 2006) and the expected 7°C increment from the Q26D mutation (Pakula and Sauer, 1990). These decrements in stability, relative to the parent host sequences, may reflect the intrinsic difficulty of encoding two Cro C-terminal conformations simultaneously in a single region of primary sequence. The sequence space that encodes potential stability-conserving pathways between these two folds may be somewhat limited. Alternatively, the lowered thermal stabilities may reflect limitations of our simple heuristic mutagenesis/design approach. Bryan and Orban have also noted a connection between loss of native stability and the potential for population of alternative folded states (Bryan and Orban, 2010). Thus, an intrinsic part of moving toward a new fold may be moving away from the stability optimum of the original fold.
In the Introduction, we noted that pathways of fold change could in principle involve one or more structural intermediates rather than proceeding in an all-or-nothing fashion. In this light, it is notable that λWDD-SASF2 shows hints of movement toward an intermediate structure. In particular, the secondary chemical shift analysis shows some evidence of a shortened β-hairpin and enhanced helical content near the end of the C-terminal region, as well as possible attenuated dynamics of the very C-terminus relative to P22-SASF2. The beginning of the C-terminal region, which includes the end of helix 3 from λ Cro and the beginning of helix 4 from P22 Cro, shows a relatively indeterminate secondary structure in λWDD-SASF2 and possible increased backbone dynamics. Neither of these observations, however, points unambiguously to an intermediate structure.
Now that full C-terminal chameleons have been designed and characterized, it is quite evident that the N-terminal halves of P22 Cro and λ Cro exert contextual influences on the structure of the C-terminal half. These influences may derive from a variety of N-terminal sequence differences. One obvious distinction is that λ Cro has a longer N-terminal sequence relative to the beginning of the conserved helix-turn-helix motif. This N-terminal tail encodes the first β-strand that interacts with the C-terminal hairpin, and the length of this region could be critical for stable formation of the β-sheet fold. Other potentially significant differences include the length of the sequence near the end of helix 3 that links the N-terminal and C-terminal halves. The length of this region could easily exert a topological influence on the folding of the C-terminal subdomain. These and other potential sequence determinants of Cro structural evolution are the subject of ongoing study.
The designed sequences could be intrinsically structurally ambivalent, or could have a strong intrinsic secondary structure preference that is overridden in one of the two contexts. When given the SASF2 sequence as input, both CSSP2 (Yoon et al., 2007) and PSIPRED (Jones, 1999) predict secondary structures similar to those experimentally observed in λWDD-SASF2 (data not shown). In the region of greatest structural difference, CSSP2 shows a high β-strand propensity (0.75 on a scale of 0 to 1) for SASF2 in the sequence corresponding to strand 2 of λ Cro (residues 40–45; IFLTIY), and a significant but lower α-helix propensity (0.54) for the overlapping sequence corresponding to helix 4 of P22 Cro (residues 36–45; EKDAIFLTIY). Interestingly, both wild-type parent sequences also show high β-strand propensity in the strand 2 region (0.71 for λ Cro and 0.70 for P22 Cro), but the helical propensity of the helix 4 region is much higher for wild-type P22 Cro (0.57) than for wild-type λ Cro (0.41). The chameleonism of SASF2 may derive from its high intrinsic β-strand tendency in the strand 2 region, which is overridden in the P22 Cro context by a combination of moderate local helical propensity plus a strong tertiary influence toward the helical fold. It is also noteworthy that the presence of highly hydrophobic sequence patterns in the strand 2 region of SASF2 favors β-strand secondary structure (West and Hecht, 1995). At the same time, increases in local sequence hydrophobicity have been shown to reduce conformational specificity in at least two proteins (Cordes et al., 2000; Hill and DeGrado, 2000), and this may be a factor favoring the chameleonism of our designed sequences, since the sequence of 39–46 for SASF2 (AIFLTIYT) is more hydrophobic than the corresponding sequence for either wild-type P22 Cro (AYRLEIVT) or wild-type λ Cro (KIFLTINA).
Chameleons—local, identical regions of sequence that can adopt different structure in different global sequence/structure contexts—of at least eight residues in length occur naturally in protein structure databases among unrelated proteins (Mezei, 1998; Guo et al., 2007). Chameleons, and even full-length sequences that switch folds, have been the subject of numerous selection and design experiments (Minor and Kim, 1996; Ambroggio and Kuhlman, 2006a,b; Alexander et al., 2007; Alexander et al., 2009). For at least two small protein domains (25–55 residues) that undergo natural fold evolution, very small sequence changes have been found to alter the fold, implying that significant regions of sequence within these proteins exhibit chameleonism (Tidow et al., 2004; Meier et al., 2007). The present work highlights the potential importance of chameleonism in the structural evolution of the Cro family (a domain of ∼60 residues), by showing that single ∼24-residue sequences, representing essentially the entire divergent region of the structure, can encode both basic structural forms found among family members. The chameleonism observed in this region also illustrates the important role of the global sequence and structure context in determining the fold.
Funding
This work was supported by the National Institute for General Medical Sciences at the National Institutes of Health (grant number R01 GM066806 to M.H.J.C.).
References
- Albright R.A., Matthews B.W. J Mol. Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- Alexander P.A., He Y., Chen Y., Orban J., Bryan P.N. Proc. Natl Acad. Sci. USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander P.A., He Y., Chen Y., Orban J., Bryan P.N. Proc. Natl Acad. Sci. USA. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggio X.I., Kuhlman B. J. Am. Chem. Soc. 2006a;128:1154–1161. doi: 10.1021/ja054718w. [DOI] [PubMed] [Google Scholar]
- Ambroggio X.I., Kuhlman B. Curr. Opin. Struct. Biol. 2006b;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Becktel W.J., Schellman J.A. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- Berjanskii M.V., Wishart D.S. Nucleic Acids Res. 2007;35:W531–537. doi: 10.1093/nar/gkm328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan P.N., Orban J. Curr. Opin. Struct. Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes M.H., Burton R.E., Walsh N.P., McKnight C.J., Sauer R.T. Nat. Struct. Biol. 2000;7:1129–1132. doi: 10.1038/81985. [DOI] [PubMed] [Google Scholar]
- Dubrava M.S., Ingram W.M., Roberts S.A., Weichsel A., Montfort W.R., Cordes M.H. Protein Sci. 2008;17:803–812. doi: 10.1110/ps.073330808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.T., Jaromczyk J.W., Xu Y. Proteins. 2007;67:548–558. doi: 10.1002/prot.21285. [DOI] [PubMed] [Google Scholar]
- Hill R.B., DeGrado W.F. Structure. 2000;8:471–479. doi: 10.1016/s0969-2126(00)00130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- LeFevre K.R., Cordes M.H. Proc. Natl Acad. Sci. USA. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., Yu H. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Meier S., Ozbek S. Bioessays. 2007;29:1095–1104. doi: 10.1002/bies.20661. [DOI] [PubMed] [Google Scholar]
- Meier S., Jensen P.R., David C.N., Chapman J., Holstein T.W., Grzesiek S., Ozbek S. Curr. Biol. 2007;17:173–178. doi: 10.1016/j.cub.2006.10.063. [DOI] [PubMed] [Google Scholar]
- Meyerguz L., Kleinberg J., Elber R. Proc. Natl Acad. Sci. USA. 2007;104:11627–11632. doi: 10.1073/pnas.0701393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezei M. Protein Eng. 1998;11:411–414. doi: 10.1093/protein/11.6.411. [DOI] [PubMed] [Google Scholar]
- Minor D.L., Jr., Kim P.S. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- Myers J.K., Pace C.N., Scholtz J.M. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlove T., Konieczka J.H., Cordes M.H. Structure. 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Newlove T., Atkinson K.R., Van Dorn L.O., Cordes M.H. Biochemistry. 2006;45:6379–6391. doi: 10.1021/bi052541c. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D.H., Tronrud D.E., Matthews B.W. J. Mol. Biol. 1998;280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- Pakula A.A., Sauer R.T. Nature. 1990;344:363–364. doi: 10.1038/344363a0. [DOI] [PubMed] [Google Scholar]
- Roessler C.G., Hall B.M., Anderson W.J., Ingram W.M., Roberts S.A., Montfort W.R., Cordes M.H. Proc. Natl Acad. Sci. USA. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzinger S., Kroon G.J., Foss T.R., Wright P.E., Dyson H.J. J. Biomol. NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- Tidow H., Lauber T., Vitzithum K., Sommerhoff C.P., Rosch P., Marx U.C. Biochemistry. 2004;43:11238–11247. doi: 10.1021/bi0492399. [DOI] [PubMed] [Google Scholar]
- Tuinstra R.L., Peterson F.C., Kutlesa S., Elgin E.S., Kron M.A., Volkman B.F. Proc. Natl Acad. Sci. USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorn L.O., Newlove T., Chang S., Ingram W.M., Cordes M.H. Biochemistry. 2006;45:10542–10553. doi: 10.1021/bi060853p. [DOI] [PubMed] [Google Scholar]
- Vuister G.W., Bax A. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- Wang A., Bolen D.W. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- West M.W., Hecht M.H. Protein Sci. 1995;4:2032–2039. doi: 10.1002/pro.5560041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Yadid I., Kirshenbaum N., Sharon M., Dym O., Tawfik D.S. Proc. Natl Acad. Sci. USA. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Welsh W.J., Jung H., Yoo Y.D. Comput. Biol. Chem. 2007;31:373–377. doi: 10.1016/j.compbiolchem.2007.06.002. [DOI] [PubMed] [Google Scholar]