Abstract
The SAGA complex is a conserved, multifunctional co-activator that has broad roles in eukaryotic transcription. Previous studies suggested that Tra1, the largest SAGA component, is required either for SAGA assembly or for SAGA recruitment by DNA-bound transcriptional activators. In contrast to Saccharomyces cerevisiae and mouse, a tra1Δ mutant is viable in Schizosaccharomyces pombe, allowing us to test these issues in vivo. We find that, in a tra1Δ mutant, SAGA assembles and is recruited to some, but not all, promoters. Consistent with these findings, Tra1 regulates the expression of only a subset of SAGA-dependent genes. We previously reported that the SAGA subunits Gcn5 and Spt8 have opposing regulatory roles during S. pombe sexual differentiation. We show here that, like Gcn5, Tra1 represses this pathway, although by a distinct mechanism. Thus, our study reveals that Tra1 has specific regulatory roles, rather than global functions, within SAGA.
Keywords: chromatin structure, co-activators, SAGA, transcription
Introduction
Transcription initiation by RNA polymerase II is a highly regulated process that involves several different classes of factors, including gene-specific activators, general transcription factors, and co-activators. Co-activators are multiprotein complexes that can possess many different activities, including histone modification and nucleosome remodelling. In addition, co-activators interact with both gene-specific activators and general transcription factors to allow transcription. Thus, co-activators have critical roles in regulating gene expression and many studies have demonstrated the importance of co-activator functions in important cellular processes (Naar et al, 2001). One well-studied, multifunctional co-activator is the SAGA complex (Spt-Ada-Gcn5-acetyltransferase). SAGA was initially discovered in Saccharomyces cerevisiae and is highly conserved throughout eukaryotes, including Schizosaccharomyces pombe, Drosophila melanogaster, mice, and humans (Koutelou et al, 2010). A combination of genetic, biochemical, and structural studies have shown that SAGA subunits are organized into several functional modules with distinct regulatory roles (reviewed in Rodriguez-Navarro (2009) and Koutelou et al (2010)). While extensive studies have elucidated both regulatory roles and the functional organization, several aspects of SAGA function remain poorly understood, including the functions of many of its components in vivo. In addition, there is still little insight into how multiple activities within a co-activator complex are coordinated in transcriptional regulation.
The SAGA subunit Tra1 and its mammalian orthologue TRRAP (Transformation/Transcription domain-Associated Protein) are integral components of two distinct histone acetyltransferase (HAT) complexes, SAGA and NuA4 (Grant et al, 1998; Saleh et al, 1998; Vassilev et al, 1998; Allard et al, 1999). TRRAP was originally identified in mammalian cells as an interacting partner for the c-Myc and E2F transcription factor oncoproteins (McMahon et al, 1998). Tra1 and TRRAP are very large proteins (around 400 kDa) and belong to the phosphoinositide 3 kinase-related kinase family, but lack some of the catalytic residues required for kinase activity. The exact roles of Tra1 and TRRAP during transcription have been challenging to study genetically, as Tra1 is essential for viability in S. cerevisiae (Saleh et al, 1998) and TRRAP is essential for early embryonic development in mice (Herceg et al, 2001). Nonetheless, studies using partial loss of function or conditional mutant alleles have implicated Tra1 and TRRAP in a number of important cellular processes, including the resistance to a variety of stress conditions, the control of mitotic checkpoints, and the maintenance of stem cell niches (reviewed in Murr et al (2007)). It has been difficult, however, to determine whether these roles are mediated through SAGA or NuA4. Finally, biochemical studies have shown direct interaction between several transcription activators and Tra1, suggesting that its primary role is to target co-activator complexes to specific promoters (Brown et al, 2001; Bhaumik et al, 2004; Fishburn et al, 2005; Reeves and Hahn, 2005). In addition, its large size, lack of kinase activity, and presence in both SAGA and NuA4 have suggested that Tra1 may serve as a scaffold for complex assembly or for recruitment to chromatin (Grant et al, 1998; Allard et al, 1999; Murr et al, 2007; Knutson and Hahn, 2011).
In order to gain new insights into SAGA function in vivo, we initiated an analysis of SAGA from the fission yeast S. pombe (Helmlinger et al, 2008), as S. pombe is highly divergent from S. cerevisiae (Wood et al, 2002). We previously reported that, although SAGA subunit composition is highly conserved in S. pombe, analysis of some SAGA mutants revealed unexpected roles for this co-activator in regulating the switch from proliferation to sexual differentiation (Helmlinger et al, 2008). Specifically, we found that Gcn5 acts as a repressor of the genes required for sexual differentiation in nutrient-rich conditions, whereas Spt8 is required for the induction of these genes when cells are starved. This study showed that SAGA can utilize distinct activities within the complex to have opposite roles in a developmental pathway, depending upon nutrient conditions.
Here, we extend our characterization of S. pombe SAGA by performing functional analysis of all viable SAGA deletion mutants, focusing on the roles of Tra1. We confirm the recent observation that Tra1 is not essential for viability in S. pombe (Calonge et al, 2010). Interestingly, in S. pombe, there are two paralogous proteins, Tra1 and Tra2 (Hayashi et al, 2007). We show that, in contrast to Tra1, Tra2 is required for viability, and that Tra1 associates specifically with SAGA while Tra2 associates specifically with NuA4. The viability of a tra1Δ mutant allowed us to address two longstanding issues about the roles of Tra1 within SAGA. First, we demonstrate that, surprisingly, Tra1 is not required for the assembly of SAGA, suggesting that it has regulatory, rather than structural roles. Second, using microarray and chromatin immunoprecipitation (ChIP) experiments, we test whether Tra1 is required for SAGA recruitment. Our results show that in a tra1Δ mutant, SAGA is still recruited to some promoters but not to others. Thus, Tra1 is required for SAGA recruitment in vivo in a gene-specific manner. Finally, we demonstrate a role for Tra1 in the S. pombe sexual differentiation pathway, where SAGA was previously shown to act as either a repressor or an activator, depending on nutrient levels (Helmlinger et al, 2008). Taken together, these studies have allowed us to define more precisely the roles of Tra1 within the SAGA complex.
Results
Deletion analysis of all SAGA subunit genes reveals important functional differences between S. pombe and S. cerevisiae
We have previously reported the purification of the S. pombe SAGA complex and found that its subunit composition is very similar to that of the S. cerevisiae, Drosophila, and mammalian complexes, emphasizing the high degree of conservation of SAGA across eukaryotes (Helmlinger et al, 2008). In order to analyse the roles of the S. pombe SAGA complex in vivo, we deleted each of the genes that encode a subunit of the complex (summarized in Supplementary Table 1), with the exception of taf5+ and taf6+, which were previously shown to be essential for viability (Yamamoto et al, 1997; Mitsuzawa and Ishihama, 2002). Our results here show that the only other essential SAGA genes are those that encode Taf9, Taf10, and Taf12 (Supplementary Figure S1). The essential nature of the TAFs is almost certainly due to their presence in TFIID (Elmlund et al, 2009).
Strikingly, there were two major differences between S. pombe and S. cerevisiae with respect to the requirement of each SAGA subunit for growth or viability (Supplementary Table 1). First, we observed that an S. pombe spt3Δ mutant is severely impaired for growth (Figure 1), in contrast to an S. cerevisiae spt3Δ mutant, which has only a modest growth impairment (Winston et al, 1984). Notably, an S. pombe spt3Δ mutant grows significantly slower than deletion mutants of genes encoding core SAGA subunits, Spt7 and Ada1 (Figure 1; Supplementary Figure S1), which are required for the integrity of the complex in S. cerevisiae (Grant et al, 1997; Sterner et al, 1999; Wu and Winston, 2002). This observation suggests that Spt3 has a more critical role in S. pombe than in S. cerevisiae, possibly outside of SAGA. Second, we made the surprising observation that a tra1Δ deletion mutant is viable and grows normally on rich medium at 32°C (Figure 1). This result was surprising as previous studies have shown that Tra1 is essential for viability in both S. cerevisiae and mice (Saleh et al, 1998; Herceg et al, 2001). The viability of S. pombe a tra1Δ mutant has also recently been described elsewhere (Calonge et al, 2010).
Figure 1.
SAGA mutants show a range of phenotypes under different stress conditions. Growth phenotypes of SAGA mutants compared with an isogenic wild-type strain. Cells were grown in liquid YES medium to stationary phase, subjected to 10-fold serial dilutions and spotted onto test plates, as indicated. The growth phenotypes observed on rich medium (YES), supplemented with 3% raffinose, 15 mM caffeine, 1 M KCl, 10 mM hydroxyurea (HU), or incubated at 38°C, as well as on minimal medium (EMM) containing 0.02 μg/ml rapamycin, or 20 μg/ml canavanine (see Supplementary Table 2 for details) are shown here. All plates were incubated at 32°C for 3 days, except caffeine, KCl, and HU plates, which were incubated for 6 days. All strains were spotted onto one plate, but arranged in two separate panels for constructing the figure. The results from one representative experiment out of four independent replicates are shown.
Analysis of the growth phenotypes of SAGA deletion mutants
To gain more information about the in vivo roles of SAGA and to identify functional modules within the complex, we tested the viable deletion mutants for growth defects under 37 different growth conditions (Figure 1; Supplementary Table 2; Supplementary Figure S2). These results show that the mutants fall into several distinct phenotypic groups and reveal differences from S. cerevisiae. The results summarized here are grouped by the functional modules within SAGA as defined by analysis in S. cerevisiae. For the three SAGA core components, Ada1, Spt7, and Spt20, only two mutants, ada1Δ and spt7Δ, were severely impaired for growth in normal conditions. In contrast, spt20Δ causes a more modest growth defect on rich medium, while showing impaired growth under a number of other conditions. For the SAGA HAT module, consisting of Gcn5, Ada2, and Ada3, deletion mutants share several similar phenotypes (Figure 1), including sensitivity to high temperature (38°C) and to high salt as previously shown for gcn5Δ (Johnsson et al, 2006). These results extend our previous analysis that showed that these three factors function together based on genome-wide expression analysis and by the demonstration that they all repress the transcription of sexual differentiation genes (Helmlinger et al, 2008). Sgf29 is an SAGA component that has been shown to directly interact with Ada3 in rat cells (Kurabe et al, 2007). Consistent with this interaction, our analysis shows that an sgf29Δ mutant has a subset of the mutant phenotypes seen in gcn5Δ, ada2Δ, and ada3Δ mutants. In S. cerevisiae, spt3Δ and spt8Δ mutants behave similar to each other, consistent with the evidence that Spt3 and Spt8 are functionally related in the recruitment of TBP. In S. pombe, however, spt3Δ and spt8Δ mutants behave very differently. In contrast to the severe growth defect of an S. pombe spt3Δ mutant, spt8Δ causes only a modest defect in growth in rich medium. Unexpectedly, an spt8Δ mutant shows pleiotropic phenotypes that are very similar to those of an spt20Δ mutant (Figure 1), suggesting that Spt8 may be functionally related to Spt20 in S. pombe. The SAGA DUB module contains four proteins, Ubp8, Sgf73, Sgf11 and Sus1, all required for Ubp8 histone deubiquitylation activity. Among these four mutants, the few mutant phenotypes observed were different for each mutant, suggesting that these four proteins are not all dedicated to Ubp8 function (Figure 1; Supplementary Table 2). Recent work has indeed shown that Sgf73 has nucleosome-binding properties, in addition to recruiting the DUB module into SAGA (Bonnet et al, 2010). Finally, we noticed several phenotypes for a tra1Δ mutant that did not fall into any of the other patterns. In addition to these distinct patterns, all SAGA mutants were sensitive to HU, albeit to different degrees, indicating a role for all SAGA activities during S-phase progression. Together, these results indicate that SAGA is not essential for viability but has important roles in specific cellular processes and several of its activities are required for adaptation to distinct stress conditions.
S. pombe has two paralogous tra genes that are differently required for viability
The most striking observation from our analysis of SAGA deletion mutants was the discovery that tra1+ is not required for viability in S. pombe, whereas it is in S. cerevisiae and mice. Furthermore, homology searches for orthologues of S. cerevisiae Tra1 or human TRRAP protein sequence identify two genes in the S. pombe genome, SPAC16F5.03c and SPAC1F5.11c, named Tra1 and Tra2 (Hayashi et al, 2007). Our additional homology searches identified Tra1 and Tra2 orthologues only in the genomes of the four Schizosaccharomyces species sequenced. A multiple sequence alignment of Tra1 and Tra2 from three Schizosaccharomyces species with their eukaryotic orthologues, Tra1 or TRRAP, suggests that an ancestral tra1 gene duplicated before speciation within the Schizosaccharomyces lineage (Supplementary Figure S3). In addition, examination of the Tra1 and Tra2 amino-acid sequences reveals that they contain the same domain structure as Tra1 or TRRAP orthologues, including a FAT domain, a phosphatidyl inositol-like kinase (PIK) domain, and a FATC domain at the C-terminal end of the protein. Similarly to other Tra1 and TRRAP orthologues, both Tra1 and Tra2 lack some of the residues critical for catalytic activity in the PIK domain (Supplementary Figure S4), suggesting that the lack of kinase activity is conserved in S. pombe Tra1 and Tra2.
The identification of two tra genes in S. pombe prompted us to ask whether tra1+ and tra2+ are functionally redundant, possibly explaining why a tra1Δ mutant is viable. To test this hypothesis, we deleted tra2+ in a diploid strain to generate tra2+/tra2Δ heterozygotes, which were then sporulated to analyse the tra2Δ phenotype. Our results show that tra2+ is essential for viability in S. pombe, as we observe a 2:2 segregation of inviability and all of the viable spores were tra2+ (Figure 2; data not shown). Thus, tra1+ cannot compensate for the loss of tra2+.
Figure 2.
The tra2+ gene is essential for viability. Progeny after sporulation and tetrad dissection of a tra2+/tra2Δ∷natMX6 heterozygous diploid is shown. The four progeny from each tetrad are labelled a–d and five representatives tetrads are shown of a total of 37 analysed. The only colonies growing are wild type, as they were all sensitive to nourseothricin (NAT), showing that a tra2Δ mutant is inviable.
S. pombe Tra1 and Tra2 are specific for SAGA and NuA4, respectively
We previously reported that S. pombe SAGA, purified by the core subunits Spt7 or Ada1, contains only Tra1 (Helmlinger et al, 2008). Recently, another study showed that the NuA4 HAT complex, purified by several different NuA4 components, contains only Tra2 (Shevchenko et al, 2008), suggesting that Tra1 is specific for SAGA and Tra2 is specific for NuA4. To test this possibility directly, we purified Tra1 and Tra2 using a tandem affinity purification (TAP) tag and analysed the co-purifying proteins by SDS–PAGE and mass spectrometry. Because TAP fusions to the 3′ ends of tra1+ and tra2+ caused mutant phenotypes, we constructed strains in which the TAP sequence was fused onto the 5′ ends of the tra1+ and tra2+ genes; these strains have wild-type phenotypes (Supplementary Figure S5). Our analysis showed that the purifications of Tra1 and Tra2 identified distinct complexes (Supplementary Table 3). Tra1 co-purified with all of the proteins found in the S. pombe SAGA complex (Helmlinger et al, 2008). In addition, several subunits of the Tel2-containing ASTRA complex co-purified with Tra1 (Supplementary Table 3) as expected (Shevchenko et al, 2008). In contrast to Tra1, Tra2 co-purified with 10 proteins, each a subunit of the NuA4 complex (Supplementary Table 3). While our analysis did not detect three NuA4 subunits, Bdf1, Alp13, and Act1, they were previously identified as subunits of the S. pombe NuA4 complex (Shevchenko et al, 2008). The NuA4 HAT subunit, Mst1, is essential for viability (Gomez et al, 2008), suggesting that Tra2 is essential because of its role in NuA4. In conclusion, our purification results demonstrate that Tra1 is a component of SAGA and ASTRA, while Tra2 is specific for NuA4. Interestingly, the related Tor1 and Tor2 proteins are each in distinct complexes, in both S. pombe and S. cerevisiae (Jacinto, 2008).
Proteomic analysis of a strain with a TAP sequence fused to the 3′ end of tra1+ showed that this fusion is defective for assembly into SAGA but not into ASTRA (Supplementary Figure S6). This behaviour is similar to a previously described purification of a C-terminally tagged Tra1 (Shevchenko et al, 2008). When we tested this tra1∷TAP strain for mutant phenotypes, we discovered that it behaves similar to a tra1Δ mutant (Supplementary Figure S5), suggesting that most of the mutant phenotypes we have observed are caused by loss of Tra1 from SAGA and not from ASTRA. This observation also suggests that the C-terminal end of Tra1 is important for its normal function, consistent with mutational analyses of the S. cerevisiae TRA1 gene, which have identified residues that are critical for Tra1 function within the C-terminal FATC domain (Hoke et al, 2010; Knutson and Hahn, 2011).
Tra1 is not required for assembly of the SAGA complex
The viability of a tra1Δ mutant allowed us to address several important aspects about the roles of Tra1 in SAGA. First, we tested whether Tra1 is required for SAGA assembly in vivo. A role for Tra1 in SAGA structure seemed likely, given its extremely large size (∼420 kDa) and previous results suggesting that Tra1 may be required for SAGA to assemble on chromatin (Memedula and Belmont, 2003). To test this possibility, we purified SAGA from a tra1Δ deletion mutant using an Ada1–TAP fusion and analysed the proteins by SDS–PAGE and mass spectrometry. We found that SAGA assembles in a tra1Δ mutant, lacking only Tra1 and none of the other subunits (Figure 3). Mass spectrometry analysis confirmed that all SAGA subunits were present in SAGA purifications from a tra1Δ mutant compared with those from wild type, although there was a reduced level of three subunits, Sgf73, Ubp8, and Sus1 (Supplementary Table 3). No Tra2 peptides were detected in the mutant SAGA complex, confirming that Tra1 and Tra2 are not interchangeable. Taken together, these results show that Tra1 does not contribute significantly to the integrity of SAGA and is thus not a core subunit of the complex. This conclusion is consistent with our genetic analysis (Figure 1), which showed that a tra1Δ mutation causes fewer phenotypes than do spt7Δ and ada1Δ mutations.
Figure 3.
Tra1 is not required for the integrity of the SAGA complex. The Ada1–TAP protein was purified from wild-type (tra1+) and tra1Δ strains. Co-purifying proteins were separated on an SDS 5–20% gradient polyacrylamide gel and visualized by silver staining. An untagged control strain is shown (‘no tag’). The positions of molecular weight markers (MW) are shown on the right. The mass spectrometry analysis of these purifications is presented in Supplementary Table 3.
Transcriptome signatures of all viable SAGA deletion mutants reveal distinct functional classes
As SAGA assembles in a tra1Δ mutant (Figure 3) and tra1Δ mutant phenotypes are distinct from other SAGA mutants (Figure 1), we wanted to determine the genome-wide effects on transcription caused by tra1Δ and to compare them with other SAGA mutants. To do this, we performed genome-wide expression analyses on all viable SAGA deletion mutants (Supplementary Table 4).
To compare the mutant profiles, we performed hierarchical clustering and found that the SAGA mutants fall into several classes (Figure 4A; Supplementary Figure S7). Three classes contain multiple SAGA mutants and are described below, whereas the tra1Δ, sgf73Δ, ubp8Δ, and sgf29Δ mutants are each in a class by themselves. Class 1 contains ada1Δ and spt7Δ mutants which, as expected from the similarity of their mutant phenotypes, have almost identical expression profiles. In this class, 1133 genes showed mRNA levels altered by >1.5-fold and 374 by >2-fold (Figure 4C and D). Class 2, as previously reported (Helmlinger et al, 2008), contains gcn5Δ, ada2Δ, and ada3Δ mutants. For this class, 178 genes have mRNA levels altered >1.5-fold and 56 by >2-fold (Figure 4C and D). Unexpectedly, Class 3 contains sgf11Δ, sus1Δ, spt20Δ, and spt8Δ mutants. In this class, 1527 genes have mRNA levels altered >1.5-fold and 520 by >2-fold (Figure 4C and D). The composition of Class 3, combined with the distinct expression profiles of ubp8Δ and sgf73Δ, was unexpected for three reasons: first, the members of the DUB module, Ubp8, Sgf11, Sus1, and Sgf73, do not all cluster together and therefore likely have distinct roles within SAGA; second, Sgf11, Sus1, Spt20, and Spt8 do cluster and thus likely regulate a similar set of genes; and third, Spt20, previously thought of as a core component of SAGA, regulates a distinct set of genes from the core subunits Ada1 and Spt7. We also determined the significance of the overlap between the lists of affected genes found in the three classes and found significant overlaps between all three. In summary, our results show that, overall, SAGA is required for normal expression of a large number of S. pombe genes, but that different modules are important for the regulation of distinct sets of genes.
Figure 4.
Gene expression signatures of all viable SAGA deletion mutants. (A, B) Hierarchical cluster analyses of 3502 genes varying in expression over 1.5-fold in at least one mutant with columns representing the different mutants indicated and rows representing genes. The mRNA levels in each mutant relative to the levels in wild-type cells are colour coded as indicated at lower right, with missing data in white. The three classes discussed in the main text are indicated at the bottom. (A) All viable SAGA deletion mutants were clustered using the Change correlation. (B) The tra1Δ, spt7Δ, and ada1Δ mutants were re-clustered using the Smooth correlation. (C, D) Venn diagrams show the degree of overlap between the three different classes of SAGA mutants. Classes were identified from the hierarchical clustering shown in (A): Class 1 consists of spt7Δ and ada1Δ mutants; Class 2 consists of ada2Δ, ada3Δ, and gcn5Δ mutants; Class 3 consists of sgf11Δ, spt20Δ, spt8Δ, and sus1Δ mutants. The total number of genes affected >1.5-fold for each data set is indicated in parenthesis. The significance of overlaps between different gene lists was calculated in GeneSpring GX7 (Agilent) by using a standard Fisher's exact test.
Finally, our results show that tra1Δ has a distinct profile from all other SAGA mutants (Figure 4A and B), with 1052 genes controlled by Spt7 and Ada1, but independently of Tra1. For the 81 genes affected in common to tra1Δ, spt7Δ, and ada1Δ mutants, we performed gene ontology (GO) analysis and found significant enrichment for genes induced in response to stress (P=0.003). This category does not overlap with the significantly enriched GO categories identified for S. cerevisiae tra1 mutants previously studied (Mutiu et al, 2007). In addition to genes that are commonly affected by tra1Δ, spt7Δ, and ada1Δ mutations, 144 other genes are affected by tra1Δ, but not by spt7Δ and ada1Δ mutations. The expression of these genes might be controlled by ASTRA or, alternatively, these genes might be aberrantly regulated by a form of SAGA lacking Tra1. We note that the levels of the ste11+ and mei2+ mRNAs are elevated in the tra1Δ mutant, suggesting a role for Tra1 in the S. pombe sexual differentiation pathway. More evidence about this role is presented in a later section.
Analysis of the contribution of Tra1 to the recruitment of SAGA to promoters
Several studies have provided strong evidence that Tra1 is the subunit of SAGA that directly interacts with transcription activators and is thus necessary for SAGA recruitment at promoters (Brown et al, 2001; Bhaumik et al, 2004; Fishburn et al, 2005; Reeves and Hahn, 2005). The viability of tra1Δ mutants in S. pombe allowed us to test this model in vivo. To test if Tra1 is required for the recruitment of SAGA, we used wild-type and tra1Δ strains to determine the physical association of SAGA with the promoters of several genes, chosen based on our microarray analysis (Supplementary Table 4). These included two genes, ssa2+ and SPBC8E4.01c+, whose transcription is SAGA dependent, but Tra1 independent; and five other genes, pgk1+, fba1+, bip1+, act1+, and adh1+, whose transcription is SAGA independent. In wild-type cells, as measured by Spt7 ChIP, SAGA bound to regions upstream of the coding sequences of all genes tested, albeit to different degrees (Figure 5). In tra1Δ mutants, SAGA recruitment was not affected at the pgk1+, fba1+, bip1+, act1+, and adh1+ promoters. In contrast, we observed a 6- to 8-fold reduction in SAGA occupancy at ssa2+ and SPBC8E4.01c+ (Figure 5), although we still observed binding of Spt7 that was significantly above background (Supplementary Figure S8A). Together, these results suggest that SAGA is recruited by distinct mechanisms at different genes. Whereas Tra1 greatly contributes to SAGA recruitment at some genes, it is clearly dispensable at others, demonstrating the existence of Tra1-independent mechanisms for SAGA recruitment.
Figure 5.
Effects of a tra1Δ mutation on SAGA recruitment at distinct genes. Spt7 occupancy was assessed by ChIP analysis of chromatin extracts from strains in which Spt7 was tagged with 13 copies of a Myc epitope, either in a wild-type (dark grey) or in a tra1Δ (light grey) background. Cells were grown to mid-log phase in minimal medium, in the presence of nutrients. Occupancy levels were quantified by real-time PCR of ‘IP’ over ‘input’ DNA and normalized to background levels detected at a non-transcribed region (mat3). Spt7 binding was specifically detected over the putative regulatory regions of pgk1+, fba1+, bip1+, act1+, adh1+, SPBC8E4.01c+, and ssa2+. The coordinates of the regions that were tested are provided in Supplementary Table 6. The background %IP values obtained from ChIP analysis of the corresponding ‘no tag’ strains are shown in Supplementary Figure S8A and B. Each column represents the mean relative %IP value±s.e. (n=4).
Tra1 is required for regulation of the S. pombe sexual differentiation pathway
We then wanted to examine the role of Tra1 at genes where a requirement for SAGA has been well characterized, so we turned to the S. pombe sexual differentiation pathway, as our microarray results suggested a role for Tra1 in this pathway. We previously showed that SAGA uses at least two of its subunits, Gcn5 and Spt8, to regulate this pathway both positively and negatively (Helmlinger et al, 2008). To follow-up on the tra1Δ microarray results, we performed quantitative RT–PCR analysis of ste11+ and mei2+ mRNA levels in a tra1Δ mutant. The ste11+ gene encodes the master regulator of the sexual differentiation pathway and mei2+ encodes a regulator of meiotic genes (reviewed in Harigaya and Yamamoto (2007)). The transcription of both genes is normally repressed during growth in rich medium and is induced upon a shift to starvation conditions. Our results show that, in the tra1Δ mutant, both genes are aberrantly derepressed in rich medium, similarly to our previous observations in a gcn5Δ mutant (Figure 6A and B; see Helmlinger et al (2008)). Consistent with these results, tra1Δ mutants ectopically mate in rich medium (Figure 6C), as was previously shown for gcn5Δ (Helmlinger et al, 2008). These results suggest that, similarly to Gcn5, Tra1 is required for repression of sexual differentiation genes in nutrient-rich conditions.
Figure 6.
Tra1 and Gcn5 repress sexual differentiation, but act through independent pathways. (A, B) Expression analysis of ste11+ (A) and mei2+ (B) by quantitative RT–PCR of RNA prepared from h90 homothallic strains. Cells were grown to mid-log phase in minimal medium in the presence of nutrients, and then shifted for 4 h to either rich medium (dark grey) or starvation medium (light grey). act1+ served as a control for normalization across RT–PCR samples. Normalized ste11+ or mei2+ mRNA levels in a wild-type strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value±s.e. (n=4). (C) Cells were grown to mid-log phase in minimal medium in the presence of nutrients, and examined under direct light microscopy for conjugation and sporulation. Mating efficiency (%) was calculated by dividing the number of zygotes or asci (each counted as two cells) by the number of total cells (at least 300 cells). Each column represents the mean value±s.e. (n=4).
We then did additional tests to determine whether Tra1 and Gcn5 repress by similar mechanisms. We first took a genetic approach based on our previous results that spt8Δ mutants are defective for ste11+ and mei2+ transcription during nutrient starvation, and that this defect is suppressed by gcn5Δ (Helmlinger et al, 2008). We tested if tra1Δ, like gcn5Δ, suppresses the spt8Δ derepression defect. To do this, we constructed a tra1Δ spt8Δ double mutant and compared it with the gcn5Δ spt8Δ double mutant with respect to ste11+ and mei2+ mRNA levels. In contrast to what was seen for gcn5Δ, tra1Δ failed to suppress spt8Δ, as ste11+ and mei2+ mRNA levels remained low in the tra1Δ spt8Δ double mutant (Figure 6A and B). This result strongly suggests that Tra1 functions in a manner distinct from Gcn5 in transcriptional repression of ste11+ and mei2+. This possibility is also supported by the observation that mating and meiosis are derepressed two-fold more in a tra1Δ gcn5Δ double mutant compared with each single mutant (Figure 6C).
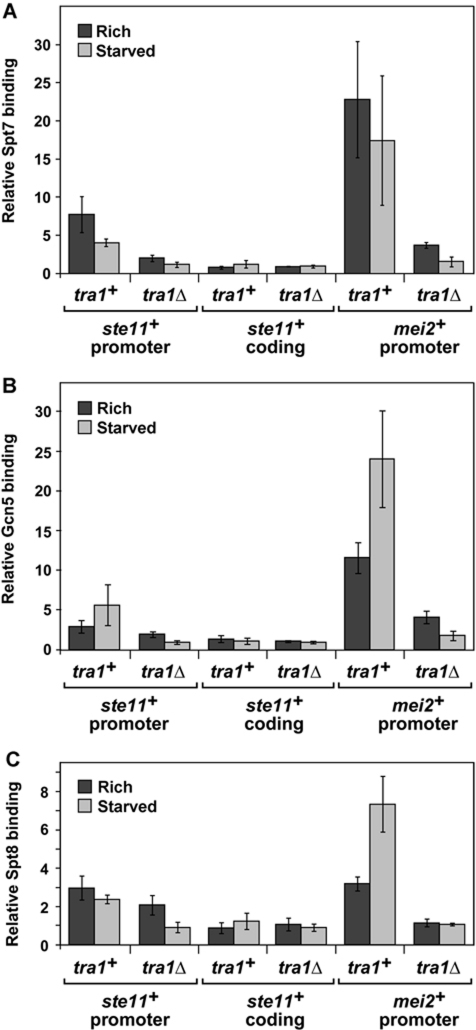
To address the respective roles of Tra1 and Gcn5 by another approach, we tested whether Tra1 is required for the recruitment of SAGA to the ste11+ and mei2+ promoters. Our previous results showed that Gcn5 is not required for SAGA recruitment at these promoters (Helmlinger et al, 2008). For Tra1, we performed ChIP analysis of three SAGA components, Spt7, Gcn5, and Spt8, comparing wild-type and tra1Δ strains. For all three SAGA components tested, their association at the ste11+ and mei2+ promoters is reduced in a tra1Δ mutant when cells were grown in either rich or starvation medium (Figure 7; Supplementary Figure S8C). We note that SAGA is still recruited to a limited degree in a tra1Δ mutant in rich medium, again suggesting Tra1-independent mechanisms of SAGA recruitment. As a control, western analyses show that steady-state levels of Spt7, Gcn5, and Spt8 are not affected in tra1Δ mutants compared with wild-type cells (Supplementary Figure S8D). Taken together with our genetic results, these ChIP results strongly suggest that Tra1 and Gcn5 repress ste11+ and mei2+ transcription by distinct mechanisms.
Figure 7.
Tra1 contributes to the recruitment of SAGA to the promoters of ste11+ and mei2+ in nutrient-rich and starved conditions. Spt7 (A), Gcn5 (B), or Spt8 (C) occupancy was assessed by ChIP analysis of chromatin extracts from strains in which each protein was myc-tagged either in a wild-type or in a tra1Δ background. Cells were grown to mid-log phase in minimal medium in the presence of nutrients, and then shifted for 4 h to either rich medium (dark grey) or starvation medium (light grey). Occupancy levels were quantified by real-time PCR of anti-myc IP over input DNA and normalized to background levels detected at a non-transcribed region (mat3), which are set at 1. The coordinates of the promoter and coding regions that were tested are provided in Supplementary Table 6. The background %IP values obtained from ChIP analysis of the corresponding ‘no tag’ strains are shown in Supplementary Figure S8C. Each column represents the mean relative %IP value±s.e. (n=4–6).
Discussion
In this work, we have analysed the growth phenotypes and genome-wide expression changes of all viable S. pombe SAGA deletion mutants, providing the most comprehensive analysis to date of regulation by SAGA in any organism. Our analysis has uncovered several new and unexpected aspects regarding SAGA function. In particular, confirming the recent finding by Calonge et al (2010), we show that Tra1 is not essential for viability in S. pombe, in marked contrast with S. cerevisiae or mammalian cells. We also show that an S. pombe Tra1 paralogue, Tra2, is essential for viability and demonstrate that Tra1 is specific for SAGA, whereas Tra2 is specific for NuA4. The latter result is in agreement with a recent proteomic analysis of chromatin modifying complexes (Shevchenko et al, 2008). We then took advantage of the viability of tra1Δ mutants to address several fundamental questions about Tra1 function. Unexpectedly, we found that Tra1 does not have a role in SAGA assembly, suggesting that, rather, it has a regulatory role within the complex. Supporting this conclusion, microarray analyses revealed that Tra1 regulates the expression of only a subset of genes regulated by the core SAGA subunits, Ada1 and Spt7, and that Tra1 is not absolutely required for the recruitment of SAGA to promoters. Importantly, our conclusions are supported by the fact that we observed a strong agreement between the phenotypic and microarray analyses of tra1Δ and the other SAGA mutants.
An important finding from our work is that Tra1 is required for the recruitment of SAGA at some promoters but not others. Our demonstration of Tra1-dependent recruitment of SAGA fits well with several in vivo and in vitro studies that showed Tra1–activator interactions (Bhaumik and Green, 2001; Brown et al, 2001; Fishburn et al, 2005; Reeves and Hahn, 2005). In addition, a recent study used ChIP assays to show that specific domains within S. cerevisiae Tra1 are required for the full recruitment of Spt7 to the HIS4 and GAL1 promoters (Knutson and Hahn, 2011). Together with our ChIP analyses using a complete tra1Δ deletion mutant in S. pombe, these results demonstrate that Tra1 is required for the recruitment of SAGA at certain promoters in vivo. Surprisingly, we also found that SAGA associates with other promoters in a Tra1-independent manner. This observation suggests that additional subunits within SAGA can recruit the complex to certain promoters. Indeed, although most studies of SAGA recruitment suggested that Tra1 is the main target of several activators, other SAGA subunits have been identified as potential targets and to contribute to SAGA recruitment, such as Spt3, Taf12, Taf6 or Ada1 (Klein et al, 2003; Fishburn et al, 2005; Qiu et al, 2005; Reeves and Hahn, 2005).
Finally, our studies have provided new insights into our understanding of the regulation of S. pombe sexual differentiation by the SAGA complex. We have shown that Tra1, like Gcn5, inhibits sexual differentiation in nutrient-rich medium by repression of ste11+ and mei2+ transcription. However, our genetic tests have suggested that Tra1 and Gcn5 regulate this pathway independently of each other, suggesting that Tra1 acts through another mechanism. Thus, we conclude that Tra1 represses the expression of genes required for sexual differentiation by a more complex mechanism than simply recruiting SAGA to their promoters. However, the observation that Tra1 is largely required for SAGA recruitment to the promoters of ste11+ and mei2+ upon starvation implies that these two genes can be induced without any detectable SAGA binding at their promoters. It is thus possible that, upon starvation, other transcriptional co-activators act redundantly with SAGA to contribute to the full induction of these genes. In S. pombe, the switch from proliferation to sexual differentiation in response to varying nutrient levels in the environment is a crucial decision because the commitment to mating and meiosis is irreversible. Our data suggest that SAGA has multiple regulatory roles in both the repression and induction of ste11+ and mei2+ transcription, highlighting the critical nature of SAGA as a regulator of this cell fate decision.
Materials and methods
S. pombe strains and genetic procedures
All S. pombe strains used are listed in Supplementary Table 5. Standard culture media (YES (Yeast Extract Supplemented) or (EMMc) Edinburgh Minimal Medium complete) and genetic manipulations were used, as described previously (Forsburg and Rhind, 2006). For all nutrient starvation experiments, cells were inoculated in nutrient-rich minimal medium (EMM+5 g/l NH4Cl and 2% glucose) and grown at 30°C to mid-log phase (∼0.5 × 107 cells/ml). Cells were then pelleted at room temperature, washed once with nutrient-starved minimal medium (EMM without NH4Cl and 0.5% glucose), inoculated in either nutrient-rich or starved medium and incubated for 4 h at 30°C. In the text, ‘nutrient starvation’ thus refers to the complete removal of the nitrogen source and to a four-fold decrease in glucose concentration in the medium. For mating assays, homothallic strains were grown as described above and the number of zygotes and asci were counted under light microscopy. Mating efficiency was calculated using the following formula: 2 × (number of zygotes and asci)/(total number of vegetative cells+2 × number of zygotes or asci). Gene deletions were constructed by replacing the respective open reading frame (ORF) with ura4+, kanMX6, or natMX6 cassettes using PCR-based gene targeting and lithium-acetate transformation, as described (Bähler et al, 1998). In each case, PCR amplification was performed using primers of 100 bases, with 80 bases to direct homologous recombination (primers listed in Supplementary Table 6). Transformants were screened by PCR analysis. Strains with epitope-tagged SAGA subunits were constructed by fusing the corresponding tag to the 3′ end of the respective gene by homologous recombination, removing the stop codon. Correct integration was verified by PCR and western blot analysis.
Western blot analysis
Protein extracts were prepared as described previously (Cheung et al, 2008) and western blot analysis was performed using the PAP antibody (peroxidase-anti-peroxidase; Sigma), an anti-Rpb1 antibody (8WG16; Covance), or an anti-Myc antibody (A14; Santa Cruz Biotechnology). Equal loading was controlled by staining the transfer membranes with Ponceau red (Sigma).
TAP and mass spectrometry analysis
Protein complexes were purified by the TAP method (Rigaut et al, 1999). Ada1 was tagged C-terminally with an HA3–TAP2 fusion epitope (from plasmid pFS209, generous gift from N Rhind). Ada1–TAP preparations were performed from 2 l YES cultures grown at 32°C to mid-log phase. The tra1+ and tra2+ ORFs were TAP tagged at their N-terminal end and thus placed under the control of the nmt1+ promoter. TAP–Tra1 and TAP–Tra2 purifications were performed from 2 l mid-log phase EMMc cultures, without thiamine, allowing overexpression of tra1+ and tra2+ (Supplementary Figure S5B). For qualitative purposes, purified proteins were separated on an SDS 5–20% gradient polyacrylamide gel (Invitrogen) and visualized by silver staining. Total protein mixtures were precipitated with 20% trichloroacetic acid (Sigma) and then in-solution digested with trypsin. In all, 10–20% was analysed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Peptides were separated across a 55-min gradient ranging from 7 to 30% ACN in 0.1% FA in a microcapillary (125 μm × 17 cm) column packed with C18 reverse-phase material (Magic C18AQ, 5 μm particles, 200 Å pore size, Michrom Bioresources, Auburn, CA) and online analysed on a hybrid LTQ-OrbitrapXL mass spectrometer (Thermo Scientific, Bremen, Germany). For each cycle, one full MS scan acquired at high mass resolution was followed by 10 MS/MS spectra on the linear ion trap from the 10 most abundant ions. MS/MS spectra were searched against the S. pombe protein sequence database using the Sequest algorithm (Eng et al, 1994). Spectral matches were first filtered to <5% false-discovery rate (FDR) at the peptide level using a target-decoy database strategy (Elias and Gygi, 2007), and subsequently to <1% FDR at the protein level. The final list of purified subunits was obtained by subtracting protein matches that were also found in an untagged control sample. We focused our analysis on the subunits of SAGA, ASTRA and NuA4 complexes, according to previously described compositions.
Microarray experiments
Total RNA was isolated using a hot-phenol protocol as described (Lyne et al, 2003). Between 10 and 20 μg of total RNA were labelled by direct incorporation of either fluorescent Cy3- or Cy5-dCTP (GE Healthcare), and the fluorescently labelled product was hybridized to S. pombe cDNA microarrays as described (Lyne et al, 2003). Microarrays were scanned using a GenePix 4000B laser scanner (Axon Instruments) and fluorescence intensity ratios were calculated with GenePix Pro (Axon Instruments). The data were normalized using a customized script (Lyne et al, 2003). At least two biological repeats were analysed for each mutant with dye swaps. To analyse the data, repeats for every given mutant were averaged. The significance of overlaps between different gene lists was calculated in GeneSpring GX7 (Agilent) by using a standard Fisher's exact test. Hierarchical clustering was performed in GeneSpring GX7 using the Change correlation (Figure 4A) or Smooth correlation (Figure 4B), with genes containing no data in ⩽50% of the conditions being discarded. All processed data are available in Supplementary Table 4 or at http://www.bahlerlab.info/.
RT–PCR analysis
Total RNA was purified using hot acidic phenol as described (Lyne et al, 2003) and contaminating DNA was removed by DNase I digestion. In all, 100 ng RNA was then reverse transcribed with an oligo-dT primer using the Invitrogen SuperScript III first-strand synthesis kit at 55°C. The thermal cycling conditions comprised an initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min. The primer sequences are listed in Supplementary Table 6.
Chromatin immunoprecipitation
ChIP experiments were performed as previously described (Hickman and Winston, 2007), with minor modifications. Briefly, cell cultures were crosslinked in 1% formaldehyde for 30 min. Cells were then broken by bead-beating, and the chromatin fraction was sheared to 200–500 bp fragments using a Bioruptor sonicator (Diagenode). For immunoprecipitations (IPs), 5 μl of anti-Myc antibody (A14; Santa Cruz Biotechnology) were incubated overnight at 4°C with the chromatin extracts and then coupled with 50 μl of protein-G-sepharose beads (GE Healthcare Life Sciences). ChIP DNA was quantified by real-time PCR, using a Stratagene MX3000P. Real-time PCR was done in triplicate for each primer set, using a standard curve that was established by serial 10-fold dilutions of a representative input DNA. Thermal cycling conditions were as described for RT–PCR analysis. For myc-tagged SAGA subunits, occupancy levels were determined by dividing the percent IP (%IP) for a myc-tagged SAGA subunit at ste11+ or mei2+ (IP/input ratio) by the %IP for a non-transcribed region from the mat3 locus (Petrie et al, 2005). To determine the specificity of enrichment of the tagged protein, the corresponding untagged control samples were included in each ChIP experiment. The primer sequences are listed in Supplementary Table 6. All tagged SAGA subunits used for ChIP analysis displayed wild-type mating phenotypes (data not shown).
Accession code
Raw microarray data are available from ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-678.
Supplementary Material
Acknowledgments
We thank Christine Kiely and Mark Hickman for critical reading of the manuscript. We are grateful to Danesh Moazed, Nick Rhind, and Chikashi Shimoda for providing plasmids or strains. DH was supported by a long-term postdoctoral Research Fellowship from the Human Frontier Science Program and by a Charles A King Trust Postdoctoral Fellowship from the Bushrod H Campbell and Adah F Hall Charity Fund. This work is supported in part by the University of Washington's Proteomics Resource (UWPR95794) to JV, by NIH grant HG3456 to SPG, by Cancer Research UK to JB, and by NIH grant GM45720 to FW.
Author contributions: DH designed and performed experiments; FW assisted experimental design and supervised the project; DH and FW wrote the manuscript; SM performed whole-genome microarray analysis overseen by JB; JV and DLS performed the MS analysis overseen by SPG. All authors reviewed and approved the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR (2004) In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev 18: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang YH, Spedale G, Atkinson RA, Romier C, Hamiche A, Pijnappel WW, Timmers HT, Tora L, Devys D, Kieffer B (2010) The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep 11: 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337 [DOI] [PubMed] [Google Scholar]
- Calonge TM, Eshaghi M, Liu J, Ronai Z, O’Connell MJ (2010) Transformation/transcription domain-associated protein (TRRAP)-mediated regulation of Wee1. Genetics 185: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F (2008) Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Linder T, Szilagyi Z, Rofougaran R, Hofer A, Hebert H, Lindahl M, Gustafsson CM (2009) Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure 17: 1442–1452 [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S (2005) Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell 18: 369–378 [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EB, Nugent RL, Laria S, Forsburg SL (2008) Schizosaccharomyces pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics 179: 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR III, Workman JL (1998) The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell 2: 863–867 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Yamamoto M (2007) Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res 15: 523–537 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Hatanaka M, Nagao K, Nakaseko Y, Kanoh J, Kokubu A, Ebe M, Yanagida M (2007) Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12: 1357–1370 [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villen J, Gygi SP, Bahler J, Winston F (2008) The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev 22: 3184–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ (2001) Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet 29: 206–211 [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Winston F (2007) Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol 27: 7414–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke SM, Irina Mutiu A, Genereaux J, Kvas S, Buck M, Yu M, Gloor GB, Brandl CJ (2010) Mutational analysis of the C-terminal FATC domain of Saccharomyces cerevisiae Tra1. Curr Genet 56: 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E (2008) What controls TOR? IUBMB Life 60: 483–496 [DOI] [PubMed] [Google Scholar]
- Johnsson A, Xue-Franzen Y, Lundin M, Wright AP (2006) Stress-specific role of fission yeast Gcn5 histone acetyltransferase in programming a subset of stress response genes. Eukaryot Cell 5: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Nolden M, Sanders SL, Kirchner J, Weil PA, Melcher K (2003) Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J Biol Chem 278: 6779–6786 [DOI] [PubMed] [Google Scholar]
- Knutson BA, Hahn S (2011) Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol Cell Biol 31: 818–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY (2010) Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabe N, Katagiri K, Komiya Y, Ito R, Sugiyama A, Kawasaki Y, Tashiro F (2007) Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc. Oncogene 26: 5626–5634 [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Memedula S, Belmont AS (2003) Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr Biol 13: 241–246 [DOI] [PubMed] [Google Scholar]
- Mitsuzawa H, Ishihama A (2002) Identification of histone H4-like TAF in Schizosaccharomyces pombe as a protein that interacts with WD repeat-containing TAF. Nucleic Acids Res 30: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z (2007) Orchestration of chromatin-based processes: mind the TRRAP. Oncogene 26: 5358–5372 [DOI] [PubMed] [Google Scholar]
- Mutiu AI, Hoke SM, Genereaux J, Hannam C, MacKenzie K, Jobin-Robitaille O, Guzzo J, Cote J, Andrews B, Haniford DB, Brandl CJ (2007) Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1. Genetics 177: 151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70: 475–501 [DOI] [PubMed] [Google Scholar]
- Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF (2005) RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol 25: 2331–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG (2005) Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Hahn S (2005) Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol 25: 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S (2009) Insights into SAGA function during gene expression. EMBO Rep 10: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR III, Lees-Miller SP, Cole MD, Brandl CJ (1998) Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J Biol Chem 273: 26559–26565 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan NJ, Shevchenko A, Stewart AF (2008) Chromatin central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol 9: R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y (1998) The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell 2: 869–875 [DOI] [PubMed] [Google Scholar]
- Winston F, Durbin KJ, Fink GR (1984) The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39: 675–682 [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Wu PY, Winston F (2002) Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol 22: 5367–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Poon D, Weil PA, Horikoshi M (1997) Molecular genetic elucidation of the tripartite structure of the Schizosaccharomyces pombe 72 kDa TFIID subunit which contains a WD40 structural motif. Genes Cells 2: 245–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.