Abstract
The bacterial flagellum is one of nature's most amazing and well-studied nanomachines. Its cell-wall-anchored motor uses chemical energy to rotate a microns-long filament and propel the bacterium towards nutrients and away from toxins. While much is known about flagellar motors from certain model organisms, their diversity across the bacterial kingdom is less well characterized, allowing the occasional misrepresentation of the motor as an invariant, ideal machine. Here, we present an electron cryotomographical survey of flagellar motor architectures throughout the Bacteria. While a conserved structural core was observed in all 11 bacteria imaged, surprisingly novel and divergent structures as well as different symmetries were observed surrounding the core. Correlating the motor structures with the presence and absence of particular motor genes in each organism suggested the locations of five proteins involved in the export apparatus including FliI, whose position below the C-ring was confirmed by imaging a deletion strain. The combination of conserved and specially-adapted structures seen here sheds light on how this complex protein nanomachine has evolved to meet the needs of different species.
Keywords: bacterial flagellar motor, electron cryotomography, motility, phylogenetic profiling, subtomogram average
Introduction
The bacterial flagellum is a paradigm in modern molecular biology. Its structural complexity, multi-phasic and strictly regulated self-assembly, mechanical capabilities, functional interdependence with the chemosensory system, and both pathogenic and ecophysiological importance have made it central to studies and discussions in both the scientific and popular literature (Berg, 2003; McCarter, 2006; Pallen and Matzke, 2006; Chevance and Hughes, 2008; Minamino et al, 2008; Sowa and Berry, 2008; Snyder et al, 2009). The flagellum consists of a motor also known as the basal body, a flexible linker termed the hook, and a filament that behaves as a helical propeller and is typically many times the length of the bacterium itself. The motor converts ion flux across the cytoplasmic membrane into a torque that rotates the flagellum. In some organisms such as Escherichia coli, counter-clockwise rotation generates thrust that propels the cell forward. Signals from chemoreceptor arrays (Briegel et al, 2009) modulate the probability of the flagellar motor reversing direction (to spin clockwise) (Hazelbauer et al, 2008), which causes the bacterium to randomly re-orient. Thus, if a bacterium detects that it is swimming towards nutrients, the chemosensory system can prolong its movement in a specific direction.
Although all bacterial flagella share similarities in structure and mechanism, extensive variation in number, placement and usage exist between species. While cells such as Vibrio cholerae and Caulobacter crescentus, for example, exhibit a single polar flagellum, others, including Salmonella enterica and E. coli distribute several or many flagella around their periphery. Under different conditions, some bacteria alternate between polar and lateral flagellar systems (McCarter, 2004). The flagella of spirochaetes do not typically pierce the outer membrane, but instead remain in the periplasm where they rotate and/or contort the cell (Murphy et al, 2006; Liu et al, 2009; Kudryashev et al, 2010). Flagellar rotation is powered either by a proton-motive force (e.g., E. coli) or by a sodium-motive force (e.g., V. cholerae). The ‘run and tumble’ swimming mode that switches between clockwise and counter-clockwise rotations is best known, but other bacteria function differently, for example by varying the speed of a unidirectional motor. The molecular architectures of the helical flagellar filaments are also diverse (Galkin et al, 2008).
The flagellar motor is constructed from >20 proteins, and its basic morphology consists of an extended axial rod with coaxial rings termed the L-, P-, S-, M- and C-rings based on their locations relative to the cell. Previous analyses of flagellar motor genes have shown that they are widely distributed throughout and conserved across many bacterial lines of descent, but it has not been clear how observed sequence variations relate to the final structure of the macromolecular complex (Pallen et al, 2005; Liu and Ochman, 2007; Snyder et al, 2009). Structural studies of the motor to date have focussed on a small number of key organisms and have been hampered by the motor's size, complexity, membrane localization and attachment to the cell envelope. These factors make motors difficult if not impossible to purify intact. X-ray crystallography has nevertheless revealed the structures of some of the isolated motor proteins, and complementary biochemical and genetic analyses have located many of them within the intact motor. Single-particle cryoEM studies have produced nanometre-resolution structures of the purified S. enterica motor, albeit without stators or the export apparatus (Thomas et al, 2006). In just the past few years, the development of electron cryotomography (ECT) has made it possible to image the structures of complete motors within intact cells in 3D to ‘macromolecular’ (several nanometres) resolution (Li and Jensen, 2009; Milne and Subramaniam, 2009). The first in situ structures have been from the thin spirochaetes Treponema primitia (Murphy et al, 2006) and Borrelia burgdorferi (Liu et al, 2009; Kudryashev et al, 2010). Those species demonstrated large peripheral structures not seen in the S. enterica single-particle reconstruction, hinting that the diversity of motor structures might be great.
Results and discussion
Data collected and overall appearance of the motors
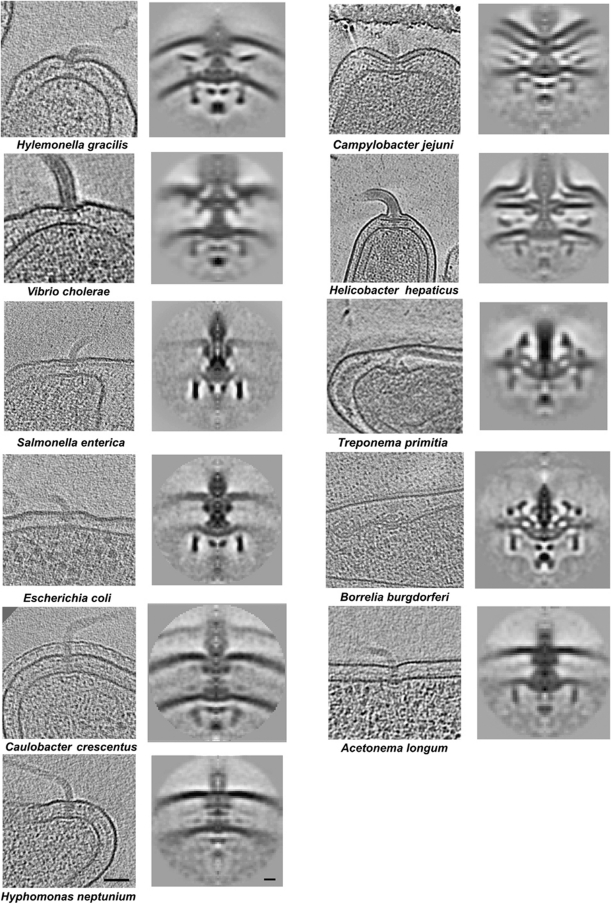
We sought to sample the structural diversity of bacterial flagellar motors. To this end, we imaged the flagellar motors from 11 phylogenetically diverse bacteria chosen for their general interest as model organisms; involvement in animal host associations or free-living lifestyles, and suitability for ECT. The selected bacteria include those with polar (C. crescentus and Campylobacter jejuni), peritrichous (S. enterica and E. coli) and periplasmic (the spirochaetes T. primitia and B. burgdorferi) flagella. Bacteria employing Na-driven motors (V. cholerae) or ‘sheathed’ flagella (V. cholerae and Helicobacter hepaticus) were also included (see Supplementary Table S1 for comprehensive details on the phylogenetics and characteristics of each motor). Using high-throughput imaging methods (Suloway et al, 2009), for each species hundreds of cryotomograms of whole cells were collected. Subtomograms containing motors were computationally extracted from the data, mutually aligned, averaged and cylindrically symmetrized to obtain resolutions of a few nanometres (see Materials and methods).
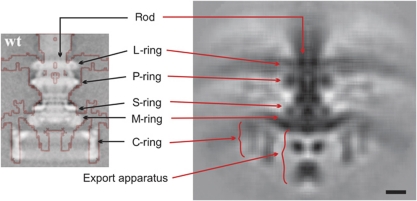
While all the motors had clear similarities, their overall appearances were strikingly diverse (Figure 1). Comparison of the S. enterica structure with a previous single-particle cryoEM reconstruction (Thomas et al, 2006) exhibited similar features at similar positions along the rod and C-ring (Figure 2, left panel), cross-validating the two approaches, but the ECT reconstruction here also showed the position and curvature of the membranes and elements of the export apparatus, albeit at lower resolution. To highlight similarities among the motors, we generated a ‘generic’ motor by aligning and averaging the axial slices of all 11 structures. As shown in Figure 2 (right panel), the core structure of the rod, L-, P-, S-, M- and C-rings, and the export apparatus, as well as their relative locations with regard to the membranes are consistent across all motors. Because the C-rings of different species have different diameters, several C-rings appear in the generic average. This is in contrast with the other parts such as the rod, the LP- and MS-ring complexes and the export apparatus, whose boundaries remain sharp, indicating high structural conservation in their structures across species. Using these conserved features as landmarks, the densities present in the 11 independent motor reconstructions were carefully compared with those in the generic and S. enterica structures and assigned to known structures where possible (Figure 3). To assist in this process and provide insight into the differences, we correlated our imaging results with genomic data for each organism, sequencing and annotating new genomes when necessary. Lists of orthologues of known flagellar proteins were hand-curated (see Supplementary Table S2) and novel, previously unrecognized flagellar motor gene products were sought using their co-occurrence with motor genes in operons.
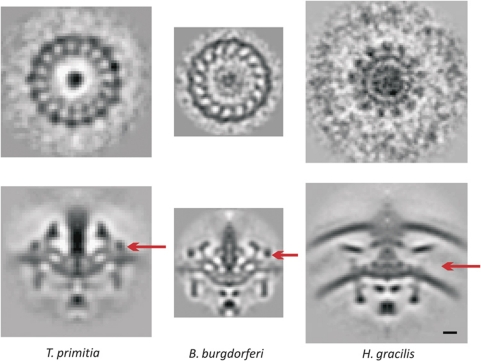
Figure 1.
Flagellar motor structures obtained by ECT and subtomogram averaging. Left column: 20-nm thick central slices through tomograms of individual cells exhibiting flagellar motors, arranged in the same order as they appear on the phylogenetic tree shown in Supplementary Figure S2. Scale bar, 50 nm. Right column: Axial slices through average reconstructions of each motor. Scale bar, 10 nm. (Note that the motor structure of T. primitia was published previously; Murphy et al, 2006.)
Figure 2.
Structure of the common core and its comparison with an earlier cryoEM single-particle reconstruction. Left: Isosurface of the S. enterica motor obtained by ECT and subtomogram averaging (red line) superimposed on an earlier single-particle reconstruction of purified basal bodies from the same organism (grey levels) (Thomas et al, 2006). Right: A generic motor structure obtained by aligning and averaging the axial slices of all 11 motors reconstructed here. Scale bar, 10 nm.
Figure 3.
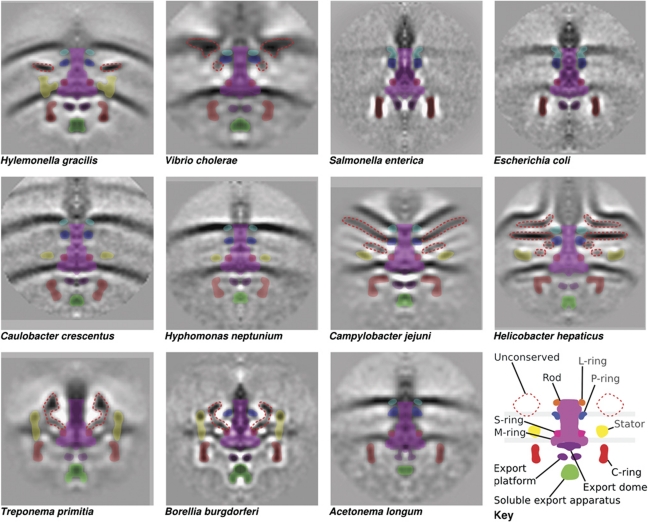
Assignment of densities. Manual segmentation of conserved (solid colours) and unconserved (dotted lines) motor components based on visual inspection. The conserved components from bottom to top are soluble export apparatus (FliH, FliI and FliJ); export platform and dome (FlhA, FlhB, FliO, FliP, FliQ and FliR); C-ring (FliG, FliM and FliN); MS-ring (FliF); stators (MotA and MotB in the H+-dependent stators or PomA and PomB in the Na+-dependent stators); rod (FliE, FlgB, FlgC, FlgF and FlgG); P-ring (FlgI); and L-ring (FlgH).
Export apparatus
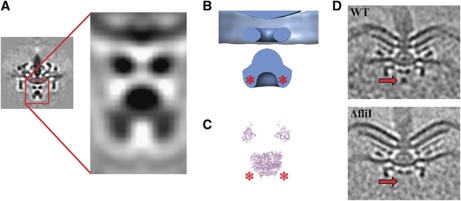
The MS-ring in the inner membrane serves as the starting point for motor assembly. As expected, homologues of the sole component protein of the MS-ring, FliF, are present in all genomes. The periplasmic S-ring is correspondingly clear in all the organisms, but the membrane-embedded M-ring is less distinct due to contrast matching with the membrane. Compared with the single-particle reconstruction of S. enterica, there are three major additional densities present in the ECT reconstructions below the MS-ring: a convex dome bulging into the cytoplasm immediately beneath the MS-ring, a torus 10 nm lower and a spherical density 10 nm below the torus (Figure 4, schematic). Based on their position, presence in the intact cells, and absence in the single-particle reconstruction, we hypothesized that these three additional densities correspond to the dedicated type III secretion system (T3SS) that exports the proteins that form the rod, hook and filament through the hollow inner bore of the assembling flagellum. This flagellar export apparatus typically consists of six transmembrane (FlhA, FlhB, FliO, FliP, FliQ and FliR) and three soluble (FliH, FliI and FliJ) proteins.
Figure 4.
Structure of the export apparatus. (A) Enlarged view of the B. burgdorferi export apparatus and (B) 3D isosurface illustrating from top to bottom the export dome, torus and spherical density. (C) Atomic models of the cytoplasmic domains of FlhA above and the hexameric F1-ATPase, a homologue of FliI, shown at the same scale as (B) to show the correspondence of their sizes to the torus and spherical density. (D) Wild-type (top) and ΔfliI (bottom) C. jejuni motors confirming that the spherical density (red arrows, present above and absent below) is FliI.
The transmembrane proteins must reside at least in part in the dome, as it appears to be the continuation of the cytoplasmic membrane across the motor. The bulging of the dome (most prominent in E. coli, C. jejuni, B. burgdorferi, T. primitia and Acetonema longum) may be necessary to accommodate the transmembrane proteins, as the circular area within the plane of the M-ring in single-particle reconstructions was judged insufficient (Suzuki et al, 2004). Both FlhA and FlhB possess C-terminal cytoplasmic domains at the end of long (∼30 amino acid) linkers (Saijo-Hamano et al, 2004, 2010; Zarivach et al, 2008; Moore and Jia, 2010; Worrall et al, 2010). The torus immediately below the dome, therefore, likely corresponds to their cytoplasmic domains, and the weak density seen between the dome and the torus in some of the reconstructions (most notably S. enterica) to the linkers. Together, the cytoplasmic domains of FlhA and FlhB have been referred to as the ‘export platform’, and may have been at least in part what was previously referred to as the ‘C rod’ in freeze-etch images (Katayama et al, 1996). Structures of homologues of these domains are available (Zarivach et al, 2008; Saijo-Hamano et al, 2010), and match the dimension of the torus reasonably well, though it remains unclear how they pack together or how many are required to complete the ring.
We reasoned that the spherical density below the torus likely corresponded to the ubiquitous ATPase FliI. FliI is thought to oligomerize into a roughly spherical cyclic hexamer ∼9.5 nm in diameter (Claret et al, 2003), as illustrated by models based on the homologous F1-ATPase (Miwa and Yoshida, 1989) using crystal structures of FliI (Imada et al, 2007) or the FliI homologue EscN from the T3SS (Zarivach et al, 2007). This is in good agreement with the ∼10 nm diameter seen for the spherical density in the reconstructions presented here (Imada et al, 2007; Zarivach et al, 2007). To test the hypothesis that this spherical density corresponds to FliI, we recorded cryotomograms of a C. jejuni strain lacking the fliI gene. C. jejuni was chosen for this test because it can be manipulated genetically and exhibits a clear density in this region. Despite the absence of fliI, the deletion strain produced sufficient motors for subtomogram averaging, consistent with a recent study in Salmonella (Paul et al, 2008), demonstrating that assembly can proceed without an absolute requirement for the export apparatus component FliI. The structure of the fliI-deletion strain was essentially identical to the wild type except for the spherical density, which was completely absent, confirming its identity (Figure 4D).
In addition to FliI, two other proteins (FliH and FliJ) are also probably part of the spherical density. FliH, a FliI regulator encoded in the genomes of all organisms but Hyphomonas neptunium, is known to bind FliI at its extreme N-terminus. If the FliI hexamer is oriented towards the membrane in the same way as its homologue, the F1-ATPase, as suggested in a recent study (Ibuki et al, 2011), the N-terminus of each FliI monomer will point towards the cytoplasm. The cytoplasmic-facing protuberances on the spherical density are, therefore, likely FliH (asterisks in Figure 4B). The poorly conserved FliJ has been shown to occupy the central pore of the FliI hexamer, and is structurally similar to the γ-subunit of the F1F0-ATPase (Ibuki et al, 2011). Although fliJ could not be detected in some organisms, its absence may be due to a difficulty in identifying the poorly conserved sequence.
The assignment of the torus as the cytoplasmic domains of FlhA and FlhB and the spherical density as the FliHIJ complex rationalize why in a recent tomographic study of detergent-treated B. burgdorferi cells, the torus but not the spherical density was visible (because the torus is covalently linked to the dome) (Liu et al, 2009). These assignments also explain why the spherical density is faint in the reconstructions of S. enterica and E. coli presented here: S. enterica and E. coli were included in the present survey because they are key model systems, but because both of them are too thick for high resolution ECT, these cells were gently lysed just before freezing (see Materials and methods), which would likely disrupt the localization of soluble proteins like FliI. The shape and prominence of the FliHIJ density in the average reconstructions may also be affected by the facts that FliI occurs as both a monomer and a hexamer and is also known to interact with the relatively distant C-ring, and may not therefore occupy a consistent location.
C-ring
Assembling around FliF (the MS-ring) and the export apparatus is the C-ring, which is involved in export, torque generation and directional switching. Accordingly, the C-ring is present in all 11 organisms and three C-ring structural genes, fliG, fliM and fliN, are conserved across all genomes. Considerable variation is evident, however, in the appearance and diameter of the C-rings, which range from 34 nm in C. crescentus to 57 nm in T. primitia (Figures 1 and 3). Consistent with the cryoEM single-particle reconstruction of the S. enterica motor (Thomas et al, 2006), all the three γ-proteobacteria V. cholerae, S. enterica and E. coli as well as the thin β-proteobacterium Hylemonella gracilis have similar cross-sections and diameters of ∼40 nm. The C-rings from H. neptunium and C. crescentus (both α-proteobacteria) are less clear (suggesting incomplete occupancy or mobility) and exhibit a distinctive conical reduction in diameters from, for example in C. crescentus, 36 nm near the inner membrane to 26 nm at the tip near the FliHIJ complex. The average diameter of the C-rings in the pathogenic ε-proteobacteria C. jejuni and H. hepaticus is 49 nm, and these display stronger densities at the membrane-proximal side of the C-ring next to the export platform immediately beneath the MS-ring. It is known that the membrane-proximal part of the C-ring is composed of FliG (Thomas et al, 2006; Liu et al, 2009); and therefore, these stronger densities may represent alternative, more stable, or higher occupancy conformations of FliG, or additional components of the motor exclusive to the ε-proteobacteria. The spirochaetes B. burgdorferi and T. primitia and the diderm firmicute A. longum possess C-rings with an even larger average diameter of 54 nm, but maintain the same cross-section as S. enterica. No correlation could be made between tomographic density and the presence of FliY, a FliN paralogue with an additional domain found in A. longum, T. primitia, H. hepaticus, C. jejuni and S. enterica.
Rod and L/P-rings
Assembling atop the S-ring is the rod, which acts as the central driveshaft through the cell envelope, transmitting torque applied to the MS-ring to the hook and filament. The rod is comprised of ubiquitous paralogous proteins that are known to form a proximal (FliE, FlgB, FlgC and FlgF) and a distal (FlgG) rod, but no new details about the arrangement of these proteins could be gleaned from the tomograms. The length of the rod was well conserved, as judged by the distinct densities seen in the generic average for the S- and P-rings and inner and outer membranes. The distance between the distal end of the S-ring and the centre of the outer membrane was 22 nm (measured in the generic average), confirming earlier measurements of the rod length on isolated flagellar hook-basal bodies (HBBs) from wild-type S. enterica (Takahashi et al, 2009).
The periplasmic P- and L-rings around the rod are thought to function as bushings through the peptidoglycan layer and lipopolysaccharide of the outer membrane, respectively. Because the rings coat the outside of the rod, both proteins (FlgH and FlgI) are exported into the periplasm via the Sec pathway instead of through the dedicated flagellar T3SS (Homma et al, 1987; Jones and Macnab, 1990). Unlike the rod proteins, however, it has previously been noted that the genes encoding the L/P-ring components are not ubiquitous, and the variation seen here calls into question the presumed roles of these proteins (Pallen et al, 2005). The reconstructions of S. enterica, E. coli, H. neptunium, C. crescentus and A. longum represent the ‘standard’ cases, where both the L- and P-ring genes are present, the P-ring is visible in the tomograms, and the L-ring is not, likely because it is embedded in the outer membrane. In H. gracilis, both genes are present, and two rings are visible underneath the outer membrane, suggesting that the L-ring may not be embedded in the membrane in this species, although it is not clear why this alternative arrangement is seen. In the sheathed flagella of H. hepaticus and V. cholerae, the outer membrane continues as a sheath around the flagellar hook and filament. The L-ring proteins in these two organisms presumably still form a complex with the P-ring, but the diverse structures in that region make assignments unclear. It is noteworthy that the H. hepaticus L-ring gene does not encode an otherwise ubiquitous cysteine residue in the vicinity of the amino terminus (Schoenhals and Macnab, 1996). This residue has been shown to be part of a signal peptide II cleavage consensus motif that is subsequently lipoylated. The situation is further complicated by the fact that this cysteine residue is nevertheless retained in the other organism with a sheathed flagellum, V. cholerae. In the spirochaetes B. burgdorferi and T. primitia, the L-ring protein FlgH is absent, as is thought due to the fact that the flagellar filaments in these organisms never cross the outer membrane (Pallen et al, 2005), but remain within the periplasm. The P-ring protein FlgI is absent only in the genome of T. primitia. This was first observed by tomography in a recent study of a B. burgforferi FlgI mutant, in which a density around the rod was absent (Liu et al, 2009). Here, we observe a similar absence of ring density on the rod of T. primitia, corresponding to FlgI. It is noteworthy that FlgI in the spirochaetes is distant from the peptidoglycan layer, at odds with its supposed function. FlgI is also found in the Leptospiraceae, another family within the spirochaete class, so it may have a special alternative function in the spirochaetes (Chevance et al, 2007).
Stator complex
Above the C-ring and surrounding the MS-ring in the inner membrane are stator complexes that are thought to extend towards and bind to the peptidoglycan layer. Through interactions with the C-ring proteins, the stators transform the flow of H+ or Na+ ions across the membrane into torque to drive the motor. Each stator complex is composed of two proteins, MotA and MotB in the H+-dependent stators (e.g., E. coli), or PomA and PomB in the Na+-dependent stators (e.g., V. cholerae), with an apparent 4:2 stoichiometry. In our averaged structures, we see distinctive rod-shaped stator ring structures above the inner membrane with connections on the other side of the membrane to the C-ring in the β-proteobacterium H. gracilis and the spirochaetes B. burgdorferi and T. primitia. As previously shown elsewhere (Murphy et al, 2006; Liu et al, 2009; Kudryashev et al, 2010) and reproduced in Figure 5, 16 copies of the stator complexes are clearly seen in the averaged tomograms of the spirochaetes B. burgdorferi and T. primitia even before rotational averaging. In contrast, the symmetry observed in the H. gracilis stator ring is 13 (Figure 5). In the α-proteobacteria (H. neptunium and C. crescentus), stator densities are visible, but they are less prominent and their symmetries could not be discerned. In the ε-proteobacteria C. jejuni and H. hepaticus, stator densities are clear, but they have a different shape in cross-section, lie nearly parallel to the cytoplasmic membrane, and extend further into the periplasm. The reason that the symmetry of these motors could not be discerned may be that the stator system is highly dynamic with rapid turnover of subunits (Leake et al, 2006) and perhaps variable conformations, causing low occupancy and heterogeneity. These cells also have single, polar flagella, and so because the rod-shaped cells freeze lying flat on the grid, no direct ‘top’ views of the motor were obtained, which makes detecting rotational symmetries more difficult. The rest of the species may not exhibit clear stator density because the periplasmic domains of the stator complex (MotA/B or PomA/B) are thin, as shown in cryoEM images of purified PomA/B in liposomes (Yonekura et al, 2006).
Figure 5.
Symmetries in the stator region. Upper row: Radial slices through the stator regions of three average motors before rotational averaging showing 16-, 16- and 13-fold symmetry, respectively. Lower row: Axial slices through the averaged and symmetrized motors with arrows showing the height at which the radial slices were taken. Scale bar, 10 nm.
Unconserved densities
In addition to this somewhat conserved structural core, many of the organisms exhibited additional, presumably proteinaceous densities that are likely to be unrecognized, novel components of the motor. The most striking is the spirochaetes' large ‘P-collar’ that forms a bowl-like structure around the rod within the stator ring, and which has been postulated to stabilize the motor (Murphy et al, 2006). To generate hypotheses about which proteins form the P-collar, we exploited the fact that genes encoding proteins involved in stable complexes are frequently encoded in genomic proximity to one-another (Huynen et al, 2000). Furthermore, as the protein is not a component of the cylindrical core of the rod, hook or filament, we assumed its secretion would be via the Sec pathway as opposed to the dedicated flagellar T3SS. Using these criteria (see Materials and methods), we narrowed our search to a candidate gene for involvement in the P-collar, flbB, first identified in a comprehensive study of B. burgdorferi flagellar genetics (Ge et al, 1997). This gene is consistently near other flagellar motor genes in spirochaetes and includes the appropriate periplasmic export signal.
While the L/P-ring complex simply surrounds the rod as it passes through the lipopolysaccharide membrane and peptidoglycan layer in the well-studied flagellar motors, in V. cholerae it appears to serve as a foundation for two additional structures. V. cholerae possesses two proteins, MotX and MotY, which form a so-called T-ring that is thought to assist in the formation and binding of the MotA/MotB stators (Terashima et al, 2006). Here, this T-ring can be seen lower and to the outside of the P-ring, to which it is connected. There is also a substantial increase in density near the presumed position of the L-ring. Hosogi et al (2011) recently described this entire density in the closely related Vibrio alginolyticus HBB as the L-ring. Based on the fact that the L-ring-encoding flgH gene is ∼360 amino acids long in both S. enterica and V. cholerae, we propose that the unusually large and oddly shaped ring structure must also include an additional, as-yet unidentified protein.
The L/P-ring complex also appears to serve as a structural anchor in the ε-proteobacteria C. jejuni and H. hepaticus. Here, the L/P-ring complex is attached to large periplasmic basal disks. In C. jejuni, an extensive disk-like density is found in the periplasm immediately below and parallel to the outer membrane around the rod and above the stators. This disk connects to the P-ring and has an outer radius of 48±9 nm, calculated from the edge of the rod along the disk. A second disk with a radius of 32±7 nm sits beneath the first and seems to be faintly connected to the M/S-ring. In H. hepaticus, a similarly extensive (32±2 nm in radius) but flat disk is found between what are probably enhanced L- and P-rings. A similarly large disk (85 nm average radius) connected to the L/P-ring complex was observed previously by EM of negatively stained flagellar motors purified from another ε-proteobacterium, Wolinella succinogenes, which is closely related to C. jejuni and H. hepaticus (Engelhardt et al, 1993). While all three of these large basal disks are likely composed of homologous proteins, bioinformatic analyses yielded no candidate proteins for this structure, likely unaided by the well-discussed genomic fragmentation and lack of synteny within the ε-proteobacteria (Eppinger et al, 2004).
It is interesting to note that the β-proteobacterium H. gracilis also exhibited a periplasmic disk above the stators, but the relationship of this disk to the larger ε-proteobacterial basal disks is unclear. The H. gracilis disk is at the same height as the P-ring but does not connect, exhibiting an inner and outer radius of 18±2 and 31±2 nm, respectively. Previously, similar disks have been observed in another β-proteobacterium, Aquaspirillum serpens (Coulton and Murray, 1978). The function of these disks remains unclear, as their occurrence does not seem to correlate with any obvious phenotype (such as sheathed flagella) or habitat (such as viscous environment), besides that they are so far exclusive to (but not required for) polar flagella. Based on their position, they may stabilize the stators to sustain larger torques.
Above the basal disk and immediately below the highly curved bend in the outer membrane of the sheathed H. hepaticus is yet another disk-like density. As mentioned above, in the other sheathed bacterium, V. cholerae, the L-ring is unusually large and odd-shaped. These structures may stabilize the outer membrane and the sheath against the rotation of the flagellum. Finally, two previous studies of negatively stained, purified C. crescentus motors gave conflicting reports about whether or not a so-called E-ring existed between the M/S- and L/P-ring complexes (Stallmeyer et al, 1989; Kanbe et al, 2005). It remains unclear whether there are additional rings here in the C. crescentus and H. neptunium reconstructions.
Conclusion
In summary, three-dimensional structures of 11 phylogenetically diverse bacterial flagellar motors were obtained in situ at a few nanometre resolutions via ECT and subtomogram averaging. The positions of key proteins of the export apparatus were determined. While the motors all exhibited a common core built from the products of conserved flagellar genes, their overall appearances were strikingly different due to variations in the structure of that core and the presence of unique peripheral densities, showing that each is a related but specially adapted nanomachine.
Materials and methods
Culture conditions
C. crescentus CB15N, E. coli RP437 and MG1655, V. cholerae TRH7000, H. hepaticus (ATCC 51449), C. jejuni (ATCC 29428), B. burgdorferi B31 (ATCC 35210), A. longum APO-1 DSM 6540 and T. primitia strain ZAS-2 cells were grown as described previously (Murphy et al, 2006; Briegel et al, 2009).
H. neptunium (ATCC 15444) was grown in 2216 marine broth (Difco) at 30°C for 24–48 h. The cell culture was incubated on ice during plunge freezing.
S. enterica subsp. enterica serovar Typhimurium str. LT2 was grown in LB plus 0.3 M sucrose and up to 10 mM MgSO4. Penicillin (466 IU/ml) was added 15 and 60 min before plunge freezing.
H. gracilis was cultivated from a rotten lily taken from a Caltech pond by placing a drop of liquid upon a 0.22-μm filter resting atop agar containing 10 mM MOPS pH 7.0, 0.5 g tryptone and 0.5 g yeast extract per litre. Colonies appeared several weeks later. The 16S rRNA was sequenced from a liquid culture and found to be identical to the type strain of H. gracilis. Cultures were grown in the above media without MOPS for 2 days and only reached an OD of 0.05. The cells were checked for motility with a light microscope. The cells were then centrifuged and concentrated 10-fold in the same media for plunge freezing onto the EM grids (Murphy, 2007).
Genome sequencing
H. gracilis (ATCC 19624) cultures were obtained from the ATCC and cultured in liquid and agar media containing 5 g peptone, 0.5 g yeast extract, 0.02 g Tween-80 and 0.1 g K2HPO4 made up to 1 l with nanopure water with optional addition of 15 g agar. Colonies were initially grown on agar plates and subsequently used to inoculate liquid cultures. The identity of the cultures was confirmed by PCR amplification and sequencing of 16S rRNA. DNA was purified from 2 l cultures by lysing cells with lysozyme followed by consecutive RnaseA, proteinase K, SDS, sodium acetate and chloroform/phenol mixture additions. DNA was washed with ethanol and dried. A paired-end genomic DNA library was prepared from this sample and 76 bp reads generated using Illumina sequencing at Caltech's Millard & Muriel Jacobs Genetics & Genomics Laboratory (http://mmjggl.caltech.edu/sequencing/). Reads were truncated to 67 bp and assembled using Velvet (Zerbino and Birney, 2008). Using a kmer size of 49, 3.6 Mb of sequence data was generated and distributed over 152 contigs with an n50 of 75 kb and a maximum contig length of 192 kb. This partially assembled genome was annotated for ORFs using the NCBI PGAAP service and flagellar genes semi-automatically annotated using reciprocal best-hit methodology and manual curation. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AEGR00000000. The version described in this paper is the first version, AEGR01000000.
The genome sequencing of A. longum APO-1 was performed at Stanford in the laboratory facility of Stephen Quake and will be described elsewhere. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AFGF00000000. The version described in this paper is the first version, AFGF01000000. Genome sequencing results for T. primitia ZAS-2 will be described elsewhere; the closed sequence for this strain has been deposited into GenBank as CP001843.
EM sample preparation, data collection and tomogram reconstruction
EM R2/2 copper/rhodium Quantifoil or lacy carbon grids were glow discharged and coated with a 3 × -concentrated solution of 10 nm colloidal gold particles (Ted Pella). A 5 × -concentrated solution of 10 nm colloidal gold was also added to the cells immediately before plunge freezing. A 4-μl droplet of the sample solution was applied to the EM grid, then blotted and plunge frozen into liquid ethane (Dubochet et al, 1988) or into a liquid ethane–propane mixture (Tivol et al, 2008) using a Vitrobot (FEI Company) (Iancu et al, 2006) or in-house plunger. To flatten the thickest cell types, E. coli and S. enterica cells were incubated with 466 IU/ml penicillin G for up to 60 min at 30°C (Eisenbach and Adler, 1981), which caused some cells to rupture when blotted just before plunge freezing. The grids were stored under liquid nitrogen until data collection.
EM images were collected using a Polara 300-kV FEG transmission electron microscope (FEI Company) equipped with an energy filter (slit width 20 eV; Gatan) on a 2k × 2k Ultrascan CCD camera or, later, a lens-coupled 4k × 4k UltraCam (Gatan). Typically, tilt series were recorded from −60° to 60° with an increment of 1° semi-automatically around 1 or 2 axes (Iancu et al, 2005) at 8–12 μm under-focus using the predictive UCSF-Tomo package (Zheng et al, 2007) or Leginon (Suloway et al, 2009). Cumulative doses of up to 200 e−/Å2 were used.
Image tilt series were generally binned by two (in X and Y) and 3D tomograms were calculated automatically by Raptor (Amat et al, 2008) using the Peach distributed computing system (Leong et al, 2005). In cases where Raptor was not available or failed, the semi-automatic IMOD software package (Mastronarde, 1997) was used. No digital filters were used to reduce noise. Image tilt series and final reconstructions were deposited into an in-house web-based database. For a more detailed protocol, see Chen et al (2010).
Subtomogram extraction, alignment, averaging and symmetrization
Bsoft (Heymann et al, 2008) and the Peach (Leong et al, 2005) distributed computing system were used for iterative subtomogram extraction, alignment, averaging and symmetrization. For V. cholerae, H. neptunium and A. longum, tomograms of different magnifications were used, and the tomograms of higher magnification were re-sampled to match the pixel size of the tomograms of lowest magnification. The positions of flagellar motors in the tomograms were marked by eye at the entry point of the flagellar filament into the outer membrane. For alignment, subtomograms centred on the motors were extracted, bandpass filtered between 200 nm and the first CTF zero and masked in reciprocal space within the missing wedge. Rotational alignments were done with 3° step sizes, as no further improvement was seen with finer steps. Subtomograms containing flagellar motors were first extracted by hand and aligned to a reference arbitrarily chosen from the data sets. The aligned motors were then averaged, and the average was rotated so that the rod axis coincided with the z-axis. This result was cylindrically symmetrized around the z-axis. Subtomograms were computationally re-extracted based on the previous alignment result, realigned to the symmetrized average motor, re-averaged and symmetrized. The whole process was iterated until the coordinates of motors stabilized. Average motors were checked computationally for symmetry about the rod before applying any symmetry. In the cases of the spirochaetes and H. gracilis, where symmetry was detected in the stator rings, the average reconstruction was symmetrized accordingly (16- and 13-fold, respectively). In all other cases, 16-fold symmetry was applied simply to improve the signal-to-noise ratio of the final motor structure. Note the cylindrical symmetrization does not affect the axial structures obtained as shown in Figure 1. All the symmetrized average motor structures have been deposited into the EMDataBank.
Tomograms were inspected and measured with IMOD (Mastronarde, 1997). 2D image thresholding and superposition was done with Adobe Photoshop (Adobe Systems Inc.). The resolution was estimated by separately averaging and symmetrizing two halves of the data set and correlating them using Fourier shell correlation with a threshold of 0.5 (Supplementary Figure S1).
Identification of FliI location
Starting with wild-type Campylobacter jejuni 81-176 SmR (DRH212) containing an rpsL allele conferring streptomycin resistance (Hendrixson et al, 2001), strain DRH2257 was constructed through in-frame deletions of fliI and marker gene astA (the enzyme arylsulphatase), which does not affect flagellar biosynthesis or motility (Joslin and Hendrixson, 2009). DRH212 and DRH2257 were grown on MH agar under microaerophilic conditions for 3 days, re-streaked and grown overnight. Colonies were resuspended in MH broth, plunge frozen and imaged as described above except that images were collected at −15 μm defocus. Subtomographic averages were generated from 21 wild-type and 19 ΔfliI motors, respectively, using PEET from the IMOD suite of programs without symmetrization (Mastronarde, 1997).
Bioinformatics analysis
To define orthologous gene families, flagellar structural protein genes were initially grouped according to the KEGG Orthology (KO) Database (Kanehisa et al, 2010). Family assignments were verified and/or revised by extensive literature searches and manual BLASTing to confirm the automated KO assignments. For those genomes not covered by the KO Database, genes were assigned to orthologous gene families by BLAST reciprocal best-hit methods against the previously curated family allocations. Non-flagellar gene family assignments were made automatically using the KO Database without further manual curation.
To predict functional linkages between known flagellar genes and novel genes, the mutual information score was calculated between operon occurrence vectors of each orthologous gene family using a previously described operon prediction method (Overbeek et al, 1999). An operon occurrence vector describes in which operons a given orthologous gene family resides: that is, for each orthologous family, a binary vector is assigned of dimension equal to the number of operons in the combined sequenced genomes used for this analysis. Thus, each gene family's operon occurrence vector will be predominantly ‘0’, with occasionally a ‘1’ for those operons in which that family occurs. For each defined novel substructure observed by tomography (e.g., the P-collar), the subset of organisms in which it is found were noted, and the mutual information between all gene families exclusive to that subset was calculated. Orthologous families predicted to be functionally linked to known flagellar genes by a high mutual information score were further filtered for periplasmic export sequences using SignalP (Emanuelsson et al, 2007).
Supplementary Material
Acknowledgments
We thank Drs Jane H Ding and Alasdair McDowall for computational and EM support, respectively; Sarkis K Mazmanian for use of the microaerobic chamber; and Igor Antoshechkin for advice on genome assembly. This work was supported by the Howard Hughes Medical Institute, the Gordon and Betty Moore Foundation Cell Center and the National Science Foundation.
Author contributions: GJJ designed the research; SC, MB, GEM, AB, ZL, JS, EIT, AM, JRL and MJD cultured cells and/or collected tomograms; SC averaged subtomograms with help from GEM; JRL and MB coordinated the sequencing and annotation of needed genomes, MB performed bioinformatics, and together with DRH identified export densities, including subtomogram averaging of the comparative wild-type and mutant C. jejuni motors; SC, MB and GJJ analysed the results and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M (2008) Markov random field based automatic image alignment for electron tomography. J Struct Biol 161: 260–275 [DOI] [PubMed] [Google Scholar]
- Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54 [DOI] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen SY, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ (2009) Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA 106: 17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, McDowall A, Dobro MJ, Briegel A, Ladinsky M, Shi J, Tocheva EI, Beeby M, Pilhofer M, Ding HJ, Li Z, Gan L, Morris DM, Jensen GJ (2010) Electron cryotomography of bacterial cells. J Vis Exp 39; doi:10.3791/1943, http://www.jove.com/details.stp?id=1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FFV, Hughes KT (2008) Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FFV, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa SI, Hughes KT (2007) The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21: 2326–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret L, Calder SR, Higgins M, Hughes C (2003) Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol 48: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulton JW, Murray RGE (1978) Cell envelope associations of Aquaspirillum Serpens flagella. J Bacteriol 136: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P (1988) Cryo-Electron Microscopy of vitrified specimens. Q Rev Biophys 21: 129–228 [DOI] [PubMed] [Google Scholar]
- Eisenbach M, Adler J (1981) Bacterial-cell envelopes with functional flagella. J Biol Chem 256: 8807–8814 [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Engelhardt H, Schuster SC, Baeuerlein E (1993) An archimedian spiral—the basal disk of the Wolinella Flagellar Motor. Science 262: 1046–1048 [DOI] [PubMed] [Google Scholar]
- Eppinger M, Baar C, Raddatz G, Huson DH, Schuster SC (2004) Comparative analysis of four Campylobacterales. Nat Rev Microbiol 2: 872–885 [DOI] [PubMed] [Google Scholar]
- Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH (2008) Divergence of quaternary structures among bacterial flagellar filaments. Science 320: 382–385 [DOI] [PubMed] [Google Scholar]
- Ge Y, Old IG, Saint Girons I, Charon NW (1997) Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J Bacteriol 179: 2289–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS (2008) Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, Akerley BJ, DiRita VJ (2001) Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol 40: 214–224 [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cardone G, Winkler DC, Steven AC (2008) Computational resources for cryo-electron tomography in Bsoft. J Struct Biol 161: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Komeda Y, Iino T, Macnab RM (1987) The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol 169: 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogi N, Shigematsu H, Terashima H, Homma M, Nagayama K (2011) Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J Struct Biol 173: 67–76 [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res 10: 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, Gan L, He Y, Jensen GJ (2006) Electron cryotomography sample preparation using the Vitrobot. Nat Protoc 1: 2813–2819 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Wright ER, Benjamin J, Tivol WF, Dias DP, Murphy GE, Morrison RC, Heymann JB, Jensen GJ (2005) A ‘flip-flop’ rotation stage for routine dual-axis electron cryotomography. J Struct Biol 151: 288–297 [DOI] [PubMed] [Google Scholar]
- Ibuki T, Imada K, Minamino T, Kato T, Miyata T, Namba K (2011) Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol 18: 277–282 [DOI] [PubMed] [Google Scholar]
- Imada K, Minamino T, Tahara A, Namba K (2007) Structural similarity between the flagellar type III ATPase Flil and F-1-ATPase subunits. Proc Natl Acad Sci USA 104: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Macnab RM (1990) Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol 172: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin SN, Hendrixson DR (2009) Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191: 2656–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe M, Shibata S, Umino Y, Jenal U, Aizawa SI (2005) Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151: 433–438 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S (1996) Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol 255: 458–475 [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F (2010) Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169: 54–61 [DOI] [PubMed] [Google Scholar]
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP (2006) Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443: 355–358 [DOI] [PubMed] [Google Scholar]
- Leong PA, Heymann JB, Jensen GJ (2005) Peach: a simple Perl-based system for distributed computation and its application to cryo-EM data processing—Ways & means. Structure 13: 505–511 [DOI] [PubMed] [Google Scholar]
- Li Z, Jensen GJ (2009) Electron cryotomography: a new view into microbial ultrastructure. Curr Opin Microbiol 12: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ (2009) Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191: 5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RY, Ochman H (2007) Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci USA 104: 7116–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (1997) Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol 120: 343–352 [DOI] [PubMed] [Google Scholar]
- McCarter LL (2004) Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7: 18–29 [DOI] [PubMed] [Google Scholar]
- McCarter LL (2006) Regulation of flagella. Curr Opin Microbiol 9: 180–186 [DOI] [PubMed] [Google Scholar]
- Milne JLS, Subramaniam S (2009) Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat Rev Microbiol 7: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K (2008) Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18: 693–701 [DOI] [PubMed] [Google Scholar]
- Miwa K, Yoshida M (1989) The alpha 3 beta 3 complex, the catalytic core of F1-ATPase. Proc Natl Acad Sci USA 86: 6484–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SA, Jia YH (2010) Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J Biol Chem 285: 21060–21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE (2007) Cryoelectron Tomography of Bacteria and Their Macromolecular Machines. Pasadena, USA: California Institute of Technology [Google Scholar]
- Murphy GE, Leadbetter JR, Jensen GJ (2006) In situ structure of the complete Treponema primitia flagellar motor. Nature 442: 1062–1064 [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N (1999) The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA 96: 2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Matzke NJ (2006) From the origin of species to the origin of bacterial flagella. Nat Rev Microbiol 4: 784–790 [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Penn CW, Chaudhuri RR (2005) Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13: 143–149 [DOI] [PubMed] [Google Scholar]
- Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT (2008) Energy source of flagellar type III secretion. Nature 451: 489–492 [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Imada K, Minamino T, Kihara M, Shimada M, Kitao A, Namba K (2010) Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol Microbiol 76: 260–268 [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Minamino T, Macnab RM, Namba K (2004) Structural and functional analysis of the C-terminal cytoplasmic domain of FIhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J Mol Biol 343: 457–466 [DOI] [PubMed] [Google Scholar]
- Schoenhals GJ, Macnab RM (1996) Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J Bacteriol 178: 4200–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LAS, Lornan NJ, Futterer K, Pallen MJ (2009) Bacterial flagellar diversity and evolution: seek simplicity and distrust it? Trends Microbiol 17: 1–5 [DOI] [PubMed] [Google Scholar]
- Sowa Y, Berry RM (2008) Bacterial flagellar motor. Q Rev Biophys 41: 103–132 [DOI] [PubMed] [Google Scholar]
- Stallmeyer MJB, Hahnenberger KM, Sosinsky GE, Shapiro L, Derosier DJ (1989) Image reconstruction of the flagellar basal body of Caulobacter crescentus. J Mol Biol 205: 511–518 [DOI] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ (2009) Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 167: 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Yonekura K, Namba K (2004) Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol 337: 105–113 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Mizuno S, Hirano T, Chevance FFV, Hughes KT, Aizawa SI (2009) Autonomous and FliK-dependent length control of the flagellar rod in Salmonella enterica. J Bacteriol 191: 6469–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M (2006) The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol Microbiol 62: 1170–1180 [DOI] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ (2006) The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188: 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ (2008) An improved cryogen for plunge freezing. Microsc Microanal 14: 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall LJ, Vuckovic M, Strynadka NCJ (2010) Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci 19: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Yakushi T, Atsumi T, Maki-Yonekura S, Homma M, Namba K (2006) Electron cryomicroscopic visualization of PomA/B stator units of the sodium-driven flagellar motor in liposomes. J Mol Biol 357: 73–81 [DOI] [PubMed] [Google Scholar]
- Zarivach R, Deng WY, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NCJ (2008) Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453: 124–127 [DOI] [PubMed] [Google Scholar]
- Zarivach R, Vuckovic M, Deng WY, Finlay BB, Strynadka NCJ (2007) Structural analysis of a prototypical ATPase from the type III secretion system. Nat Struct Mol Biol 14: 131–137 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA (2007) UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157: 138–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.