Abstract
Mammalian centromeric cohesin is protected from phosphorylation-dependent displacement in mitotic prophase by shugoshin-1 (Sgo1), while shugoshin-2 (Sgo2) protects cohesin from separase-dependent cleavage in meiosis I. In higher eukaryotes, progression and faithful execution of both mitosis and meiosis are controlled by the spindle assembly checkpoint, which delays anaphase onset until chromosomes have achieved proper attachment to microtubules. According to the so-called template model, Mad1–Mad2 complexes at unattached kinetochores instruct conformational change of soluble Mad2, thus catalysing Mad2 binding to its target Cdc20. Here, we show that human Sgo2, but not Sgo1, specifically interacts with Mad2 in a manner that strongly resembles the interactions of Mad2 with Mad1 or Cdc20. Sgo2 contains a Mad1/Cdc20-like Mad2-interaction motif and competes with Mad1 and Cdc20 for binding to Mad2. NMR and biochemical analyses show that shugoshin binding induces similar conformational changes in Mad2 as do Mad1 or Cdc20. Mad2 binding regulates fine-tuning of Sgo2's sub-centromeric localization. Mad2 binding is conserved in the only known Xenopus laevis shugoshin homologue and, compatible with a putative meiotic function, the interaction occurs in oocytes.
Keywords: Mad1, Mad2, MIM, SAC, shugoshin
Introduction
In all eukaryotes, accurate chromosome segregation during both mitosis and meiosis is fundamental for the propagation and inheritance of stable genomes. Errors in this process lead to aneuploidies and have dire consequences such as cell death, cancer, infertility or genomic disorders like Down syndrome. At the time of their replication in S-phase, sister chromatids become paired by a DNA-embracing, ring-shaped protein complex termed cohesin (Uhlmann and Nasmyth, 1998; Gruber et al, 2003; Ivanov and Nasmyth, 2005; Haering et al, 2008). At centromeres, this linkage persists up to anaphase of mitosis or meiosis II. Then, the kleisin subunit of cohesin, Scc1 in mitosis or Rec8 in meiosis, is cleaved by a giant cysteine endopeptidase, separase and sister chromatids are pulled towards opposite ends of the dividing cell (Uhlmann et al, 1999, 2000).
Cohesin on chromosome arms is removed earlier. In higher eukaryotic mitosis, it gets displaced in prophase by a proteolysis-independent process that requires phosphorylation of the cohesin subunit Scc3SA2 (Losada et al, 1998; Waizenegger et al, 2000; Sumara et al, 2002; Hauf et al, 2005). Similarly, segregation of homologous chromosomes is triggered by separase-dependent cleavage of arm cohesin in anaphase of meiosis I and requires phosphorylation of Rec8 (Brar et al, 2006; Kudo et al, 2009; Katis et al, 2010). During these early attacks, centromeric cohesin is protected by shugoshins, which might provide physical shielding but, most importantly, recruit the ubiquitous protein phosphatase 2A (PP2A), thus counteracting phosphorylation of cohesin at centromeres (Kitajima et al, 2004, 2006; Riedel et al, 2006; Tang et al, 2006). In mammals, shugoshin-1 (Sgo1) and shugoshin-2 (Sgo2) protect centromeric cohesin throughout early mitosis and meiosis I, respectively (Tang et al, 2004; McGuinness et al, 2005; Lee et al, 2008; Llano et al, 2008). However, despite this apparent division of labour, mammalian Sgo2 is expressed in the soma, where it has been associated with chromosome alignment, cohesin protection or tension sensing (Kitajima et al, 2006; Gomez et al, 2007; Huang et al, 2007; Tanno et al, 2010).
The anaphase promoting complex or cyclosome (APC/C) is a multisubunit ubiquitin ligase that triggers anaphase by targeting for proteasomal destruction the separase inhibitor securin and the cyclin-dependent kinase 1 regulatory subunit cyclin B1 (Irniger et al, 1995; King et al, 1995; Cohen-Fix et al, 1996; Zou et al, 1999). APC/C activity is regulated by the conserved spindle assembly checkpoint (SAC) that monitors the attachment of spindle microtubules (MTs) to the kinetochores of chromosomes. Kinetochores, which are unattached or do not experience tension due to improper interaction with MTs, emit a ‘wait-anaphase’ signal that arrests cells at metaphase, thereby giving them time to correct the error. To this end, the SAC targets Cdc20, an essential co-activator of the APC/C (Fang et al, 1998; Schott and Hoyt, 1998). The SAC relies on a template-like conformational activation of its key factor Mad2, which is catalysed by a kinetochore-bound complex of Mad1 and Mad2 (De Antoni et al, 2005). Mad2 belongs to the HORMA-domain-containing family of proteins (Aravind and Koonin, 1998) and exhibits two native conformations (Luo et al, 2004) commonly referred to as Mad2open and Mad2closed (De Antoni et al, 2005). Activation of Mad2 depends on a short-lived interaction between Mad2open from a cytosolic pool with Mad2closed stably bound to kinetochore-localized Mad1. This instructs a conformational change in Mad2open facilitating its conversion to Mad2closed, which binds Cdc20 and thereby inhibits ubiquitylation of APC/C targets like securin and cyclin B1 (De Antoni et al, 2005; Vink et al, 2006; Mapelli et al, 2007; Simonetta et al, 2009). Mad1 and Cdc20 bind to identical sites on Mad2, which is why they compete for Mad2-binding in vitro. Yet, Mad1 is essential to load Mad2 onto Cdc20 in vivo. What seems contradictory at first is elegantly explained by the above template model.
Once the Mad2closed–Cdc20 complex has formed, two additional SAC components, BubR1 (Mad3) and Bub3, are recruited to form the heterotetrameric mitotic checkpoint complex or MCC (Sudakin et al, 2001). When all chromosomes achieve proper attachment to the mitotic spindle, the SAC gets inactivated and the MCC is disassembled leading to APC/CCdc20-mediated ubiquitylation of securin and cyclin B1 (Clute and Pines, 1999; Reddy et al, 2007; Stegmeier et al, 2007). Inactivation of the SAC in terms of the Mad1–Mad2 template depends on both dynein-mediated transport of kinetochore-bound Mad1–Mad2 complexes to spindle poles and p31comet, which mimics Mad2open and binds to Mad2closed–Mad1, thus inhibiting further conformational activation of soluble Mad2open (Howell et al, 2001; Xia et al, 2004; Mapelli et al, 2006; Yang et al, 2007). In recent years, it has become clear that the SAC is also crucial for the faithful execution of the meiotic cell division (Wassmann et al, 2003b; Homer et al, 2005; Niault et al, 2007; McGuinness et al, 2009).
Aside its cohesin-protective function, shugoshin has been implicated in the tension-sensitive branch of the SAC (Indjeian et al, 2005). While S. pombe shugoshin likely fulfils this task by localizing the kinase Aurora B to centromeres (Kawashima et al, 2007; Vanoosthuyse et al, 2007), the molecular mechanism of Saccharomyces cerevisiae shugoshin in SAC signalling remains elusive. Mad1 and Cdc20 have long been regarded as the only factors harbouring a Mad2-interaction motif (MIM). Only a recent report of Mad2 inhibiting the mitotic kinesin MKlp2 suggests that Mad2 has additional targets (Lee et al, 2010).
In the present study, we identify Mad2 as a novel interaction partner of human Sgo2. We show that human Sgo2 is dispensable for somatic cell division, in agreement with Pendas and colleagues (Llano et al, 2008) who have demonstrated an exclusively meiotic role of Sgo2. We provide extensive biochemical evidence that human Sgo2 interacts with Mad2 in a manner that closely resembles the interactions of Mad1 and Cdc20 with Mad2. We demonstrate that this Mad2 binding is shared by the only known Xenopus laevis shugoshin homologue and, thus, represents a conserved property of vertebrate shugoshin. Consistent with a possible meiotic role of this interaction, a complex of endogenous shugoshin and Mad2 exists in X. laevis oocytes.
Results
Specific association of shugoshin and Mad2
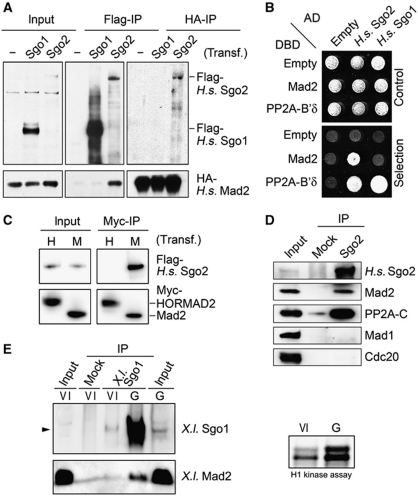
In vertebrate mitosis, the protection of centromeric cohesion and the generation of the ‘wait-anaphase’ signal at kinetochores correlate well in timing. Together with the functional links established between shugoshin family members and the SAC in yeasts (Indjeian et al, 2005; Kawashima et al, 2007; Vanoosthuyse et al, 2007), this prompted us to look for a possible interaction of human shugoshins with known SAC components. To this end, Flag-tagged Sgo1 or -2 were overexpressed together with various HA-tagged SAC factors in HEK293T cells. Subsequent reciprocal affinity purifications from corresponding lysates revealed that Sgo2 but not Sgo1 interacted with Mad2 (Figure 1A). This interaction was independently confirmed by yeast-two-hybrid analysis. Both human shugoshins bound PP2A, as expected, but only Sgo2 also interacted with Mad2 (Figure 1B).
Figure 1.
Mad2 is a novel interaction partner of human Sgo2 but not Sgo1. (A) Human Sgo2 but not Sgo1 interacts with Mad2 in vivo. HEK293T cells were transfected with plasmids to express HA–Mad2 alone or HA–Mad2 together with Flag-Sgo1 or Flag-Sgo2. Cells were arrested in mitosis by treatment with nocodazole (200 ng/ml) for 16 h, lysed and proteins were immunoprecipitated using the different tags. Immunoprecipitates were analysed by immunoblotting and detection of the tags. (B) Yeast-two-hybrid assay using combinations of plasmids encoding the indicated fusion proteins (AD: GAL4 transactivation domain, DBD: GAL4 DNA-binding domain). Growth on non-selective SC-Leu-Trp and on selective SC-His-Leu-Trp plates demonstrate equal plating and interaction, respectively. (C) Human Sgo2 is not a general interactor of HORMA-domain-containing proteins. Anti-Myc immunoprecipitates from mitotic HEK293T cells transiently expressing Flag-H.s. Sgo2 and either Myc–H.s. Mad2 (‘M’) or Myc–HORMAD2 (‘H’) were analysed by immunoblotting and detection of the tags. (D) Existence of the H.s. Sgo2–Mad2 complex at endogenous protein levels. HEK293T cells arrested in mitosis were lysed and Sgo2 was extracted from chromatin in high salt buffer (400 mM NaCl). Sgo2 was immunoprecipitated and probed for associated proteins by western blot. Unspecific IgG (mock) served as negative control. (E) Endogenous X.l. Sgo1 and Mad2 interact in meiosis. Lysates from stage VI or GVBD oocytes were subjected to IP using anti-X.l. Sgo1 antibodies or unspecific IgG (mock) as control. Immobilized material was analysed by immunoblotting and detection as indicated. GVBD (germinal vesicle breakdown) was assessed by visual inspection and Cdk1 activity assay (H1).
Mad2 is a member of a protein family sharing a common structural motif called the HORMA (Hop1/Rev7/Mad2) domain (Aravind and Koonin, 1998). Hence, we tested whether the interaction with human Sgo2 is a specific aspect of Mad2 or a property of the HORMA domain per se. Human Mad2 and HORMAD2, another HORMA-domain protein, which has been implicated in the regulation of DNA double-strand breaks and the synaptonemal complex during meiosis (Wojtasz et al, 2009), were probed for their ability to bind Sgo2 in a co-immunoprecipitation (co-IP) experiment. While immobilized Mad2 efficiently co-precipitated human Sgo2, no binding could be detected between Sgo2 and HORMAD2 (Figure 1C). Thus, the interaction of human Sgo2 with Mad2 requires specific aspects of Mad2 not common to all HORMA-domain proteins.
Next, we asked whether the Sgo2–Mad2 interaction took place also between endogenous proteins, that is without overexpression of any binding partner. When Sgo2 was immunoprecipitated from lysates of mitotically arrested HEK293T or HeLa cells, not only its established binding partner PP2A co-purified but Mad2 did as well (Figure 1D and data not shown). This interaction was again specific because Mad2 did not associate with mock IgG beads. Thus, human Sgo2 and Mad2 bind each other in vivo. Moreover, since Mad1 or Cdc20 were undetectable in the Sgo2 immunoprecipitate, this association was not bridged by these two known binding partners of Mad2.
To address whether other vertebrate shugoshins also associate with Mad2, we additionally characterized murine Sgo1 and -2 as well as the only known shugoshin from X. laevis. Indeed, the Mad2-binding ability is conserved in M.m. Sgo2 and X.l. Sgo1 but not M.m. Sgo1, as judged by IP or yeast-two-hybrid assays (Supplementary Figure S1A–C). Because of the important meiotic functions of mammalian Sgo2, we next asked whether a shugoshin–Mad2 complex could be isolated from meiotic tissue. For reasons of accessibility of the required biological material, we turned to X. laevis. Endogenous X.l. Sgo1 was immunoprecipitated from oocytes that were arrested in prophase of meiosis (stage VI) or had been induced by progesterone to enter meiosis I (GVBD stage). Subsequent immunoblotting revealed (1) strong upregulation of X.l. Sgo1 upon entry into meiosis and (2) the specific co-purification of endogenous Mad2 with X.l. Sgo1 (Figure 1E). Further biochemical analyses revealed that despite their relatively low homology in sequence and length, human Sgo2 and Xenopus Sgo1 behaved indistinguishable in regard to Mad2 (see below).
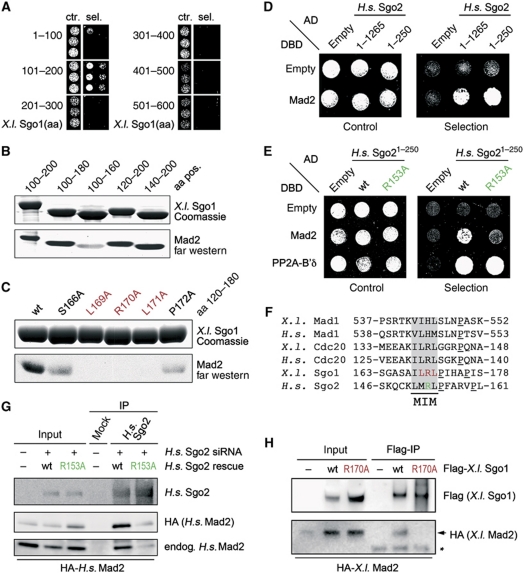
Mapping of the Mad2-binding site in shugoshin identifies a bona fide MIM
To gain further insight into structural requirements of the shugoshin–Mad2 interaction, we sought to map the Mad2-binding site within X.l. Sgo1. First, different shugoshin fragments were tested for interaction with Mad2 in a yeast-two-hybrid assay. Because of the lack of structural information, Sgo1 was divided into parts of roughly 100 amino acids (aa). We observed a specific interaction of X.l. Sgo1101–200 but not of the other shugoshin fragments with Mad2 (Figure 2A). To further narrow down the Mad2-binding region, we deleted additional amino acids at both the N- and the C-terminus of the X.l. Sgo1101–200 fragment. Corresponding recombinant proteins were analysed in a far-western assay for Mad2 binding (Figure 2B). While a fragment comprising aa 100–180 was still able to bind Mad2, a further C-terminal deletion of 20 aa (100–160) strongly reduced Mad2 interaction. In contrast, N-terminal deletions of the Sgo1 fragment (120–200, 140–200) did not impair Mad2 binding. Thus, the region between aa 160 and 180 of X.l. Sgo1 is critical for association with Mad2. Next, we introduced a series of single Ala mutations within this region and characterized them again by far-western assay (Figure 2C). Mutation of either of the residues 169–171 abolished Mad2 binding while changing the flanking residues 166 or 172 weakened the interaction. Likewise, mutating Arg 170 to Ala fully abrogated the ability of X.l. Sgo1 to interact with Mad2 in a yeast-two-hybrid assay (Supplementary Figure S1C).
Figure 2.
Shugoshin interacts with Mad2 via a conserved Mad1-/Cdc20-like MIM. (A) Mapping of the Mad2-binding site in shugoshin by yeast-two-hybrid. Indicated AD-tagged fragments of X. laevis Sgo1 were assayed for interaction with DBD-tagged X.l. Mad2 (aa=amino acids; ctr.=control: SC-Leu-Trp; sel.=selection: SC-Leu-Trp-His). (B) Fine-mapping of the Mad2-binding site in shugoshin by far-western assay. Membrane-immobilized X.l. Sgo1 polypeptides were incubated with native RGSH6-tagged Mad2, which was then detected by immunoblotting against its tag. (C) Creation of Mad2-binding deficient shugoshin mutants. Wild-type (wt) or point mutant X.l. Sgo1 fragments comprising amino-acid (aa) residues 120–180 were analysed by Mad2-far western as in (B). (D) Mad2 binds close to H.s. Sgo2's N-terminus. A yeast-two-hybrid assay was performed similar to (A). (E) A single amino-acid exchange (R153A) abolishes Mad2-binding capability of H.s. Sgo2 in a yeast-two-hybrid assay (experiment as in (A)). (F) Mad2-binding shugoshins contain a conserved Mad2-Interaction Motif (MIM). Amino-acid sequence alignment of Mad2-binding sites in shugoshin, Mad1 and Cdc20 from Xenopus and human origin. (G) A single point mutation in H.s. Sgo2 (R153A) interferes with Mad2 binding in vivo. HEK293T cells were transfected with the indicated siRNA and plasmids. Anti-Sgo2 immunoprecipitates from respective lysates were analysed by immunoblotting and detection. (H) Specific interaction of overexpressed X.l. Sgo1 and Mad2 in stage VI X. laevis oocytes. Oocytes were injected into their nuclei with plasmids encoding Flag-X.l. Sgo1 wild type or R170A and/or HA–Mad2. Extracts from 50 nuclei per sample were subjected to anti-Flag IP followed by immunoblotting and detection using the indicated antibodies (arrow: HA–Mad2, asterisk: IgG light chain).
We were able to extend these findings to the human Sgo2–Mad2 complex. An N-terminal 250 amino-acid fragment corresponding to roughly one fifth of H.s. Sgo2 was sufficient for Mad2 binding as exemplified by yeast-two-hybrid assay (Figure 2D). Furthermore, changing Arg 153 of human Sgo2 to Ala was sufficient to severely impair Mad2 binding without affecting expression levels or interaction with the PP2A subunit B’δ (Figure 2E; Supplementary Figure S1D). We additionally tested the R to A mutations within the context of full-length proteins. Wild-type (wt) and mutant forms of human Sgo2 and Xenopus Sgo1 were transiently overexpressed in HEK293T cells or stage VI frog oocytes, respectively, and then immunopurified. In both systems, the co-precipitation of Mad2 was specifically observed for wt shugoshin but severely impaired in the point mutant (Figure 2G and H).
Interestingly, in both X.l. Sgo1 and H.s. Sgo2, the predicted coiled-coil domain, which mediates PP2A binding, and the arginine, which is critical for Mad2 binding, are separated by 30–35 aa. Thus, binding of Mad2 to shugoshin might not interfere with simultaneous PP2A–shugoshin interaction. This speculation was indeed confirmed by in vitro reconstitution of a heterotrimeric complex, in which the interaction of Mad2 with PP2A-B’δ was bridged by recombinant X.l. Sgo1 (Supplementary Figure S2A).
The established Mad2-binding partners Mad1 and Cdc20 share a short MIM that is characterized by one basic amino acid surrounded by several hydrophobic residues (Luo et al, 2002; Sironi et al, 2002). A sequence alignment revealed that the Mad2-binding sites identified in X.l. Sgo1 and H.s. Sgo2 were intriguingly similar to the MIMs from Xenopus and human Mad1 and Cdc20 (Figure 2F). Together with our mutational analyses, this observation strongly argues that H.s. Sgo2 and X.l. Sgo1 each bind Mad2 via a bona fide MIM. Consistently, MIM-spanning peptides of H.s. Sgo2 (KRISKQCKLMRLPFAR) and X.l. Sgo1 (SAILRLPIH) bound to Mad2 with a KD of 0.69 and 0.60 μM, respectively, as determined by isothermal titration calorimetry (ITC; Supplementary Figure S2B and data not shown). This strong interaction was confirmed by 1D proton NMR, in which a Mad2 solution was titrated with a X.l. Sgo1 peptide of 25 residues (Supplementary Figure S2C). Here, signals for NHε indole side chains of tryptophans of Mad2 between 10.0 and 11.4 p.p.m. splitted upon shugoshin peptide addition. The complete disappearance of the Trp peaks of free Mad2 at a 1:1 molar ratio of both binding partners indicated quantitative complex formation and a KD of ⩽1 μM.
The ITC experiments also suggested a Mad2:shugoshin ratio of 0.55 and 1.33, respectively, which is most consistent with a 1:1 stoichiometry of the complex. To test this prediction, a complex of recombinant Mad2 and a X.l. Sgo1 fragment (aa 108–207) was tandem affinity purified via a different tag on each binding partner (Supplementary Figure S2D). Then, Mad2 and shugoshin were separated by SDS–PAGE and quantified by densitometry of Coomassie-stained protein bands to relative amounts of 1.13 and 1.0, respectively, assuming equal staining behaviour. We conclude that one molecule of Mad2 associates with one molecule of shugoshin. As the heterodimeric complex was assembled from bacterially expressed proteins and purified to near homogeneity (Supplementary Figure S2E), this binding is direct and does not require other factors.
Shugoshin binds Mad2 in a Mad1/Cdc20-like manner
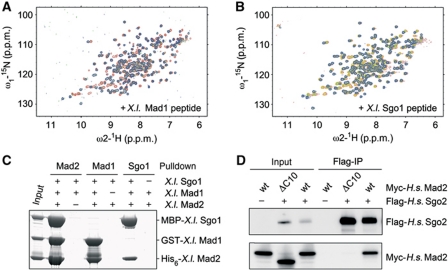
The identification of a MIM in members of the shugoshin protein family suggested that the shugoshin–Mad2 complex might also share other characteristics with the known Mad1–Mad2 and Cdc20–Mad2 complexes. We therefore tested a series of predictions associated with this hypothesis.
Ligand binding by Mad2 as it occurs in active SAC signalling leads to a global structural rearrangement of Mad2 (Luo et al, 2002). We therefore asked whether a Mad2-binding shugoshin peptide would induce similar changes. To address this question, we used two dimensional [1H,15N]-HSQC NMR spectroscopy to monitor structural changes occurring in [15N]-labelled Mad2 upon titration with unlabelled Mad1- or X.l. Sgo1 peptides. As expected, binding of a Mad1 peptide altered a large number of Mad2’s chemical shifts (Figure 3A). Importantly, addition of the X.l. Sgo1 peptide induced chemical shifting of essentially the same residues (Figure 3B). This strongly argues that both peptides trigger the same conformational rearrangement in Mad2 and, hence, bind to identical sites. Furthermore, at roughly equimolar concentrations of Mad2 and shugoshin peptide, the NMR spectra displayed double peaks, which correspond to the signals of the free- and the peptide-bound Mad2, thus revealing slow chemical exchange between both species. Once again, this underscores the strong affinity of the X.l. Sgo1 peptide towards Mad2.
Figure 3.
Shugoshin and Mad1 bind to the same site on Mad2. (A) A 2D [1H,15N]-HSQC NMR spectrum of [15N]-labelled Mad2 was acquired in the absence (red) or in the presence of 0.8 mol equivalents (green) or 5 mol equivalents (blue) of the X.l. Mad1 peptide ‘TKVIHLSLN’. (B) 2D NMR experiment performed essentially as in (A) using the X.l. Sgo1 peptide ‘SAILRLPIH’. The spectrum for Mad2 alone is shown in red. Spectra in the presence of 0.4, 0.8 or 5 mol equivalents of X.l. Sgo1 peptide are shown in yellow, green and blue, respectively. Note that the final positions of the Mad2 signals were slightly different. This is due to the different sequences of the added peptides and, hence, different chemical environment and does not indicate different binding sites. (C) Mutually exclusive binding of Mad1 or X.l. Sgo1 to Mad2. A mixture of GST-X.l. Mad1485–586, MBP-X.l. Sgo1108–207 and His6-X.l. Mad2 (input) was passed over glutathione-, amylose- or Ni2+-NTA beads. Proteins retained on the different affinity resins after washing were identified by SDS–PAGE and Coomassie staining. To control for unspecific binding in each case, the protein carrying the respective tag was omitted. (D) Constitutively open Mad2 (Mad2-ΔC10) does not bind to H.s. Sgo2. Anti-Flag immunoprecipitates from transfected HEK293T cells expressing the indicated proteins were analysed by immunoblotting and detection of the tags.
If it were indeed true that shugoshin and Mad1 bind Mad2 in the same manner, then their binding to Mad2 should be mutually exclusive. We tested this hypothesis in an in vitro assay using differentially tagged recombinant versions of Xenopus Mad1, Sgo1 and Mad2. We combined all three proteins and subsequently re-isolated each of them in separate pull-down assays (Figure 3C). While Mad1 and Sgo1 both co-purified with Mad2, as expected, an affinity-purified preparation of Mad1 contained Mad2 but no Sgo1. Vice versa Mad1 was absent from a Sgo1–Mad2 complex purified by pulling on Sgo1. Thus, a heterotrimeric Mad1–Mad2–Sgo1 complex does not exist. Instead, Mad1 and Sgo1 compete for a closely related if not identical binding site on Mad2. The same result was obtained when Cdc20 was used in place of Mad1 in a corresponding competition experiment (Supplementary Figure S2G).
A specific trait of the Mad1–Mad2 and Cdc20–Mad2 interactions is that in both complexes Mad2 is in its closed conformation (Luo et al, 2002; Sironi et al, 2002). In contrast, a Mad2 mutant, which lacks the C-terminal 10 aa and is locked in its open conformation (Mad2-ΔC10), is unable to engage in these interactions (Luo et al, 2000; Sironi et al, 2001). To test whether these rules also apply to shugoshin–Mad2, we assessed the shugoshin-binding capability of Mad2 wt versus -ΔC10. Immunoprecipitates of human Sgo2 from transfected HEK293T cells were probed for the presence of co-expressed Mad2. In contrast to wt Mad2, the constitutively ‘open’ deletion mutant indeed failed to interact with Sgo2 (Figure 3D; see also Supplementary Figure S2A and D for X.l. Sgo1). The same loss of Sgo2 binding was exhibited by a dominant-negative Mad2 mutant, in which Asp residues mimic constitutive phosphorylation and compromise Mad1 binding (Supplementary Figure S2F; Wassmann et al, 2003a).
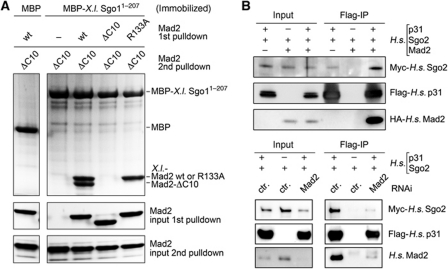
According to the template model, free Mad2open is recruited to Mad1–Mad2closed complexes at unattached kinetochores by direct but transient interaction with Mad2closed (De Antoni et al, 2005). To address whether, likewise, Mad2open could associate with Shugoshin–Mad2 complexes, we first incubated an immobilized fragment (aa 1–207) of X.l. Sgo1 with wt Mad2, Mad2-ΔC10 or Mad2-R133A, a mutant that is incapable of homotypic interaction (Sironi et al, 2001). As expected, wt Mad2 and the R133A mutant bound to shugoshin, while the constitutively ‘open’ Mad2-ΔC10 did not (Figure 4A). After washing away unbound Mad2, all samples were incubated with Mad2-ΔC10 in a second step. Finally, the shugoshin beads were washed again before bound material was analysed by SDS–PAGE and Coomassie staining. Interestingly, Mad2-ΔC10 was able to bind to the beads in the second round but only if the immobilized shugoshin had been pre-charged with wt Mad2. This result recapitulates with shugoshin what has so far only been shown for Mad1 and Cdc20 (De Antoni et al, 2005; Mapelli et al, 2006; Yang et al, 2008), namely the existence of a shugoshin–Mad2closed–Mad2open complex.
Figure 4.
Shugoshin-bound Mad2 is in the closed conformation. (A) A heterotrimeric shugoshin–Mad2closed–Mad2open complex. Amylose beads charged with MBP-X.l. Sgo11–207 were incubated with wild-type (wt) or mutant (ΔC10 or R133A) Mad2, extensively washed and then incubated with Mad2-ΔC10. Beads were washed again and SDS eluates analysed by PAGE and Coomassie staining. (B) p31comet as a probe for the Mad2closed conformer bound to H.s. Sgo2. Transfected HEK293T cells expressing Flag-p31, Myc–Sgo2 and/or HA–Mad2 as indicated were subjected to anti-Flag IP followed by immunoblotting and detection of the tags. In one case, endogenous Mad2 was partially knocked down by siRNA addition to the calcium phosphate transfection mix and additional liposome-based siRNA transfection as exemplified by Mad2 western.
To provide additional in vivo evidence that shugoshin-associated Mad2 is indeed in its closed conformation, we explored the fact that the SAC antagonist p31comet mimics Mad2open and specifically binds only to Mad2closed (Xia et al, 2004; Mapelli et al, 2006; Yang et al, 2007). We therefore analysed if p31comet was associated with human Sgo2 and whether this interaction depended on the presence of Mad2. Indeed, H.s. Sgo2 co-purified with p31comet from transfected HEK293T cells (Figure 4B). Importantly, the interaction was enforced by overexpression of Mad2 (top panels) and weakened by siRNA-mediated Mad2 knockdown (lower panels). Thus, shugoshin-associated Mad2 mediates the recruitment of p31comet and, hence, must be in its closed conformation.
Human Sgo2 is dispensable for faithful mitosis
Yen and co-workers reported that Sgo2-depleted HeLa cells undergo delayed anaphase with persisting spindle defects and lagging chromosomes (Huang et al, 2007). However, they did not detect sister chromatid cohesion defects in response to their siRNA (henceforth called oligo ‘Y’). In contrast, Watanabe and colleagues reported partial loss of sister chromatid cohesion, a much greater prolongation of mitosis and cell death upon transfection of a different Sgo2 siRNA (oligo ‘W’) at high concentration (Tanno et al, 2010).
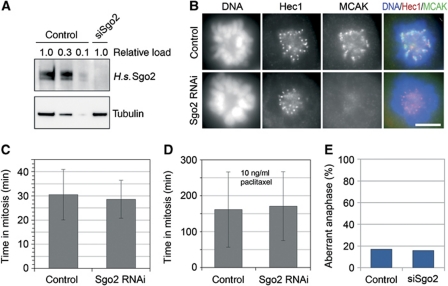
When compared side by side in HeLa cells, both siRNAs depleted Sgo2 equally well (Supplementary Figure S3A). Surprisingly, a dramatic prolongation of mitosis and induction of apoptosis was observed only for oligo ‘W’ but not ‘Y’ (Supplementary Figure S3B and C). This result led us to refrain from using oligo ‘W’ in subsequent, more detailed analyses of putative Sgo2 functions in mitosis. Instead, we used oligo ‘Y’ to deplete >90% of Sgo2 from HeLa cells (Figure 5A). Consequently, all centromeric signals for MCAK were lost (Figure 5B). Consistent with our first siRNA experiment but contradicting the findings by Huang et al (2007), this did not lead to a significant lengthening of mitosis as judged by phase-contrast microscopy. The time from cell rounding until cleavage furrow ingression averaged 30.5 (±10.5) min for mock-treated and 28.5 (±7.5) min for Sgo2-less cells (Figure 5C). Even when challenged by a low dose (10 ng/ml) of the spindle toxin paclitaxel, control and Sgo2-depleted cells exhibited identical extensions of mitosis to about 160 min (Figure 5D). In contrast to cells without Mad2, those without Sgo2 did also not display an override of a short-term mitotic SAC-mediated arrest triggered by the spindle toxins nocodazole or vinblastine (Supplementary Figure S4A). Furthermore, the ability to stay mitotically arrested upon prolonged treatment with paclitaxel or nocodazole was unaffected by knockdown of Sgo2 (Supplementary Figure S4B). In our hands, HeLa cells stably expressing histone-H2B-GFP also did not suffer from an increased rate of aberrant anaphases in the absence of Sgo2 as judged by scoring lagging chromosomes and anaphase bridges (Figure 5E). Finally, chromosome spreads revealed that Sgo2 was dispensable for maintenance of sister chromatid cohesion in prometaphase-arrested cells, while Sgo1 was clearly required (Supplementary Figure S4C). Collectively, these experiments show that >90% of Sgo2 can be depleted from human cells without causing any significant phenotype. Consistent with the study by Llano et al (2008), our findings therefore strongly suggest that Sgo2 has no crucial role during mitosis. We propose that the most significant functions of mammalian Sgo2—and hence also of its interactions—are limited to meiosis.
Figure 5.
Sgo2 is not required for mitotic progression. (A) H.s. Sgo2 is efficiently depleted by RNAi. Mock- and Sgo2 siRNA-treated HeLa cells were harvested 55 h after transfection (treatment with 200 ng/ml nocodazole for final 16 h). Western blots of cell lysates were probed by immunoblotting for Sgo2 and α-tubulin. Different amounts of control lysate (‘relative load’) were analysed for a semi-quantitative estimate of Sgo2 depletion efficiency. (B) MCAK mislocalizes in Sgo2-less cells. HeLa cells were transfected with Sgo2 siRNA or control treated, fixed 48 h thereafter and processed for immunofluorescence microscopy using antibodies against Hec1 and MCAK (scale bar=10 μm). (C) Sgo2-less cells progress normally through mitosis. HeLa cells were transfected with Sgo2 siRNA or control treated for a total time of 55 h. Following release from a single thymidine block, cells were analysed by live cell imaging (error bars: s.d.; n>100). (D) Sgo2-less cells exhibit a normal SAC response. A live cell imaging experiment was conducted essentially as in (B), with the exception that 10 ng/ml paclitaxel was added to the culture medium before mitotic entry (error bars: s.d.; n>70). (E) Sgo2-less cells do not suffer from lagging chromosomes. A HeLa cell line stably expressing histone-H2B-eGFP was either control treated or transfected with Sgo2 siRNA for a total time of 55 h. Cells were released from a single thymidine block and subjected to live cell fluorescence microscopy (n>60).
Mad2 binding is required for focused localization of Sgo2 to centromeres
Despite the lack of obvious mitotic Sgo2 functions, the dynamic localization of Sgo2 in meiosis II is nevertheless closely mirrored in mitosis (Gomez et al, 2007). We therefore used immunofluorescence microscopy to localize human Sgo2 relative to the outer kinetochore marker Hec1 in different stages of mitosis. Consistent with earlier reports (Gomez et al, 2007), Sgo2 localized to centromeres in late pro- and prometaphase and then re-localized to inner kinetochores in metaphase (Supplementary Figure S5A). When Sgo2 first localized to chromatin in early prophase, interestingly, the signals were also split into two dots per chromosome. Thus, Sgo2 changes its chromosomal localization twice in early mitosis, namely from inner kinetochores to centromeres and back. Temporal correlation between the late re-localization of Sgo2 and the formation of tension across amphitelically attached sister kinetochores spurred the idea that Sgo2 might somehow be physically pulled outward by the attaching MTs (Gomez et al, 2007). The failure of Sgo2 to leave the centromeres in nocodazole-arrested cells is consistent with this model. However, we noted that in paclitaxel-treated cells Sgo2 travelled from centromeres to kinetochores at least partially (Supplementary Figure S5B). Thus, attachment of MTs might suffice for Sgo2 re-localization and the exertion of pulling force might largely be dispensable. In summary, the above results suggest that both the dynamics and mechanisms of Sgo2 (re-)localization are more complex than previously anticipated.
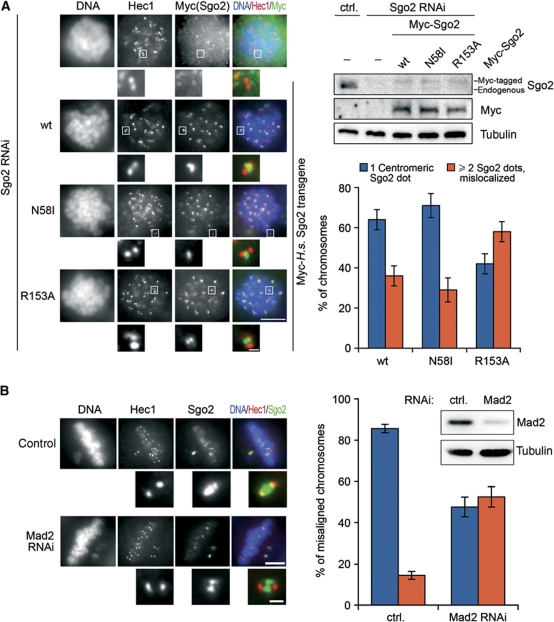
Following this characterization, we next investigated whether Mad2 binding would influence Sgo2's centromeric localization in prometaphase-arrested cells. As Sgo2 dimerizes via its N-terminal coiled coil (Xu et al, 2009), this required prior depletion of endogenous Sgo2 by RNAi and expression of Myc6-tagged Sgo2-R153A from a transfected plasmid encoding an siRNA-resistant mRNA. As controls, we alternatively expressed Myc6-tagged versions of either wt Sgo2 or Sgo2-N58I, a point mutant unable to interact with PP2A (Supplementary Figure S6B). At first glance, all versions of recombinant Sgo2 seemed to localize properly to centromeres in nocodazole-treated HeLa cells (Figure 6A) and to rescue the centromeric localization of MCAK (Supplementary Figure S6C). However, on close inspection we noted that the Sgo2-R153A signal was frequently split into two dots perpendicular to the axis defined by the Hec1-labelled kinetochores (Figure 6A). In contrast, Sgo2-wt and -N58I almost always localized as a single focus on a theoretical line connecting the two Hec1 dots. If the mislocalization of Sgo2-R153A were the specific result of its compromised Mad2-binding capability, then wt Sgo2 should display the same mislocalization in the absence of Mad2. To test this prediction, we conducted immunofluorescence microscopy of endogenous Sgo2 in cells partially depleted of Mad2 by short-term RNAi. As it is difficult to unambiguously identify sister kinetochores within the bulk of congressed chromosomes, cells were additionally treated with a low dose of nocodazole to induce misalignment of individual chromosomes. Indeed, 50% of the Sgo2 signals on these isolated chromosomes were split into two dots perpendicular to the inter-kinetochore axis, while Sgo2 showed normal centromere localization in 85% of mock-depleted metaphase cells (Figure 6B). When the same cells were additionally treated for 3 h with the proteasome inhibitor MG132 to ensure arrest in metaphase, the Sgo2 signals were mostly found split into four dots, two flanking each kinetochore (Supplementary Figure S6C and D). At this stage, Sgo2 in control cells had frequently re-localized towards kinetochores giving rise to the typical two signals. We conclude that Mad2 binding is important for the proper localization of human Sgo2 within sub-centromeric regions.
Figure 6.
Fine-tuning of Sgo2 localization by Mad2 binding. (A) Splitting of the centromeric signal for a Mad2-binding deficient Sgo2 mutant. GL2 (ctrl.) or Sgo2 siRNA-treated cells were additionally transfected either with empty vector (−) or with plasmids encoding siRNA-resistant Myc-tagged Sgo2-wt, -N58I or -R153A. Cells were arrested in mitosis with nocodazole and analysed by fluorescence microscopy as labelled on top (left panels) or by western blot using Sgo2-, Myc- and α-tubulin antibodies (upper right panels). Taking into account the relatively high transfection efficiency, we estimate that Sgo2 transgenes were not expressed above physiological levels. Scale bars are 10 μm in the large panels and 0.5 μm in the close-up images of kinetochores/centromeres. The graph shows a quantification of the observed phenotypes (three independent experiments, n>1000 chromosomes each, error bars: s.d.). (B) Centromeric Sgo2 immunofluorescence signals split into two upon depletion of Mad2. HeLa cells were transfected with either control (ctrl.=GL2) or Mad2 siRNA and cultured for 24 h. A low concentration of nocodazole (10 ng/ml) was added and 3 h thereafter cells were analysed by Mad2- and α-tubulin western blots (upper right panels) or fixed and stained for the indicated markers (left panels). Individual misaligned chromosomes from >160 metaphase cells and two independent experiments were selected and quantitatively analysed for centromeric Sgo2 localization (graph on right; error bars: s.d.). Scale bars are as in (A).
Discussion
Here, we report that human Shugoshin-2 (H.s. Sgo2) binds to the essential SAC protein Mad2. Moreover, we provide exhaustive biochemical evidence that H.s. Sgo2 binds to the same site of Mad2 and induces similar conformational changes in Mad2 as do the checkpoint component Mad1 and the APC/C co-factor Cdc20. Underscoring the relevance of this observation, we find this Mad2-binding behaviour to be conserved not only in murine Sgo2 but also in X. laevis Sgo1—the only shugoshin known for this species to date. Existence of a stable interaction between X.l. Sgo1 and Mad2 was somewhat surprising because synteny analyses clearly identify X.l. Sgo1 as an orthologue of mammalian Sgo1, which is unable to interact with Mad2 (Figure 1A). Xenopus egg extracts, which are routinely used to study the first embryonic cell cycle, exhibit a functional prophase pathway upon entry into mitosis (Sumara et al, 2002). Thus, the absence of X.l. Sgo1 should result in precocious separation of sister chromatids if X.l. Sgo1 would be functionally redundant with mammalian Sgo1. Yet, mitotic chromosomes replicated in X.l. Sgo1-depleted extract merely exhibit an increased inter-kinetochore distance but otherwise retain pairing of their sister chromatids (Rivera and Losada, 2009). Including Mad2-binding ability as an additional indicator, it is tempting to speculate that, following gene duplication, reciprocal specializations have evolved for Sgo1 and Sgo2 in amphibians versus mammals. The strong upregulation of X.l. Sgo1 levels in oocytes upon entry of meiosis is consistent with this speculation (Figure 1E).
We could show the Mad2 interaction for shugoshins from different species and by using a variety of techniques. Yet, we were unable to detect co-localization by immunofluorescence microscopy, for example a localization of Mad2 to centromeres at prometaphase. However, this does not exclude that Mad2 is recruited to a fraction of centromeric shugoshin in amounts that are below the detection limit. It does also not exclude that this small Mad2–shugoshin population might nevertheless exert an important function in meiosis. In fact, under certain conditions even kinetochore-localized Mad2 is difficult to detect although it still mediates a robust cell-cycle arrest. For example, Mad2 becomes largely undetectable at kinetochores when cells are treated with paclitaxel or siRNAs against Bub1 or Hec1. Yet, these cells retain a functional, Mad2-dependent SAC (Waters et al, 1998; Martin-Lluesma et al, 2002; Johnson et al, 2004; BM, MO and OS, unpublished data).
In our hands, depletion of Sgo2 from HeLa cells gave rise to obvious phenotypes only for one siRNA (Tanno et al, 2010) but not for another (Huang et al, 2007). For subsequent, more detailed characterizations, we relied on the latter because in both cases Sgo2 was efficiently depleted as judged by anti-Sgo2 western and IF analysis of Sgo2 and MCAK. Furthermore, HeLa cells were unaffected also by transfection of a third siRNA (number 3 in Huang et al, 2007), which also depleted Sgo2 by >90% (data not shown). But why do Yen and co-workers see mitotic phenotypes upon depletion of Sgo2 from human cells (Huang et al, 2007), while our further analyses with the same oligos failed to reveal overt mitotic defects? Differences in knockdown efficiencies can be excluded (compare Figure 3A of Huang et al, 2007 with Figure 5A of this work). Instead, functional variations between different HeLa cell cultivates, which will certainly exist after decades of separation, might explain this discrepancy. However, depletion of Sgo2 from other human cell lines—HCT-116 and U2OS—also goes unpunished (BM, OS, unpublished observations), indicating that mammalian Sgo2 is usually dispensable for mitosis. Llano et al (2008) reported that mitosis is normal in murine Sgol2−/− cells. Our data underscore this finding and extend it to cultured human cells.
Given the highly similar properties of Mad1–Mad2 and Cdc20–Mad2 complexes, our biochemical experiments could not decide a priori whether shugoshin might be a Mad1-like upstream checkpoint component or a Cdc20-like downstream checkpoint target or both. Our functional analyses showed that human cells lacking Sgo2 retain full SAC competence in mitosis. In contrast, preventing Mad2–Sgo2 interaction by mutation of Sgo2 or depletion of Mad2 resulted in a mislocalization of Sgo2 in mitotic cells, suggesting that Sgo2 might constitute an unexpected Cdc20-like downstream target of the SAC. This interpretation would be consistent with the roles of the kinases Bub1 and Aurora B to ensure proper centromeric enrichment of shugoshins (Kitajima et al, 2004; Tang et al, 2004; Huang et al, 2007; BM, MO and OS, unpublished data).
While the Mad1–Mad2 complex is present throughout the cell cycle (Chen et al, 1999), binding of Cdc20 to Mad2 correlates with mitosis and an active SAC (Li et al, 1997; Wassmann and Benezra, 1998). Comparative IP of Sgo2 from prometaphase- versus S-phase arrested cells revealed that Mad2, like PP2A but unlike MCAK, is a constitutive binding partner of Sgo2 in a somatic cell cycle (Supplementary Figure S7). Thus, with respect to this cell cycle dependence of Mad2 binding, Sgo2 behaves more like the upstream SAC component Mad1. In fact, the Sgo2–Mad2 complex might also act as a template-like source of a ‘wait-anaphase’ signal. While we can clearly show that this is not the case in mitosis, Sgol2 knockout mice might have a potential SAC defect in meiosis. Here, sister chromatid cohesion is prematurely lost in anaphase of meiosis I when Sgo2 is absent. A corollary of the presence of single chromatids in early meiosis II would be the constitutive activation of the tension-sensitive branch of the SAC. Yet, indicative of a leaky arrest, a high percentage of spermatids exit meiosis II and differentiate into highly aneuploid albeit morphologically normal sperm cells (Llano et al, 2008). An effective SAC has been demonstrated in male meiosis (Nagaoka et al, 2011), supporting the notion that Sgo2 might have a function in meiotic SAC integrity. It will therefore be interesting to see whether replacing endogenous Sgo2 with a Mad2-binding deficient mutant might result in a compromised SAC in meiosis II.
A functional SAC is also important during female meiosis I where Mad2 regulates the timing of the first polar body extrusion. In the absence of Mad2, meiosis I is significantly shortened (Homer et al, 2005). According to current models, a diffusible SAC signal is generated at kinetochores and constitutively inactivated in the cytosol (Doncic et al, 2005), leading to a gradient of APC/C inhibitory activity. It is conceivable that Mad1 alone might be unable to sustain SAC activity in oocytes considering the large cytoplasm to chromatin ratio. Thus, the Sgo2–Mad2 complex could support or relay the meiotic SAC signal. Interestingly, Mad2 is no longer readily detectable at kinetochores about 2 h before anaphase onset of meiosis I (Wassmann et al, 2003b) and it is currently not clear how APC/C activity is restrained during this period of time. As we can detect Shugoshin–Mad2 binding in meiotic tissue, a potential influence of Sgo2 on meiosis I duration should, therefore, be addressed in future studies.
While Sgo2 is essential during the first meiotic division, it has to be somehow inactivated in meiosis II to allow separase-dependent cleavage of centromeric cohesin and subsequent segregation of sister chromatids. According to one model, bipolar attachment of chromosomes to the spindle in meiosis II leads to re-localization of Sgo2 to kinetochores, deprotecting centromeric cohesin (Gomez et al, 2007). However, there are indications that re-localization might not be sufficient for the inactivation of meiotic shugoshin. In fission yeast, for example, enforced bipolar attachment during meiosis I in a rec12Δmoa1Δ strain does not cause premature loss of sister chromatid cohesion (Yokobayashi and Watanabe, 2005). It is therefore tempting to speculate that Mad2 might act as a negative regulator of the cohesion-protective function of Sgo2.
Materials and methods
Bacterially expressed proteins and interaction studies thereof
Unless specified otherwise (see below), bacterial expression and native purifications were conducted as follows: At OD600 nm=0.5–1.0, a culture of Escherichia coli Rosetta 2 cells (Novagen), which had been transformed with the corresponding expression plasmid, was supplemented with IPTG to 1 mM. After shaking for additional 3 h at 37°C, cells were harvested by centrifugation. All further steps were performed at 4°C. Using a microfluidizer (Avestin), cells were lysed in PBHS (10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, 537 mM NaCl) plus 10 mM imidazole, 5 mM β-mercaptoethanol (for His-tagged proteins) or plus 10 mM DTT (all others). Lysates were centrifuged at 40 000 g for 30 min and soluble tagged proteins were batch purified from the supernatants by overnight incubation with the appropriate affinity resin. For MBP-Tev3-, GST- and N-terminally His6-tagged proteins, this was immobilized amylose (New England Biolabs), -glutathione or -Ni2+-ions (Qiagen), respectively. From here on, procedures differed depending on the individual experiment. Mad2 was purified as described previously (Sironi et al, 2002) followed by gel filtration using a Superdex 75 column (GE Healthcare) and dialysis against Mad2 buffer (20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA). To probe interaction between X.l. Sgo1 and Mad2, 5 μg of immobilized MBP-Tev3-Sgo1 (aa 1–207) was rotated for 4 h with 50 μg of purified His-tagged X.l. Mad2-wt, -ΔC10 or -R133A. The amylose beads were then washed in PBHS plus 10 mM DTT, 0.2% Triton X-100. To further test binding of Mad2open (Mad2-ΔC10) to pre-assembled Sgo1–Mad2 complexes, all samples were incubated with 50 μg of His6-Mad2-ΔC10 as before, washed in Mad2 buffer, eluted in SDS sample buffer and finally analysed by PAGE on precast gradient gels (Serva, Heidelberg) with subsequent Coomassie staining. To demonstrate mutually exclusive binding of Xenopus Mad1 (or Cdc20) and Sgo1 to Mad2, the three differentially tagged proteins (or fragments thereof) were individually purified, competitively eluted from the corresponding affinity matrix and mixed in equal amounts. The mixture was then split and dialysed either against Mad2 buffer (for MBP- and GST-pulldowns) or against 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 20 mM imidazole, 0.1 mM DTT (for His6-pulldown) before they were combined with amylose-, glutathione- or Ni2+-NTA beads. After 3 h of rotation, beads were washed in PBHS plus 1 mM DTT, 0.05% Triton X-100 and analysed as described above. For tandem affinity purification of Xenopus Sgo1–Mad2 complexes, 0.2 mg of purified MBP-Tev3-Sgo1 fragments (aa 99–207 or 108–207) immobilized on 25 μl of amylose beads were combined with 140 μg each of purified, dialysed His-tagged X.l. Mad2-wt, -ΔC10 or -R133A. After 3 h of rotation, the beads were washed extensively with PBHS plus 10 mM DTT and once with 1 × PBS plus 0.5 mM DTT, 0.05% Tween 20. Then, the beads were rotated overnight with GST-tagged TEV-protease. The resulting eluates were adjusted to 0.4 M NaCl and 20 mM imidazole, combined with 15 μl Ni2+-NTA agarose and rotated for another 3 h. The Ni2+-NTA beads were washed with PBHS plus 0.5 mM DTT, 0.05% Tween 20 and 30 mM imidazole before being eluted with imidazole (250 mM) containing buffer and analysed as described above.
To probe for the existence of a trimeric PP2A–Shugoshin–Mad2 complex, 5 μg of purified and dialysed His6-Mad2 (wt or ΔC10) were immobilized on 10 μl of Ni2+-NTA agarose. To these, 100 μl extract from Myc–PP2A-B’δ overexpressing HEK293T cells (in 20 mM Tris–HCl (pH 7.7), 400 mM NaCl, 10 mM NaF, 20 mM β-glycerophosphate, 5 mM MgCl2, 0.1% Triton X-100, 5% glycerol, 10 mM imidazole) were added in the presence or absence of 10 μg MBP-Tev3-X.l. Sgo1 (aa 1–207). Following incubation for 4 h at 4°C and extensive washing of the beads, immobilized proteins were resolved by SDS–PAGE using precast gradient gels (Serva, Heidelberg). Mad2 and X.l. Sgo1 were visualized by Coomassie staining, while PP2A-B’δ was detected by immunoblotting using anti-Myc antibodies. For far-western blots, fragments of X.l. Sgo1 were tagged N-terminally with ketosteroid isomerase and C-terminally with six histidine residues. Corresponding pET31 derivatives were expressed in E. coli Rosetta 2 DE3 as described above. Cells were suspended in 8 M urea, 100 mM NaH2PO4, 10 mM Tris–HCl (pH 7.6), 10 mM imidazole, 5 mM β-mercaptoethanol and lysed by sonification. Lysates were centrifuged at 20 000 g for 30 min and His-tagged proteins were purified by Ni2+-NTA pulldown. Beads were washed in 8 M urea, 100 mM NaH2PO4, 10 mM Tris–HCl (pH 7.6), 20 mM imidazole, 5 mM β-mercaptoethanol and eluted in SDS sample buffer. Eluted proteins were separated by SDS–PAGE and blotted onto a nitrocellulose membrane. The membrane was blocked in 1 × PBS, 5% milk powder and then incubated (4°C, overnight) with a solution of RGS-His6-tagged X.l. Mad2 (10 μg/ml in 1 × PBS, 1% BSA). Subsequently, Sgo1-bound Mad2 was detected by standard western blot using mouse-anti-RGS-His6.
Immunoprecipitation
Transiently transfected cells were harvested and resuspended in LP2 (20 mM Tris–HCl (pH 7.7), 100 mM NaCl, 10 mM NaF, 20 mM β-glycerophosphate, 5 mM MgCl2, 0.1% Triton X-100, 5% glycerol supplemented with complete protease inhibitor cocktail (Roche)). To extract endogenous human Sgo2, NaCl was adjusted to 400 mM before IP. Lysates were prepared using a Dounce homogenizer, incubated for 30 min on ice and then centrifuged for 1 h at 16 000 g. Depending on the tag, recombinant proteins were immunoprecipitated from the resulting supernatants by anti-Flag, anti-Myc or anti-HA affinity matrices. For IP of endogenous human Sgo2, anti-Sgo2 polyclonal antibody coupled to protein G-sepharose beads was used. IPs were carried out in batch procedure and at 4°C for 4–15 h. Beads were recovered by passing the suspensions over Mobicols (MoBiTec), washed with lysis buffer and eluted in SDS sample buffer. Eluates were finally analysed by immunoblotting. X.l. oocyte nuclear extracts were prepared by lysing 50 nuclei per sample in 200 μl of LP2 and centrifugation at 16 000 g for 30 min. Whole oocytes (250 per sample) were lysed in 1 ml XB (Murray, 1991), with additional 300 mM NaCl, protease inhibitors and 10 μM cytochalasin B. After centrifugation at 15 000 g for 15 min in a swing-out rotor, the central layer was recovered for IPs. Samples were subsequently processed as above.
NMR methods
All NMR spectra were acquired at 300 K on a Bruker DRX 500 MHz spectrometer. Typically, NMR samples contained up to 0.1 mM of protein in 50 mM KH2PO4, 50 mM Na2HPO4, 150 mM NaCl, 5 mM DTT, pH 7.4. Water suppression was carried out using the WATERGATE sequence. NMR data were processed using the Bruker program Xwin-NMR version 3.5. NMR ligand binding experiments were carried out in an analogous way to those previously described (Stoll et al, 2001). In all experiments, 500 μl of 0.1 mM X.l. Mad2 in PBS buffer containing 10% D2O and 1 mM stock solutions of the peptides in PBS were used. Mad1 or shugoshin peptides were added stepwise to the Mad2 solution leading to molar ratios of protein:peptide ranging between 10:1 and 0.2:1. After each step of peptide addition 1D proton or 2D [1H,15N]-HSQC spectra were recorded.
Nuclear injection of Xenopus oocytes
Female frogs were injected with 50 U PMSG 5 days before ovarial surgery. Frogs were anaesthetized in 0.1% ethyl-aminobenzoate methane sulphonate salt (Sigma) in water. Ovary tissue was harvested via a small cut in the abdomen closed afterwards by suture with separate stitches for muscle wall and skin. Oocytes (2 ml ovary tissue per 10 ml buffer) were defolliculated 10–12 h by collagenase treatment (0.5 mg/ml collagenase NB4G (Serva)) in buffer OR1 (5.0 mM Hepes-KOH, 82.5 mM NaCl, 2.5 mM KCl, 1.0 mM MgCl2, 1.0 mM Na2HPO4, pH 7.6). 10 nl water or plasmid DNA dissolved in water (0.1–0.4 mg DNA/ml) were injected directly into the nucleus of hand-selected stage VI oocytes followed by incubation at 18°C for 20 h in OR3 (5.0 mM Hepes-KOH, 82.5 mM NaCl, 2.5 mM KCl, 1.0 mM MgCl2, 1.0 mM Na2HPO4, 1 mM CaCl2, 50 μg/ml Gentamicin, pH 7.6).
Isolation of oocyte nuclei
Nuclei of injected or uninjected oocytes were harvested as described in detail previously (Gall and Wu, 2010), collected in pools of 50 nuclei and used either directly or frozen in liquid nitrogen and stored at −80°C.
Induction of meiosis in Xenopus oocytes
Meiosis was induced in oocytes by addition of water-soluble progesterone (Sigma-Aldrich, 10 μg/ml in OR3). Meiosis resumption (germinal vesicle breakdown) was visible as white spot formation at the animal pole and defined as time point t=0 of meiotic progression. Histone H1 kinase assays were performed as described previously (Gorr et al, 2006).
Supplementary Material
Since the publication of this paper, the authors have noticed that the panels of figure S7B are interchanged. This has been corrected in this file as of 14 June 2011.
Acknowledgments
We thank Thomas U Mayer for the MCAK antibody, Adrian Salic and Markus Moser for cDNA. We are grateful to Jutta Hübner, Monika Ohlraun and Alexander Strasser for technical assistance, Kay Hofmann for bioinformatic advice. We thank Stefan Heidmann for critical reading of the manuscript and all Stemmann laboratory members for helpful discussions. This work was supported by German Cancer Aid (Deutsche Krebshilfe e.V., Grant 107327) and Minna James Heineman Foundation (Minerva).
Author contributions: BM, MO and OS conceived and designed the experiments. BM and MO carried out the experiments except for the data shown in Figure 3A and B, Supplementary Figure S2B and C (KR, UR and TH), Supplementary Figure S2D and E (OS) and 1E and 2H (DH and BM). BM and OS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravind L, Koonin EV (1998) The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci 23: 284–286 [DOI] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A (2006) Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature 441: 532–536 [DOI] [PubMed] [Google Scholar]
- Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG (1999) The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell 10: 2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1: 82–87 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10: 3081–3093 [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A (2005) The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol 15: 214–225 [DOI] [PubMed] [Google Scholar]
- Doncic A, Ben-Jacob E, Barkai N (2005) Evaluating putative mechanisms of the mitotic spindle checkpoint. Proc Natl Acad Sci USA 102: 6332–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW (1998) The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev 12: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Wu Z (2010) Examining the contents of isolated Xenopus germinal vesicles. Methods 51: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez R, Valdeolmillos A, Parra MT, Viera A, Carreiro C, Roncal F, Rufas JS, Barbero JL, Suja JA (2007) Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep 8: 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr IH, Reis A, Boos D, Wuhr M, Madgwick S, Jones KT, Stemmann O (2006) Essential CDK1-inhibitory role for separase during meiosis I in vertebrate oocytes. Nat Cell Biol 8: 1035–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer HA, McDougall A, Levasseur M, Yallop K, Murdoch AP, Herbert M (2005) Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev 19: 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ (2007) Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol 177: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW (2005) The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307: 130–133 [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K (1995) Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81: 269–278 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849–860 [DOI] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS (2004) Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci 117 (Part 8): 1577–1589 [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y (2007) Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW (1995) A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81: 279–288 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y (2004) The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52 [DOI] [PubMed] [Google Scholar]
- Kudo NR, Anger M, Peters AH, Stemmann O, Theussl HC, Helmhart W, Kudo H, Heyting C, Nasmyth K (2009) Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J Cell Sci 122 (Part 15): 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y (2008) Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 10: 42–52 [DOI] [PubMed] [Google Scholar]
- Lee SH, McCormick F, Saya H (2010) Mad2 inhibits the mitotic kinesin MKlp2. J Cell Biol 191: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R (1997) MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA 94: 12431–12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E, Gomez R, Gutierrez-Caballero C, Herran Y, Sanchez-Martin M, Vazquez-Quinones L, Hernandez T, de Alava E, Cuadrado A, Barbero JL, Suja JA, Pendas AM (2008) Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev 22: 2400–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G (2000) Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol 7: 224–229 [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H (2002) The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell 9: 59–71 [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H (2004) The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol 11: 338–345 [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, Musacchio A (2006) Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J 25: 1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, Musacchio A (2007) The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell 131: 730–743 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabe AM, Helmhart W, Kudo NR, Wuensche A, Taylor S, Hoog C, Novak B, Nasmyth K (2009) Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol 19: 369–380 [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW (1991) Cell cycle extracts. Methods Cell Biol 36: 581–605 [PubMed] [Google Scholar]
- Nagaoka SI, Hodges CA, Albertini DF, Hunt PA (2011) Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol 21: 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niault T, Hached K, Sotillo R, Sorger PK, Maro B, Benezra R, Wassmann K (2007) Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS ONE 2: e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW (2007) Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446: 921–925 [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61 [DOI] [PubMed] [Google Scholar]
- Rivera T, Losada A (2009) Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma 118: 223–233 [DOI] [PubMed] [Google Scholar]
- Schott EJ, Hoyt MA (1998) Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics 148: 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, Novak B, Musacchio A, Ciliberto A (2009) The influence of catalysis on mad2 activation dynamics. PLoS Biol 7: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A (2002) Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J 21: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A (2001) Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J 20: 6371–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER III, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ (2007) Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446: 876–881 [DOI] [PubMed] [Google Scholar]
- Stoll R, Renner C, Hansen S, Palme S, Klein C, Belling A, Zeslawski W, Kamionka M, Rehm T, Muhlhahn P, Schumacher R, Hesse F, Kaluza B, Voelter W, Engh RA, Holak TA (2001) Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry 40: 336–344 [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H (2006) PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell 10: 575–585 [DOI] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H (2004) Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 101: 18012–18017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y (2010) Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev 24: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K (1998) Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Prykhozhij S, Hardwick KG (2007) Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol Biol Cell 18: 1657–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, De Antoni A, Massimiliano L, Ciliberto A, Faretta M, Salmon ED, Musacchio A (2006) In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr Biol 16: 755–766 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Wassmann K, Benezra R (1998) Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc Natl Acad Sci USA 95: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Liberal V, Benezra R (2003a) Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J 22: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Niault T, Maro B (2003b) Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol 13: 1596–1608 [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 141: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5: e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H (2004) Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J 23: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W (2009) Structure and function of the PP2A-shugoshin interaction. Mol Cell 35: 426–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, Yu H, Luo X (2008) Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol 6: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X (2007) p31comet blocks Mad2 activation through structural mimicry. Cell 131: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y (2005) The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817 [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285: 418–422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Since the publication of this paper, the authors have noticed that the panels of figure S7B are interchanged. This has been corrected in this file as of 14 June 2011.