Abstract
Rotation and switching of the bacterial flagellum depends on a large rotor-mounted protein assembly composed of the proteins FliG, FliM and FliN, with FliG most directly involved in rotation. The crystal structure of a complex between the central domains of FliG and FliM, in conjunction with several biochemical and molecular-genetic experiments, reveals the arrangement of the FliG and FliM proteins in the rotor. A stoichiometric mismatch between FliG (26 subunits) and FliM (34 subunits) is explained in terms of two distinct positions for FliM: one where it binds the FliG central domain and another where it binds the FliG C-terminal domain. This architecture provides a structural framework for addressing the mechanisms of motor rotation and direction switching and for unifying the large body of data on motor performance. Recently proposed alternative models of rotor assembly, based on a subunit contact observed in crystals, are not supported by experiment.
Keywords: flagella, co-crystal, rotor
Introduction
The flagellar motor of bacteria is a rotary device (Berg and Anderson, 1973) energized by the membrane ion gradient (Larsen et al, 1974; Glagolev and Skulachev, 1978). Its mechanism has been studied for >30 years and a great deal is known about the performance of the motor under various circumstances (for a review, see Sowa and Berry, 2008). The identities and stoichiometries of components that form the various substructures in the flagellum are also well established (Macnab, 2003). The stators consist of the membrane proteins MotA and MotB, which form complexes with subunit composition MotA4MotB2 (Sato and Homma, 2000; Kojima and Blair, 2004). Each motor has several independent stator complexes that function to conduct the energizing ions (Blair and Berg, 1990) and harness ion movement to rotation. In a current model, protons move on and off a conserved aspartate residue in MotB (Zhou et al, 1998b), to drive conformational changes that apply torque to the rotor (Kojima and Blair, 2001). The rotor and stator engage in electrostatic interactions that involve conserved charged residues in a cytoplasmic domain of MotA and in the C-terminal domain of FliG (Lloyd and Blair, 1997; Zhou and Blair, 1997; Zhou et al, 1998a; Yakushi et al, 2006).
Flagellar rotation and direction is controlled by a large protein assembly on the rotor called the switch complex (Yamaguchi et al, 1986a). It is formed from the proteins FliG, FliM and FliN, each present in many copies, and corresponds structurally to the C-ring of the flagellar basal body (Francis et al, 1992, 1994; Zhao et al, 1996a, 1996b; Thomas et al, 2006; Figure 1). The lower part of the switch complex is formed from FliN and the FliM C-terminal domain (FliMC); FliN is organized in doughnut-shaped tetramers that alternate with the FliMC domains in an array at the membrane distal region of the C-ring (hereafter referred to as the ‘bottom’) (Brown et al, 2005; Paul and Blair, 2006; Thomas et al, 2006; Sarkar et al, 2010b). The switch complex functions in flagellar assembly as well as in rotation (Yamaguchi et al, 1986b). FliN interacts with components of the type III secretion apparatus housed in the basal body (Gonzalez-Pedrajo et al, 2006; McMurry et al, 2006; Paul et al, 2006) and may facilitate assembly by assisting in the delivery of protein subunits that form exterior parts of the structure (the filament and hook). FliN is also critical for direction switching and contains a binding site for the signalling molecule phospho-CheY that promotes clockwise (CW) rotation (Sarkar et al, 2010a). The thinner side-wall of the C-ring, above the FliN4FliMC array, is formed from the FliM middle domain (FliMM; Park et al, 2006; Brown et al, 2007). Mutational analyses indicate that FliM has a large role in direction switching (Sockett et al, 1992) and its N-terminal domain, which is predicted to have an extended conformation, binds phospho-CheY (Welch et al, 1993; Lee et al, 2001). FliG is proximal to the membrane (hereafter referred to as the ‘top’ position) and comprises three domains (Irikura et al, 1993; Lloyd et al, 1996; Lee et al, 2010), each with distinct functions: The N-terminal domain (FliGN) interacts with the FliF protein which forms the MS-ring, the middle domain (FliGM) interacts with FliM, and the C-terminal domain (FliGC) contains a set of conserved charged residues that interact with charged residues in the cytoplasmic domain of MotA (Lloyd and Blair, 1997; Zhou et al, 1998a; Yakushi et al, 2006).
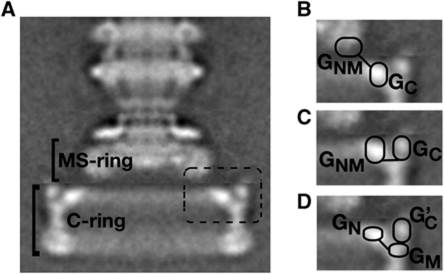
Figure 1.
Hypotheses for FliG organization in the flagellar rotor. (A) The flagellar basal body of wild-type Salmonella (Thomas et al, 2001). The dashed box indicates the region shown magnified in the other panels. (B) FliG-domain arrangement discussed by Thomas et al (2006). (C) Hypothesis of Brown et al (2007). (D) Arrangement based on a FliGM–FliGC contact observed in crystals. The contact is postulated to involve either two different FliG subunits (Lee et al, 2010) or a single FliG subunit (Minamino et al, 2011).
Current electron microscopic reconstructions of the flagellar basal body are highly detailed and provide strong constraints on the overall shape of the switch complex (Figure 1; Thomas et al, 2006). Whereas it is clear that FliG must lie at the top of the C-ring to enable interaction with the stator, presently there is no consensus regarding the assignment of specific FliG domains to the features observed in electron micrographs. Thomas and co-workers suggested that FliGC might correspond to the inner lobe of density at the top of the C-ring, with the other parts of FliG falling in the bottom part of the MS-ring (Figure 1B). Brown and co-workers favour an assignment of FliGC to the outer lobe of density at the top of the C-ring, and the N-terminal and middle domains of FliG to the inner lobe (Figure 1C). Lee et al (2010) and Minamino et al (2011) take this a step further and propose that both the middle- and C-terminal domains of FliG lie in the outer part of the C-ring, with the N-terminal domain alone accounting for the inner lobe (Figure 1D). Their models are based on an interaction between FliGC and FliGM observed in FliG crystals, which was judged to be biologically relevant on the grounds that it involves surfaces with conserved hydrophobic character and occurred in more than one crystal form. Experimental tests of the crystal contact-based model have not been reported.
The known features of switch-complex organization have been deduced from crosslinking and mutational experiments guided by structures of the individual components (Park et al, 2006; Brown et al, 2007; Sarkar et al, 2010b). The lower part of the C-ring has been studied most fully; systematic disulphide crosslinking studies of this region produced a structural model that fits well with the EM reconstructions (Thomas et al, 2006), and that additionally revealed a subunit movement that occurs upon CW/CCW direction switching (Sarkar et al, 2010b). Similar information on the upper part of the switch complex is needed to provide a structural framework for understanding motor rotation and switching. Uncertainties regarding the arrangement of FliG in particular must be addressed, as it is the component that functions directly in rotation. Herein, we report a range of experiments that culminate in a specific, firmly grounded model of FliG and FliM organizations. The crystal structure of a complex between major domains of FliG and FliM is described, together with several biochemical and mutational experiments to probe their arrangement within the flagellar motor. The results support an architecture like that proposed by Brown et al (2007). The crystal contact-based models (Lee et al, 2010; Minamino et al, 2011) are not supported; several results indicate that the FliGM–FliGC contact observed in crystals does not occur within the flagellum and that FliGC interacts directly with FliMM instead. The new structural model provides a long-sought framework for addressing molecular details of the motor mechanism.
Results and discussion
Structure of a FliGM:FliMM complex
In the model of Thomas et al (2006) only the C-terminal domain of FliG resides within the C-ring, whereas the FliGN and FliGM domains lie within the lower part of the MS-ring (Figure 1B). This model would thus preclude a direct interaction between FliGM and FliM, because FliM is believed to lie within the C-ring. However, our previous binding studies revealed an interaction between FliGM and FliM, occurring through a conserved ‘EHPQR’ surface motif on FliGM and a conserved ‘GGXG’ motif in the middle domain of FliM (Mathews et al, 1998; Brown et al, 2002, 2007). To examine this interaction further, we determined the crystal structure of a complex of the FliGM and FliMM domains, using thermostable proteins from Thermotoga maritima. The T. maritima proteins show high sequence similarity to those of Escherichia coli, including conservation of hydrophobic character at core positions, and in some cases can partially complement E. coli mutants (Lloyd et al, 1999; Brown et al, 2002, 2005). Thus, the T. maritima proteins provide relevant structural models for the proteins from the otherwise better-characterized enteric species.
The co-crystal structure (Figure 2) shows FliMM and FliGM domains essentially like the previously described individual structures (Brown et al, 2002; Park et al, 2006), except that a helix at the C-terminus of FliGM is shortened and packed more closely against the body of the domain than in the previous FliGMC structure (Brown et al, 2002) or in the just-reported structure of a 3-residue deletion variant of FliGMC (Minamino et al, 2011). The close-packed conformation of this helix is stabilized in part by contacts with FliMM in the complex (Figure 2E), and is similar to what is observed in the Aquifex aeolicus FliG structure (Lee et al, 2010). Most of the FliGM:FliMM interface is formed from the EHPQR motif of FliG and the GGXG motif and adjacent regions of FliM, as was suggested by the binding and mutational studies (Mathews et al, 1998; Brown et al, 2007). These motifs generate an interface from the inter-digitation of two surface loops that link helices on both proteins. Within the contact, FliM Met131, which immediately precedes the FliM GGXG motif, inserts into a hydrophobic patch composed of FliG Val172, Val176 and Val133 (T. maritima numbering is used here in the discussion of the T. maritima protein structures). Gln129 of the FliG EHPQR motif hydrogen bonds to the peptide backbone of the FliM GGXG motif, and FliG-His127 contacts FliM-Tyr124 and hydrogen bonds to FliM-Asp128. FliM-Asp128, whose conformation is also stabilized by interaction with the GGXG motif, further sets the interface orientation by forming a highly conserved salt bridge with FliG-Arg161. Interactions between conserved residues at the top of FliM α1′ (Thr144 and Ile146) and the end of FliGM αA (Phe 122 and Glu 126) also contribute to the association. In total, the FliMM–FliGM contact buries ∼750 Å2 surface area per subunit, with a calculated free energy of association ΔG=−6.3 kcal mol−1, hydrophobic surface specificity of 0.32 and surface complementarity of 0.51 (Lawrence and Colman, 1993; Krissinel and Henrick, 2007). These parameters reflect an interface of medium affinity (ca. >μM) often characteristic of binding partners that associate and dissociate as part of their function (Park et al, 2004).
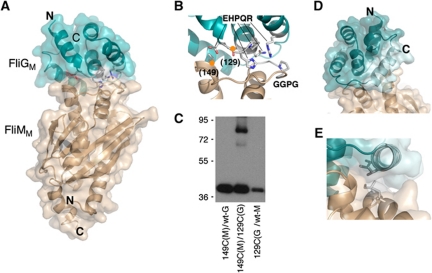
Figure 2.
Structure of the T. maritima FliMM:FliGM complex. (A) Overall shape of the complex. The N- and C-termini of FliMM are oriented towards the bottom in this view; these parts of FliM are directed towards the bottom of the C-ring in the flagellar basal body (Park et al, 2006; Sarkar et al, 2010b). (A stereo version of this figure is provided in Supplementary data.) (B) The FliGM:FliMM interface. The EHPQR residues of FliG and the GGXG motif of FliM are indicated. Orange circles mark positions where Cys residues were introduced to confirm the interaction by crosslinking. Numbers are for the E. coli protein. Unbiased electron density for the EHPQR and GGXG motifs is shown in Supplementary data. (C) Crosslinking through the introduced Cys residues. Crosslinking was induced using Cu-phenanthroline. (D) Packing of the helix near the C-terminus of FliGM against the body of the domain. The helix is shown in lighter colour. In the previous crystal structure of FliGMC (Brown et al, 2002), this helix is detached from the domain and makes extensive inter-subunit crystal contacts instead. (E) Hydrophobic contacts between the helix and FliMM that stabilize the close-packed conformation of the helix.
As a further check that the interaction seen in the co-crystal structure is relevant to the protein arrangement in the motor, Cys residues were introduced in the E. coli FliG and FliM proteins, at positions that are close in the crystal structure (FliG-129 and FliM-149, in E. coli numbering), and oxidative crosslinking was induced in cells. The Cys-substituted proteins crosslinked in high yield (Figure 2C). Taken together with the binding and mutational studies, this result indicates that the FliGM–FliMM arrangement observed in the crystal resembles that actually occurring in the motor. Switch-complex models with FliGM in the MS-ring, and thus removed from FliM (Figure 1B), are therefore unlikely.
Interaction between FliM and FliGC
The binding study that identified the FliM–FliGM interaction also gave evidence of a binding interaction between FliM and FliGC. The interaction involves a conserved hydrophobic patch on the surface of FliGC opposite the stator-interaction site (Brown et al, 2007). Mutations in the FliGC hydrophobic patch, like mutations in the FliGM EHPQR motif, were found to weaken the FliG–FliM binding. Stock and co-workers (Lee et al, 2010) re-interpreted these findings to mean that FliG binds FliM through a surface composed jointly from the FliGM and FliGC domains, which were assumed to associate together in the motor in the same way as occurs in the FliG crystals. The FliGM:FliMM structure shows, however, that FliGM has a FliM-binding surface distinct from the surface that associates with FliGC in the crystal contact (Figure 2; Supplementary Figure S1). The disruption of FliM binding by the hydrophobic-patch mutations is, thus, more readily explained in terms of a direct interaction between FliM and FliGC (Brown et al, 2007), and appears incompatible with models based on the FliGM–FliGC crystal contact (Lee et al, 2010; Minamino et al, 2011), where FliGM intervenes between FliGC and FliM. The hydrophobic-patch mutations were studied in the context of the full-length FliG protein, however, which might complicate the interpretation in terms of the individual domain interactions. To characterize the FliM–FliGC interaction more directly, we expressed FliGC (consisting of residues 185–331) as a separate domain and tested its binding to FliM in a pull-down assay. The separately expressed FliGC domain showed clear binding to FliM (Figure 3). Using collections of surface-residue mutations, the interaction was mapped to the hydrophobic patch of FliGC and the GGXG motif and adjacent regions on FliMM (Figure 3; Supplementary Figure S2). The FliM mutations that weakened the FliGC–FliMM interaction also disrupted function, as assayed either by motility in soft agar or by export of flagellin (Supplementary Figure S2). The co-crystal structure shows that the GGXG motif also binds FliGM; thus, essentially the same part of FliM is involved in interactions with both FliGM and FliGC (Supplementary Figure S2) and loss of function in the GGXG mutants might reflect disruption of either or both contacts.
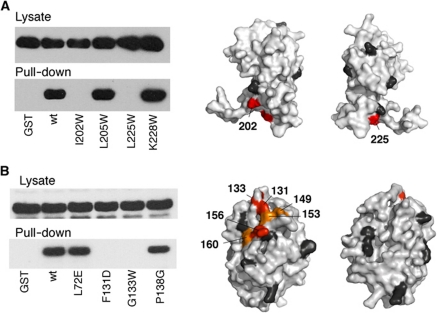
Figure 3.
Interaction between FliM and FliGC detected in GST pull-down assays. (Representative results are shown; see Supplementary Figure S2 for additional data.) Blots were probed with anti-FliM antibody. (A) Effects of FliGC mutations on the binding to FliM. Positions where mutations eliminated binding are coloured red; black indicates positions where mutations had no effect. (B) Effects of FliM mutations on binding to FliGC. Colouring as in panel (A), plus orange to indicate positions where binding was weakened.
We used disulphide crosslinking to verify the occurrence of the FliMM–FliGC interaction in cells and to obtain constraints on its geometry. Single-Cys residues were introduced at seven positions in the E. coli FliGC protein in the vicinity of the hydrophobic patch, and six positions in the E. coli FliMM protein near the GGXG motif, and all the pairwise combinations were studied. Oxidative crosslinking was induced in cells and products were examined on immunoblots. Several Cys pairs gave reproducible, moderately strong FliG–FliM crosslinking (Figure 4A; Supplementary Tables 3 and 4). A model for the FliMM:FliGC complex, constructed by bringing into proximity the highest yielding pairs (Figure 4B), places the hydrophobic patch of FliGC in contact with residues on FliMM with conserved strongly hydrophobic (Val127, Phe131 and Val153) or partially hydrophobic (Thr149) character (amino-acid residues and numbering are for the FliM protein of E. coli). One of the high-yielding Cys pairs also exhibited intergenic suppression: the Cys replacement at FliG residue 225 caused a complete loss of motility that was substantially rescued by the Cys replacement at residue 149 of FliM (Supplementary Figure S3). The size and shape of the FliMM:FliGC assembly provide an acceptable match to features observed in the upper part of the C-ring in EM reconstructions (Supplementary Figure S3B).
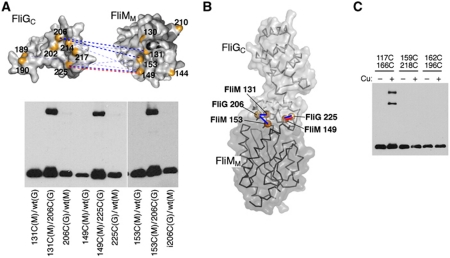
Figure 4.
Crosslinking experiments to probe the FliMM–FliGC relationship. (A) Positions of Cys replacements and summary of the crosslinking results. Dotted blue lines connect Cys pairs of residues that formed disulphide crosslinks, with the thickness of the line indicating relative yield. Representative gels are shown below; blots were probed with anti-HA antibody. The red dashed line connects a Cys pair that, in addition to crosslinking, showed mutational suppression (see the text and Supplementary Figure S3A). (B) Model for the FliMM–FliGC assembly based on the crosslinking results. The highest yield Cys pairs are indicated. (C) Tests of the crystal contact-based model for FliG organization. The 117/166 Cys pair in FliGM was shown previously to crosslink efficiently (Lowder et al, 2005) and is included as a positive control. The 159/218 and 162/196 Cys pairs are in close proximity in the crystal contact model (Lee et al, 2010; see Supplementary Figure S4 for an illustration). These failed to crosslink, using either Cu-phenanthroline (shown) or iodine (data not shown).
FliGN and FliGM are in proximity
The binding and crosslinking results establish that FliGC interacts directly with FliMM, and thus argue against the FliGC–FliGM interaction that has been postulated on the basis of crystal contacts (Lee et al, 2010; Minamino et al, 2011). Complicated architectures involving both types of interaction might still be imagined, and so we introduced Cys pairs at positions that are in close proximity in the crystal contact models and tested for disulphide crosslinking in cells. Two Cys pairs were made, both of which are predicted to allow close approach of the sulphur atoms (van der Waals distance or nearer; see Supplementary Figure S4). One pair (residues 162/196; E. coli numbering) retained about half of wild-type function in a soft agar motility assay, while the other (159/218) functioned at about 10% of wild type. Neither Cys pair showed detectable crosslinking in cells, using either Cu-phenanthroline (Figure 4C) or iodine as oxidizing agents (data not shown). The bifunctional reagent bis-maleimidohexane (BMH) that can bridge more distant thiols was also tried, and also showed no crosslinking through these positions (data not shown).
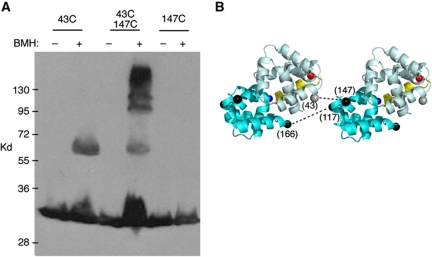
These results indicate that FliMM, and not FliGM, is located under FliGC in the outer part of the C-ring, in accordance with the proposal of Brown et al (2007). FliGM must then occupy a more-inward location, nearer FliGN (as in Figure 1C). The FliGN and FliGM domains are widely separated in the crystal structure of A. aeolicus FliG, but might adopt a more-compact conformation in the motor where the protein can engage in its normal interactions with FliM and FliF. In the A. aeolicus structure, both FliGN and FliGM, as well as the helix joining them, display sizable hydrophobic surfaces that appear to be stabilized by crystal contacts (Supplementary Figure S5). To test whether FliGN and FliGM actually lie near each other in the motor, we constructed three double-Cys mutants (31/146, 43/147 and 50/147), each with a replacement near an edge of FliGM and an edge of FliGN, and examined crosslinking in cells using the bifunctional reagent BMH. These Cys pairs are distant (Cβ-to-Cβ distances in the >30 Å range) in the rotor model of Lee et al (Supplementary Figure S4). The 43/147 Cys pair was crosslinked by BMH to form both dimer and trimer products. The corresponding single-Cys mutants either failed to crosslink (position 147) or formed dimer but none of the larger multimers (position 43) (Figure 5A). Previously identified crosslinks between FliGM and FliGM provide constraints on the relative orientation of the FliGM domains (Lowder et al, 2005). If FliGN is positioned in an appropriate orientation near FliGM, the 43–157 crosslink can be accounted for while simultaneously satisfying the previous FliGM–FliGM constraints (Figure 5B). We conclude that the FliGN and FliGM subdomains are not widely separated as observed in the A. aeolicus crystal structure, but are in relatively close proximity in the flagellar motor of E. coli.
Figure 5.
Proximity of FliGN to FliGM. (A) Crosslinking of position 43 in FliGN to position 147 in FliGM by bis-maleimidohexane. Crosslinking was carried out at 23°C for 10 min. (B) A hypothetical arrangement of the FliGN and FliGM domains that could account for the observed FliGN–FliGM and FliGM–FliGM crosslinking. The FliGN domain (residues 5–89) is pale-cyan and FliGM (residues 104–184) is cyan. The segment linking the domains (residues 90–103) is yellow and the positions to which it would connect (carboxy-terminus of FliGN and amino-terminus of FliGM) are red and blue. The relative orientation of the FliGM domains is based on a previous study (Lowder et al, 2005), which identified positions giving efficient FliGM–FliGM crosslinking; one such pair (117–166) is shown. The orientation of FliGN, which is intended to be approximate only, is based on the observed FliGN–FliGM crosslink (A) and constraints imposed by the inter-domain connection (the length of the connecting helix and the positions it must connect). Spheres indicate positions of Cβ positions, (grey in FliGN and black in FliGM). Residue numbers are for the E. coli protein. For previously identified instances of crosslinking, including the indicated 117/166 Cys pair, see Lowder et al (2005).
Organization of torque-generating elements of the rotor
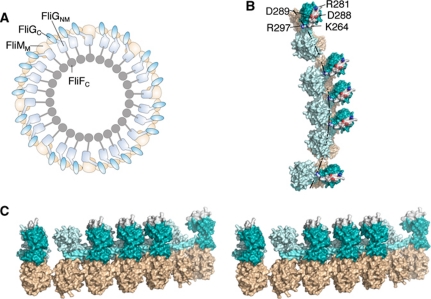
FliG binds to the MS-ring protein FliF (Oosawa et al, 1994; Kihara et al, 2000; Grunenfelder et al, 2003) and the available evidence indicates that both FliG and FliF are present in about 26 copies per motor (Jones et al, 1990; Francis et al, 1992; Sosinsky et al, 1992; Thomas et al, 2001, 2006; Suzuki et al, 2004). FliM is believed to be present in more, about 34, copies per motor (Thomas et al, 1999, 2006; Young et al, 2003). A subunit arrangement that can accommodate the different FliG and FliM copy numbers has been proposed (Brown et al, 2007). The dual FliM–FliG interactions that have been characterized here are key elements in the model. The FliMM domains are proposed to occur in two kinds of structural setting. Most are in an approximately vertical orientation, forming the outer wall of the C-ring and interacting with the hydrophobic patch of FliGC. A subset of FliMM domains, typically 8 or 9 (equal to the number of FliM subunits present in excess over FliG subunits), are tilted slightly inward where they interact with FliGM instead (Figure 6). FliG subunits are, therefore, also of two kinds; most are supported by a single FliM that is positioned under FliGC and binds through the hydrophobic patch, while a subset (again about 8 or 9) is bound to two FliM subunits and is thus supported through both the middle- and C-terminal domains (Figure 6A).
Figure 6.
Structural model for the upper part of the C-ring. (A) Overall plan of FliG and FliM organizations. The arrangement is similar to that proposed by Brown et al (2007), with adjustments to reflect more-current information on FliG structure. FliM is light brown and FliG is cyan. (B) More detailed view of a section of the rotor. Colouring is as in (A), but with the three parts of FliG (FliGNM, linking helix and FliGC) coloured with increasing intensity, and the active-site ridge shown in atom colours to highlight the conserved charged residues that interact with the stator (Zhou et al, 1998a). The dashed line indicates the hypothesized path of the stator (relative to the rotor) as the motor turns (see the text). (C) Stereo-view (crossed-eye) of a section of the rotor. The view is in a roughly radial direction (out-to-in). The active-site ridge on FliGC is coloured white.
The results here, combined with results in the previous structural study of FliMM (Park et al, 2006), allow us to develop this structural model in detail. To construct the model explicitly, FliMM domains were first positioned as they would be in a 34-member ring, in the relative orientation determined by crosslinking experiments of Park et al (2006). FliG domains (either FliGC or FliGNM, as dictated by the model) were positioned on top of the FliMM domains, in the orientations determined in the FliMM:FliGM co-crystal structure (Figure 2) or by FliMM:FliGC crosslinking (Figure 4). The structure of the complex when placed in the rotor simultaneously satisfies crosslinking constraints between adjacent subunits of FliMM and FliGM. The helix that joins FliGM and FliGC was extended straight to residue 193 (just before the Gly-Gly linker that joins it to the C-terminal domain), as observed in the FliGMC crystal structure (Brown et al, 2002). The subsets of FliMM domains that are associated with FliGNM were then tilted inward. This tilt was sufficient to bring the end of the linking helix close to the Gly-Gly linker of the adjacent FliGC domain, to which it connects in the model. This matching of termini required no other assumptions but did depend on the linking helix assuming the roughly tangential orientation observed in the present co-crystal structure, rather than the roughly radial orientation observed in the previous T. maritima FliGMC structure (Brown et al, 2002). The other FliGNM domains (those not bound to FliMM) were oriented similarly and were positioned to maintain, as nearly as possible, the same relationship with the attached FliGC. Like the other elements in the assembly, the FliGNM domains occur in slightly varied situations, in this case consisting of close groups of 3 or 4 separated by slightly larger gaps at the position of the inward-tilted FliMM (Figure 6). All of the FliGNM domains, including those not bound to FliMM, would also be held in place by attachment to FliF. The protein subsets in different environments might have different stabilities within the structure, and consistent with this, Delalez et al (2010) recently reported that about two thirds of the FliM subunits in the motor are in relatively rapid exchange whereas the rest are more stably bound.
EM reconstructions indicate that features in the outer part of the C-ring have ∼34-fold symmetry, whereas the inner lobe has roughly 26-fold symmetry (Supplementary Figure S6). In the structural model, the symmetry transition occurs between FliGNM (assigned to the inner lobe) and FliGC (assigned to the outer lobe) (Figure 6). The linking helix connects to FliGNM through an extended segment (residues 162–168 in E. coli numbering) that is relatively non-conserved, and to FliGC through the aforementioned Gly-Gly motif. Either of these linkages might provide flexibility to accommodate the symmetry mismatch between FliG and FliM (see Supplementary Figure S9 for an illustration).
The structural model developed here agrees well with the electron microscopic reconstructions (Thomas et al, 2001, 2006). The unified FliGNM domain has a size matching the inner lobe of density at the top of the C-ring (Supplementary Figure S7). The model also accounts for the effects of a deletion/fusion mutation removing large parts of FliG and FliF (Supplementary Figure S8). The ∼26-fold symmetry observed for the inner lobe in the reconstructions (Supplementary Figure S6) is well explained by an individual FliGNM domain in each lobe. FliGC can satisfactorily account for the outer lobe at the top of the C-ring (see Supplementary Figure S3 for shape comparison). While FliG is present in only about 26 copies, the 34-fold symmetry observed for the outer part of the C-ring arises because the FliGC domains are held in position by the underlying FliMM domains, which are present in 34 copies. The gaps in the upper edge of the C-ring (where FliMM is tilted inward and FliGC is absent) would have been obscured by the symmetry averaging of the EM reconstructions. The electron density in the outer lobes should reflect roughly three-fourths occupancy, and consistent with this, the outer lobe is substantially less intense than the inner lobe in most reconstructions (Figure 1; Supplementary Figure S3; Thomas et al, 2001, 2006).
To evaluate the consistency of our molecular model with the EM reconstructions, we built full representations of the upper C-rings composed from FliG and FliMM and then averaged the outer lobes over 34-fold symmetry and the inner lobes over 26-fold symmetry. Electron density for the protomer subunits was then calculated to 20 Å resolution and compared with the EM maps (Supplementary Figure S6). Our model reasonably recapitulates the general shape of the electron density in the outer and inner C-ring lobes and the greater weighting of electron density in the inner lobe. In contrast, the FliG organization proposed by Lee et al (2010) does not fit the density or shape of the inner lobe as well, nor does it account for the varied symmetries in the C-ring (Supplementary Figure S6). Their model would also result in overlap between adjacent FliM subunits when the FliMM domain is docked onto FliGM according to the co-crystal structure (Supplementary Figure S6). The model of Minamino and co-workers (2011) was not developed in sufficient detail to allow for detailed comparison with the EM reconstructions. We emphasize, however, that neither of the crystal contact-based models appears consistent with the present binding, crosslinking and structural results.
FliGC interacts with the stator protein MotA (Zhou et al, 1998a), and models for the rotation mechanism are typically focussed on the FliGC–MotA interface. In the structural model developed here, FliGC is absent in several positions around the rotor. Extensive physiological measurements have so far not given evidence of halting motor performance, provided the membrane is normally energized; even motors operating with a single stator unit (the full motor has about 10; Block and Berg, 1984; Blair and Berg, 1988; Reid et al, 2006) can rotate smoothly under high load (Reid et al, 2006) or rapidly under low load (Yuan and Berg, 2008). One of the charged residues of FliG that is important for rotation and that interacts with the stator (Arg 297 in the protein of E. coli) is at the inner edge of FliGC, close to FliGNM. We propose that the stator complexes are centred roughly above this position, where they could interact with both FliGC and FliGNM (Figure 5B). Interactions with more-inward parts of the rotor (the FliGNM domains) might be important for propelling the rotor through the gap positions. An interaction with the FliGNM domains might also be more consistent with the measured size of rotational steps, which average about 1/26th of a revolution (Sowa et al, 2005). While the FliGNM domains have not previously been implicated in motor rotation in the same way as FliGC, presently there is no evidence against their involvement, and the occurrence of some Mot− (immotile but flagellate) mutations near the C-terminus of FliF, in segments that are known to bind to FliGN (Grunenfelder et al, 2003), would be in accordance with this proposal.
NMR experiments gave evidence of a FliG–FliM interaction different from any found here (Dyer et al, 2009). That interaction involves surfaces of FliG and FliM that, if brought together, would orient the charged ridge of FliG downward and away from the stator. Such an interaction appears unlikely to occur in the fully assembled motor but may nevertheless occur, and have a useful role, in the cell. In a FliG molecule with its middle domain bound to FliM, the C-terminal domain could be re-oriented (by rotations in the Gly-Gly linker) to interact with FliMM in the way observed in the NMR experiments. This more-compact conformation might provide a means of stabilizing the protein before its assembly into the C-ring. The FliGM–FliGC interaction observed in FliG crystals could have a similar role, helping to shield hydrophobic surfaces from inappropriate interactions until the normal interaction partners become available in the later stages of C-ring assembly.
In a recent proposal for the motor mechanism, rotation occurs as one part of the stator presses inward against angled surfaces of the rotor, and as another part, engaged through electrostatic interactions, moves tangentially (Blair, 2009). The structural information here is consistent with the essential elements of that hypothesis, provided the stator is positioned to allow interactions with both FliGC and FliGNM, as proposed above (Supplementary Figure S10). Most importantly, the present structural model for the torque-generating elements should provide a useful framework for addressing the molecular details of rotation and switching.
Materials and methods
Protein preparation
Coding sequences for T. maritima FliM residues 1–249 (FliMNM, which contains the CheY-binding peptide and CheC-like domain) and FliG residues 117–195 (FliGM195, which includes the middle domain and the segment linking it to FliGC) were PCR cloned into the vector peT28a (Novagen) and expressed with a 6-histidine (His) tag in E. coli strain BL21-DE3 (Novagen) in LB broth with kanamycin selection (25 μg/ml). The proteins were purified on Nickel-NTA columns and the His tags were removed by thrombin digestion. The proteins were combined and run on a Superdex-200 sizing column (Pharmacia), followed by pooling of fractions and concentration (Centriprep; Amicon) in GF buffer (50 mM Tris pH 7.5, 150 mM NaCl and 4.5 mM DTT). The complex of FliMNM and FliGM was co-eluted a second time on Superdex-200 column and further concentrated for crystallization trials.
Crystallization and data collection
Multiple initial conditions for growing FliGM195/FliMNM complex crystals were found in commercial screening solutions (Hampton). The crystals with the best morphology appeared in a 2-ml drop (1:1 mixture of protein in GF buffer and reservoir) from a sealed well under vapour diffusion against a reservoir of 0.1 M MES pH 6.5, 10% dioxane and 1.6 M ammonium sulphate (Hampton Research). Diffraction data were collected under 100 K nitrogen stream at Cornell High-Energy Synchrotron Source (A1) on a CCD detector (Quantum-210, Area Detector System). The data sets were reduced and scaled using HKL200 (Otwinowski and Minor, 1997).
Structure determination and refinement
The FliGM195/FliMM complex structure was determined by molecular replacement (MR) with PHASER (McCoy et al, 2007) using as a model the RCSB deposited coordinates PDB codes 2HP7 (T. maritima FliM) and 1LKV (T. maritima FliG). Two FliMM domains and one FliGM domain were found by MR; the second FliGM was placed manually in the residual electron density. Several residues of FliGM195 (helix E) were removed from the initial model and rebuilt manually in XFIT (McRee, 1992). The final model was refined with the program CNS amidst cycles of manual model building (Brunger et al, 1998). Given the 3.5 Å resolution, only grouped B-factor refinement was applied. The model consists of two FliMM and two FliGM units that form an antiparallel dimer in the asymmetric unit through association of the FliMM α1 and α1’ helices and the truncated FliMM C-termini with the opposing FliGM. Data collection and refinement statistics are summarized in Supplementary Table 1.
Protein interface analysis
Protein interfaces were analysed by the Protein Interfaces, Surfaces and Assemblies service PISA at European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), authored by Krissinel and Henrick (2007). Conservation of surface-exposed residues on FliMM and FliGMC was mapped with the Consurf server (Ashkenazy et al, 2010) and interface complementarity was evaluated with SC (Lawrence and Colman, 1993).
Strains
E. coli strains and plasmids used are listed in Supplementary Table 2.
Site-directed mutagenesis and assays of motility
Mutagenesis was performed using the QuikChange method (Stratagene) with oligonucleotides synthesized in core facilities of the University of Utah. Mutations were confirmed by sequencing. For assays of function, strains with deletions in the relevant genes were transformed with wild-type or mutant plasmids, and motility in soft agar, swimming in liquid, and flagellation were measured as described previously (Tang and Blair, 1995). Motility plates contained tryptone broth and 0.27% bacto agar, appropriate antibiotic(s) and IPTG at concentrations of 0, 40 and 100 μM to allow function to be tested over a range of expression levels. Plates were incubated at 32°C and swarm diameters were measured at regular intervals. Rates were determined from plots of diameter versus time.
Binding assays
Binding of FliM to FliGC was measured using a pull-down assay with GST fused to residues 185–331 of FliG. Proteins were expressed separately in two strains, using plasmid pHT100 (Tang et al, 1996) derivatives to express the GST fusions to FliGC, pDB72 (Tang et al, 1996) to express FliM (or its variants) and pKP41 to express FliN. For most experiments, FliN was coexpressed with FliM because FliM alone is prone to aggregation (Mathews et al, 1998). Control experiments used GST only, expressed from plasmid pHT100. Most binding experiments used strain RP3098, a ΔflhDC mutant that expresses no flagellar genes from the chromosome (Tang et al, 1996).
Cells were cultured overnight at 32°C in 40 ml TB or LB containing appropriate antibiotics and 400 μM IPTG for expressing GST (pHT100) or GST-fused FliGC constructs. FliM and its mutant variants (pDB72) and FliN (pKP41) were cultured at the same condition containing appropriate antibiotics. IPTG (40 μM) was used to induce expression of FliM and 10 μM Na-salicylate to induce expression of FliN. Cells were harvested and resuspended in lysozyme-containing phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 5 mM EDTA and 0.2 mM APMSF (4-amidinophenylmethanesulphonyl fluoride)) and 0.1% CHAPS. Following a 1-h incubation on ice, the cells were further disrupted by sonication, debris was pelleted (16 000 g, 40 min, 4°C), and 50 ml of the supernatant was stored for use in estimating the amount of FliM present before addition of affinity beads. The rest (∼1 ml) was transferred to a clean tube, mixed with 150 μl of a 50% slurry of glutathione Sepharose 4B (Pharmacia) prepared according to the manufacturer's directions, and incubated for 1 h at room temperature with gentle rotation to allow binding. The Sepharose beads were then pelleted by a 1-min microcentrifuge spin, and washed twice with 1 ml of phosphate-buffered saline, each time pelleting again with a brief spin. The beads were then incubated with 50 μl of elution buffer (50 mM reduced glutathione in 50 mM Tris–HCl (pH 8.0)) for 10 min at room temperature with gentle rotation to release the GST–FliGC and associated proteins. Beads were then pelleted and the supernatant was collected for analysis by SDS–PAGE and immunoblotting using anti-FliM antibody.
Crosslinking
Initial crosslinking experiments were carried out using the catalyst Cu [1,10-phenanthroline]3. Plasmids expressing the Cys-substituted FliG and FliM proteins were co-transformed into the fliGM deletion strain DFB247. FliG-only, single-Cys mutants were transformed into the fliG deletion strain DFB225. Cells were cultured at 37°C for 4–5 h in LB medium containing required antibiotics and then diluted 100-fold into LB broth (containing antibiotics) and grown overnight with 50 μm IPTG at 37°C. Using A600 readings to estimate culture density, equal numbers of cells from each culture were transferred to a centrifuge tube, pelleted (3000 g, 10 min) and resuspended in 200 μl of motility buffer (0.067 M sodium chloride, 0.01 M potassium phosphate pH 7.0 and 10−4 M EDTA), then divided into two 100 μl fractions. Crosslinking reagent (11 μl in 50% ethanol) was added to one sample, and non-crosslinked controls received just the 50% ethanol. The crosslinking reagent contained 4 mM CuSO4 and 16 mM 1,10-phenanthroline, and was freshly prepared from a 1-M stock of phenanthroline in 95% ethanol and a 400-mM stock of CuSO4 in water. Samples were rotated gently for 5 min at room temperature. Reactions were quenched after 5 min by addition of N-ethylmaleimide (2.2 μl from a 1-M stock in 95% ethanol) and EDTA (12.6 μl from a 0.5-M stock). Cells were then mixed with non-reducing gel-loading buffer, boiled and used for electrophoresis.
Some crosslinking experiments used the bifunctional reagent BMH. In all, 100 μl of cells was mixed with 2 μl of 50 mM BMH (dissolved in dimethyl sulphoxide and stored at −20°C), and incubated at room temperature for 10 min. Reactions were quenched with N-ethylmaleimide (2 μl from a 1-M stock in 95% ethanol). Control samples received just DMSO. Cells were mixed with reducing gel-loading buffer, boiled, and used for electrophoresis.
SDS–PAGE and immunoblotting
Protein samples were separated on 10% SDS–PAGE gels and transferred onto nitrocellulose using a semidry transfer apparatus (Bio-Rad). Rabbit polyclonal antibody against FliM was prepared as described previously (Tang and Blair, 1995; Tang et al, 1995) and used at 1500-fold dilution. Haemagglutinin-tagged FliG was detected using mouse anti-HA antibody at 1000-fold dilution (Covance, USA). Bands were visualized using the Super Signal West Picoluminol system (Pierce) and X-ray film.
Supplementary Material
Acknowledgments
This work was supported by grants GM64664 and GM087260Z from the National Institutes of Health. We thank members of the Blair and Crane laboratories for discussions. KP gratefully acknowledges support from Dale A Stringfellow Graduate Fellowship in Microbiology and GGB acknowledges support under NIH fellowship F31 GM078789.
Author contributions: KP, GGB, AMB, BRC and DB designed research; KP, GGB and AMB performed experiments; KP, GGB, AMB, BRC and DB analysed data; and KP, BRC and DB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucl Acids Res 38 (Web Server issue): W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Anderson RA (1973) Bacteria swim by rotating their flagellar filaments. Nature 245: 380–382 [DOI] [PubMed] [Google Scholar]
- Blair DF (2009) Structure and mechanism of the flagellar rotary motor. In Pili and Flagella: Current Research and Future Trends, Jarrell K (ed), 8, p 238 Caister Academic Press: Norfolk [Google Scholar]
- Blair DF, Berg HC (1988) Restoration of torque in defective flagellar motors. Science 242: 1678–1681 [DOI] [PubMed] [Google Scholar]
- Blair DF, Berg HC (1990) The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60: 439–449 [DOI] [PubMed] [Google Scholar]
- Block SM, Berg HC (1984) Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309: 470–472 [DOI] [PubMed] [Google Scholar]
- Brown PN, Hill CP, Blair DF (2002) Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J 21: 3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Mathews MAA, Joss LA, Hill CP, Blair DF (2005) Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol 187: 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Terrazas M, Paul K, Blair DF (2007) Mutational analysis of the flagellar rotor protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol 189: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Delalez NJ, Wadhams GH, Rosser G, Xue Q, Brown MT, Dobbie IM, Berrie RM, Leake MC, Armitage JP (2010) Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acad Sci USA 107: 11347–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer CM, Vartanian AS, Zhou H, Dahlquist FW (2009) A molecular mechanism of bacterial flagellar motor switching. J Mol Biol 388: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM (1992) Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA 89: 6304–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Sosinsky GE, Thomas D, DeRosier DJ (1994) Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Glagolev AN, Skulachev VP (1978) The proton pump is a molecular engine of motile bacteria. Nature 272: 280–282 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pedrajo B, Minamino T, Kihara M, Namba K (2006) Interactions between C-ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol 60: 984–998 [DOI] [PubMed] [Google Scholar]
- Grunenfelder B, Gehrig S, Jenal U (2003) Role of the cytoplasmic C-terminus of the FliF motor protein in flagellar assembly and rotation. J Bacteriol 185: 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM (1993) Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol 175: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Macnab RM, Okino H, Aizawa S-I (1990) Stoichiometric analysis of the flagellar hook-(basal body) complex of Salmonella typhimurium. J Mol Biol 212: 377–387 [DOI] [PubMed] [Google Scholar]
- Kihara M, Miller GU, Macnab RM (2000) Deletion analysis of the flagellar switch protein FliG of Salmonella. J Bacteriol 182: 3022–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF (2001) Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40: 13041–13050 [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF (2004) Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43: 26–34 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Larsen SH, Adler J, Gargus JJ, Hogg RW (1974) Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA 71: 1239–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein-protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D (2010) Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-Y, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L-S, Kustu S, Berry EA, Wemmer DE (2001) Crystal structure of an activated response regulator bound to its target. Nat Struct Biol 8: 52–56 [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Blair DF (1997) Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol 266: 733–744 [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Tang H, Wang X, Billings S, Blair DF (1996) Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol 178: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SA, Whitby FG, Blair DF, Hill CP (1999) Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400: 472–475 [DOI] [PubMed] [Google Scholar]
- Lowder BJ, Duyvesteyn MD, Blair DF (2005) FliG subunit arrangement in the flagellar rotor probed by targeted crosslinking. J Bacteriol 187: 5640–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM (2003) How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100 [DOI] [PubMed] [Google Scholar]
- Mathews MAA, Tang HL, Blair DF (1998) Domain analysis of the FliM protein of Escherichia coli. J Bacteriol 180: 5580–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J App Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry JL, Murphy JW, Gonzalez-Pedrajo B (2006) The FliN-FliH interaction mediates localization of Flagellar export ATPase FliI to the C ring complex. Biochemistry 45: 11790–11798 [DOI] [PubMed] [Google Scholar]
- McRee DE (1992) XtalView: a visual protein crystallographic software system for X11/Xview. J Mol Graph 10: 44–47 [Google Scholar]
- Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV, Namba K (2011) Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol 9: e1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Oosawa K, Ueno T, Aizawa S-I (1994) Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol 176: 3683–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Beel BD, Simon MI, Bilwes AW, Crane BR (2004) In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci USA 101: 11646–11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR (2006) Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci USA 103: 11886–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Blair DF (2006) Organization of FliN subunits in the flagellar motor of E. coli. J Bacteriol 288: 2502–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Harmon J, Blair DF (2006) Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J Bacteriol 188: 5240–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SW, Leake MC, Chandler JH, Low CJ, Armitage JP, Berry RM (2006) The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA 103: 8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar MK, Paul K, Blair D (2010a) Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci USA 107: 9370–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar MK, Paul K, Blair DF (2010b) Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem 285: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Homma M (2000) Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275: 5718–5722 [DOI] [PubMed] [Google Scholar]
- Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM (1992) Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol 174: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky GE, Francis NR, DeRosier DJ, Wall JS, Simon MN, Hainfeld J (1992) Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc Natl Acad Sci USA 89: 4801–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y, Berry RM (2008) Bacterial flagellar motor. Q Rev Biophys 41: 103–132 [DOI] [PubMed] [Google Scholar]
- Sowa Y, Rowe AD, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM (2005) Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437: 916–919 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yonekura K, Namba K (2004) Structure of the rotor of the bacterial flagellar motor revealed by electron cryo-microscopy and single-particle image analysis. J Mol Biol 337: 105–113 [DOI] [PubMed] [Google Scholar]
- Tang H, Billings S, Wang X, Sharp L, Blair DF (1995) Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol 177: 3496–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Blair DF (1995) Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol 177: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Braun TF, Blair DF (1996) Motility protein complexes in the bacterial flagellar motor. J Mol Biol 261: 209–221 [DOI] [PubMed] [Google Scholar]
- Thomas D, Morgan DG, DeRosier DJ (2001) Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J Bacteriol 183: 6404–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ (2006) The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188: 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR, Morgan DG, DeRosier DJ (1999) Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA 96: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S-I, Eisenbach M (1993) Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA 90: 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF (2006) Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J Bacteriol 188: 1466–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones CJ, Macnab RM (1986a) Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol 168: 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab RM (1986b) Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol 166: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HS, Dang H, Lai Y, DeRosier DJ, Khan S (2003) Variable symmetry in Salmonella typhimurium flagellar motors. Biophys J 84: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Berg HC (2008) Resurrection of the flagellar rotary motor near zero load. Proc Natl Acad Sci USA 105: 1182–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Amsler CD, Matsumura P, Khan S (1996a) FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol 17: 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Pathak N, Jaffe H, Reese TS, Khan S (1996b) FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol 261: 195–208 [DOI] [PubMed] [Google Scholar]
- Zhou J, Blair DF (1997) Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol 273: 428–439 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lloyd SA, Blair DF (1998a) Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA 95: 6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF (1998b) Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180: 2729–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.