Abstract
Mitochondrial outer membrane permeabilization is a watershed event in the process of apoptosis, which is tightly regulated by a series of pro- and anti-apoptotic proteins belonging to the BCL-2 family, each characteristically possessing a BCL-2 homology domain 3 (BH3). Here, we identify a yeast protein (Ybh3p) that interacts with BCL-XL and harbours a functional BH3 domain. Upon lethal insult, Ybh3p translocates to mitochondria and triggers BH3 domain-dependent apoptosis. Ybh3p induces cell death and disruption of the mitochondrial transmembrane potential via the mitochondrial phosphate carrier Mir1p. Deletion of Mir1p and depletion of its human orthologue (SLC25A3/PHC) abolish stress-induced mitochondrial targeting of Ybh3p in yeast and that of BAX in human cells, respectively. Yeast cells lacking YBH3 display prolonged chronological and replicative lifespans and resistance to apoptosis induction. Thus, the yeast genome encodes a functional BH3 domain that induces cell death through phylogenetically conserved mechanisms.
Keywords: ageing, BH3 domain, mitochondrial apoptosis, MOMP, yeast
Introduction
Within the mitochondrial pathway of apoptosis, the release of apoptogenic factors from the intermembrane space and the dissipation of the mitochondrial transmembrane potential (Δψm) is controlled by pro- and anti-apoptotic members of the BCL-2 protein family, each characteristically possessing one to four BCL-2 homology domains (BH1–4) (Green and Kroemer, 2004; Antignani and Youle, 2006). The pro-apoptotic branch of the BCL-2 family includes the multidomain proteins BAX and BAK, as well as BH3-only proteins like BID or BIM (Kelekar and Thompson, 1998). Upon induction of apoptosis, several BH3-only proteins as well as the multidomain protein BAX are targeted to mitochondria where they induce mitochondrial outer membrane permeabilization (MOMP), thereby initiating the regulated disintegration of the cell (Green and Kroemer, 2004; Antignani and Youle, 2006).
BCL-2 family members are thought to activate phylogenetically ancient pathways in yeast, since the basic framework of metazoan cell death execution is conserved and functional in this model organism (Madeo et al, 1997, 2002; Ludovico et al, 2002; Wissing et al, 2004; Buttner et al, 2007; Eisenberg et al, 2007; Carmona-Gutierrez et al, 2010). Human BAX and other BCL-2 family members have been widely studied in yeast (Ligr et al, 1998; Priault et al, 1999; Khoury and Greenwood, 2008) based on the premise that yeast cells would constitute an environment devoid of endogenous BCL-2 proteins and hence simplify the interpretation of functional analyses.
Here, we report the unexpected finding that the yeast genome encodes a BH3 domain-containing protein (Ybh3p) that regulates the mitochondrial pathway of apoptosis in a phylogenetically conserved manner.
Results
Yeast Ynl305cp contains a BH3 domain
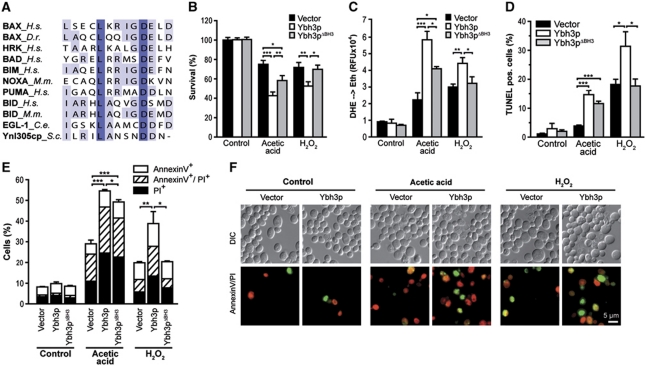
Analysis of the amino-acid sequence of Ynl305cp unveiled a putative BH3 domain within the C-terminus of this yeast protein, which displays a high degree of similarity to BH3 domains from higher eukaryotes (Figure 1A) (Kelekar et al, 1997). Consequently, we designated this protein ‘yeast BH3-only protein’ (Ybh3p). We generated yeast cells overexpressing Ybh3p under the control of an inducible promoter and examined its effects on clonogenic survival. The overexpression of Ybh3p per se did not affect the survival of yeast cells in resting conditions within 24 h. However, Ybh3p sensitized cells to the lethal effect of moderate doses of H2O2 and acetic acid (Figure 1B). Death mediated by Ybh3p was accompanied by a burst of reactive oxygen species (ROS), as assessed by the ROS-driven conversion of non-fluorescent dihydroethidium (DHE) to fluorescent ethidium (Figure 1C). A Ybh3p mutant lacking the C-terminal putative BH3 domain (Ybh3pΔBH3) was markedly less effective in inducing ROS accumulation (indicated by DHE oxidation) and loss of clonogenic survival than full-length Ybh3p (Figure 1B and C). Death induced by H2O2 completely depended on the presence of the BH3 domain. Instead, death upon acetic acid treatment was only partly inhibited by deletion of the BH3 domain, pointing to an additional lethal function within this protein (Figure 1B and C). Similar results were obtained upon overexpression of Ybh3p and Ybh3pΔBH3 in cells deleted of endogenous YBH3 (Supplementary Figure S1A–C). Additionally, elevated levels of Ybh3p but not of Ybh3pΔBH3 triggered cell death upon high doses of rapamycin (Supplementary Figure S1D), a treatment known to activate the mitochondrial pathway of apoptosis (Paglin et al, 2005).
Figure 1.
Yeast Ynl305cp contains a bona fide BH3 domain. (A) Sequence comparison of BH3 domains of indicated BCL-2 family members and Ynl305cp. Following sequences were used: BAX (Homo sapiens, Q07812), BAX (Danio rerio, AF231015), HRK (H. sapiens, NP_003797), BAD (H. sapiens, CAG46757), BIM (H. sapiens, O43521), NOXA (also known as PMAIP1, Mus musculus, BAA95781), PUMA (also known as BBC3, H. sapiens, Q9BXH1), BID (H. sapiens, P55957) BID (M. musculus, P70444), EGL-1 (Caenorhabditis elegans, O61667) and Ynl305cp (S. cerevisiae, AAT92698). (B, C) Survival determined by clonogenicity (B) and quantification of ROS accumulation (DHE → Eth) (C) of Ybh3p- or Ybh3pΔBH3-overexpressing cells or corresponding vector control treated or not with 120 mM acetic acid or 0.6 mM H2O2 (mean±s.e.m., n=6). (D) Quantification of DNA fragmentation using TUNEL staining of Ybh3p- or Ybh3pΔBH3-overexpressing cells or corresponding vector control after treatment with or without 120 mM acetic acid or 0.6 mM H2O2, respectively. In each experiment, 30 000 cells were analysed using flow cytometry (mean±s.e.m., n=4). (E) Quantification of phosphatidylserine externalization and loss of membrane integrity using AnnexinV/PI-co-staining of Ybh3p- or Ybh3pΔBH3-overexpressing cells or corresponding vector control after treatment with or without 120 mM acetic acid or 0.6 mM H2O2, respectively. In each experiment, 30 000 cells were analysed using flow cytometry (mean±s.e.m., n=4). (F) Representative micrographs of AnnexinV (green)/PI (red)-co-staining of cells overexpressing Ybh3p or harbouring the empty vector after treatment with or without 120 mM acetic acid or 0.6 mM H2O2, respectively. *P<0.05, **P<0.01 and ***P<0.001.
To characterize the nature of cell death triggered by Ybh3p, we quantified phenotypic changes indicative of apoptosis. While apoptotic DNA fragmentation was detected by TUNEL staining (Figure 1D), combined Annexin V/propidium iodide (PI) staining was used to detect cell surface exposure of phosphatidylserine, an early apoptotic event (Annexin V+), and loss of membrane integrity, indicating necrotic cell death (PI+). This allows for the discrimination between early apoptotic (Annexin V+/PI−), late apoptotic/secondary necrotic (Annexin V+/PI+) and necrotic (Annexin V−/PI+) death (Figure 1E and F). Ybh3p-facilitated cell death was accompanied by an increase in both apoptotic and necrotic markers (Figure 1D–F). Upon treatment with acetic acid or H2O2, Ybh3p enhanced phosphatidylserine externalization and DNA fragmentation. At the same time, the percentage of AnnexinV−/PI+ cells doubled upon Ybh3p overexpression, suggesting an increase in necrotic cell death (Figure 1E and F). Depending on the death stimulus, deletion of the BH3 domain prevented (H2O2) or reduced (acetic acid) the occurrence of apoptotic and necrotic cell death markers (Figure 1D–F). To further elucidate the apoptotic pathway deployed by Ybh3p, we overexpressed Ybh3p in the absence of identified key players of yeast apoptosis, including the metacaspase Yca1p, the serine protease Nma111p (HTRA2/Omi), the apoptosis-inducing factor Aif1p and the mitochondrial endonuclease Nuc1p (ENDOG) (Supplementary Figure S2A). Ybh3p-mediated cell death was not affected by the deletion of any of these genes, suggesting that Ybh3p triggers a cell death pathway independent of Yca1p, Aif1p, Nma111p or Nuc1p. Of note, Ybh3p, Yca1p and Nuc1p all induced comparable death rates when overexpressed under the same conditions (Supplementary Figure S2B and C).
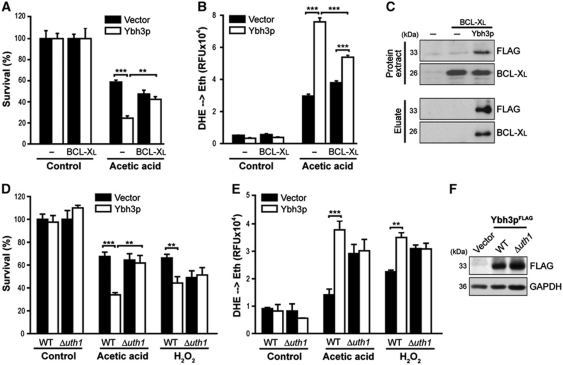
Cell killing mediated by pro-apoptotic BCL-2 family members like BAX and BAK can be antagonized by anti-apoptotic members like BCL-2 or BCL-XL. Consistently, death and ROS accumulation (evidenced by oxidation of DHE) induced by Ybh3p were inhibited by co-expression of human BCL-XL (Figure 2A and B). Furthermore, immunoprecipitation demonstrated Ybh3p to interact with BCL-XL (Figure 2C). The mitochondrial outer membrane protein Uth1p, a regulator of ageing, oxidative stress responses and mitochondrial biogenesis (Kennedy et al, 1995; Bandara et al, 1998; Camougrand et al, 2004) has been identified as a molecular determinant for mammalian BAX-induced yeast death (Camougrand et al, 2003). Uth1p was found to be dispensable for targeting human BAX to yeast mitochondria, yet was required for BAX-induced ROS production, MOMP (Camougrand et al, 2003) and mitophagy (Kissova et al, 2006). Similarly, we found that Ybh3p lost its capacity to facilitate cell killing and ROS generation in the absence of Uth1p (Figure 2D and E), although deletion of UTH1 had no effect on the stability of Ybh3p (Figure 2F). Overexpression of Uth1p did not influence the lethal effects of Ybh3p (Supplementary Figure S2D and E). In conclusion, these data suggest that Ybh3p acts via a mitochondrial cell death pathway similar to that activated by mammalian BAX.
Figure 2.
BCL-XL interacts with Ybh3p and inhibits Ybh3p-mediated death. (A, B) Survival determined by clonogenicity (A) and quantification of ROS accumulation using DHE → Eth conversion (B) of cells overexpressing Ybh3p alone or co-expressing Ybh3p and BCL-XL and the corresponding vector controls upon treatment with or without 120 mM acetic acid. Survival was normalized to untreated control cultures (mean±s.e.m., n=5). (C) Protein extracts of cells expressing BCL-XL either alone or in combination with FLAG-tagged Ybh3p as well as of cells harbouring both empty vectors were used to immunoprecipitate FLAG-tagged Ybh3p using agarose beads coupled with a monoclonal anti-FLAG antibody. Immunoprecipitates (eluates) as well as whole protein extracts were analysed via immunoblot for BCL-XL and Ybh3p. (D, E) Survival determined by clonogenicity (D) and quantification of ROS accumulation using DHE → Eth conversion (E) of WT and Δuth1 cells overexpressing Ybh3p or harbouring the empty vector upon treatment with or without 120 mM acetic acid or 0.6 mM H2O2. Survival was normalized to untreated isogenic vector control (mean±s.e.m., n=6). (F) Immunoblot analysis of WT and Δuth1 cells for FLAG-tagged Ybh3p. **P<0.01 and ***P<0.001.
Yeast BH3 peptide promotes apoptotic changes on mammalian cells and mitochondria
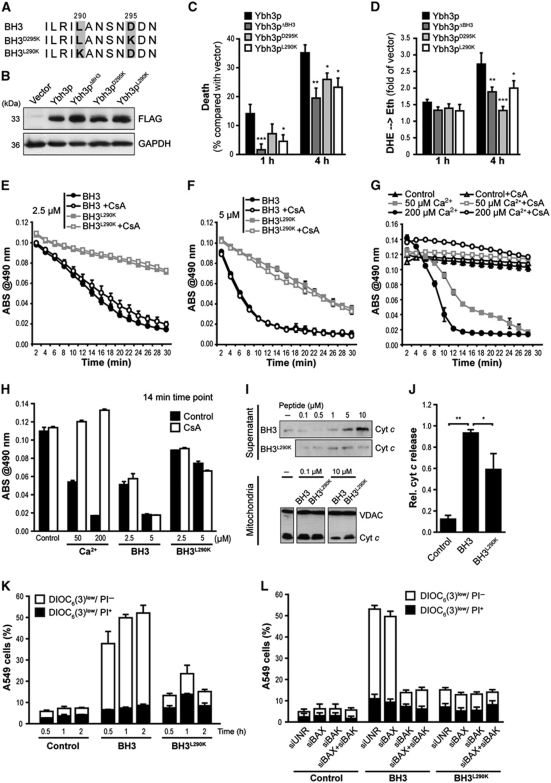
To further evaluate the contribution of the putative BH3 domain to the pro-apoptotic activity of Ybh3p, we generated constructs harbouring separate single point mutations in two highly conserved residues (Ybh3pL290K and Ybh3pD295K) (Figure 3A). Although expressed at similar levels, both point mutants were less active in inducing ROS production and cell death than native Ybh3p (Figure 3B–D). We next determined whether the BH3 domain of yeast Ybh3p can permeabilize mammalian mitochondria and kill mammalian cells. Incubation of isolated mouse liver mitochondria with the native BH3 (but not BH3L290K) peptide induced mitochondrial swelling (Figure 3E–H) and caused rapid release of cytochrome c (cyt c) from mitochondria (Figure 3I and J). We verified equal mitochondrial uptake of the BH3 and BH3L290K peptides (Supplementary Figure S3A). Potential mechanisms behind BH3 domain-mediated MOMP are highly debated (Green and Kroemer, 2004; Antignani and Youle, 2006). One controversial question is whether the permeability transition pore (PTP), a complex of proteins that consists of cyclophilin D as essential subunit, the adenine nucleotide translocase (ANT) as regulatory component and additional factors (Baines et al, 2005; Nakagawa et al, 2005; Schinzel et al, 2005; Baines, 2009), is required for BAX-induced apoptosis (Marzo et al, 1998; Shimizu et al, 1999; Kumarswamy and Chandna, 2009). While pre-incubation of mitochondria with the PTP-inhibitor cyclosporine A (CsA) severely reduced mitochondrial swelling promoted by Ca2+ (Figure 3G and H), BH3 domain-induced swelling was unaffected (Figure 3E, F and H), indicating that the yeast BH3 domain can permeabilize murine mitochondria through a CsA-insensitive mechanism.
Figure 3.
The yeast BH3 domain induces apoptotic changes in isolated mitochondria as well as in mammalian and yeast cells. (A) Amino-acid sequence alignment of the native yeast BH3 domain and the BH3D295K and BH3L290K point mutants. (B) Immunoblot analysis of WT yeast cells for FLAG-tagged Ybh3p, Ybh3pΔBH3, Ybh3pD295K or Ybh3pL290K. (C, D) Death determined by clonogenicity (C) and quantification of ROS accumulation using DHE → Eth conversion (D) of yeast cells overexpressing Ybh3p, Ybh3pΔBH3, Ybh3pD295K or Ybh3pL290K after treatment with 120 mM acetic acid for 1 or 4 h. Death rates were calculated by setting the survival of vector control cells to 100% and thus 0% death (mean±s.e.m., n=8). (E–H) Swelling of mouse liver mitochondria as indicated by loss of absorbance (ABS) at 490 nm. Mitochondria were pre-incubated or not with 5 μM CsA, followed by the administration of 2.5 μM (E) or 5 μM (F) of native (BH3) or mutated (BH3L290K) peptide or indicated concentrations of Ca2+(G). To permit direct comparison, swelling after 14 min under all conditions analysed was depicted in (H). Representative experiments are shown, with data representing mean±s.e.m. of three replicates. (I, J) Immunoblot analysis of supernatants and mitochondrial pellets (I) from mouse liver mitochondria incubated with the indicated concentration of native (BH3) or mutated (BH3L290K) peptide using an antibody specific for cyt c. For supernatants, the signal intensities of cyt c were quantified and normalized to VDAC intensities of deployed mitochondria (J) (mean±s.e.m., n=3). (K) Non-small cell lung cancer (A549) cells were incubated with 30 μM of the native (BH3) or mutated (BH3L290K) peptide, followed by the flow cytometric assessment of Δψm dissipation (DiOC6(3)low) and loss of plasma membrane integrity (PI+) (mean±s.e.m., n=5). (L) A549 cells transfected with a control siRNA (siUNR) or with siRNA targeting BAX, BAK or with a combination of these were treated with 30 μM of the native (BH3) or mutated (BH3L290K) peptide followed by the flow cytometric assessment of Δψm dissipation (DiOC6(3)low) and loss of plasma membrane integrity (PI+). Data represent mean±s.e.m. and one representative experiment is shown with n=3. *P<0.05, **P<0.01 and ***P<0.001.
Next, human non-small cell lung cancer cells (A549) were incubated with the yeast BH3 peptide fused to an Antennapedia plasma membrane translocation domain, which allows the peptide to enter cells. Addition of this fusion peptide resulted in Δψm dissipation and cell death depending on the conserved L290 residue within the BH3 domain (Figure 3K). Cellular uptake of the BH3 and BH3L290K peptides was similar (Supplementary Figure S3B). Notably, knockdown of BAX by small-interfering RNA (siRNA) did not affect apoptotic changes triggered by the BH3 peptide, while knockdown of BAK completely inhibited Δψm dissipation (Figure 3L). Thus, the native yeast BH3 peptide triggers apoptosis of human cancer cells by a mechanism that strictly requires the presence of BAK.
Upon apoptosis induction, the yeast BH3 protein localizes to mitochondria
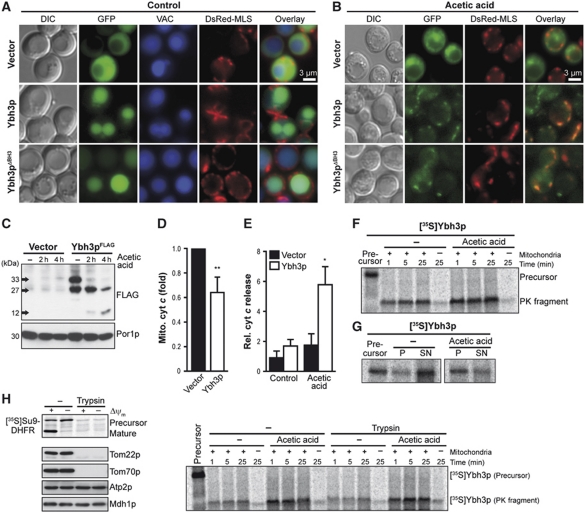
Upon apoptotic insults, several pro-apoptotic members of the BCL-2 protein family are targeted to mitochondria where they induce MOMP and eventually cellular demise (Green and Kroemer, 2004; Antignani and Youle, 2006; Lovell et al, 2008). Similarly, GFP-tagged Ybh3p translocated from predominantly vacuolar sites in healthy cells to discrete cytoplasmic foci upon acetic acid treatment, partly co-localizing with a fluorescent protein (DsRed) targeted to mitochondria (Figure 4A and B). The distribution of GFP-Ybh3p was indistinguishable from that of GFP-Ybh3pΔBH3, indicating that the yeast BH3 domain, though necessary for Ybh3p-mediated cell death (Figure 1B and C), is dispensable for the subcellular redistribution of Ybh3p (Figure 4A and B). Immunoblot analysis of the mitochondrial fractions from cells overexpressing C-terminally tagged Ybh3pFLAG revealed the presence of a 33 kDa protein (full-length Ybh3p) and a smaller fragment of 27 kDa, both of which were progressively degraded in cells treated with acetic acid leading to a FLAG-immunoreactive molecule of 12 kDa after 4 h (Figure 4C). Whether this represents a specific cleavage product or is the result of unspecific proteolytic degradation remains elusive yet. The mitochondrial integration of Ybh3p coincided with the release of cyt c from yeast mitochondria (Figure 4D; Supplementary Figure S4A). Consistently, the cytosol from cells overexpressing Ybh3p was enriched in cyt c upon treatment with acetic acid (Figure 4E; Supplementary Figure S4B).
Figure 4.
Ybh3p translocates to mitochondria and is imported in a TOM-receptor-independent way. (A, B) Fluorescence microscopy of yeast cells expressing GFP-tagged Ybh3p or Ybh3pΔBH3 or harbouring the vector control for 16 h before (A) and after (B) treatment with acetic acid. Vacuoles were counterstained using Celltracker Blue. For visualization of mitochondria, the mitochondrial marker DsRed Su1-69 (DsRed-MLS) was co-expressed. (C) Immunoblot analysis of mitochondrial fractions harvested from cells overexpressing FLAG-tagged Ybh3p without or with acetic acid treatment for 2 and 4 h before subcellular fractionation. (D) The cyt c content of mitochondrial fractions harvested from cells overexpressing Ybh3p compared with isogenic vector control after 4 h of acetic acid treatment determined by immunoblot analysis and subsequent quantification of cyt c signal compared with porin signal (Por1p, mitochondrial marker) (mean±s.e.m., n=6). (E) The cyt c content in supernatants after release of cytosolic content via short permeabilization. Cells overexpressing Ybh3p or harbouring the empty vector were pre-treated or not with 120 mM acetic acid for 2 h prior to permeabilization followed by immunoblot analysis. Cyt c signal in supernatants was normalized to cyt c content in whole cell extracts (mean±s.e.m., n=5). (F, G) 35S-labelled Ybh3p was incubated with isolated yeast mitochondria, followed by treatment with proteinase K (F) or carbonate extraction (G) and analysis by NuPAGE and autoradiography. For carbonate extraction, supernatants (SN) and pellets (P) were analysed. Cells were grown in the presence or absence of acetic acid prior to isolation of mitochondria. (H) Yeast mitochondria isolated from cells grown in the presence or absence of 90 mM acetic acid were incubated with 35S-labelled Su9-DHFR for 30 min or with 35S-labelled Ybh3p for the indicated time followed by treatment with proteinase K and analysis by NuPAGE and autoradiography. Isolated mitochondria were incubated with or without trypsin prior to the import reaction. In addition, mitochondria were analysed for Tom22p, Tom70p, Atp2p and Mdh1 using immunoblotting. Where indicated the membrane potential (Δψ) was dissipated prior to the import reaction. See also Supplementary Figure S4. *P<0.05 and **P<0.01.
In organello mitochondrial import studies using radioactively labelled [35S]Ybh3p revealed that Ybh3p was incorporated into the membrane of isolated mitochondria (as shown by a proteinase K resistant fragment) (Figure 4F). This import seemed more efficient into mitochondria isolated from cells that had been pre-treated with acetic acid and were hence undergoing apoptosis. Additionally, carbonate extraction to reveal the portion of membrane-embedded Ybh3p demonstrated that the membrane integration of Ybh3p was elevated upon apoptosis induction (Figure 4G). Remarkably, shaving of mitochondria with trypsin to remove outer mitochondrial TOM receptors (e.g. Tom22p and Tom70p) did not affect the level of integrated Ybh3p (Figure 4H), while it completely abrogated the import of [35S]Su9-DHFR, a protein that requires TOM receptors for its import into mitochondria (Figure 4H). Thus, the integration of Ybh3p into mitochondria occurs independently of mitochondrial surface proteins (such as TOM receptors). In summary, these results demonstrate that Ybh3p can target mitochondria of both healthy and stressed cells, yet it is preferentially imported into mitochondria of cells destined to die.
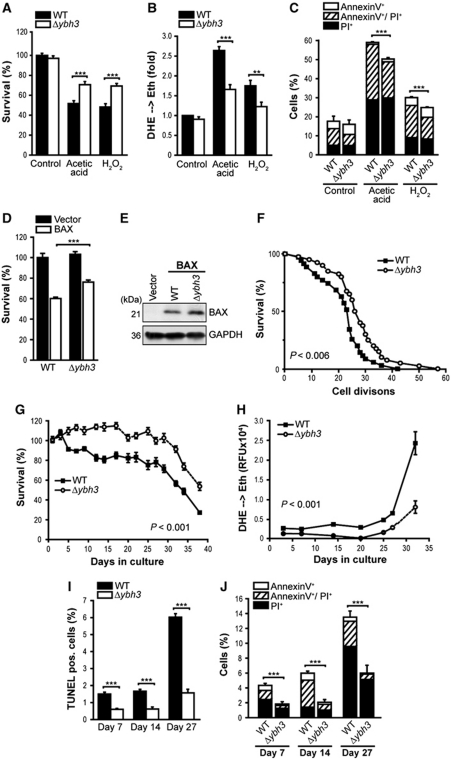
Deletion of YBH3 protects against cell death induced by H2O2, acetic acid, mammalian BAX expression and ageing
The finding that overexpressed Ybh3p sensitized cells to apoptotic stimuli prompted us to analyse the apoptotic response of cells devoid of YBH3 (Δybh3). Consistent with the effects of its overexpression, the deletion of YBH3 protected cells against various apoptotic insults (Figure 5; Supplementary Figure S5). Upon treatment with H2O2 or acetic acid, Δybh3 cells displayed reduced death, DHE-detectable ROS accumulation and apoptotic phosphatidylserine externalization (Figure 5A–C), yet were undistinguishable from control cells with respect to the proportion of necrotic Annexin V−/PI+ cells (Figure 5C). Thus, Ybh3p regulates apoptotic cell death. The effect of YBH3 deletion upon acetic acid treatment was further compared with that of YCA1, NUC1 and TOR1 (target of rapamycin) (Supplementary Figure S5A and B). While deletion of YBH3 and TOR1, which has previously been shown to protect against a variety of stresses (Wei et al, 2008; Almeida et al, 2009), yielded the strongest cytoprotective effects, deletion of YCA1 caused a strong decrease in ROS production (as indicated by DHE oxidation). Deletion of NUC1, which reportedly enhances necrotic death on fermentative media (Buttner et al, 2007), sensitized cells to death induction. Assessment of survival upon treatment with various death inducers showed that deletion of YBH3 protected from heat shock, rapamycin and Cu2+, but had no effect on death induced by dithiothreitol (DTT) or amiodarone (AM) (Supplementary Figure S5C).
Figure 5.
Deletion of YBH3 protects against H2O2, acetic acid, BAX expression and ageing. (A, B) Survival determined by clonogenicity (A) and quantification of ROS accumulation (DHE → Eth) (B) of wild-type (WT) and Δybh3 cells treated or not with acetic acid or H2O2 (mean±s.e.m., n=8). (C) Flow cytometric quantification of phosphatidylserine externalization and loss of membrane integrity using AnnexinV/PI co-staining of cells described in (A) (mean±s.e.m., n=4). (D) Survival determined by clonogenicity upon heterologous expression of murine BAX in WT and Δybh3 cells. Viability was determined after 20 h of BAX expression (mean±s.e.m., n=8). (E) Immunoblot analysis of WT and Δybh3 cells expressing BAX. (F) Replicative lifespan of WT and Δybh3 cells. A representative experiment is shown with n⩾40. Indicated P-value (calculated using Tarone-Ware) refers to median lifespan of Δybh3 compared with WT. (G, H) Survival determined by clonogenicity (G) and quantification of ROS accumulation using DHE → Eth conversion (H) of WT and Δybh3 cells during chronological ageing on glycerol-containing media. Colony forming units of WT cells at day 1 were set to 100%. One representative ageing experiment is shown, with data representing mean±s.e.m. of six independent cultures. (I, J) Flow cytometric quantification of TUNEL staining (I) and AnnexinV/PI-co-staining (J) of chronologically aged WT and Δybh3 cells at indicated days during ageing (mean±s.e.m., n=4). See also Supplementary Figure S5. **P<0.01 and ***P<0.001.
Interestingly, mammalian BAX has been demonstrated to require the presence of tBID for efficient mitochondrial integration and subsequent membrane permeabilization (Lovell et al, 2008; Leber et al, 2010). Therefore, we addressed whether Ybh3p cooperates with heterologously expressed BAX to promote lethality and expressed murine BAX in yeast cells that were either deficient or proficient for Ybh3p. Despite being expressed at similar levels, BAX was more efficient in killing wild-type cells than Δybh3 cells (∼40 and ∼25% dead cells in WT and Δybh3, respectively) (Figure 5D and E).
Cellular ageing across species is causally associated with increased apoptosis (Purdom and Chen, 2003; Herker et al, 2004; Brust et al, 2010). We thus evaluated the contribution of Ybh3p to the termination of chronological lifespan (which corresponds to the ageing of post-mitotic metazoan cells) and replicative lifespan (which reflects ageing of mitotic metazoan cells, including stem cells). YBH3 deletion resulted in a significant extension of replicative lifespan (Figure 5F). Moreover, during chronological ageing on a non-fermentable carbon source (glycerol), which amplifies mitochondrial mass (Stevens, 1981) and, therefore, might increase the impact of mitochondrial death effectors, Δybh3 cells exhibited improved survival and reduced ROS levels when compared with wild-type cells (Figure 5G and H). In addition, the deletion of YBH3 efficiently protected against apoptotic and (upon prolonged ageing) necrotic death as measured by staining with Annexin V/PI and TUNEL (Figure 5I and J). Re-introduction of Ybh3p (FLAG-tagged or GFP-tagged) but not that of Ybh3pΔBH3 could reverse the protective effect of YBH3 deletion during chronological ageing on glycerol (Supplementary Figure S5D–G). These data posit Ybh3p as the first known pro-apoptotic yeast protein that regulates both the replicative and chronological lifespans.
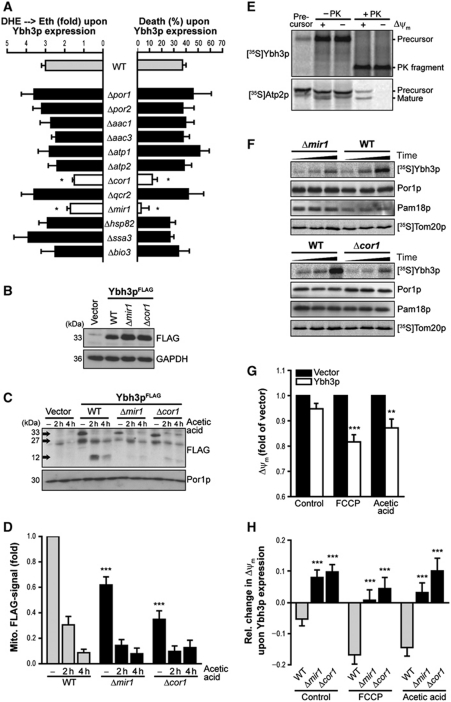
Ybh3p-mediated breakdown of mitochondrial membrane potential and cell death depend on Mir1p and Cor1p
To identify regulators of Ybh3p-mediated apoptosis, we immunoprecipitated FLAG-tagged Ybh3p and used mass spectrometry to reveal the identities of co-purified proteins. The majority of isolated proteins were mitochondrial, including an ANT isoform as putative regulator of the PTP (Supplementary Table S1). We determined the capacity of Ybh3p to induce ROS production and lethality in deletion mutants of all proteins that co-purified with FLAG-tagged Ybh3p. Among these, Cor1p, a core subunit of the ubiqinol-cyt c oxidoreductase complex, and Mir1p, a mitochondrial phosphate carrier, were indispensable for the cytocidal activity of Ybh3p (Figure 6A). Immunoblot analyses of mitochondrial translocation of Ybh3pFLAG revealed that the mitochondrial relocalization of Ybh3p was compromised in both Δmir1 and Δcor1 cells (Figure 6C and D) despite comparable expression levels of Ybh3p in wild-type, Δmir1 and Δcor1 cells (Figure 6B–D). In this context, we were unable to detect Ybh3p in the cytosolic fraction (Supplementary Figure S6A).
Figure 6.
Mir1p and Cor1p facilitate mitochondrial import of Ybh3p as well as Ybh3p-mediated breakdown of mitochondrial membrane potential and subsequent cell death. (A) Death and ROS accumulation of wild-type (WT) cells and indicated deletion mutants overexpressing Ybh3p upon treatment with 120 mM acetic acid. Percentage of death as well as fold of DHE → Eth conversion mediated by overexpression of Ybh3p compared with corresponding isogenic vector control was plotted (mean±s.e.m., n=6–8). (B) Immunoblot analysis of Ybh3pFLAG expression in WT, Δmir1 and Δcor1 cells. (C, D) Representative immunoblot (C) and quantification of mitochondrial full-length Ybh3pFLAG normalized to porin (Por1p, mitochondrial loading control) (D) in mitochondrial fractions isolated from WT, Δmir1 and Δcor1 cells overexpressing Ybh3pFLAG treated 120 mM with acetic acid for 2 and 4 h or left untreated (mean±s.e.m., n=12). (E) [35S]-labelled precursor proteins were incubated with yeast mitochondria followed by treatment with proteinase K (+ PK). Plus and minus signs denote whether Δψm had been dissipated prior to the import reaction. (F) Yeast mitochondria isolated from WT, Δmir1 and Δcor1 cells were incubated with [35S]-labelled Ybh3p for 1, 5 and 25 min or with [35S]-labelled Tom20p for 1, 4 and 20 min followed by carbonate extraction (pH 10.5) and analysis by NuPAGE and autoradiography. In addition, mitochondrial carbonate resistant pellets from the Ybh3p import reactions were analysed for Por1p and Pam18p as markers for outer and inner membrane using immunoblotting. (G) Measurement of Δψm using TMRM in yeast cells overexpressing Ybh3p or harbouring the empty vector. Cells were treated or not with acetic acid or FCCP before flow cytometric analysis. TMRM fluorescence upon Ybh3p overexpression compared with similarly treated vector control was indicated (mean±s.e.m., n=12). (H) Change of Δψm using flow cytometric assessment of TMRM fluorescence upon Ybh3p overexpression in WT, Δmir1 and Δcor1 cells treated with acetic acid or FCCP or left untreated. Fold change compared with isogenic vector control was plotted (mean±s.e.m., n=12). See also Supplementary Figure S6 and Supplementary Table S1. *P<0.05, **P<0.01 and ***P<0.001.
In vitro translation, coupled with mitochondrial import studies, demonstrated that the disruption of Δψm did not impede the insertion of Ybh3p into mitochondrial membranes, although it did abrogate the import of the ATPase subunit Atp2p, which served as a positive control (Figure 6E). Thus, an intact Δψm is neither required for mitochondrial targeting nor for subsequent import of Ybh3p.
Mitochondria isolated from Δmir1 and Δcor1 cells were tested for their capability to incorporate radioactively labelled [35S]Ybh3p and subjected to carbonate extraction to reveal the amount of membrane-embedded Ybh3p. In the absence of either Mir1p or Cor1p, the efficiency of Ybh3p membrane insertion was markedly decreased, while the import of [35S]Tom20p was unaffected, suggesting a rather specific role of Mir1p and Cor1p instead of a general import function (Figure 6F). While overexpression of Cor1p alone or in combination with Ybh3p did not alter cell survival, Mir1p overexpression clearly sensitized cells to death upon acetic acid treatment (Supplementary Figure S6B and C). Notably, the effects of Mir1p and Ybh3p amplified each other, as combined high levels of both proteins aggravated death and ROS accumulation. Additionally, the protective effect of YBH3 deletion was completely abrogated in cells devoid of Mir1p (Supplementary Figure S6D), suggesting that Ybh3p and Mir1p act in the same cell death pathway. The death of cells with defects in this signalling pathway can no longer be prevented by deletion of YBH3.
Mammalian BAX is known to directly affect the Δψm during apoptosis (Shimizu et al, 1999). To assess a similar role for Ybh3p, Δψm was examined using the Δψm-sensitive dye TMRM in Ybh3p-overexpressing cells treated with acetic acid or FCCP, a protonophore that causes Δψm dissipation. Even in the absence of apoptotic stimuli, a slight Δψm decrease was detectable in Ybh3p-overexpressing cells (Figure 6G), and this effect was exacerbated by acetic acid or FCCP (Figure 6G). In both Δmir1 and Δcor1 cells, Ybh3p completely failed to dissipate Δψm (Figure 6H). Thus, Ybh3p-induced Δψm dissipation correlates with the efficiency of its integration into mitochondria.
Knockdown of the human orthologues of Cor1p (QCR1) or Mir1p (PHC) reduces BAX aggregation and attenuates apoptosis in U2OS cells
Based on the conserved nature of Mir1p and Cor1p, we hypothesized that the orthologues of these proteins might regulate BAX-mediated apoptosis in human cells. The human orthologue of yeast Cor1p is QCR1 (UQCR1), a nuclear-encoded component of mitochondrial complex III, and that of Mir1p is SLC25A3 (solute carrier family 25, member 3), also known as phosphate carrier (PHC). Interestingly, PHC, a component of the ATP ‘synthasome’ (which also includes ANT and the FOF1 ATPase) (Alcala et al, 2008) has been previously implicated as a regulator of apoptosis in human cells because it physically interacts with the human cytomegalovirus-encoded BCL-2 analogue vMIA (Poncet et al, 2006) and triggers apoptosis upon overexpression (Alcala et al, 2008). Knockdown of QCR1 or PHC by siRNAs (Figure 7H) was performed on a human osteosarcoma cell line (U2OS) that was stably transfected with GFP-tagged human BAX. Treatment of these cells with the anti-cancer agent mitoxantrone (MTX) caused the redistribution of GFP-BAX from a diffuse (cytosolic) to a punctate (mitochondrial) pattern, as determined by fluorescence video microscopy and automated image analysis of GFP granularity (Figure 7A and B). Knockdown of either QCR1 or PHC significantly reduced the mitochondrial redistribution of BAX. In addition, transfection with siRNAs specific for either QCR1 or PHC, but not with an unrelated control siRNA (siUNR), led to a clear reduction in caspase activity (Figure 7C and D) and to an inhibition of MTX-induced Δψm dissipation, as determined with two distinct Δψm-sensitive dyes, CMTMRos and DIOC6(3) (Figure 7E–G). Notably, MTX-induced Δψm dissipation occurred well before the rupture of the plasma membrane, as determined by DIOC6(3)/PI co-staining (Figure 7G). Thus, depletion of the human orthologues of Cor1p and Mir1p interrupted pro-apoptotic mitochondrial signalling in human cells, paralleled by a reduction in the mitochondrial translocation of BAX.
Figure 7.
Knockdown of the human orthologues of Cor1p (QCR1) and Mir1p (PHC) reduces BAX aggregation and attenuates apoptosis in U2OS cells. (A, B) U2OS cells stably expressing GFP-BAX were transfected with siRNAs against human COR1 (siQCR1) or MIR1 (siPHC) or with an siRNA against an unrelated sequence (siUNR) for 48 h and subsequently treated with MTX or left untreated for 12 h. Nuclei were counterstained with Hoechst 33342 and subjected to automated image acquisition and analysis. Representative images (A) and quantitative analysis of GFP-BAX aggregation (B) are shown (mean±s.e.m., n=8). (C, D) U2OS transfected with siRNA and treated with 1 μM MTX or left untreated were immunostained with anti-cleaved caspase3 antibody followed by anti-rabbit Alexa488 conjugated antibody. Upon counterstaining with Hoechst 33342, automated image acquisition was performed. Representative images (C) and quantitative analysis (D) of cleaved-caspase3/Alexa488 signals are shown (mean±s.e.m., n=4). (E, F) U2OS cells treated as described in (C) were analysed for Δψm using the membrane potential-sensitive dye CMTMRos and counterstained with Hoechst 33342. Representative images (E) and quantitative analyses (F) are shown (mean±s.e.m., n=8). (G) U2OS cells treated as described in (C) were stained with PI and the membrane potential-sensitive dye DiOC6(3) and subjected to flow cytometric quantification. The percentage of cells displaying loss of mitochondrial membrane potential alone (DiOC6(3)low/PI−) or in combination with plasma membrane rupture (DiOC6(3)low/PI+) is plotted (mean±s.e.m., n=3). (H) Validation of the siRNA-mediated knockdown using specific antibodies for PHC, QCR1 and actin. ***P<0.001.
Discussion
Within the last decade, key components of the apoptotic machinery and complex apoptotic processes have been shown to be conserved between animals and yeast. Still, yeast has been assumed to lack orthologues of BCL-2 protein family members. Here, we identified a yeast protein that contains a BH3 domain (Ybh3p) and shares functional characteristics with pro-apoptotic members of the mammalian BCL-2 family. Overexpression of Ybh3p sensitizes yeast cells to apoptotic stimuli, while its knockout reduces cell death and prolongs replicative as well as chronological lifespan. Like several pro-apoptotic BCL-2 family members, Ybh3p translocates to mitochondria where it triggers Δψm dissipation and subsequent cell death. These consequences can be prevented by BCL-XL, which interacts with Ybh3p. Though mitochondrial targeting of BAX has been shown to constitute a determining factor in mitochondrion-dependent cell death (Antonsson et al, 1997; Antignani and Youle, 2006), the underlying mechanism remains highly controversial (Green and Kroemer, 2004; Antignani and Youle, 2006). One view suggests that BAX itself is capable of perforating the mitochondrial outer membrane and artificial lipid bilayers (Antonsson et al, 1997; Schlesinger et al, 1997; Martinou and Green, 2001), while others suggest that BAX acts on mitochondria by activating the latent activity of pre-existing channel proteins, thereby altering their diameter and specificities. Conflicting results have been obtained as to whether ANT or voltage-dependent anion channel (VDAC) would be required (Marzo et al, 1998; Shimizu et al, 1999) or not (Harris et al, 2000; Shimizu et al, 2000; Baines et al, 2007) for BAX-induced cell death. Collectively, our data demonstrate a physical interaction between Ybh3p and components of the PTP as well as with a subunit of the FOF1ATPase. However, Ybh3p continued to execute death even in the absence of these proteins, and its BH3 domain induced swelling of isolated mouse mitochondria in a CsA-insensitive manner. Mitochondrial import of Ybh3p neither required an intact Δψm nor specific TOM receptors. Instead, two Ybh3p-interacting proteins, Mir1p and Cor1p, were found to be required for efficient mitochondrial integration of Ybh3p, subsequent Δψm dissipation, ROS accumulation and cell death. Similarly, their human orthologues, PHC and QCR1, mediated the recruitment of BAX to mitochondria in human cells. PHC has been shown to interact with VDAC as well as with ANT, and binding between PHC and ANT has been reported to be stabilized upon apoptosis induction (Alcala et al, 2008). Thus, depending on apoptotic activators and conditions, PHC might even modulate PTP function in specific scenarios.
Ybh3p was first described as a member of the family of BAX-inhibitor 1 (BI-1) proteins, which operate as anti-apoptotic proteins in the endoplasmic reticulum of all phyla, including animals, plants and yeast (Xu and Reed, 1998; Chae et al, 2003). The cytoprotective function of BI-1 proteins has been mapped to their C-termini, which harbour functionally important residues, including positively charged (arginine and lysine) and negatively charged (glutamate and aspartate) amino acids (Supplementary Figure S7). However, systematic comparisons between Ybh3p and BI-1 proteins from other phyla indicate that Ybh3p does not possess such an ampholytic cluster of charged amino acids in its C-terminus and differs in respect to hydrophobicity and coiled-coil probability (Supplementary Figure S7). Instead, Ybh3p contains a BH3 domain (Figure 1A; Supplementary Figure S7). In addition, the yeast protein Ybh3p displays similarity to the so-called transmembrane BI-1 motif containing (TMBIM) superfamily of proteins, which probably misled their categorization into the family of BI-1 proteins (Chae et al, 2003). Interestingly, Hu et al (2009) recently suggested that the TMBIM family exhibits rather weak similarity. The authors identified the lifeguard proteins (LFG1–LFG5) as a distinct and ubiquitous eukaryotic sub-family within this superfamily. They suggested that a single LFG4-like ancestor present in plants, fungi and protozoa has branched into five sub-families (LFG1–LFG5) in animals and classified the yeast protein Ybh3p as an LFG4. Notably, these sub-families do not contain BI-1 proteins (Hu et al, 2009). Sequence comparison of the C-termini of the human proteins RECS1 (LFG3 sub-family) and LFG (belonging to the LFG2 sub-family) using the BH3 consensus from Prosite indicated the presence of a putative C-terminal BH3 domain (Supplementary Figure S7). Although distinct regulatory apoptotic functions have been assigned to several members of this LFG family, a possible involvement of the BH3-like domain has not been investigated. Our results suggest that the only BH3 domain that has thus far been identified in the yeast proteome is contained in a protein that, in higher eukaryotes, has distinct (and perhaps even opposed) functions. It is tempting to speculate that Ybh3p could operate as the sole member of its family in yeast, while animals would have developed a more sophisticated multi-component regulatory system involving multiple BCL-2 family proteins. Notably, Ybh3p interacts with BCL-XL and its BH3 peptide requires BAK to mediate apoptosis of human cells, arguing in favour of crucial cell death scenarios conserved across phyla.
In summary, we have identified a yeast BH3-only protein that mediates mitochondrion-dependent apoptosis. Moreover, we have demonstrated that the mitochondrial proteins Mir1p and Cor1p are pivotal for Ybh3p-facilitated cell death in yeast. Similarly, their mammalian orthologues PHC and QCR1 are required for targeting BAX to mitochondria in human cells. Our data suggest that the pro-apoptotic modus operandi of Ybh3p is phylogenetically conserved and that yeast can be employed for the further in-depth analysis of this evolutionary ancient cell death pathway.
Materials and methods
Yeast strains and molecular biology
Experiments were carried out in Saccharomyces cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and respective null mutants, obtained from Euroscarf. Strains were grown at 28°C and 145 r.p.m. on SC medium containing 0.17% yeast nitrogen base (Difco), 0.5% (NH4)2SO4 and 30 mg/l of all amino acids (except 80 mg/l histidine and 200 mg/l leucine), 30 mg/l adenine and 320 mg/l uracil with 2% glucose, 2% galactose or 2% glycerol, respectively. To confirm the phenotype of Δybh3 (Δynl305c) from Euroscarf, self-generated mutants obtained using the vector pUG72 (Gueldener et al, 2002) to amplify a gene-specific URA3-knockout cassette were tested. All primers and restriction enzymes used are listed in Supplementary Table S2. To construct YBH3-FLAG and YBH3ΔBH3-FLAG in pESC-his (Stratagene) and pUG35FLAG (GFP has been replaced by FLAG in pUG35; Niedenthal et al, 1996) and YBH3-GFP and YBH3ΔBH3-GFP in pUG36-ura (Niedenthal et al, 1996), inserts were amplified by PCR using genomic DNA from BY4741 as template. For generation of the point mutants YBH3L290K and YBH3D295K, antisense primers harbouring these mutations (Supplementary Table S2) were used for amplification by PCR. For expression of BCL-XL, the plasmid pRS416-BCL-XL (gift from A Szallies) was used. For pulldown analysis, murine BCL-XL has been cloned into pESC-ura (Stratagene) using previously published pCM252-BCL-XL as template (Polcic et al, 2005). To construct FLAG-tagged UTH1, MIR1 and COR1 in pESC-ura (Stratagene), inserts were amplified using genomic DNA from BY4741 as template and adequate primers (Supplementary Table S2). The double mutant Δmir1Δybh3 was obtained by deletion of YBH3 in Δmir1 cells using the vector pUG72 (Gueldener et al, 2002) to amplify a YBH3-specific URA3-knockout cassette. For overexpression of FLAG-tagged Yca1p and Nuc1p, previously published constructs in pESC-his were used (Madeo et al, 2002; Buttner et al, 2007).
Yeast survival plating and tests for cell death markers
Clonogenic survival plating was performed as described before (Madeo et al, 2002; Herker et al, 2004). For overexpression studies, cultures were inoculated from fresh overnight cultures to OD600 of 0.1, grown for ∼5 h to OD600 of 0.4 and shifted to galactose media for promoter induction. Then, cells were either (i) treated with 0.6 mM H2O2 or 0.5 μg/ml rapamycin immediately and grown for 20 h or (ii) grown for ∼16 h (∼8·107 cells/ml) and treated with 120 mM acetic acid for 4 h or (iii) left untreated for 20 h. Aliquots were collected to perform survival plating. Notably, at least three different clones were tested for overexpression in survival assays to rule out clonogenic variation of the effects. For calculation of death rates, the isogenic vector control was set to 100% survival under respective conditions. For deletion studies with acetic acid (120 mM), H2O2 (1.0 mM), AM (20 μM), DTT (5 mM) and CuSO4 (0.5 mM), cultures were inoculated from fresh overnight cultures to OD600 0.1 (∼1·106 cells/ml), grown to early stationary phase (∼8·107 cells/ml) and treated with respective death stimuli for 4 h or left untreated. Same cells were subjected to 15 min of heat shock at 46°C. For rapamycin treatment, cultures were inoculated to OD600 0.2 and grown for 4 h before incubation with 0.5 μg/ml rapamycin for 14 h. Aliquots were collected to perform survival plating. For chronological ageing experiments, cultures were inoculated to OD600 of 0.1 and aliquots were collected to perform survival plating at the indicated time points. Tests for apoptotic (TUNEL and Annexin V staining) and necrotic (PI staining) markers as well as for markers of oxidative stress (DHE staining) were performed as described (Buttner et al, 2007). For flow cytometric (BD FACSAria) quantifications, 30 000 cells were evaluated and analysed with BD FACSDiva software. For the assessment of mitochondrial membrane potential (Δψm), cells were stained with tetramethylrhodamine methyl ester (TMRM, Molecular Probes, Invitrogen), which accumulates in mitochondria driven by their transmembrane potential. Approximately 1 × 107 cells were incubated with 10 μg/ml TMRM for 40 min at 28°C while shaking. Cells were cytofluorometrically analysed after treatment with or without 120 mM acetic acid or 10 μM FCCP (Sigma) for 7 or 15 min. Background fluorescence without dye was analysed for each strain and subtracted. All statistical analyses were performed using one-way ANOVA (or two-way ANOVA for ageing experiments) followed by a Tukey post hoc test.
Yeast replicative lifespan determination
The replicative lifespan measurements were performed as described previously (Laun et al, 2001). All lifespans were determined on defined SC-glucose media for a cohort of at least 45 cells. Standard deviations of the median lifespans were calculated according to Kaplan–Meier statistics. To determine whether two given lifespan distributions are significantly different at the 98% confidence level, Breslow, Tarone-Ware and log-rank statistics were used. All statistical calculations were performed using the software package SPSS 15.0 (SPSS Inc.).
Epifluorescence microscopy
For intracellular localization studies of the fusion proteins Ybh3p-GFP and Ybh3pΔBH3-GFP in living cells, cultures were inoculated from fresh overnight cultures to OD600 of 0.1, grown for ∼20 h, treated with 120 mM acetic acid for 30 min or left untreated and analysed using epifluorescence microscopy. The vacuole was counterstained using CellTracker Blue CMAC (Molecular Probes, Invitrogen) at a concentration of 100 μM in 10 mM HEPES for 20 min and mitochondria were visualized using a DsRed Su1-69 protein, encoded by the vector pYX142 under the control of a TPI promoter (Jakobs et al, 2003). For epifluorescence microscopy, a Zeiss Axioskop microscope with adequate filters (dsRed, GFP and DAPI, Zeiss) was used.
Yeast cell fractionation, cyt c release and immunoblotting
Preparation of whole cell extracts as well as of mitochondrial and cytosolic fractions was performed as previously described (Madeo et al, 2002; Wissing et al, 2004). For analysis of cyt c release, cells were treated or not with acetic acid and subjected to spheroblastation followed by short permeabilization and immunoblotting to detect cyt c in whole cell extracts and supernatants. For details regarding culturing of cells, cell fractionation, cyt c release and immunoblotting, please see the Extended Experimental Procedures.
In organello import of preproteins into mitochondria
For the detailed description of the purification of mitochondria, in organello import studies, or carbonate extraction, please see Extended Experimental Procedures. Briefly, acetic acid treatment was carried out for 2 h by addition of 90 mM acetic acid prior to the isolation of mitochondria. Cells were harvested and homogenized, and highly purified mitochondria were isolated by differential centrifugation (Meisinger et al, 2006). Preproteins were radiolabelled by in vitro transcription and translation in the presence of [35S]methionine and incubated with isolated mitochondria. Analysis was performed by SDS–PAGE or NuPAGE Novex (Invitrogen) and digital autoradiography (PhosphorImager, Molecular Dynamics). For the removal of outer membrane receptor proteins, mitochondria were treated with 25 μg/ml trypsin prior to the import reaction.
Pulldown assay and LC-MS/MS analysis
To investigate a potential interaction between Ybh3p and BCL-XL, cells expressing either BCL-XL alone or co-expressing BCL-XL and FLAG-tagged Ybh3p were lysed and subjected to pulldown analysis using agarose beads coupled to a monoclonal anti-FLAG antibody (ANTI-FLAGTM M2 Affinity Gel, SIGMA) and subsequent immunoblotting using a specific BCL-XL antibody (Abcam). The same beads were used for the purification of FLAG-tagged Ybh3p and potential direct and indirect interactors. For the identification of co-purified proteins, specific protein bands where excised after separation on SDS–PAGE, tryptically digested and analysed by LC-MS/MS analysis. For details on pulldown assay, LC-MS/MS analysis and general MS/MS search parameters, please see the Extended Experimental Procedures.
Analysis of apoptotic changes in isolated mouse liver mitochondria
For the assessment of mitochondrial swelling, mitochondria isolated from mouse liver and resuspended in swelling buffer, as previously described (Petit et al, 1998), were pre-incubated or not with 5 μM CsA for 5 min, followed by the administration of the yeast BH3 peptides (GL Biochem Ltd.) or Ca2+ at the indicated concentrations. Immediately after the addition of the peptides, absorbance at 490 nm was followed at ∼2 min intervals. To analyse the release of cyt c, mitochondria isolated from mouse liver and resuspended in swelling buffer were incubated with the indicated concentrations of native (BH3) or mutated (BH3L290K) peptide for 20 min at RT. Mitochondrial suspensions were centrifuged at 10 000 r.p.m. for 10 min at 4°C, and supernatants and mitochondrial pellets (lysed with 50 μl of lysis buffer) were subjected to immunoblot analysis with antibodies specific for VDAC1 and cyt c (Cell Signaling Technology) and the appropriate horseradish peroxidase-labelled secondary antibody (SouthernBiotech). For validation of mitochondrial uptake, mitochondria were incubated with 10 μM FITC-labelled peptides for 20 min and FITC intensities were analysed using flow cytometry (FACScan, BD) and normalized to baseline fluorescence of FITC-label peptides quantified using a fluorescence reader (FluoStar Optima BMG).
Cell culture and staining procedures
For cell cultures, transfection and siRNA sequences as well as image acquisition, please see the Extended Experimental Procedures. U2OS cells stably expressing GFP-BAX were transfected with siRNA for 48 h and subsequently treated with 1 μM MTX or left untreated for 12 h. U2OS cells were reseeded in 96-well black-walled imaging plates (BD Falcon) 24 h before the drug treatment. The cells were fixed with 3.7% PFA for 30 min and nuclei were counterstained with 2 μg/ml Hoechst 33342 for additional 10 min before automated image acquisition and subsequent image analysis. For Δψm assessment, U2OS cells transfected with the siRNAs for 48 h were treated with 1 μM MTX or left untreated for 12 h. The Δψm was visualized by staining the cells with 150 nM CMTMRos (Molecular Probes, Invitrogen) for 30 min before fixation, Hoechst 33342 counterstaining and image acquisition. For quantification of caspase3 cleavage, fixed cells were permeabilized with 0.1% Triton X 100 and immunostained with anti-cleaved caspase3 antibody (Cell Signaling Technology) followed by anti-rabbit Alexa488-coupled secondary antibody (Invitrogen) both diluted in 2% BSA (w:v in PBS) according to the manufacturer's instructions. Subsequently, nuclei were counterstained with 2 μg/ml Hoechst 33342 for 10 min.
Cytofluorometric analyses of U2OS and A549 cells
U2OS cells transfected with indicated siRNAs were analysed for Δψm dissipation using DiOC6(3) (40 nM, Molecular Probes, Invitrogen) and loss of plasma membrane integrity using PI (1 μg/ml, Sigma) after treatment with or without MTX. Non-small cell lung cancer cells (A549 cells) were incubated for the indicated time with the native (BH3) or mutated (BH3L290K) peptide at the concentration of 30 μM, followed by the assessment of Δψm loss using the same concentrations of DiOC6(3) and PI. To estimate the amount of peptide delivered into the cell, A549 cells were incubated with 30 μM FITC-labelled peptides and FITC intensities were flow cytometrically analysed after 0.5, 1 and 1.5 h. Cytofluorometric analyses were performed using an FACSCalibur equipped with Cell Quest Pro software (Becton Dickinson).
Supplementary Material
Acknowledgments
This work was supported by the Austrian Science Fund FWF (Grants T414-B09 to SB, S9304-B05 to FM and DC-G, S9302-B05 to MB, LIPOTOX to FM and PR, and W1226-B18 to FM and LH), the European Commission (Apo-Sys to GK, FM and TE, ArtForce and ChemoRes to GK, Project MIMAGE to MB), Ligue contre le cancer (équipe labellisée), Agence Nationale de la Recherche, Institut National pour le Cancer and Cancéropôle Ile-de-France (to GK) and GRK1478 (to F-NV and CM).
Author contributions: FM, SB and GK planned the project and wrote the manuscript. FM, SB, GK, CM and LG analysed the data. SB, DR, F-NV, LG, BM, TE, OK, LH, DC-G, PR, PL, CK and GR performed the experimental work. FM, SB, GK, CM, MB, and K-UF designed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alcala S, Klee M, Fernandez J, Fleischer A, Pimentel-Muinos FX (2008) A high-throughput screening for mammalian cell death effectors identifies the mitochondrial phosphate carrier as a regulator of cytochrome c release. Oncogene 27: 44–54 [DOI] [PubMed] [Google Scholar]
- Almeida B, Ohlmeier S, Almeida AJ, Madeo F, Leao C, Rodrigues F, Ludovico P (2009) Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics 9: 720–732 [DOI] [PubMed] [Google Scholar]
- Antignani A, Youle RJ (2006) How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol 18: 685–689 [DOI] [PubMed] [Google Scholar]
- Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod JJ, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou JC (1997) Inhibition of Bax channel-forming activity by Bcl-2. Science 277: 370–372 [DOI] [PubMed] [Google Scholar]
- Baines CP (2009) The molecular composition of the mitochondrial permeability transition pore. J Mol Cell Cardiol 46: 850–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434: 658–662 [DOI] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD (2007) Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol 9: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara PD, Flattery-O’Brien JA, Grant CM, Dawes IW (1998) Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr Genet 34: 259–268 [DOI] [PubMed] [Google Scholar]
- Brust D, Hamann A, Osiewacz HD (2010) Deletion of PaAif2 and PaAmid2, two genes encoding mitochondrial AIF-like oxidoreductases of Podospora anserina, leads to increased stress tolerance and lifespan extension. Curr Genet 56: 225–235 [DOI] [PubMed] [Google Scholar]
- Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F (2007) Endonuclease G regulates budding yeast life and death. Mol Cell 25: 233–246 [DOI] [PubMed] [Google Scholar]
- Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S (2003) The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol Microbiol 47: 495–506 [DOI] [PubMed] [Google Scholar]
- Camougrand N, Kissova I, Velours G, Manon S (2004) Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res 5: 133–140 [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F (2010) Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ 17: 763–773 [DOI] [PubMed] [Google Scholar]
- Chae HJ, Ke N, Kim HR, Chen S, Godzik A, Dickman M, Reed JC (2003) Evolutionarily conserved cytoprotection provided by Bax inhibitor-1 homologs from animals, plants, and yeast. Gene 323: 101–113 [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Buttner S, Kroemer G, Madeo F (2007) The mitochondrial pathway in yeast apoptosis. Apoptosis 12: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G (2004) The pathophysiology of mitochondrial cell death. Science 305: 626–629 [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MH, Vander Heiden MG, Kron SJ, Thompson CB (2000) Role of oxidative phosphorylation in Bax toxicity. Mol Cell Biol 20: 3590–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F (2004) Chronological aging leads to apoptosis in yeast. J Cell Biol 164: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Smith TF, Goldberger G (2009) LFG: a candidate apoptosis regulatory gene family. Apoptosis 14: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW (2003) Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J Cell Sci 116: 2005–2014 [DOI] [PubMed] [Google Scholar]
- Kelekar A, Chang BS, Harlan JE, Fesik SW, Thompson CB (1997) Bad is a BH3 domain-containing protein that forms an inactivating dimer with Bcl-XL. Mol Cell Biol 17: 7040–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelekar A, Thompson CB (1998) Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol 8: 324–330 [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR Jr, Zhang J, Guarente L (1995) Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80: 485–496 [DOI] [PubMed] [Google Scholar]
- Khoury CM, Greenwood MT (2008) The pleiotropic effects of heterologous Bax expression in yeast. Biochim Biophys Acta 1783: 1449–1465 [DOI] [PubMed] [Google Scholar]
- Kissova I, Plamondon LT, Brisson L, Priault M, Renouf V, Schaeffer J, Camougrand N, Manon S (2006) Evaluation of the roles of apoptosis, autophagy, and mitophagy in the loss of plating efficiency induced by Bax expression in yeast. J Biol Chem 281: 36187–36197 [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Chandna S (2009) Putative partners in Bax mediated cytochrome-c release: ANT, CypD, VDAC or none of them? Mitochondrion 9: 1–8 [DOI] [PubMed] [Google Scholar]
- Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M (2001) Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol 39: 1166–1173 [PubMed] [Google Scholar]
- Leber B, Lin J, Andrews DW (2010) Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene 29: 5221–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH (1998) Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett 438: 61–65 [DOI] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW (2008) Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135: 1074–1084 [DOI] [PubMed] [Google Scholar]
- Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M (2002) Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell 13: 2598–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 139: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU (2002) A caspase-related protease regulates apoptosis in yeast. Mol Cell 9: 911–917 [DOI] [PubMed] [Google Scholar]
- Martinou JC, Green DR (2001) Breaking the mitochondrial barrier. Nat Rev Mol Cell Biol 2: 63–67 [DOI] [PubMed] [Google Scholar]
- Marzo I, Brenner C, Zamzami N, Jurgensmeier JM, Susin SA, Vieira HL, Prevost MC, Xie Z, Matsuyama S, Reed JC, Kroemer G (1998) Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science 281: 2027–2031 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y (2005) Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature 434: 652–658 [DOI] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12: 773–786 [DOI] [PubMed] [Google Scholar]
- Paglin S, Lee NY, Nakar C, Fitzgerald M, Plotkin J, Deuel B, Hackett N, McMahill M, Sphicas E, Lampen N, Yahalom J (2005) Rapamycin-sensitive pathway regulates mitochondrial membrane potential, autophagy, and survival in irradiated MCF-7 cells. Cancer Res 65: 11061–11070 [DOI] [PubMed] [Google Scholar]
- Petit PX, Goubern M, Diolez P, Susin SA, Zamzami N, Kroemer G (1998) Disruption of the outer mitochondrial membrane as a result of large amplitude swelling: the impact of irreversible permeability transition. FEBS Lett 426: 111–116 [DOI] [PubMed] [Google Scholar]
- Polcic P, Su X, Fowlkes J, Blachly-Dyson E, Dowhan W, Forte M (2005) Cardiolipin and phosphatidylglycerol are not required for the in vivo action of Bcl-2 family proteins. Cell Death Differ 12: 310–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, Le Bras M, Briere JJ, Jalil A, Le Moigne R, Brenner C, Hahn G, Wittig I, Schagger H, Lemaire C, Bianchi K, Souquere S, Pierron G, Rustin P, Goldmacher VS, Rizzuto R et al. (2006) Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J Cell Biol 174: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M, Camougrand N, Chaudhuri B, Schaeffer J, Manon S (1999) Comparison of the effects of bax-expression in yeast under fermentative and respiratory conditions: investigation of the role of adenine nucleotides carrier and cytochrome c. FEBS Lett 456: 232–238 [DOI] [PubMed] [Google Scholar]
- Purdom S, Chen QM (2003) p66(Shc): at the crossroad of oxidative stress and the genetics of aging. Trends Mol Med 9: 206–210 [DOI] [PubMed] [Google Scholar]
- Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ (2005) Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci USA 102: 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger PH, Gross A, Yin XM, Yamamoto K, Saito M, Waksman G, Korsmeyer SJ (1997) Comparison of the ion channel characteristics of proapoptotic BAX and antiapoptotic BCL-2. Proc Natl Acad Sci USA 94: 11357–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Narita M, Tsujimoto Y (1999) Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399: 483–487 [DOI] [PubMed] [Google Scholar]
- Shimizu S, Shinohara Y, Tsujimoto Y (2000) Bax and Bcl-xL independently regulate apoptotic changes of yeast mitochondria that require VDAC but not adenine nucleotide translocator. Oncogene 19: 4309–4318 [DOI] [PubMed] [Google Scholar]
- Stevens L (1981) Regulation of the biosynthesis of putrescine, spermidine and spermine in fungi. Med Biol 59: 308–313 [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD (2008) Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 4: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F (2004) An AIF orthologue regulates apoptosis in yeast. J Cell Biol 166: 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Reed JC (1998) Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell 1: 337–346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.