Abstract
Mitochondrial morphology is controlled by two opposing processes: fusion and fission. Drp1 (dynamin-related protein 1) and hFis1 are two key players of mitochondrial fission, but how Drp1 is recruited to mitochondria and how Drp1-mediated mitochondrial fission is regulated in mammals is poorly understood. Here, we identify the vertebrate-specific protein MIEF1 (mitochondrial elongation factor 1; independently identified as MiD51), which is anchored to the outer mitochondrial membrane. Elevated MIEF1 levels induce extensive mitochondrial fusion, whereas depletion of MIEF1 causes mitochondrial fragmentation. MIEF1 interacts with and recruits Drp1 to mitochondria in a manner independent of hFis1, Mff (mitochondrial fission factor) and Mfn2 (mitofusin 2), but inhibits Drp1 activity, thus executing a negative effect on mitochondrial fission. MIEF1 also interacts with hFis1 and elevated hFis1 levels partially reverse the MIEF1-induced fusion phenotype. In addition to inhibiting Drp1, MIEF1 also actively promotes fusion, but in a manner distinct from mitofusins. In conclusion, our findings uncover a novel mechanism which controls the mitochondrial fusion–fission machinery in vertebrates. As MIEF1 is vertebrate-specific, these data also reveal important differences between yeast and vertebrates in the regulation of mitochondrial dynamics.
Keywords: Drp1, hFis1, mitochondrial fusion and fission, SMCR7L, MIEF1/MiD51
Introduction
Mitochondria have critical roles in many cellular processes, including energy metabolism, intracellular calcium homeostasis and apoptosis. Mitochondrial morphology is dynamic and controlled by a balance between mitochondrial fusion and fission (Okamoto and Shaw, 2005; Chan, 2006; Kiefel et al, 2006). When mitochondrial fusion is reduced, mitochondria become fragmented due to ongoing fission. Conversely, when the balance shifts towards fusion, mitochondria elongate resulting in a tubular interconnected network. Mitochondrial dynamics controls not only the morphology but also the function of mitochondria, thereby impacting on a wide range of cellular processes. Dysfunction of mitochondrial dynamics has been implicated in ageing and a variety of human diseases (Liesa et al, 2009; Schafer and Reichert, 2009) including neurodegenerative diseases (Knott et al, 2008; Mattson et al, 2008; Reddy et al, 2009; Wang et al, 2009), diabetes (Zorzano et al, 2009), cardiovascular disease (Hom and Sheu, 2009) and cancer (Modica-Napolitano and Singh, 2004; Grandemange et al, 2009).
The molecular mechanisms controlling mitochondrial fusion and fission are beginning to be unravelled, and genes important for these processes have been identified (Okamoto and Shaw, 2005; Hoppins et al, 2007; Merz et al, 2007; Berman et al, 2008; Liesa et al, 2009). The dynamin-like GTPases, mitofusin 1 (Mfn1) and mitofusin 2 (Mfn2), have a critical role in the fusion process. Mfn1 and Mfn2 are localized to the outer mitochondrial membrane, and are believed to interact in trans to promote mitochondrial fusion (Koshiba et al, 2004). OPA1 (optic atrophy 1), another dynamin-like GTPase, is localized to the inner mitochondrial membrane and mediates the fusion of the inner membranes (Delettre et al, 2000). On the fission side, Drp1 (dynamin-related protein 1) has a central role (Lackner and Nunnari, 2010), and mice lacking Drp1 die at an early embryonic stage (Ishihara et al, 2009; Wakabayashi et al, 2009). Drp1 is also a dynamin-like GTPase and is primarily distributed in the cytoplasm, but shuttles between the cytoplasm and the mitochondrial surface (Smirnova et al, 2001). At the mitochondrial surface, Drp1 is assembled into a higher-order complex, and is thought to wrap around the mitochondria to induce fission via its GTPase activity (Liesa et al, 2009). hFis1 is a fission-promoting integral mitochondrial outer membrane protein and believed to serve as a receptor for recruitment of Drp1 to mitochondria (James et al, 2003; Yoon et al, 2003). In yeast, Fis1p (the hFis1 orthologue) interacts with Dnm1p (the Drp1 orthologue) and recruits Dnm1p to the mitochondrial surface through one of the two adaptor proteins, Mdv1p or Caf4p (Mozdy et al, 2000; Tieu and Nunnari, 2000; Tieu et al, 2002; Griffin et al, 2005). However, orthologues of Mdv1p and Caf4p have not been identified in vertebrates (Westermann, 2010a). Moreover, increased or reduced levels of hFis1 do not affect the amount of mitochondrial-associated Drp1 (Suzuki et al, 2003; Lee et al, 2004), suggesting that additional factors potentially contribute to the recruitment of Drp1 to the mitochondrial surface. One Drp1-recruiting factor, mitochondrial fission factor (Mff), has recently been identified (Otera et al, 2010), but the process of how Drp1 becomes recruited to mitochondria and regulates mitochondrial fission is likely to be more complex and warrants further investigation (Hoppins et al, 2007; Santel and Frank, 2008; Liesa et al, 2009; Lackner and Nunnari, 2010).
In this report, we have characterized a novel vertebrate-specific integral mitochondrial outer membrane protein, designated MIEF1 (mitochondrial elongation factor 1). Ectopic expression of the protein results in extensive mitochondrial elongation and perinuclear clustering, whereas depletion of the protein induces mitochondrial fragmentation. MIEF1 interacts with Drp1 and triggers translocation of cytoplasmic Drp1 to mitochondria, but despite this fact, MIEF1 acts as a suppressor to sequester Drp1 and impedes Drp1-mediated fission, leading to a mitochondrial fusion phenotype. MIEF1 also interacts with hFis1, separate from its interaction with Drp1, and elevated hFis1 levels reverse the MIEF1-induced fusion phenotype. Our data provide novel insights into how cytoplasmic Drp1 is recruited to mitochondria and how Drp1-mediated fission is regulated in vertebrates. In addition to inhibiting Drp1, MIEF1 also actively promotes fusion, in a manner distinct from mitofusins. Our finding that the vertebrate-specific protein MIEF1 interacts with Drp1 and hFis1, which are highly conserved in yeast and vertebrates, also reveals important differences between yeast and vertebrates with regard to the underpinning mechanisms for regulation of mitochondrial morphology.
Results
MIEF1—a novel integral mitochondrial outer membrane protein
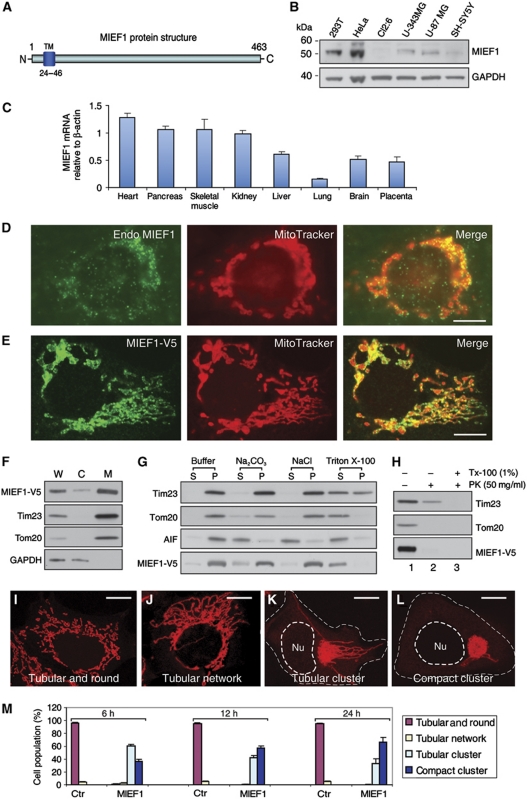
To identify proteins with potential roles in controlling mitochondrial morphology, we searched an intracellular protein localization database (http://www.lifedb.de/lifedb/) that contains information about the intracellular localization of a large number of GFP-tagged fusion proteins (Simpson et al, 2000). We identified a mitochondrial protein, which we designated MIEF1, as ectopic expression of the protein triggered extensive mitochondrial elongation (see below). MIEF1 is encoded by the SMCR7L gene and contains 463 amino-acid residues with an N-terminal transmembrane (TM) domain (Figure 1A). MIEF1 is highly conserved in all vertebrate species analysed (Supplementary Figure S1), but was not found in yeast, invertebrates and plants.
Figure 1.
The protein structure, expression and subcellular localization of MIEF1. (A) MIEF1 is an integral outer mitochondrial membrane protein with an N-terminal TM domain. (B) Endogenous MIEF1 in various cell lines was immunoblotted using anti-MIEF1 antibody. (C) Real-time PCR analysis of MIEF1 expression in normal human adult tissues. Levels of MIEF1 mRNA were determined relative to β-actin. Data were from three independent experiments. (D, E) Both endogenous MIEF1 and exogenous MIEF1-V5 were localized to mitochondria, stained with anti-MIEF1 antibody (green) and MitoTracker Red (red). (F) The distribution of MIEF1-V5, Tim23, Tom20 and GAPDH was analysed in whole-cell lysate (W), cytosolic fraction (C) and mitochondrial fraction (M). (G) Mitochondrial fractions prepared from 293T cells expressing MIEF1-V5 were resuspended in the mitochondrial buffer (buffer) alone as control, or in buffers containing 0.1 M Na2CO3 (pH 11.5), 1 M NaCl or 1% Triton X-100 followed by centrifugation, and the membrane pellets (P) and supernatant fractions (S) were immunoblotted with indicated antibodies. (H) Mitochondrial fractions were digested with PK in the absence (lane 2) or presence (lane 3) of 1% Triton X-100 (Tx-100) for 30 min or with mock control (lane 1) and analysed for MIEF1-V5, Tim23 and Tom20. (I–L) Mitochondrial morphology in 293T cells transfected with empty vector (I, J) and MIEF1-V5 (K, L) was analysed by confocal microscopy after double staining with MitoTracker and anti-V5 antibody (not shown). Outlines of the nucleus (Nu) and the cell are drawn in dash line. (M) Percentages (mean±s.e.m.) of 293T cells with indicated mitochondrial morphologies at indicated time points post-transfection for empty vector (Ctr) transfected cells (n=495 for 6 h; n=558 for 12 h; n=568 for 24 h) and for MIEF1-V5 transfected cells (n=427 for 6 h; n=735 for 12 h; n=776 for 24 h). Data were from three independent experiments. Bars, 10 μm.
Western blotting showed that endogenous MIEF1 with a molecular mass of ∼52 kDa was expressed at various levels in a number of human cell lines (Figure 1B). Expression of MIEF1 mRNA was relatively high in adult human heart, skeletal muscle, pancreas and kidney (Figure 1C). Endogenous MIEF1, as well as a C-terminally V5-tagged MIEF1 (MIEF1-V5), was localized to mitochondria (Figure 1D and E). In agreement with this, MIEF1-V5, together with the two mitochondrial membrane proteins Tom20 and Tim23, was predominantly confined to the mitochondrial fraction (Figure 1F). Mitochondrial fractions isolated from cells expressing MIEF1-V5 were incubated under conditions that resulted in release of soluble and peripheral membrane-associated proteins from the outer and inner membranes. MIEF1-V5, like Tom20 and Tim23, was predominantly retained in the mitochondrial membrane pellet, whereas the intermembrane space protein AIF was found in the supernatant fraction (Figure 1G). This suggests that MIEF1 is an integral mitochondrial membrane protein. To determine the membrane topology of MIEF1, proteinase K (PK) treatment resulted in the disappearance of MIEF1-V5 and the outer membrane protein Tom20 from mitochondrial fractions, whereas the inner membrane protein Tim23 was protected, unless Triton X-100 was added (Figure 1H). Taken together, the data indicate that MIEF1 is anchored to the mitochondrial outer membrane with its C-terminal region facing the cytoplasm (Figure 1A).
MIEF1 promotes mitochondrial elongation and fusion
The basic mitochondrial morphology in non-transfected or empty vector transfected 293T cells was a mixed reticulum with tubular and round forms (Figure 1I), with only a few cells displaying a tubular network of mitochondria (Figure 1J, quantified in Figure 1M). In contrast, overexpression of MIEF1 induced extensive mitochondrial elongation in most (∼90%) of the cells, with frequent mitochondrial tubular clusters with long tubules emanating from the perinuclear clusters (Figure 1K) or with a more compact cluster of mitochondria (Figure 1L). The compact cluster phenotype over time became more dominant, at the expense of the tubular cluster phenotype, which initially after transfection was more prevalent (quantified in Figure 1M).
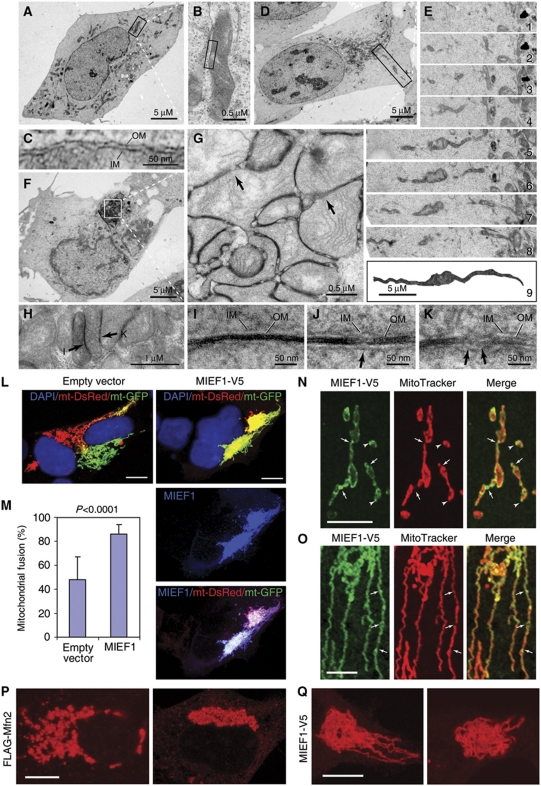
At the electron microscopy (EM) level, mitochondria in non-transfected cells appeared as multiple short tubules, and were distributed throughout the entire cytoplasm (Figure 2A–C). In contrast, in cells overexpressing MIEF1, both elongated tubular (Figure 2D and E) and compact cluster (Figure 2F–H) morphologies were observed, corresponding to the phenotypes observed in Figure 1K and L, respectively. Reconstruction of serial ultrathin sections revealed one single extremely long mitochondrion (Figure 2E), which was never observed in non-transfected cells. In areas of compact clusters of mitochondria, the two outer membranes between tethered mitochondria often became obscured due to accumulation of electron-dense material (Figure 2G, H and I), but were also found in the process of fusion (Figure 2G and J, arrows) or completely fused (Figure 2G, H and K, arrows).
Figure 2.
MIEF1 promotes mitochondrial elongation and fusion. (A) Electron micrograph of mitochondria in a non-transfected cell. (B) The mitochondrion boxed in (A) at high magnification. (C) A locally magnified mitochondrion boxed in (B). (D) Electron micrograph in a MIEF1-V5 transfected cell, with an elongated tubular cluster of mitochondria (corresponding to the image in Figure 1K). An elongated mitochondrion in the box was followed from the perinuclear region to the cell surface. (E) Serial ultrathin sections (E1–8) and a reconstruction (E9) from the serial sections of the giant mitochondrion boxed in (D). (F) Electron micrograph of mitochondria in a MIEF1-V5 transfected cell with a compact cluster of mitochondria in the perinuclear region (corresponding to the image in Figure 1L). (G) The magnified compact cluster of mitochondria boxed in (F). Arrows indicate mitochondrial membranes in the fusion process. (H) A locally magnified compact cluster of mitochondria. The areas pointed by arrows were further magnified in (I, K). (I–K) Locally magnified electron micrographs of the mitochondrial membrane structure in compact clusters illustrate how the two outer membranes between tethered mitochondria become indistinguishable (I) and undergo fusion (J, K, arrows). IM, the inner membrane; OM, the outer membrane. (L) Cocultured 293T cells expressing mito-GFP or mito-DsRed were transfected with empty vector or MIEF1-V5 and fused with PEG. Polykaryons were analysed by confocal microscopy and mitochondrial fusion was measured by colocalization of mito-GFP or mito-DsRed. (M) Mitochondrial fusion was quantified by using the colocalization model of LAS AF software. At least 20 polykaryons were analysed in each of two independent experiments. Bars, 10 μm. (N) Confocal images show accumulation of MIEF1-V5 as punctate structures at the connection sites between two mitochondrial units (arrows) and at the tips of mitochondrial tubules (arrowheads). (O) Mitochondria had undergone MIEF1-V5-induced elongation. Distinct foci of MIEF1-V5 were observed (arrows). Bars, 5 μm. (P, Q) Mitochondrial morphology in 293T cells expressing FLAG-Mfn2 or MIEF1-V5 was observed by confocal microscopy after double staining with MitoTracker and either anti-V5 or anti-FLAG antibody (not shown). Bars, 10 μm.
To further assess the role of MIEF1 in mitochondrial fusion, we used an in vivo cell fusion assay (Liesa et al, 2008; Kamp et al, 2010). Two sets of 293T cells transiently transfected with mito-GFP or mito-DsRed, respectively, were cocultured and transfected with either MIEF1-V5 or empty vector, and cell fusion was induced by polyethylene glycol (PEG). After subsequent culturing in the presence of cycloheximide, mitochondrial fusion was analysed by measuring the extent of mito-GFP and mito-DsRed colocalization. Polykaryons expressing MIEF1-V5 showed significantly higher incidence of mitochondrial fusion as compared with control (Figure 2L and M), suggesting that MIEF1 promotes mitochondrial fusion.
In keeping with its role in regulating mitochondrial fusion, MIEF1 distribution was punctate on the mitochondrial surface, at the connection sites between two adjacent mitochondrial units (Figure 2N, arrows) and at the tips of mitochondrial tubules (Figure 2N, arrowheads) in cells expressing lower amounts of MIEF1-V5. In mitochondria that underwent MIEF1-V5-induced elongation, MIEF1 coated the outer mitochondrial membrane more evenly with fewer distinct foci of MIEF1 observed (Figure 2O, arrows).
The mitochondrial phenotype caused by MIEF1 overexpression was distinct from that caused by Mfn2 overexpression. Elevated Mfn2 expression induced perinuclear aggregation (Figure 2P), in keeping with an earlier report (Huang et al, 2007), but the aggregation consisted of individual small mitochondria in a tightly apposed grape-like structure rather than an aggregation of tubular elements observed in clusters following MIEF1 overexpression (Figure 2Q). This indicates that MIEF1 induces mitochondrial fusion in a manner distinct from Mfn2. To further corroborate this notion, we examined the effect of MIEF1-V5 expression on mitochondrial morphology in cells depleted of Mfn2 by RNA interference (RNAi). Knockdown of Mfn2 resulted in mitochondrial fragmentation (Figure 3A; Supplementary Figure S2), consistent with previous reports (Eura et al, 2003; Chen et al, 2005). Expression of MIEF1-V5 reversed the Mfn2 RNAi-induced fission phenotype and resulted in mitochondrial fusion (Figure 3B and C), suggesting that Mfn2 is not essential for MIEF1-induced mitochondrial fusion.
Figure 3.
Mfn2 is not essential for MIEF1-induced mitochondrial fusion and MIEF1 exhibits oligomerization. (A) Confocal images show fragmented mitochondria in HeLa cells depleted of Mfn2 by siRNA (siMfn2–3). (B) Expression of MIEF1-V5 reverses Mfn2 RNAi-induced fragmentation, resulting in mitochondrial elongation in Mfn2-depleted HeLa cells. Bars, 10 μm. (C) Percentages (mean±s.e.m.) of cells with indicated mitochondrial morphologies in Mfn2 RNAi-treated HeLa cells in the presence or absence of exogenous MIEF1-V5. At least 200 cells in several fields were counted in two independent experiments. (D) HeLa cells were transfected with two MIEF1-specific siRNAs as indicated, or with a scrambled control siRNA (Ctr-siRNA), and analysed by immunoblotting with anti-MIEF1 antibody. (E) Representative confocal images of mitochondrial morphologies in HeLa cells transfected with Ctr-siRNA, siMIEF1-1 or siMIEF1-2 were stained with MitoTracker. Bar, 20 μm. (F) Percentages (mean±s.e.m.) of cells with indicated mitochondrial morphologies after transfection with siRNAs as indicated. At least 500 cells in several fields were counted in three independent experiments. (G) Cell extracts of 293T cells expressing MIEF1-V5 were separated by SDS-gel electrophoresis in the presence (+) or absence (−) of DTT followed by western blot with anti-V5 antibody. Under non-reducing conditions, a dimer with the molecular mass of ∼110 kDa was seen in addition to the ∼56 kDa monomer of MIEF1-V5. (H) 293T cells expressing MIEF1-V5 were chemically crosslinked with (+) or without (−) FA, and cell lysates were immunoblotted with anti-MIEF1 antibody. Several high molecular weight bands were seen under FA crosslinking conditions. (I) 293T cells were cotransfected with HA-MIEF1 and either MIEF1-V5 or empty vector, and cell lysates were subjected to IP with anti-V5 agarose followed by immunoblotting with indicated antibodies. (J) Schematic representation of deletion mutants of MIEF1 fused to a C-terminal V5-tag. (K) 293T cells expressing the indicated deletion mutants of MIEF1 were chemically crosslinked with (+) or without (−) FA, followed by immunoblotting with anti-V5 antibody.
Depletion of endogenous MIEF1 leads to mitochondrial fragmentation
To further elucidate the role of MIEF1, knockdown of endogenous MIEF1 was performed by RNAi. Reduction of endogenous MIEF1 levels by two different siRNAs, which reduced MIEF1 protein levels by >80 and 90%, respectively (Figure 3D), led to a significant increase in the proportion of cells with partially fragmented mitochondria (76.1±6.8% and 80.1±5.4%, respectively, P<0.0001), compared with control cells (4.2±2.0%) (Figure 3E and F). This demonstrates that loss of MIEF1 leads to mitochondrial fragmentation.
MIEF1 undergoes oligomerization
Next, we asked whether MIEF1 oligomerizes. Under reducing conditions, the V5-tagged MIEF1 appeared as a monomer with a molecular mass of ∼56 kDa, whereas under non-reducing conditions, it appeared as two bands, with a molecular mass of ∼110 and ∼56 kDa, respectively (Figure 3G). When cells expressing MIEF1 were chemically crosslinked, several high molecular weight bands were detected, while under non-crosslinking conditions only a single band was observed (Figure 3H). Furthermore, two differently tagged MIEF1 proteins could be coimmunoprecipitated (Figure 3I).
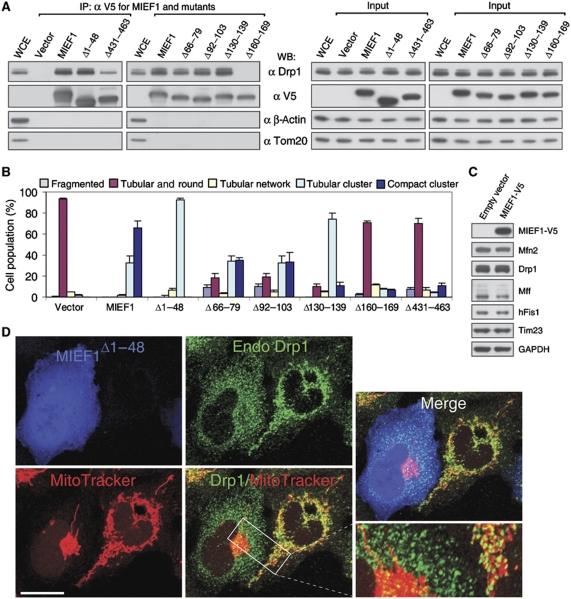
To map the region in MIEF1 important for dimerization, we analysed a set of MIEF1 deletion mutants for their ability to form dimers (summarized in Figure 3J and K). An N-terminal deletion mutant lacking the first 48 amino-acid residues (MIEF1Δ1−48) retained the capacity to dimerize, whereas a mutant lacking the first 195 residues (MIEF1Δ1−195) could not dimerize. In sum, these data show that MIEF1 can oligomerize and that residues 49 to 195 are required for MIEF1 dimerization.
MIEF1 interacts with Drp1 and recruits cytoplasmic Drp1 to mitochondria
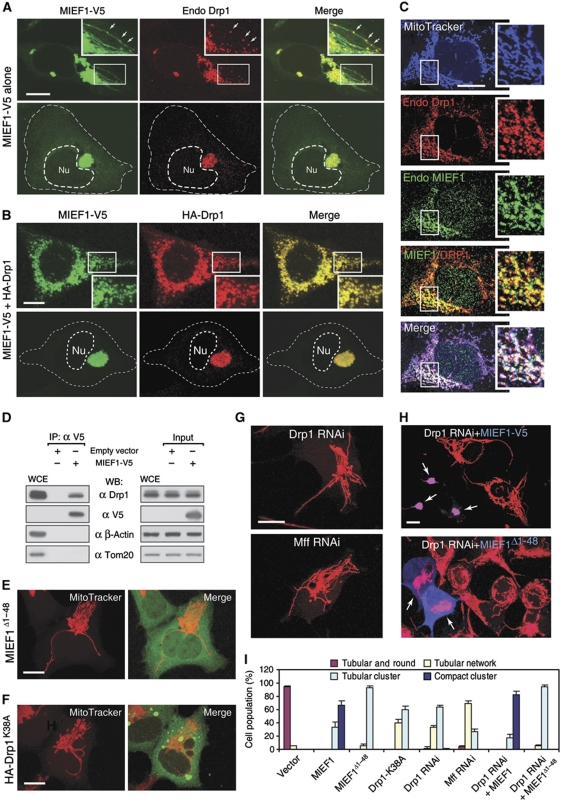
We observed that MIEF1 was colocalized with both exogenous HA-Drp1 and endogenous Drp1 on mitochondria (Figure 4A–C). We next tested whether MIEF1 interacts with Drp1, and endogenous Drp1 was indeed coimmunoprecipitated with MIEF1-V5 (Figure 4D). A functional interaction between MIEF1 and Drp1 was further supported by the fact that the MIEF1Δ1−48 mutant, which was diffusely distributed in the cytoplasm because of the deletion of the TM domain (Figure 4E), still induced an extensive tubular cluster phenotype of mitochondria. This was reminiscent of the mitochondrial phenotype caused by inhibition of Drp1 activity with a dominant negative mutant Drp1K38A lacking the GTPase activity (Figure 4F; Yoon et al, 2001). The mitochondrial tubular cluster phenotype induced by wild-type MIEF1 (Figure 1K) or by MIEF1Δ1−48 (Figure 4E) was similar to that induced by Drp1K38A (Figure 4F), or by depletion of Drp1 or Mff by RNAi (Figure 4G; Supplementary Figure S2). Importantly, Drp1 silencing did not affect the mitochondrial phenotype induced by MIEF1 and MIEF1Δ1−48 (Figure 4H, arrows; see Figure 4I for a summary of the mitochondrial phenotypes). Taken together, these data suggest that MIEF1 interacts with Drp1 and inhibits Drp1-mediated mitochondrial fission.
Figure 4.
MIEF1 interacts and colocalizes with Drp1 on mitochondria. (A) Confocal images show that MIEF1-V5 was colocalized with endogenous Drp1 (endo Drp1) in the tubular cluster (upper panel) and in the compact cluster of mitochondria (lower panel) in 293T cells expressing MIEF1-V5. Insets represent magnifications of the boxed areas. MIEF1 and Drp1 were colocalized in punctate structures on mitochondria (arrows). (B) Exogenously expressed MIEF1-V5 and HA-Drp1 were colocalized on mitochondria of 293T cells. Outlines of the nucleus (Nu) and the cell in (A, B) are drawn in dash lines. (C) Confocal images show the colocalization of endogenous MIEF1 and Drp1 as punctate structures on mitochondria. Cells were triple stained using MitoTracker (blue) followed by anti-MIEF1 (red) and anti-Drp1 (green) antibodies. Insets represent magnifications of the boxed areas. (D) After transfection with empty vector or MIEF1-V5, cell lysates of 293T cells were subjected to IP with anti-V5 agarose, and the precipitated complexes were analysed by western blot (WB) with indicated antibodies. WCE, whole cell extract from non-transfected cells. (E, F) Confocal images show mitochondrial morphology in 293T cells expressing either MIEF1Δ1−48 or HA-Drp1K38A. (G) Confocal images of mitochondria stained with MitoTracker in 293T cells depleted of Drp1 or Mff by siRNA (siDrp1-1, siMff-1). (H) Confocal images of mitochondria stained with MitoTracker in Drp1 RNAi-treated 293T cells expressing MIEF1-V5 or MIEF1Δ1−48 stained with anti-V5 antibody (blue, arrows). (I) Percentages (mean±s.e.m.) of cells with indicated mitochondrial morphologies in 293T cells transfected with empty vector (n=497), MIEF1-V5 (n=776), MIEF1Δ1−48 (n=409), HA-Drp1K38A (n=289), Drp1 RNAi (n=319), Mff RNAi (n=303), Drp1 RNAi+MIEF1-V5 (n=301) and Drp1 RNAi+MIEF1Δ1−48 (n=119). Data were determined from two or three independent experiments. Bars, 10 μm.
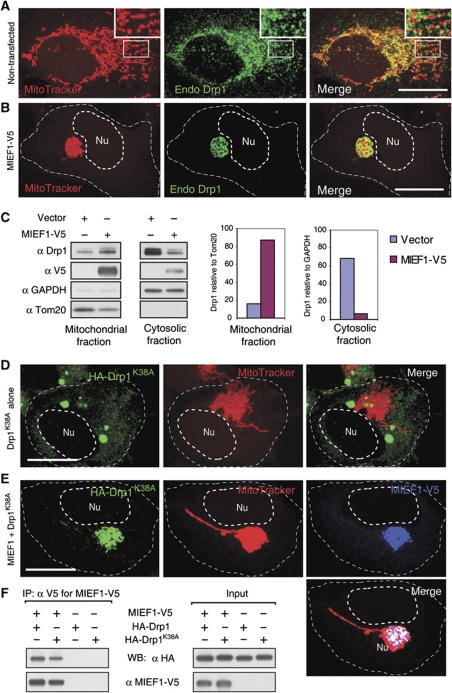
How Drp1 becomes localized to mitochondria has remained enigmatic (Okamoto and Shaw, 2005; Liesa et al, 2009), and the interaction between MIEF1 and Drp1 led us to further address whether MIEF1 was able to recruit Drp1 to mitochondria. In non-transfected 293T cells, Drp1 was both diffusely distributed in the cytoplasm and associated with mitochondria (Figure 5A), in agreement with a previous report (Smirnova et al, 2001). In contrast, in cells expressing MIEF1-V5, the majority of endogenous Drp1 was recruited from the cytoplasm to mitochondria (Figure 5B), where it colocalized with MIEF1 (see Figure 4A and B). In keeping with this, a significant increase of Drp1 levels in the mitochondrial fraction and a corresponding decrease in the cytosolic fraction in cells expressing MIEF1-V5 was observed (Figure 5C). Moreover, MIEF1-mediated mitochondrial recruitment of Drp1 was more pronounced for the Drp1K38A mutant, which showed extensive cytoplasmic localization in cells expressing Drp1K38A alone (Figure 5D) but showed a distinct mitochondrial localization following cotransfection with MIEF1 (Figure 5E). Drp1K38A, like wild-type Drp1, was readily coimmunoprecipitated with MIEF1 (Figure 5F). Collectively, these data indicate that MIEF1 recruits cytoplasmic Drp1 to mitochondria in a manner that does not require the GTPase activity of Drp1.
Figure 5.
MIEF1 recruits cytoplasmic Drp1 to mitochondria. (A) Confocal images show the distribution of endogenous Drp1 (endo Drp1) in non-transfected 293T cells. Insets represent magnifications of the boxed areas. (B) Confocal images show that endogenous Drp1 was recruited to mitochondria from the cytoplasm in 293T cells overexpressing MIEF1-V5. (C) Subcellular fractionation followed by immunoblotting show an increase of Drp1 in the mitochondrial fraction and a decrease of Drp1 in the cytosolic fraction in 293T cells overexpressing MIEF1-V5. Tom20 and GAPDH were used as markers for mitochondrial and cytosolic fractions, respectively. (D) Overexpression of the GTPase-inactive Drp1K38A alone resulted in aggregated structures (green) of HA-Drp1K38A in the cytoplasm. 293T cells were double stained with MitoTracker and anti-HA antibody. (E) Cotransfection with MIEF1-V5 recruited HA-Drp1K38A to mitochondria and MIEF1-V5 and Drp1K38A colocalized in mitochondrial clusters. 293T cells were cotransfected, and stained with MitoTracker followed by immunostaining with anti-V5 (blue) and anti-HA (green) antibodies. (F) Cell lysates of 293T cells transfected with indicated plasmids were subjected to IP with anti-V5 agarose, and the precipitated complexes were analysed by western blot (WB) with indicated antibodies. Outlines of the nucleus (Nu) and the cell in (B, D and E) are drawn in dash line. Bars, 20 μm.
Coimmunoprecipitation (co-IP) with a set of MIEF1 deletion mutants identified two regions in MIEF1 important for its interaction with Drp1 (summarized in Figure 6A; Supplementary Figure S3). Removal of the C-terminal domain (MIEF1Δ431−463) reduced but did not abolish binding to Drp1, whereas removal of amino-acid residues 160 to 169 (MIEF1Δ160−169) led to a complete loss of binding ability to Drp1 (Figure 6A). In keeping with the reduced and abolished Drp1-binding ability, most cells expressing MIEF1Δ431−463 or MIEF1Δ160−169 exhibited relatively normal mitochondrial morphology (Figure 6B; Supplementary Figure S3), whereas the MIEF1Δ66−79, MIEF1Δ92−103, MIEF1Δ130−139 and MIEF1Δ1−48 mutants, which retained the ability to bind to Drp1 (Figure 6A), produced fusion phenotypes (Figure 6B; Supplementary Figure S3). In addition, elevated levels of MIEF1 did not affect levels of Drp1, hFis1, Mff, Mfn2 and Tim23 in the cells (Figure 6C). In line with this, MIEF1Δ1−48, which lacks the TM domain but retains the ability to bind to Drp1 and to induce mitochondrial elongation (Figure 4E and I), sequestered Drp1 in the cytoplasm (Figure 6D). This suggests that MIEF1–Drp1 interaction, rather than MIEF1's mitochondrial localization, is crucial for the MIEF1-induced fusion phenotype, and in keeping with this, the mitochondrial phenotype induced by the cytoplasmic MIEF1Δ1−48 mutant was more similar to the phenotypes induced by the Drp1K38A mutant, or by RNAi-mediated silencing of the fission components Drp1 or Mff, as compared with the wild-type MIEF1 phenotype. However, mitochondrial localization of MIEF1 was required for further inducing a compact cluster of mitochondria (see Figures 4E–H and 6B).
Figure 6.
MIEF1's interaction with Drp1 rather than its mitochondrial localization is crucial for the MIEF1-induced mitochondrial fusion phenotype. (A) Cell lysates of 293T cells transfected with empty vector, wild-type and various mutants of MIEF1-V5, as indicated, were immunoprecipitated with anti-V5 agarose, and the precipitated complexes were analysed by western blot (WB). WCE, whole cell extract from non-transfected cells. Note that the MIEF1Δ160−169 mutant does not bind Drp1 and that the MIEF1Δ431−463 mutant binds only weakly. (B) Percentages (mean±s.e.m.) of cells with indicated mitochondrial morphologies in 293T cultures transfected with empty vector (n=582), wild-type MIEF1-V5 (n=815), MIEF1Δ1−48 (n=302) MIEF1Δ66−79 (n=455), MIEF1Δ92−103 (n=481), MIEF1Δ130−139 (n=517), MIEF1Δ160−169 (n=663) as well as MIEF1Δ431−463 (n=495) plasmids. Data were from three independent experiments. (C) Immunoblotting analysis of Drp1, hFis1, Mff, Mfn2 and Tim23 in 293T cells transfected with empty vector and MIEF1-V5. (D) Confocal images show that overexpression of MIEF1Δ1−48, which was cytoplasmically located due to lack of the TM domain but retained the ability to bind Drp1, resulted in reduced distribution of Drp1 in mitochondria compared to in the adjacent non-transfected 293T cells. Bar, 20 μm.
Mff, hFis1 and Mfn2 are not required for MIEF1-mediated recruitment of Drp1 to mitochondria
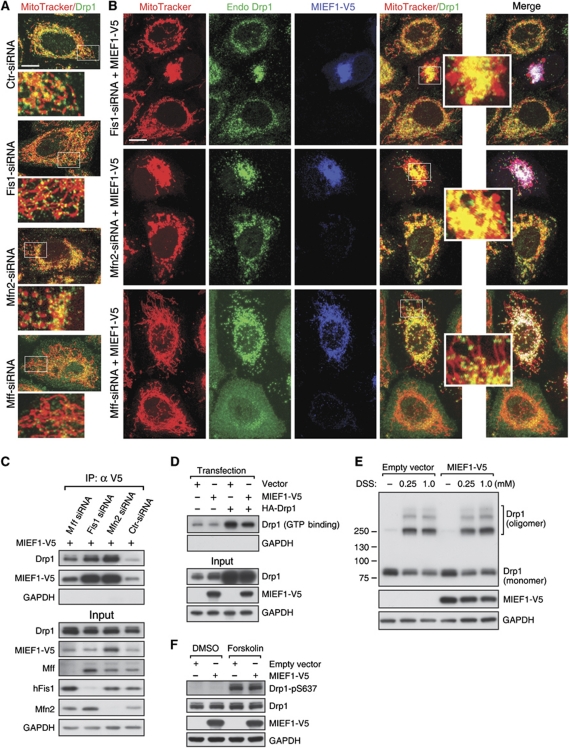
We next tested whether Mff, hFis1 or Mfn2 were required for MIEF1-induced recruitment of Drp1 to mitochondria. Depletion of hFis1 or Mfn2 alone did not produce a significant relocalization of Drp1 to mitochondria, whereas silencing of Mff reduced mitochondrial localization of Drp1 (Figure 7A; Supplementary Figure S2), in line with a recent report (Otera et al, 2010). In contrast, when MIEF1-V5 was ectopically expressed, Drp1 was recruited to mitochondria, irrespective of whether hFis1, Mfn2 or Mff were silenced (Figure 7B). In keeping with this, the binding between MIEF1-V5 and Drp1 was retained also under conditions when hFis1, Mfn2 and Mff were depleted (Figure 7C). These data indicate that Mff, hFis1 and Mfn2 are not required for MIEF1-induced recruitment of Drp1 to mitochondria.
Figure 7.
hFis1, Mfn2 and Mff are not required for MIEF1-mediated recruitment of Drp1 to mitochondria. (A) Distribution of endogenous Drp1 in control and HeLa cells depleted of hFis1, Mfn2 or Mff by RNAi (siFis1–3, siMfn2–3 or siMff-1), followed by confocal microscopy using MitoTracker Red staining and immunostaining with anti-Drp1 (green) antibody. (B) Distribution of endogenous Drp1 in hFis1, Mfn2 or Mff RNAi-treated HeLa cells expressing MIEF1-V5. Insets in (A, B) are magnifications of the boxed areas. Bars, 10 μm. (C) HeLa cells depleted of hFis1, Mfn2 or Mff by RNAi were further transfected with MIEF1-V5 and subjected to IP with anti-V5 agarose, and the precipitated complexes were analysed by western blot with indicated antibodies. (D) GTP binding of both endogenous Drp1 and exogenous HA-Drp1 was analysed by GTP-agarose pull down in 293T cells transfected with MIEF1-V5 alone, or cotransfected with MIEF1-V5 and HA-Drp1. (E) Immunoblotting to illustrate the oligomerization of endogenous Drp1 in 293T cells transfected with empty vector or MIEF1-V5 and crosslinked with DSS at indicated concentrations. (F) 293T cells transfected with empty vector or MIEF1-V5 were after 24 h incubated in the presence or absence of forskolin (20 μM), and cell extracts were immunoblotted using Phospho-DRP1 (Ser637) antibody and indicated antibodies.
MIEF1 reduces Drp1's GTP-binding activity
The potential effect of MIEF1 on Drp1's GTP-binding ability, oligomerization and phosphorlyation was assessed. The GTP binding of both endogenous Drp1 and exogenously expressed HA-Drp1 was reduced by MIEF1 overexpression (Figure 7D). In contrast, the extent of oligomerization of endogenous Drp1 was not affected by expression of MIEF1-V5 (Figure 7E). Using a phosphorylation-specific (Ser637) Drp1 polyclonal antibody, we showed that the low basal level of Drp1-Ser637 phosphorylation, which was, in keeping with previous reports (Chang and Blackstone, 2007; Cribbs and Strack, 2007; Han et al, 2008), augmented by the protein kinase A activator forskolin (Figure 7F), was not affected by MIEF1 overexpression, neither in the presence nor in the absence of forskolin (Figure 7F).
To test whether the MIEF1-induced relocalization of Drp1 to mitochondria depends on the phosphorylation status of Drp1, we expressed wild-type Myc-Drp1, the phosphorylation-deficient Myc-Drp1S637A mutant or the phosphomimetic Myc-Drp1S637D mutant in the presence or absence of MIEF1-V5. Expression of wild-type Drp1, Myc-Drp1S637A or Myc-Drp1S637D revealed a predominantly cytoplasmic distribution (Supplementary Figure S4A), whereas coexpression with MIEF1-V5 led to recruitment of wild-type Drp1, Myc-Drp1S637A and Myc-Drp1S637D to mitochondria, where they colocalized with MIEF1-V5 (Supplementary Figure S4B). Co-IP showed that MIEF1 binds to wild-type Myc-Drp1, Myc-Drp1S637A and Myc-Drp1S637D (Supplementary Figure S4C). In sum, these data show that MIEF1 reduces Drp1's GTP-binding activity, but does not affect oligomerization or phosphorylation of Drp1, and conversely, the phosphorylation status of Drp1-Ser637 is not critical for its interaction with MIEF1.
MIEF1 interacts with hFis1
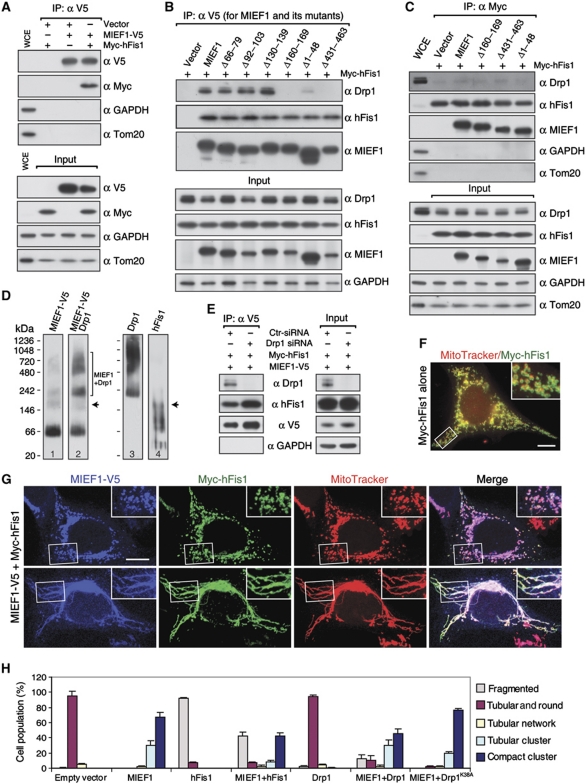
hFis1 serves as a potential mitochondrial receptor for Drp1 and is important for promoting fission (Yoon et al, 2003). We therefore examined whether MIEF1 interacts with hFis1. Co-IP revealed that hFis1 was co-immunoprecipitated both with wild-type MIEF1 and with the mutants MIEF1Δ431−463 and MIEF1Δ160−169, which only weakly or not at all bind Drp1 (Figure 8A and B). Furthermore, hFis1 binding was observed with the cytoplasmic MIEF1Δ1−48 mutant (Figure 8B). These data indicate that the interaction between MIEF1 and hFis1 is independent of MIEF1's interaction with Drp1 and its mitochondrial localization.
Figure 8.
The interaction of MIEF1 with hFis1 is independent of its interaction with Drp1 and overexpression of hFis1 partially reverses the MIEF1-induced mitochondrial fusion phenotype. (A) Cell lysates of 293T cells cotransfected with indicated plasmids were subjected to IP with anti-V5 agarose, and the precipitated complexes were immunoblotted with indicated antibodies. (B) MIEF1-V5 deletion mutants with no or reduced Drp1 binding (MIEF1Δ160−169 and MIEF1Δ431−463) and the cytoplasmic MIEF1Δ1−48 mutant had no effect on the binding between MIEF1 and hFis1. Cell lysates of 293T cells cotransfected with Myc-hFis1 and indicated MIEF1-V5 wild-type and mutant plasmids were subjected to IP with anti-V5 agarose and the precipitated complexes were immunoblotted with indicated antibodies. (C) Cell lysates of 293T cells cotransfected with Myc-hFis1 and indicated MIEF1-V5 wild-type and mutant plasmids were immunoprecipitated with anti-Myc agarose and the precipitated complexes were immunoblotted with indicated antibodies. (D) Native complexes of MIEF1, Drp1 and hFis1 were determined by NGE. Lysates from cells transfected with MIEF1-V5 and crosslinked with DSS (1 mM) were subjected to NGE followed by immunoblotting with indicated antibodies. Note: Lane 2 is the same blot as lane 1 that was reprobed with anti-Drp1 antibody after immunoblotting with anti-V5 antibody, and lanes 3 and 4 were probed using anti-Drp1 and hFis1 antibodies, respectively. (E) Cell lysates from Drp1 RNAi-treated HeLa cells cotransfected with MIEF1-V5 and Myc-hFis1 were subjected to IP with anti-V5 agarose, and the precipitated complexes were immunoblotted with indicated antibodies. (F) Confocal images of 293T cells transfected with Myc-hFis1 alone. Inset represents magnification of the boxed area. (G) Confocal images of 293T cells cotransfected with MIEF1-V5 and Myc-hFis1, and stained with MitoTracker followed by immunostaining with anti-V5 (blue) and anti-Myc (green) antibodies. Insets represent magnifications of the boxed areas. Bars, 10 μm. (H) Percentages (mean±s.e.m.) of cells with indicated mitochondrial morphologies in 293T cultures either transfected with empty vector (n=570), MIEF1-V5 (n=671), HA-Drp1 (n=405) and Myc-hFis1 (n=706) plasmid alone, or cotransfected with MIEF1-V5+HA-Drp1 (n=501), MIEF1-V5+HA-Drp1K38A (n=585) and MIEF1-V5+Myc-hFis1 (n=546) plasmids. Data were from three independent experiments.
Drp1 and hFis1 were shown to coprecipitate in IP experiments (Figure 8C), in keeping with a previous report (Yu et al, 2005), but the hFis1–Drp1 binding was much weaker than that between hFis1 and MIEF1. The avidity of the binding between hFis1 and Drp1 was not affected by coexpression of MIEF1 or MIEF1 mutants (Figure 8C), and overexpression of hFis1 did not affect the distribution of Drp1 between cytoplasm and mitochondria (Supplementary Figure S5). Non-denaturing (native blue) gel electrophoresis revealed that MIEF1 and Drp1 form complexes at ∼220 kDa and ∼480–720 kDa (Figure 8D, lanes 1 and 2), whereas MIEF1-V5 and hFis1 form an ∼200 kDa complex (Figure 8D, compare lane 1 to 4, arrows). In contrast, there was almost no high molecular weight complexes formed between Drp1 and hFis1. Co-IP further showed that MIEF1-V5 retained the ability to bind Myc-hFis1 in cells depleted of Drp1, indicating that Drp1 was not required for MIEF1–hFis1 interaction (Figure 8E). Similarly, hFis1 was not required for MIEF1–Drp1 interaction (Figure 7C). Taken together, these data suggest that MIEF1's interaction with Drp1 is independent of MIEF1's interaction with hFis1.
The MIEF1-mediated mitochondrial fusion phenotype is partially reversed by elevated hFis1 expression
To address the relationship between Drp1, hFis1 and MIEF1 further, we tested how different levels of Drp1 and hFis1 influence the MIEF1-induced fusion phenotype. Expression of hFis1 alone triggered mitochondrial fragmentation in most transfected cells (93%) (Figure 8F and H), in agreement with a previous report (Yoon et al, 2003). Coexpression of MIEF1 and hFis1 strongly reduced the proportion of cells with mitochondrial tubular and compact cluster phenotypes seen by MIEF1 transfection alone, and ∼43% of cells coexpressing MIEF1 and hFis1 exhibited fragmented mitochondria (Figure 8G and H).
Overexpression of Drp1 alone did not affect the morphology of mitochondria (Figure 8H; Supplementary Figure S6A), consistent with a previous report (Smirnova et al, 1998). Coexpression of MIEF1 and Drp1 only to a limited extent reverted the MIEF1-induced fusion phenotype (Figure 8H; Supplementary Figure S6B, upper panel). However, a small proportion of cells with fragmented mitochondria was also observed, a phenotype not seen when MIEF1 and Drp1 were expressed individually (Figure 8H; Supplementary Figure S6B, lower panel). Taken together, these data show that elevated levels of hFis1 partially reverse the MIEF1-induced fusion phenotype.
MIEF1 affects the sensitivity of cells to apoptotic stimuli and the autophagic activity
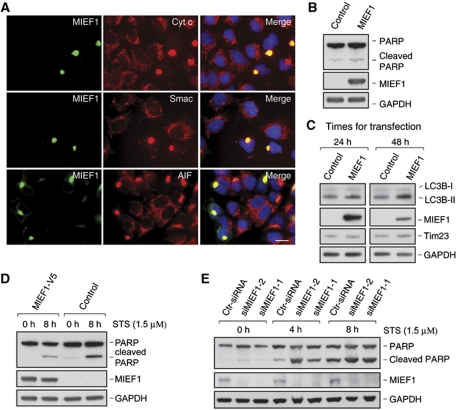
To begin to gain insights into the biological function of MIEF1, we explored whether MIEF1 regulated cell death pathways, as mitochondrial dynamics is important for the control of apoptosis (Suen et al, 2008). Expression of MIEF1 did not induce release of proapoptotic factors, such as cytochrome c, Smac/Diablo and AIF, from mitochondria (Figure 9A), nor did it affect the level of cleaved PARP (Figure 9B). In contrast, we observed an increase in the production of LC3B-II at 48 h after transfection (Figure 9C), indicating an elevated autophagic activity. We also observed that MIEF1 overexpression led to a decrease of cleaved PARP that was induced by staurosporine (STS) treatment (Figure 9D). Conversely, RNAi-mediated silencing of MIEF1 led to an enhanced production of cleaved PARP in response to STS (Figure 9E). These observations suggest that MIEF1 affects the sensitivity of cells to apoptotic stimuli and the autophagic activity.
Figure 9.
MIEF1 affects the sensitivity of cells to apoptotic stimuli and the activity of autophagy. (A) Confocal images show that MIEF1-V5 overexpression did not induce release of cytochrome c (cyt c), Smac/Diablo or AIF from mitochondria in 293T cells transfected with MIEF1-V5. Cells were immunostained with the indicated antibodies and nuclei were stained with DAPI. Bar, 20 μm. (B) PARP cleavage was analysed by immunoblotting of extracts from 293T cells transfected with empty vector (control) and MIEF1-V5. (C) The levels of LC3B-I and LC3B-II were analysed by immunoblotting of extracts from 293T cells transfected with empty vector (control) and MIEF1-V5. (D) PARP cleavage was analysed in extracts of HeLa cells transfected with MIEF1-V5 or empty vector and treated with STS (1.5 μM) for indicated time points. (E) PARP cleavage was analysed in extracts of HeLa cells transfected with control siRNA (Ctr-siRNA) or siMIEF1-1 and -2 and treated with STS (1.5 μM) for indicated time points.
Discussion
In this report, we characterize a novel vertebrate-specific mitochondrial outer membrane protein MIEF1, which regulates mitochondrial morphology. Overexpression of MIEF1 shifts the balance towards a fusion phenotype, characterized by extensively elongated mitochondrial tubules and perinuclear tubular clusters, whereas knockdown of MIEF1 leads to mitochondrial fragmentation. MIEF1 binds to and recruits Drp1 to the mitochondrial surface. Despite the fact that MIEF1 recruits Drp1 to mitochondria, it negatively affects the fission-promoting capacity of Drp1 by sequestering and inhibiting Drp1 activity, resulting in the mitochondrial fusion phenotype. To our knowledge, MIEF1 is the first Drp1 suppressor protein identified in vertebrates that efficiently impedes Drp1-mediated fission. Multiple lines of evidence support that MIEF1 has a key role in inhibition of Drp1-mediated mitochondrial fission. Firstly, MIEF1–Drp1 binding and the MIEF1-mediated recruitment of Drp1 to mitochondria do not require hFis1, Mff and Mfn2 (Figure 7), as well as are independent of Drp1 GTPase activity (Figure 5) or phosphorylation status. MIEF1 can efficiently bind to wild-type Drp1 as well as the Drp1S637A and Drp1S637D mutants (Supplementary Figure S4). Moreover, MIEF1 overexpression does not affect the phosphorylation level of Drp1 (Figure 7). Secondly, MIEF1 overexpression reduces the GTP-binding levels of both endogenous Drp1 and exogenous HA-Drp1 (Figure 7). This suggests that MIEF1 may affect the GTPase activity of Drp1 via a reduction of its GTP-binding ability. Thirdly, the MIEF1 mutant MIEF1Δ1−48, which is cytoplasmically localized but retains Drp1-binding ability and sequesters Drp1 in the cytoplasm, causes a mitochondrial fusion phenotype similar to that induced by Drp1K38A or by knockdown of Drp1 or Mff mRNA by RNAi (Figures 4 and 6). Conversely, the MIEF1 mutant MIEF1Δ160−169, which is membrane tethered but does not bind to Drp1, does not cause the mitochondrial fusion phenotype (Figure 6; Supplementary Figure S3). Collectively, the findings presented in this work shed light on the longstanding question of how Drp1 translocates from the cytoplasm to mitochondria and how Drp1-mediated fission is regulated in vertebrates (Hoppins et al, 2007; Santel and Frank, 2008; Liesa et al, 2009; Westermann, 2010b). Consistent with our findings, another group characterized the same cDNA of MIEF1(MiD51) and published their findings during the editorial processing of this work. They showed that MIEF1/MiD51 recruits Drp1 to mitochondria and that overexpression of MIEF1/MiD51 induces mitochondrial fusion, confirming a role for MIEF1 in inhibition of Drp1-mediated mitochondrial fission (Palmer et al, 2011). The function of MIEF1 is distinct from the recently characterized vertebrate-specific Mff protein (Gandre-Babbe and van der Bliek, 2008; Otera et al, 2010). While Mff, like MIEF1, recruits Drp1 to mitochondria, overexpression of Mff leads to a fission phenotype, whereas Mff knockdown promotes fusion. We therefore suggest that MIEF1 and Mff have opposite effects on mitochondrial fission. MIEF1 has a key role in inhibiting Drp1-induced mitochondrial fission, while Mff has a role in promoting Drp1-induced mitochondrial fission. Therefore, Mff and MIEF1 positively and negatively regulate Drp1-mediated mitochondrial fission in vertebrates, respectively.
Mitochondrial fusion is thought to be initiated by Mfn-mediated mitochondrial tethering. Tethering of adjacent mitochondria is required for bringing membranes into close apposition for consequent membrane fusion events, where the GTPase activity of Mfn may function downstream of mitochondrial tethering to mediate full fusion (Koshiba et al, 2004). However, it seems plausible that MIEF1, in addition to blocking Drp1 fission activity, also actively promotes fusion. Notably, overexpression of MIEF1 can induce a high proportion of cells to exhibit a compact cluster of mitochondria, while inhibition of mitochondrial fission via MIEF1Δ1−48 or Drp1K38A, or via depletion of either Drp1 or Mff does not induce a highly compact cluster phenotype (see Figure 4). This suggests that MIEF1 has the capacity to bring the mitochondrial membranes into close apposition and ultimately facilitates the process of membrane fusion as revealed by EM in Figure 2. Moreover, the data from an in vivo cell fusion assay reveal that MIEF1 overexpression promotes mitochondrial fusion. Conversely, depletion of endogenous MIEF1 results in partially fragmented mitochondria. Two lines of evidence indicate that MIEF1 promotes fusion in a manner independent of the fusion-promoting GTPase Mfn2. Firstly, the mitochondrial fusion phenotypes caused by overexpression of MIEF1 and Mfn2 are not identical, and secondly, MIEF1 is capable of inducing mitochondrial fusion also under conditions when Mfn2 expression is knocked down by RNAi. The accumulation of MIEF1 into discrete puncta at the connection sites of adjacent mitochondrial units, along with the observation that MIEF1 undergoes oligomerization by self-association, indicates that interactions between MIEF1 proteins on opposing mitochondrial membranes may contribute to the fusion process.
hFis1 is a fission-promoting protein, which was proposed to serve as a receptor for recruitment of Drp1 to mitochondria (James et al, 2003; Yoon et al, 2003). In yeast, Fis1p (the hFis1 homologue) interacts with and recruits Dnm1p (the Drp1 homologue) to mitochondria through the yeast-specific adaptor proteins Mdv1p or Caf4p (Mozdy et al, 2000; Tieu and Nunnari, 2000; Griffin et al, 2005). The fact that elevated expression of hFis1, in contrast to MIEF1, does not relocalize Drp1 to mitochondria argues against a direct role for hFis1 in the recruitment process in vertebrates. Furthermore, hFis1 localizes evenly throughout the mitochondrial surface (Suzuki et al, 2003), whereas MIEF1 and Drp1 colocalize as discrete puncta extensively. MIEF1 shows a robust interaction with hFis1, in contrast to a much weaker interaction observed between hFis1 and Drp1. The MIEF1–hFis1 interaction is likely to be of functional significance, as elevated hFis1 partially reverts the MIEF1-induced fusion phenotype. While it may be tempting to speculate that MIEF1, because it can interact with Drp1 and hFis1 through non-overlapping domains, would serve as a bridging protein between Drp1 and hFis1, our data indicate that the MIEF1–Drp1 and MIEF1–hFis1 interactions are independent. Firstly, the binding between hFis1 and Drp1 is not affected by elevated levels of MIEF1 or MIEF1 mutants. Secondly, depletion of hFis1 or Drp1 by RNAi does not affect MIEF1 interaction with Drp1 and hFis1, respectively. Thirdly, the native gel electrophoresis (NGE) shows that the complexes containing MIEF1 and Drp1 differ in size from those containing MIEF1 and hFis1. Assuming that MIEF1 independently interacts with hFis1 and Drp1, it may be argued that the relative levels of hFis1 and MIEF1 regulate Drp1-mediated mitochondrial fission, such that high levels of MIEF1 promote a MIEF1–Drp1 interaction, which inhibits Drp1-mediated fission, leading to mitochondrial fusion, whereas high levels of hFis1 reverse the MIEF1-induced fusion phenotype, thus leading to mitochondrial fission.
The characterization of MIEF1 also highlights that mitochondrial morphology is differently regulated in vertebrates and yeast. We suggest that the molecular machinery controlling mitochondrial dynamics has evolved such that two of the central components, the Dmn1p/Drp1 and Fis1p/hFis1 proteins, are highly conserved in both yeast and vertebrates, while the interacting proteins, that is MIEF1 and Mff versus Mdv1p and Caf4p, are quite evolutionarily and functionally diverged.
In conclusion, the characterization of MIEF1 sheds new light on how the molecular machinery controlling mitochondrial morphology in mammals is organized and also highlights important differences between yeast and mammals in the composition of this machinery. Given the importance of mitochondrial dynamics for many cellular processes, the discovery of a novel key regulator protein in the process may provide further options to experimentally steer the process, which may be of importance in understanding the role of mitochondrial dynamics in cancer, diabetes, cardiovascular and neurodegenerative diseases.
Materials and methods
Cell cultures
293T and HeLa cells were grown in Dulbecco's modified Eagle's medium with 10% FCS. The three human malignant glioma cell lines, U-343 MG, U-343 MGa Cl2:6 (Cl2:6) and U-87 MG, and the neuroblastoma cell line SH-SY5Y were grown in Eagle's minimum essential medium with 10% FCS.
Generation of expression constructs
The cDNA with the full-length ORF of SMCR7L (MIEF1) (NM_019008) was amplified by PCR using the Human Fetal Brain BD™ Marathon-Ready cDNA (Clontech) as template, and cloned into the pcDNA3.1/V5-His-TOPO vector (Invitrogen) to produce a C-terminally V5-His-tagged MIEF1 (MIEF1-V5). The MIEF1-V5 plasmid was verified by sequencing. We also generated an N-terminally HA-tagged MIEF1 plasmid in pcDNA3.1 (HA-MIEF1). All deletion mutants of MIEF1 diagrammed in Supplementary Figure S7 were constructed in the pcDNA3.1/V5-His-TOPO vector (Invitrogen) using standard PCR techniques. Internal deletion mutants of MIEF1 shown in Supplementary Figure S7 were constructed using a previously described method (Ho et al, 1989). All deletion constructs were verified by sequencing.
Subcellular fractionation
Subcellular fractionation of 293T cells was carried out as described (Zhao et al, 2009), or using a mitochondria isolation kit (Pierce) according to the manufacturer's instructions. Proteinase K (PK) digestion and membrane protein analyses were performed as described (Zhao et al, 2009).
RNAi silencing of MIEF1 expression
Two sets of siRNA duplexes specific for human SMCR7L (MIEF1) mRNA: siMIEF1-1 (sense 5′-GCCAAGCAAGCUGCUGUGGACAUAU-3′), siMIEF1-2 (sense 5′-GGAGCAGAACC-UGUGGUCAUGUAUU-3′) and a scrambled siRNA duplex (12935-400) with similar GC content recommended by the manufacturer were purchased from Invitrogen.
In vivo protein–protein crosslinking and co-IP
For formaldehyde (FA) in vivo crosslinking, transfected cells were washed with PBS buffer, and proteins were crosslinked by incubating cells in PBS containing 1% FA for 10 min at room temperature as described (Hajek et al, 2007). In vivo crosslinking with disuccinimidyl suberate (DSS; Pierce) was performed as described (Zhu et al, 2004) with some modifications. Co-IP experiments were carried out as described (Hajek et al, 2007) with a slight modification.
PEG cell fusion assay
PEG-mediated cell fusion experiments were performed as described (Liesa et al, 2008; Kamp et al, 2010) with slight modifications. The polykaryons were analysed by Leica TCS SP5 confocal microscopy system. Mitochondrial fusion was indicated by the percent of mito-GFP and mito-DsRed colocalization.
GTP-agarose pull-down assay
GTP-binding assays were performed as described (De Palma et al, 2010) with slight modifications.
Native gel electrophoresis
NGE was performed using the NativePAGE Novex Bis-Tris Gel System according to the manufacturer's protocols (Invitrogen). The NativeMarkTM Unstained Protein Standard (Invitrogen) was used as size marker.
Statistical analysis
The unpaired Student's t-test was applied to evaluate differences between experimental groups. P-values ⩽0.05 were considered statistically significant.
More detailed information on the materials and methods mentioned above and all additional materials and methods used are described in Supplementary data.
Supplementary Material
Acknowledgments
We thank Dr Alexander M van der Bliek (Department of Biological Chemistry, UCLA School of Medicine) for providing HA-Drp1 and HA-Drp1K38A; Dr Shigehise Hirose (Department of Biological Science, Tokyo Institute of Technology) for providing FLAG-mMfn2; Dr Yisang Yoon (Department of Anesthesiology and of Pharmacology and Physiology, University of Rochester School of Medicine and Dentistry) for the Myc-hFis1; Dr Craig Blackstone (Cellular Neurology Unit, NINDS, National Institutes of Health, Bethesda) for Myc-Drp1, Myc-Drp1S637A, Myc-Drp1S637D; and Dr Christian Haass and Dr Frits Kamp (DZNE-German Center for Neurodegenerative Diseases, Adolf-Butenandt-Institute, Biochemistry, Ludwig-Maximilians-University) for mito-GFP and mito-DsRed plasmids. This work was supported by grants from the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Cancer Society in Stockholm, Knut och Alice Wallenbergs Stiftelse, Karolinska Institutet and the Swedish Research Council (VR-M, VR-Linné and DBRM).
Author contributions: JZ conceived the project, designed research. ZJ, SJ, TL, XW, MQ, NT, OS and PU performed the experiments and data analysis. JZ, UL and MN wrote the manuscript. OS, SJ, TL and XW discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Berman SB, Pineda FJ, Hardwick JM (2008) Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ 15: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC (2006) Mitochondrial fusion and fission in mammals. Ann Rev Cell Dev Biol 22: 79–99 [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282: 21583–21587 [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280: 26185–26192 [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma C, Falcone S, Pisoni S, Cipolat S, Panzeri C, Pambianco S, Pisconti A, Allevi R, Bassi MT, Cossu G, Pozzan T, Moncada S, Scorrano L, Brunelli S, Clementi E (2010) Nitric oxide inhibition of Drp1-mediated mitochondrial fission is critical for myogenic differentiation. Cell Death Differ 17: 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26: 207–210 [DOI] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K (2003) Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem 134: 333–344 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandemange S, Herzig S, Martinou JC (2009) Mitochondrial dynamics and cancer. Seminars Cancer Biol 19: 50–56 [DOI] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC (2005) The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol 170: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek P, Chomyn A, Attardi G (2007) Identification of a novel mitochondrial complex containing mitofusin 2 and stomatin-like protein 2. J Biol Chem 282: 5670–5681 [DOI] [PubMed] [Google Scholar]
- Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M (2008) CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol 182: 573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Hom J, Sheu SS (2009) Morphological dynamics of mitochondria—a special emphasis on cardiac muscle cells. J Mol Cell Cardiol 46: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J (2007) The machines that divide and fuse mitochondria. Ann Rev Biochem 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Huang P, Yu T, Yoon Y (2007) Mitochondrial clustering induced by overexpression of the mitochondrial fusion protein Mfn2 causes mitochondrial dysfunction and cell death. Eur J Cell Biol 86: 289–302 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11: 958–966 [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC (2003) hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem 278: 36373–36379 [DOI] [PubMed] [Google Scholar]
- Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, Eimer S, Winklhofer KF, Haass C (2010) Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J 29: 3571–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefel BR, Gilson PR, Beech PL (2006) Cell biology of mitochondrial dynamics. Int Rev Cytol 254: 151–213 [DOI] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E (2008) Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci 9: 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC (2004) Structural basis of mitochondrial tethering by mitofusin complexes. Science (New York, NY) 305: 858–862 [DOI] [PubMed] [Google Scholar]
- Lackner LL, Nunnari J (2010) Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem Biol 17: 578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ (2004) Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell 15: 5001–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Borda-d’Agua B, Medina-Gomez G, Lelliott CJ, Paz JC, Rojo M, Palacin M, Vidal-Puig A, Zorzano A (2008) Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS One 3: e3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A (2009) Mitochondrial dynamics in mammalian health and disease. Physiol Rev 89: 799–845 [DOI] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A (2008) Mitochondria in neuroplasticity and neurological disorders. Neuron 60: 748–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz S, Hammermeister M, Altmann K, Durr M, Westermann B (2007) Molecular machinery of mitochondrial dynamics in yeast. Biol Chem 388: 917–926 [DOI] [PubMed] [Google Scholar]
- Modica-Napolitano JS, Singh KK (2004) Mitochondrial dysfunction in cancer. Mitochondrion 4: 755–762 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet 39: 503–536 [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Mao P, Manczak M (2009) Mitochondrial structural and functional dynamics in Huntington's disease. Brain Res Rev 61: 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S (2008) Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life 60: 448–455 [DOI] [PubMed] [Google Scholar]
- Schafer A, Reichert AS (2009) Emerging roles of mitochondrial membrane dynamics in health and disease. Biol Chem 390: 707–715 [DOI] [PubMed] [Google Scholar]
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S (2000) Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep 1: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM (1998) A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 143: 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ (2008) Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N (2003) The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J Mol Biol 334: 445–458 [DOI] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol 151: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, Nunnari J (2002) The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol 158: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol 186: 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X (2009) The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer's disease. J Neurochem 109(Suppl 1): 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B (2010a) Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol 21: 542–549 [DOI] [PubMed] [Google Scholar]
- Westermann B (2010b) Mitochondrial fusion and fission in cell life and death. Nat Rev 11: 872–884 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA (2003) The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 23: 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, McNiven MA (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell 12: 2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fox RJ, Burwell LS, Yoon Y (2005) Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci 118: 4141–4151 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin SB, Tomilin N, Castro J, Shupliakov O, Lendahl U, Nister M (2009) The novel conserved mitochondrial inner-membrane protein MTGM regulates mitochondrial morphology and cell proliferation. J Cell Sci 122: 2252–2262 [DOI] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C (2004) Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem 279: 35967–35974 [DOI] [PubMed] [Google Scholar]
- Zorzano A, Liesa M, Palacin M (2009) Role of mitochondrial dynamics proteins in the pathophysiology of obesity and type 2 diabetes. Int J Biochem Cell Biol 41: 1846–1854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.