Abstract
Parkin is an E3-ubiquitin ligase belonging to the RBR (RING–InBetweenRING–RING family), and is involved in the neurodegenerative disorder Parkinson's disease. Autosomal recessive juvenile Parkinsonism, which is one of the most common familial forms of the disease, is directly linked to mutations in the parkin gene. However, the molecular mechanisms of Parkin dysfunction in the disease state remain to be established. We now demonstrate that the ubiquitin-like domain of Parkin functions to inhibit its autoubiquitination. Moreover pathogenic Parkin mutations disrupt this autoinhibition, resulting in a constitutively active molecule. In addition, we show that the mechanism of autoregulation involves ubiquitin binding by a C-terminal region of Parkin. Our observations provide important molecular insights into the underlying basis of Parkinson's disease, and in the regulation of RBR E3-ligase activity.
Keywords: ligase, Parkinson's disease, RING, ubiquitin
Introduction
Post-translational modification of a target protein by ubiquitin signals a variety of cellular outcomes including proteasomal degradation, endosomal sorting, endocytosis and DNA repair. The conjugation process is usually effected by a cascade of enzymes; an E1 that activates the C-terminus of the ubiquitin moiety; an E2 conjugating enzyme that accepts the activated ubiquitin and co-ordinates with a third enzyme, the E3-ubiquitin ligase. The E3-ubiquitin ligase provides the specificity by determining which substrates will be ubiquitinated (Pickart and Eddins, 2004; Kerscher et al, 2006; Dye and Schulman, 2007). E3 ligases fall broadly into two classes: the HECTs that contain a catalytic cysteine responsible for the formation of a thioester intermediate, and the RING-type E3s that are thought to act as scaffolding proteins, enhancing the transfer of ubiquitin from E2 to target. The RING E3s are the larger class, comprising ∼90% of known E3s, and have varied domain structures. The components required for ubiquitination are diverse and varied; however, little is known about how the machinery itself is regulated. Although ubiquitin ligases have been largely thought to be constitutively active, it has recently been demonstrated that both HECT and RING-type E3 ligases are subject to regulation through substrate or E3 phosphorylation, or by utilisation of adaptor proteins (Kee and Huibregtse, 2007; Duda et al, 2008; Saha and Deshaies, 2008), as well as by autoinhibition of catalytic activity in the case of the HECT-type E2, Smurf2 (Wiesner et al, 2007).
Parkin, the product of the parkin gene, has E3-ubiquitin-ligase activity (Shimura et al, 2000; Zhang et al, 2000). Parkinson's disease is a neurodegenerative disorder characterised by the loss of dopaminergic neurons from the substantia nigra, and the presence of Lewy bodies, which are pathogenic aggregated inclusion bodies rich in ubiquitin and α-synuclein (Jenner and Olanow, 1998). Autosomal recessive juvenile Parkinsonism (AR-JP) is one of the most common familial forms of the disease and has been directly linked to mutations in the parkin gene (Kitada et al, 1998). In addition, heterozygous parkin mutations have also been discovered in cases of late-onset sporadic Parkinsonism, raising the possibility of its involvement in the pathogenesis of sporadic PD. Recent studies suggest a pivotal role for Parkin and PINK1 (a kinase also mutated in AR-JP) (Valente et al, 2004) in the selective degradation of mitochondria, although the mechanisms by which this occurs are still unclear (Geisler et al, 2010; Matsuda et al, 2010; Narendra et al, 2010; Vives-Bauza et al, 2010). These studies also report a requirement for activation of Parkin, whereas historically, Parkin has been considered constitutively active due to its autoubiquitination (Shimura et al, 2000; Zhang et al, 2000). In addition to its established role in Parkinsonism, Parkin is also a putative tumour suppressor (Poulogiannis et al, 2010; Veeriah et al, 2010). Parkin is a 465-residue protein that contains two RING motifs linked by a cysteine-rich in-between-RING (IBR) motif, a newly identified zinc co-ordinating motif termed RING0, and an N-terminal ubiquitin-like domain (Ubl) (Figure 1A) (Hristova et al, 2009). Parkin binds several E2s including UbcH7 and UbcH8, and the UbcH13/Uev1a E2 heterodimer that is thought to be responsible for the catalysis of K63-linked ubiquitin chains (Olzmann et al, 2007). Parkin has also been shown to be capable of inducing monoubiquitination (Hampe et al, 2006; Moore et al, 2008), multiple monoubiquitination (Matsuda et al, 2006), K48-linked polyubiquitination, and K63-linked polyubiquitination (Doss-Pepe et al, 2005; Lim et al, 2005).
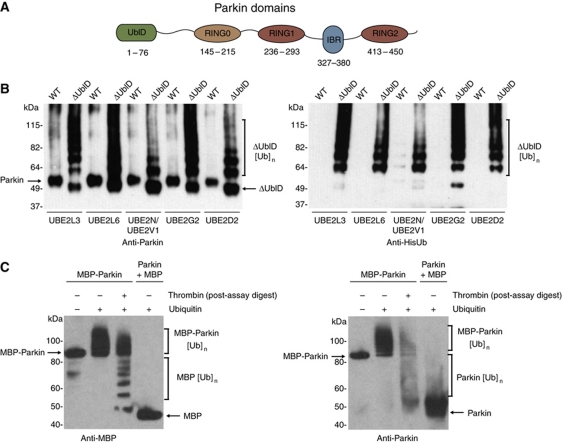
Figure 1.
The ubiquitin-like domain of Parkin inhibits its autoubiquitination. (A) Schematic representation of Parkin molecule. The ubiquitin-like (UblD), RING0 (R0), RING (R1, R2) and in-between-RING (IBR) domains are indicated. The numbering for each domain is as per the human protein. (B) Western blot analysis of autoubiquitination assays of WT Parkin (WT) and ΔUbl-Parkin (ΔUblD) reveal inhibition of Parkin autoubiquitination by the Ubl domain. Formation of ubiquitin conjugates is detected with α-Parkin (left) and α-His-Ub (right) and indicated with a bracket. The E2 used in each experiment is indicated. (C) Western blot analysis of autoubiquitination assays of MBP-Parkin, showing competence for autoubiquitination only in the fused protein. Thrombin was added after the reaction has been stopped, leading to the cleavage of ubiquitinated MBP-Parkin. Detection of conjugates by MBP and Parkin-specific antibodies reveals that most of the ubiquitin moieties are on the MBP tag (lane 3). Lane 1 has no ubiquitin. Brackets indicate ubiquitination.
Pathogenic mutations occur throughout the primary sequence and there have been many studies linking these mutations to changes in Parkin stability and solubility (Wang et al, 2005; Hampe et al, 2006; Schlehe et al, 2008). Reviews of the distribution and type of mutations occurring in the parkin gene have revealed that there are hotspots for the mutations including exon rearrangements that affect the RING domains, and that most of the missense mutations occur in the C-terminal RING-IBR-RING domains (Hedrich et al, 2004; Tan and Skipper, 2007). However, there is also a cluster of point mutations and small deletions that occur in exon 2. Exon 2 gives rise to the Ubl domain, a domain at the extreme N-terminus of Parkin that shares 30% sequence identity with human ubiquitin (Figure 1A). The functional importance of this domain remains to be established although there have been recent studies implicating it in substrate recognition, SH3-domain binding, proteasome association and regulation of cellular Parkin levels (Finney et al, 2003; Sakata et al, 2003; Fallon et al, 2006; Trempe et al, 2009). Certain disease-causing mutations in the Ubl domain can result in its unfolding (Safadi and Shaw, 2007; Tomoo et al, 2008). It has been demonstrated that the Ubl domain is not necessary for the E3-ligase activity of Parkin in vitro as a C-terminal fragment comprising the IBR-RING2 domains is capable of autoubiquitination (Matsuda et al, 2006). It has also been reported that this domain can be cleaved in vivo by caspases (Kahns et al, 2003). We set out to establish the molecular function of the Ubl domain in the regulation of Parkin’s E3 activity. We found evidence for an autoinhibitory role of the Ubl domain in regulating the autoubiquitination activity of Parkin. Furthermore, we show that pathogenic mutations disrupt this autoinhibitory function of the Ubl domain, providing a molecular rationale for pathogenic Parkin mutations.
Results
The ubiquitin-like domain of Parkin inhibits its autoubiquitination
Given the fundamental regulatory roles played by E3s, it seems likely that they should be tightly regulated. Many enzymes responsible for post-translational modifications are themselves regulated by intramolecular interactions, including kinases and acetyltransferases (Hubbard, 2002; Stavropoulos et al, 2008). Recent reports have indicated a need for Parkin activation at the mitochondria, suggesting an intrinsic or extrinsic regulation of Parkin activity must exist (Matsuda et al, 2010; Narendra et al, 2010). One possibility is that the Ubl domain of Parkin might inhibit its autoubiquitination activity through an intramolecular interaction. To test this hypothesis, we generated a deletion mutant (ΔUblD) lacking the Ubl domain and assayed its autoubiquitination activity. In contrast to wild-type (WT) Parkin, ΔUblD autoubiquitinates and forms high molecular weight ubiquitinated species (Figure 1B). Given the ability of Parkin to use several E2s, we examined whether Parkin autoinhibition is restricted to a particular E2. We found that ΔUbl-Parkin autoubiquitinates with Ube2L3 (UbcH7) and Ube2L6 (UbcH8), Ube2N/Ube2V1 (UbcH13/Uev1a), Ube2G2 (Ubc7) and Ube2D2 (UbcH5b) (Figure 1B). Furthermore, WT Parkin did not autoubiquitinate in the presence of any of the tested E2s (Figure 1B). Similar results were obtained at pH 7.4 and pH 8.0, and no activity was observed in the absence of E2 (Supplementary Figure S1A and B). These data suggest that the Ubl domain of Parkin functions to inhibit its intrinsic autoubiquitination activity.
Perturbation of Parkin tertiary structure disrupts Ubl-mediated inhibition
In contrast to our observations, numerous studies, both in vivo and in vitro, have shown that Parkin is readily autoubiquitinated (Shimura et al, 2000; Chung et al, 2004; Hampe et al, 2006). We considered the possibility that bacterially expressed Parkin may not be active due to a lack of an uncharacterised modification. To test this, we also produced Parkin in insect cells, and found that it too was inactive (Supplementary Figure S2A). Previous studies have been performed using N-terminally tagged Parkin, while our experiments are carried out using, recombinant WT human Parkin purified to homogeneity after removal of a His-smt3 (SUMO) fusion tag. The His-smt3 tag enables cleavage of the fusion protein immediately before the N-terminal methionine, generating WT Parkin with no overhang or leader sequence. We therefore wondered if these apparent differences in the activity of Parkin were the consequence of the N-terminal tag interfering with the intramolecular interaction within the protein. To explore this possibility, we tested the activity of tagged versions of Parkin. We found that MBP WT Parkin is capable of undergoing autoubiquitination (Figure 1C). Furthermore, thrombin cleavage following a ubiquitination assay revealed both the cleaved tag and Parkin itself are ubiquitinated. In contrast, MBP is not ubiquitinated when added in isolation to purified untagged Parkin (Figure 1C, lane 4). The same activity was observed with a C-terminal thioredoxin tag (Supplementary Figure S2A). In order to further explore this, we left the His-smt3 tag on Parkin and carried out the assay in the absence and presence of the smt3 protease, Ulp1. Once the smt3 tag is cleaved, the protein loses autoubiquitination activity (Supplementary Figure S2B). These data show that artificially tagging Parkin promotes the protein to be active against itself and the tag, and that cleavage of the SUMO tag inhibits autoubiquitination.
The most straightforward explanation for our observations is that Parkin exists in an autoinhibited state that involves its N-terminal Ubl domain. Further support for this hypothesis comes from longer incubation of the assay, which will be expected to affect the stability of any intramolecular interaction, we found that Parkin was autoubiquitinated (Supplementary Figure S2C). Taken together, these data suggest that Parkin is susceptible to destabilisation and becoming active, and that the dynamics of the intramolecular interaction within the molecule may influence the extent of autoinhibition. Furthermore, these observations suggest the ubiquitin-ligase activity of WT Parkin is latent, and requires various secondary factors to induce its autoubiquitination.
Ligation of the Ubl domain to ΔUblD restores the autoinhibition
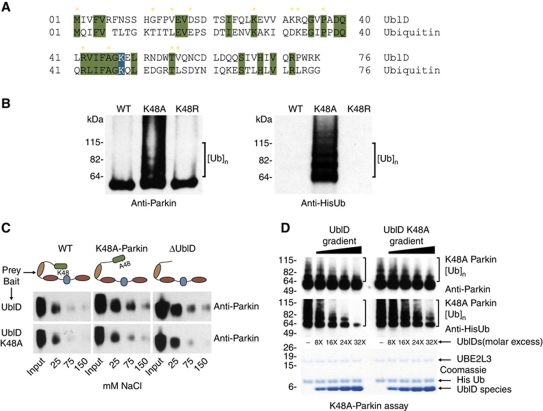
To further validate the autoinhibitory role of the Ubl domain, we reattached the Ubl domain to a constitutively active ΔUblD species. Using expressed protein ligation (EPL; see Materials and methods and Supplementary Figure S3A for details), we produced the Ubl domain with a C-terminal intein tag, and residues 95–465 of Parkin (95CParkin) with an N-terminal cysteine. Ligation of these fragments yielded full-length Parkin-EPL, likely to resemble the tertiary structure of WT Parkin as well as its autoubiquitination profile. Antibodies specific to different domains of Parkin were used to differentiate between Parkin-EPL and 95C. As expected, the C-terminal fragment (95C) readily autoubiquitinated (Supplementary Figure S3B). The Ubl-specific antibody does not detect the 95C species, but readily detects the Parkin-EPL species. Absence of any higher molecular weight bands suggests that autoubiquitination does not occur with Parkin-EPL. A positive control containing a Parkin-Ubl mutation (K48A) described later does retain activity. Thus, Parkin-EPL resembles WT Parkin in its autoubiquitination profile thereby establishing the Ubl domain as an autoinhibitory module.
Pathogenic mutations in the Ubl domain relieve autoinhibition
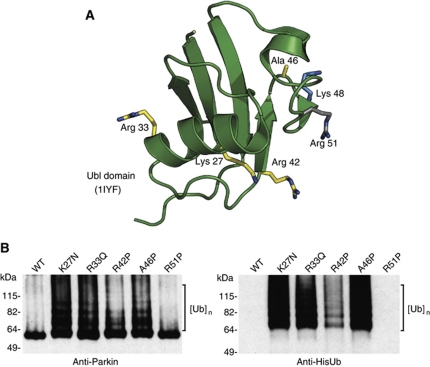
Pathogenic mutations occur throughout the primary sequence of Parkin, although there are a number of important hotspots, including a cluster of point mutations and small deletions within the Ubl domain (Terreni et al, 2001; Hedrich et al, 2002, 2004; Hoenicka et al, 2002; Tan and Skipper, 2007). Furthermore, a number of disease-causing mutations in Ubl domain result in its unfolding (Safadi and Shaw, 2007; Tomoo et al, 2008). Based on our observations, we examined whether four pathogenic point mutations, K27N, R33Q, R42P and A46P, in the Ubl domain affect the autoinhibition of Parkin (Figure 2A). We found that all four mutants were capable of autoubiquitination, while the WT was not (Figure 2B). We also generated what we predicted to be a ‘silent’ mutation, R51P, based on the structure of the Ubl domain (Sakata et al, 2003). We selected this mutation because the arginine is in a loop rather than a secondary structure element and is not conserved between species. This mutation behaves as the WT protein (Figure 2B). These data suggest that the consequence of the pathogenic mutations within the Ubl domain is the disruption of autoinhibition and activation of Parkin autoubiquitination.
Figure 2.
Pathogenic mutations in the Ubl domain relieve autoinhibition. (A) NMR structure of human Parkin showing the location of each of the pathogenic mutations in yellow, Lys48 in blue, and the site of the silent mutation in grey (PDB code 1iyf (Sakata et al, 2003)). (B) Western blot analysis of Parkin autoubiquitination shows that pathogenic point mutations render Parkin active for autoubiquitination while R51P does not. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. Ube2L3 is the E2 in the assay.
We also investigated whether the Ubl domain regulated Parkin in cells by transfecting WT or pathogenic mutants of Parkin, along with His-ubiquitin. We pulled down His-ubiquitin conjugates from the cells in the presence and absence of the proteasome inhibitor, MG132. Overall levels of WT Parkin but not mutants were similar irrespective of MG132 treatment. Furthermore, we found that a greater proportion of the transfected Parkin was ubiquitinated in the pathogenic and ΔUblD mutant forms of Parkin, when stabilised by the presence of MG132, than WT (Supplementary Figure S4). These data suggest that the Ubl domain also regulates Parkin ubiquitination in cells, and that Ubl mutants are more active.
Lysine 48 is critical for the intramolecular inhibition of Parkin
Interestingly, several of the known pathogenic point mutations in the Ubl domain are sites of sequence conservation with ubiquitin (Figure 3A). Furthermore, the Ubl domain has a lysine residue at position 48, which is completely conserved with ubiquitin. In ubiquitin, this lysine is the point of ubiquitin chain building and is responsible for generating the proteasome recognition signal (Chau et al, 1989). Therefore, we generated a K48A-Parkin species and assayed it for autoubiquitination. We found that this mutant was also capable of autoubiquitination (Figure 3B). The K48A mutation does not cause unfolding of the Ubl domain and yet behaves the same way as the mutations that do unfold the domain (Safadi and Shaw, 2007; Tomoo et al, 2008). The most straightforward explanation is that K48 is important for maintaining the inhibitory intramolecular interaction within Parkin. To explore this possibility, we mutated lysine 48 to arginine, maintaining the electrostatic nature of the residue. We found this mutant is not capable of autoubiquitination, behaving as WT protein (Figure 3B), suggesting that the charged nature of the residue is critical for the interaction. To explore this further, we performed Parkin pull-down assays using the WT and Ubl-K48A domains as bait to examine if the two domains interact (Figure 3C). We found that while the WT Ubl domain interacts weakly with Parkin, it is able to bind both Parkin-K48A and ΔUblD. Moreover, the Ubl-K48A domain is unable to interact with WT, Parkin-K48A or ΔUblD. We also reasoned that a WT Ubl domain would be able to diminish Parkin autoubiquitination in the K48A-Parkin mutant. To test this hypothesis, we titrated increasing concentrations of WT Ubl domain in the K48A-Parkin autoubiquitination assay and found that K48A-Parkin autoubiquitination is reduced. Adding K48A-Ubl to K48A-Parkin did not inhibit K48A-Parkin autoubiquitination to the same extent (Figure 3D; Supplementary Figure S5). Our findings show the Ubl domain of Parkin is responsible for the intramolecular interaction and autoinhibition of Parkin, with K48 being a critical point of interaction.
Figure 3.
Lysine 48 is required for the intramolecular interaction. (A) Sequence alignment of the Ubl domain and ubiquitin. Conserved residues are highlighted in green with K48 highlighted in blue. Yellow stars indicate pathogenic Parkin mutations. (B) Western blot analysis of Parkin autoubiquitination shows K48A renders Parkin active for autoubiquitination while K48R does not. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. (C) Western blot analysis of full-length Parkin species pulled down by either an intact Ubl domain (top panel) or mutant K48A Ubl domain (lower panel), showing that WT Ubl domain interacts with K48A-Parkin and ΔUbl-Parkin at physiological salt concentrations, while K48A-Ubl domain does not. Input and salt concentrations are indicated. (D) Western blot analysis of K48A-Parkin autoubiquitination shows that increasing concentrations of WT Ubl domain reduces K48A-Parkin autoubiquitination to a greater extent than K48A-Ubl domain. Conjugates are detected using Parkin and His antibodies and are indicated by brackets. Excess concentrations are indicated as a ratio of Ubl species to K48A-Parkin, and a coomassie-stained gel of the input is shown beneath.
The Ubl domain interacts with ΔUblD
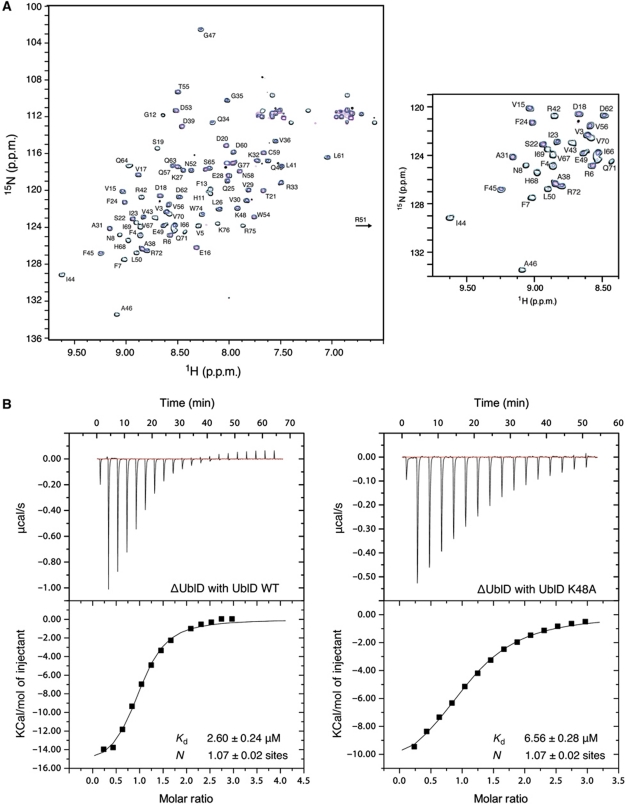
In order to confirm the interaction between the Ubl domain and the rest of Parkin, we performed NMR titration experiments using 15N-labelled Ubl domain and titrating it with residues 77–465 from Parkin (ΔUblD). In the absence of ΔUblD, the 1H–15N HSQC spectrum of the Ubl domain is disperse with narrow line widths similar to previously reported work (Figure 4A). Upon increasing concentrations of ΔUblD, many peaks in the Ubl domain gradually decrease in intensity while others exhibit significant broadening. These observations are consistent with slow exchange and the six-fold increase in apparent mass for the Ubl domain expected from an interaction between the two fragments.
Figure 4.
The Ubl domain interacts with ΔUbl-Parkin. (A) 600 MHz 1H–15N HSQC spectra showing the interaction between 15N-labelled Ubl domain in the absence (black contours) and presence of 0.33 (cyan), 0.67 (blue) and 1.0 (magenta) equivalents of ΔUblD. Residue assignments are indicated for the Ubl domain beside each peak. The spectra show the disappearance of many peaks in the Ubl domain spectrum as a function of ΔUblD concentration, indicative of an interaction with the ΔUblD fragment. To the right, a magnified view of part of the spectra is shown. (B) ITC curves showing binding of WT UblD (left) and K48A-UblD (right) to ΔUblD-Parkin. Dissociation constants and stoichiometry of the interaction are indicated.
To further quantify the interaction between the N-terminal UblD and the C-terminal ΔUblD, we conducted isothermal titration calorimetry (ITC) assays. WT UblD binds ΔUblD with a dissociation constant (Kd) of ∼2.5 μM, while K48A-UblD binds with greater than two-fold lower affinity (Kd=∼6.5 μM) (Figure 4B). These results indicate that WT Parkin exists in a closed conformation mediated through an intramolecular interaction between the Ubl domain and the C-terminal 388 residues of Parkin.
WT Parkin is more thermostable and protease resistant than mutant Parkin
In order to confirm that the differences in activity of Parkin WT versus mutants are due to the intramolecular interaction, we performed two independent stability assays.
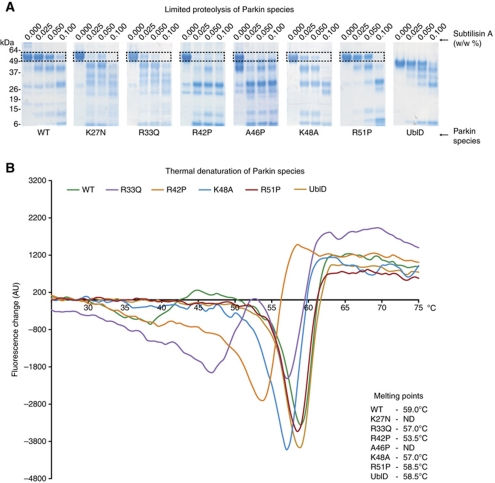
We subjected WT and mutant Parkins to limited proteolysis by subtilisin (Figure 5A). All of the pathogenic point mutants of Parkin underwent proteolysis at lower concentrations of subtilisin than WT protein, while R51P-Parkin, the mutation we predicted to be ‘silent’ that also displays no activity in vitro, was as resistant to proteolysis as WT. We then performed thermal denaturation assays by differential scanning fluorimetry (DSF) and found that WT, R51P-Parkin and ΔUblD had melting points of 59°C, 58.5°C and 58.5°C respectively, while R33Q-Parkin and K48A-Parkin were less stable, with Tm of 57°C, and R42P-Parkin less stable again with a Tm of 53.5°C (Figure 5B). A46P-Parkin and K27N-Parkin were so unstable that we were unable to obtain useful readings in all four replicates. Taken together, these data indicate that the WT Parkin is more stable than the pathogenic point mutations, and therefore more folded and less susceptible to proteolytic digest. Interestingly, ΔUblD is as thermostable as WT Parkin, suggesting that the Ubl domain could be protected from unfolding by the C-terminus of Parkin, consistent with both our hypotheses and our observations. A coomassie-stained gel of each input species is included to demonstrate that each species is purified to homogeneity (Supplementary Figure S6).
Figure 5.
WT Parkin is more stable than pathogenic mutants. (A) Proteolytic digest of Parkin species in the presence of increasing percentage of subtilisin (w/w). The boxed area indicates the degradation of all full-length species of Parkin. (B) Difference scanning fluorimetry of Parkin species. Each thermal denaturation curve is coloured according to the key, and the melting point (Tm) of each protein is noted in the figure.
Mechanism of Parkin autoinhibition
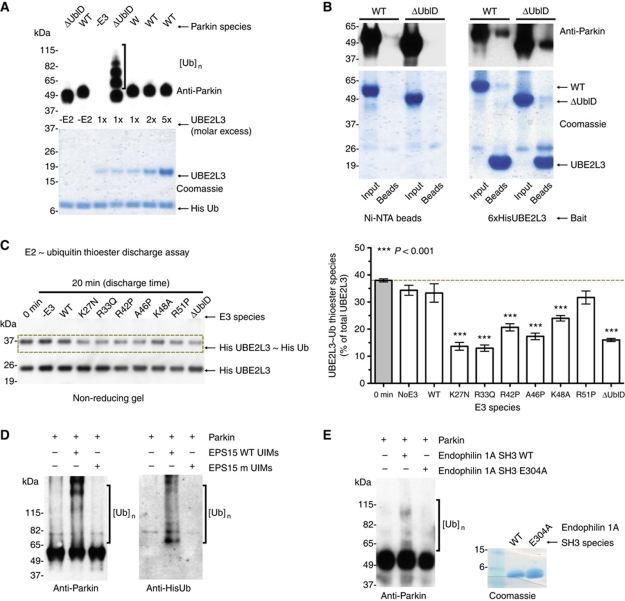
The mechanism of autoinhibition could result from the occlusion of an E2-binding site by an intramolecular interaction with the Ubl domain. In order to explore this possibility, we incubated WT inactive Parkin with increasing concentrations of E2. Ube2L3 was unable to overcome the autoinhibition, even at the highest concentrations used (Figure 6A). To determine whether there was any difference in E2 binding between WT and ΔUblD, we performed pull-down assays using 6xHis Ube2L3 as bait. The levels of WT Parkin retained were comparable to that of ΔUblD (Figure 6B). Taken together, these results show that E2 interaction and intramolecular inhibition occur independently.
Figure 6.
Ubl-binding partners but not E2 interactions ameliorate autoinhibition. (A) Coomassie and western blot analysis of Parkin autoubiquitination shows that increasing concentrations of E2s does not overcome the autoinhibition of Parkin autoubiquitination. Parkin antibodies are used for detection, and molar excess of E2 species is indicated. (B) Pull-down analysis of Parkin and ΔUblD-Parkin with Ube2L3. A beads-only control is shown (left) and Parkin retention by the E2 is analysed by coomassie staining and western blot. (C) E2∼Ub thioester discharge assay of each Parkin species. A representative western blot of His-Ube2L3 and His-Ube2L3∼His-Ub levels upon discharge induced by indicated Parkin species. Quantitation of absolute thioester levels (3 × repeats) is graphically represented (mean±s.e.m., right). Raw data were analysed using one-way ANOVA followed by Dunnett’s multiple comparison post-test using ‘0 min’ absolute thioester level (grey) as control (***P<0.001). (D) Western blot analysis of Parkin autoubiquitination in the presence of WT and mutant UIMs from the Parkin interactor EPS15. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. (E) Western blot analysis of Parkin autoubiquitination in the presence of WT and mutant SH3 domain from the Parkin interactor, endophilin 1A. Ubiquitin conjugates are detected using Parkin antibody and are indicated by brackets.
One possibility for the lack of WT Parkin activity is that it is unable to support the discharge of the ubiquitin-thioester formed on E2. In order to test this, we set up a pulse-chase assay whereby Ube2L3 was charged with ubiquitin for 10 min. Apyrase was then added to quench the charging, and was taken as a zero time point for reference. Parkin species were then added or not and the reactions were incubated for 20 min. Figure 6C shows that relative to no E3, WT and R51P-Parkin share a similar capacity for thioester discharge, while the mutant Parkins discharge the thioester-ubiquitin from E2 to a greater extent. A complete blot and a reducing gel blot are included in the Supplementary data to demonstrate the Parkin-Ub-species generated by the active Parkins and the loss of the thioester-ubiquitin from E2 under reducing conditions (Supplementary Figure S7).
We hypothesised that an alternative mechanism of inhibition could be to maintain Parkin in an ‘off’ position that would be relieved upon partner protein binding. To investigate this, we incubated Parkin with the ubiquitin-interacting motifs (UIMs) of the known Parkin interactor, EPS15. The Ubl domain of Parkin recognises the UIMs resulting in Parkin-mediated monoubiquitination of EPS15 and multi-monoubiquitination of Parkin (Fallon et al, 2006). Autoubiquitination of WT Parkin was observed when excess UIMs were added, while mutant UIM peptides (Ala to Gln) did not enable Parkin autoubiquitination (Figure 6D). The ability of WT Parkin to autoubiquitinate in the presence of the EPS15 UIMs but not with mutant UIMs suggests an interaction between EPS15 and Parkin’s Ubl domain opens the molecule and allows Parkin activity. In order to further probe this relief of autoinhibition by an interacting partner, we also assayed WT Parkin in the absence and presence of the SH3 domain from endophilin A1 (Trempe et al, 2009). This is a non-UIM-based Parkin interactor but has been shown to interact with the Ubl domain of Parkin. WT Parkin was competent for autoubiquitination in the presence of the SH3 domain but not with the E304A mutant shown to abolish binding to the Ubl domain (Trempe et al, 2009; Figure 6E). These data suggest that binding of an interacting partner can activate Parkin and opens up the possibility of Parkin effectors.
The C-terminus of Parkin harbours a Parkin UblD/Ub-binding (PUB) site
To further develop the mechanism of Parkin autoregulation, we hypothesised that there could be an as-yet-unidentified Ubl-interacting motif in the C-terminal portion of Parkin. To test this hypothesis, we generated a peptide array of Parkin ΔUblD, which was then probed with fluorescently labelled Ubl domain (Supplementary Figure S8). As the Ubl domain shares 32% sequence identity with ubiquitin, we also probed the arrays with fluorescently labelled ubiquitin (Supplementary Figure S8). We found two major hits for each protein, only one of which was shared between them. These data suggest an overlapping but not identical binding site for both ubiquitin and UblD in the C-terminus of Parkin.
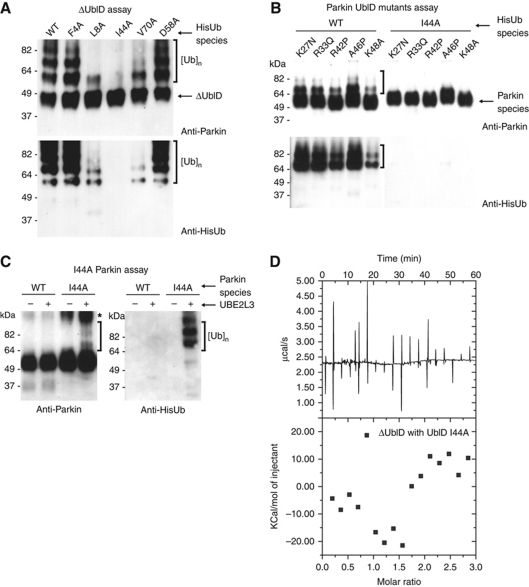
Almost all ubiquitin-binding domains bind ubiquitin through its central hydrophobic patch (referred to as the ‘Ile44’ patch) comprising Leu8–Ile44–Val70. Intriguingly, both the Ubl domain and ubiquitin have Ile44 and Val70, and the area surrounding Ile44 (including K48) is completely conserved (Figure 3A). We therefore hypothesised that the mechanism for autoinhibition could be the blocking of a ubiquitin-binding site by the Ubl domain. To test this hypothesis, we performed an autoubiquitination assay with ΔUblD using ubiquitin hydrophobic patch mutants. Surprisingly, ΔUblD was not able to autoubiquitinate in the presence of I44A ubiquitin, and had reduced activity with L8A and V70A ubiquitin (Figure 7A). To further investigate, we repeated this experiment with I44A-ubiquitin with each of the UblD pathogenic point mutants. Each of the point mutants was active with WT ubiquitin, but inactive with I44A-ubiquitin (Figure 7B). We also hypothesised that if the Ubl domain occludes a ubiquitin-binding site, I44A-Parkin should be active. To test this, we purified I44A-Parkin and assayed it for autoubiquitination. In contrast to WT, I44A-Parkin was active for autoubiquitination (Figure 7C). In addition, ITC experiments revealed ΔUblD affinity for the I44A-Ubl domain is negligible compared with WT (Figures 4B and 7D). Taken together, these data suggest that the mechanism of Parkin autoinhibition is occlusion of a Parkin UblD/ubiquitin-binding (PUB) site in the C-terminal portion of Parkin by the Ubl domain.
Figure 7.
Mechanism of Parkin autoinhibition. (A) Western blot analysis of ΔUblD autoubiquitination with mutant ubiquitin species. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. (B) Western blot analysis of Parkin point mutants with WT (left) and I44A (right) ubiquitin. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. (C) Western blot analysis of I44A-Parkin autoubiquitination. Ubiquitin conjugates are detected using Parkin and His antibodies and are indicated by brackets. (D) ITC curve depicting lack of interaction between I44A-UblD and ΔUblD.
Discussion
Ubiquitin ligases are a large family of proteins that are crucial for regulating essential cellular processes. As such, it is not surprising that they themselves would be subject to tight regulation. Indeed, a recent study of the HECT-type E3 Smurf, revealed autoinhibition of Smurf ligase activity through the C2 domain (Wiesner et al, 2007). However, with the notable exception of Ubr1 in the N-end rule pathway (Du et al, 2002), no such autoregulation has been observed in the RING E3s, although three groups recently reported regulation of the cullin–RING ligase (CRL) complex by the ubiquitin-like protein, Nedd8 (Duda et al, 2008; Saha and Deshaies, 2008; Yamoah et al, 2008). Parkin is an intriguing E3 as it has several putative substrates, is capable of multiple modes of ubiquitination, interacts with several E2s, and mutations in the parkin gene are causative in AR-JP. Therefore, Parkin serves as a good model for understanding regulation and misregulation of E3s. We have characterised the role of the Ubl domain in regulating the autoubiquitination activity of Parkin. We found that the Ubl domain of Parkin inhibits its autoubiquitination activity and that cleavage of the domain and pathogenic mutations with the Ubl domain disrupt this inhibition.
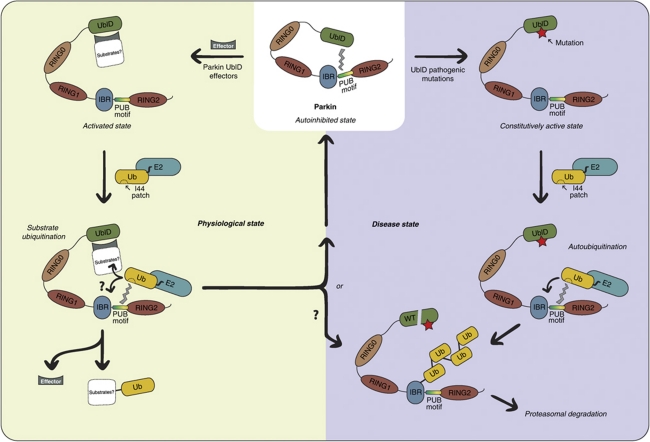
We present evidence towards a mechanism for Parkin autoregulation that involves effector binding to the Ubl domain (Figure 8). This presents an analogy to the CRLs, with the Ubl recruiting adaptors, analogous to the F-box proteins, which would in turn recruit substrates. There is some support for this model in previous studies isolating endogenous Parkin as a large, heteromeric complex (Van Humbeeck et al, 2008), and demonstrating Parkin interaction and function with CRL components (Staropoli et al, 2003). Our data also suggest a mechanism for Parkin regulation that involves unmasking a previously unidentified ubiquitin-interacting region in the C-terminus of Parkin. This suggests that the Ubl domain needs to be prised from the ubiquitin-interacting region before Parkin can connect to the charged E2 via both E2 and ubiquitin. Some support for this model comes from a very recent study showing the related RBR homologue, HHARI, forms a catalytic intermediate with ubiquitin via a thioester (Wenzel et al, 2011). Although they were unable to establish experimentally that the same is true for Parkin, our data provide a complementary means to achieve a semi-catalytic intermediate through association with the ubiquitin moiety on the E2 (Figure 8).
Figure 8.
Model for Parkin activation. In the physiological state (left), Parkin is autoinhibited and activated by effectors, which may in turn bind true Parkin substrates. The C-terminal RING regions recruit E2, with an additional interaction between the thioester charged ubiquitin on E2 and the PUB motif in Parkin. This may enhance substrate ubiquitination and/or Parkin autoubiquitination. In the disease state (right), a constitutively active Parkin autoubiquitinates and is targeted for proteasomal degradation.
We also found that fusion tags can disrupt the autoinhibition, highlighting the importance of investigating E3 function in its WT form. Previous studies performed with N-terminal tags and controlled with tag alone, but not Parkin alone, have concluded that autoubiquitination is a normal function of Parkin. Our data demonstrate that, in fact, Parkin autoubiquitination in the absence of substrate is a consequence of pathogenic mutations, and can be artificially stimulated in a non-physiological manner by N- or C-terminal tags. In addition to our study, Matsuda et al (2006) observed MBP ubiquitination in an assay using MBP-IBR-R2, and the E2, Ubc5, is capable of ubiquitinating GST when it is fused (Cooper et al, 2004; Matsuda et al, 2006). Additionally, multiple studies show that immunoprecipitation of endogenous Parkin reveals one band rather than Parkin ladders (West et al, 2003; Biasini et al, 2004; LaVoie et al, 2005; Joch et al, 2007; Van Humbeeck et al, 2008; Rakovic et al, 2010). In contrast, many studies showing immunoprecipitation of multiple higher molecular weight species of Parkin use overexpression of both Parkin (usually N-terminally tagged) and ubiquitin, contributing to a probable exaggeration of ‘in vivo’ ligase activity (Imai et al, 2000; Shimura et al, 2000; Zhang et al, 2000; Ko et al, 2010). Our data, coupled with those from endogenous studies suggest that autocatalytic ubiquitination and degradation of WT Parkin does not happen or is actively prevented in a normal cellular situation. Our studies indicate a need for caution when interpreting data from overexpressed, tagged or non-WT proteins, both in vivo and in vitro. The confusing and often conflicting reports in the literature regarding the activity of pathogenic Parkin mutations are likely due to an unintended disruption of the Ubl-mediated regulation of Parkin activity.
Several pathogenic mutations occur within the Ubl domain of Parkin. A recent study showed that four missense mutants of Parkin were rapidly degraded by the proteasome in a cellular overexpression assay (Henn et al, 2005). The mutants were only detectable in the presence of the proteasome inhibitor, MG132, supporting what we observe in vitro, in that the mutations in the Ubl domain render Parkin active for autoubiquitination and degradation. Intriguingly, a mutation that disrupts the Ubl domain fold (R42P), and one that remains intact (K48A) both result in an ‘active for autoubiquitination’ form of Parkin. It has previously been suggested that Parkin may be physiologically processed to release the Ubl domain (Schlossmacher et al, 2002). Parkin has also been shown to be cleaved by caspases after induction of apoptosis (Kahns et al, 2003), although it is unclear what the consequence of this cleavage is for the ligase activity of Parkin. Given our data, we would propose this would activate Parkin for autoubiquitination. Interestingly, there is a truncated isoform of Parkin arising from an internal start codon, which lacks the Ubl domain (Henn et al, 2005). The stability and functional significance of this isoform remains unclear, although an in vivo study revealed that HA-tagged Parkin lacking the Ubl domain had autoubiquitination activity, demonstrating that this domain is not necessary for Parkin autoubiquitination in vivo (Finney et al, 2003). Some studies have shown that ubiquitination of certain putative substrates is impaired in the absence of the Ubl domain (Corti et al, 2003; Huynh et al, 2003), although it is difficult to establish whether this is due to reduced substrate binding, or reduced activity of Parkin towards particular substrates. Both UIMs of EPS15 have been shown to be required for interaction with the Ubl domain (Fallon et al, 2006; Safadi and Shaw, 2010). Our data with the UIMs of EPS15 and SH3 domain of endophilin A1 show that partner binding can relieve the inhibition, hinting at a substrate or effector-based mechanism of activation of Parkin. There is also conflicting data on Parkin substrates. Despite so many putative substrates being identified, there is as yet no substrate that has been absolutely and independently verified in vitro and in vivo to be a direct substrate of Parkin (Corti and Brice, 2007; Dawson and Dawson, 2010). In addition, several recent studies have suggested that phosphorylation of Parkin can activate it against certain putative substrates, indicating a requirement for Parkin effectors in the cell (Matsuda et al, 2010; Narendra et al, 2010; Sha et al, 2010). It is unlikely that the only role for the Ubl of Parkin is to inhibit autoubiquitination, as substrate binding and proteasome association may be important functions. Indeed, a recent study reveals poorly folded Ubl domain mutants lose the ability to bind the proteasome (Safadi et al, 2011). However, we speculate that a possible mode of pathogenesis of the mutations in the Ubl domain of Parkin may be to misregulate Parkin by relieving inhibition of autoubiquitination. The consequence of the ubiquitination is unclear, but there are at least two potential outcomes: Parkin may become hyperactive and indiscriminately modify substrate(s), or autoubiquitination may lead to proteasomal degradation and therefore inactivation of Parkin. Given that the current consensus is that Parkin mutations are ‘loss-of-function’, it is more likely to be the latter (Figure 8). If this is the case, the Ubl domain could be a viable therapeutic target, either as a rescue therapy to increase the stability of Parkin without targeting the proteasome or to specifically disrupt the domain in order to destabilise Parkin.
The human genome encodes several proteins that contain Ubl domains. As some of these proteins are recruited to the proteasome and bind in a Ubl domain-dependent manner, it was thought that the Ubl domain is a generic proteasome-binding domain (Schauber et al, 1998; Wilkinson et al, 2001). However, there are Ubl domain containing proteins that do not associate with the proteasome (Schulze et al, 2005). Another E3-ligase component, Elongin B, has a Ubl domain, the function of which is unclear. It is possible that a generic function for Ubl domains is to regulate E3-ligase activity. Consistent with this suggestion, a bioinformatics study uncovered a high incidence of predicted Ubl domains in human ubiquitin-specific proteases (Zhu et al, 2007). Intriguingly, USP7 is predicted to have four Ubl domains C-terminal to the catalytic protease core, and has been found to bind a viral ring finger protein (ICPO). This binding protects ICPO from autoubiquitination (Zhu et al, 2007), suggesting a potential hijacking of the Ubl domains of USP7 to prevent autoubiquitination-mediated destruction of the viral protein.
In summary, our study shows that the tertiary arrangement of Parkin domains influences its activity. Parkin has >15 putative substrates, at least three of which contain domains that interact with the Ubl domain of Parkin (Tsai et al, 2003; Fallon et al, 2006). As well as the mutations within the Ubl, mutations throughout Parkin may also disrupt the tertiary arrangement, potentially exposing substrate, partner or E2∼Ub-binding sites. Our data suggest that the Ubl domain has a critical role both in maintaining the integrity of the autoinhibited conformation of Parkin and in regulating the type of ubiquitin modification. Finally, pathogenic mutations in the Ubl domain influence the intramolecular inhibitory activity of Parkin.
Materials and methods
Expression and purification of proteins
park2 gene was inserted into pET SUMO protein expression systems (Invitrogen). Ubl point mutants (K27N, R33Q, R42P, A46P, K48A/R and R51P) and Parkin truncations (ΔUbl and 95C) were generated using the Phusion® Mutagenesis Kit (Finnzymes). Restriction-free cloning (van den Ent and Lowe, 2006) was used to insert Ubl, UBE2L3 and UBE2L6 into pRSF Duet-1 (Merck) and thioredoxin from pThioHis A vector (Invitrogen) at the C-terminus of 6xHis-smt3-Parkin vector. All other E2s were purchased from Boston Biochem. pTXB1 (New England Biolabs) served as template for Mxe GyrA Intein-CBD sequence, inserted C-terminal of residue 94 to give a 6xHis-smt3 Ubl (1–94)–Intein-CBD fusion construct. A G94A mutation was made to improve autocatalytic cleavage of the C-terminal intein.
The SH3 domain from endophilin (residues 291–352) and E304A mutant (Trempe et al, 2009) were expressed as His-smt3 fusions, purified by Nickel-affinity chromatography followed by overnight cleavage with Ulp1 at 4°C and subsequent anion exchange chromatography. Purified proteins were diluted to 1 mg/ml and flash frozen at −80°C.
All 6xHis-smt3-tagged constructs were transformed into Escherichia coli BL21 (DE3) gold (Stratagene) and grown in Luria broth supplemented with 500 μM zinc chloride at 37°C until OD600 reached 0.4. Expression was induced with 25 μM IPTG for 12 h at 16°C. Harvested cells were sonicated in 50 mM Tris–HCl pH 7.5, 500 mM NaCl, 250 μM TCEP and bound to Ni-NTA beads (Qiagen). Beads were washed, equilibrated with 200 mM NaCl buffer and incubated overnight at 4°C with Ulp1 protease to cleave fusion protein. Flow through was purified through gel filtration. Pure protein was flash frozen with 10% glycerol. Expression of 6xHis-tagged protein was induced with 0.5 mM IPTG at OD600 0.6 followed by batch affinity purification as above. Proteins were eluted with 300 mM imidazole and further purified by gel filteration. MBP-Parkin, a kind gift from Professor K Tanaka, Tokyo Metropolitan Institute of Medical Science, was purified as described (Matsuda et al, 2006). All proteins were at least 99% pure as judged by Coomassie blue staining and quantification of FPLC traces.
In vitro autoubiquitination assays
Auto-ubiquitination reaction protocols were adapted from Matsuda et al (2006). In all, 25 μl reactions were conducted in reaction buffer: 50 mM Tris pH 8.0 or 7.4, 2 mM dithiothreitol (DTT), 5 mM MgCl2, 4 mM ATP and 5% glycerol. In all, 15 nM of human recombinant E1 (Boston Biochem), 0.5 μM of UBE2L3 (unless stated otherwise), 5 μM Ubiquitin species (Boston Biochem) were mixed with 0.77–1 μM of E3 species in the above reaction buffer and incubated at 37°C for 1 h (unless stated otherwise). Reactions were either stopped with SDS-loading buffer or cleaved overnight at 4°C.
Antibodies
Indicated volumes of autoubiquitination reactions were subjected to western blotting: 2 μl for anti-MBP (1/5000, New England Biolabs), 5 μl for anti-6xHis (1/2500, GE Healthcare), and 1.5 μl for anti-Parkin (1/5000, 1A1, IBL). Pettingill Technology Ltd raised polyclonal rabbit antibodies against purified Ubl domain, the anti-UblD (1/5000) was used to analyse Parkin-EPL autoubiquitination reactions. FK1 (1/1000) and FK2 (1/2000) antibodies were obtained from Enzo Life Sciences. Anti-rabbit HRP (1/1000) and anti-mouse (1/1000) HRP secondary antibodies were obtained from Dako. Anti-mouse IgM HRP (1/5000) and Strep-tactin HRP (1/4000) antibodies were obtained from Abcam and IBA, respectively.
Expressed protein ligation
The 6xHis-smt3 95C, 6xHis-smt3 Ubl (1–94)–Intein-CBD and 6xHis-smt3 K48A-Ubl (1–94)–Intein-CBD constructs were expressed, affinity purified and cleaved as described above. The K48A-Ubl and Ubl (1–94)–Intein-CBD fusions were further cleaved overnight at 4°C with 10 mM 2-mercaptoethanesulfonic acid (MESNA; Sigma) to give Ubl (1–94) with a C-terminal reactive species. Uncleaved species and Intein-CBD was separated using Chitin resin (NEB) and Ubl (1–94) species were further purified using gel filtration. Chemical ligation of Ubl (1–94) species with Cys95C (10:1 ratio) was carried out overnight at 16°C in the presence of 10 mM MESNA followed by gel filtration to separate unligated Ubl (1–94) species. Autoubiquitination assays were conducted with freshly purified Parkin-EPL and 95C species alongside the 95C truncation.
Cell transfections and ubiquitination assays
Parkin species were cloned in pcDNATM–DEST 40 mammalian expression vectors (Invitrogen) following Gateway® technology (Invitrogen). His-Ubiquitin mammalian construct was a kind gift from Dr Axel Behrens (London Research Institute). Human embryonic kidney 293 cells, maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, were transfected with 2 μg each of indicated vectors using Effectene® (Qiagen). After transfections, cells were maintained for 48 h including a 10-h treatment with 10 μM proteasome inhibitor MG132 (Calbiochem) where indicated. Cells were lysed and sonicated in cold lysis buffer (PBS, 1% Nonidet P-40, complete EDTA-free protease inhibitor cocktail (Roche)). Lysates were clarified by centrifugation and 50–100 μg of soluble lysates were analysed by western blotting using anti-Parkin and anti-actin (1/400, Abcam) antibodies. His–Ub species from 4 to 5 mg of soluble lysates were affinity purified using Ni-NTA magnetic agarose beads (Qiagen) as described (Campanero and Flemington, 1997).
Protein stability assays
Proteolytic digests were carried out using 10 μg Parkin species per 20 μl reaction in 50 mM Hepes, pH 7.4, 200 mM NaCl, 250 μM TCEP. Subtilisin was added to w/w percentages of 0.025, 0.05 and 0.1%. Reactions were carried out at 4°C for 3 h, and stopped with the addition of SDS-loading buffer and boiled. In all, 15 μl of each sample, was run on an 8–16% SDS–PAGE gel and stained with Coomassie blue.
Thermal denaturation experiments were performed using DSF. In all, 5 μg of each protein was added to 90 μl reaction buffer (50 mM Hepes pH 7.4, 200 mM NaCl, 250 μM TCEP), and 5 μl of 50 × Sypro Orange (Invitrogen) fluorescent dye was added to a final reaction volume of 100 μl. Each experiment was repeated four times in a 96-well plate in a Bio-Rad iQ5 thermal cycler, with a temperature gradient from 4°C to 100°C at steps of 0.5°C per minute. The results are displayed as the differential of the fluorescence in arbitrary units divided by the differential of the temperature, plotted against temperature. The minimum of each curve indicates the melting point (Tm) of each protein, representing the inflection point of the fluorescence curve.
Interaction assays
In all, 40–50 μl Ni-NTA beads were saturated with bait species; 6xHis-Ubl (WT or K48A), 6xHisE2 (UBE2L3 or UBE2L6) and incubated (2–3 h at 4°C) with 0.5 mg of indicated prey Parkin species; WT, K48A or ΔUblD for interactions with Ubl baits and WT and ΔUblD for E2 baits, in 50 mM Tris pH 7.5, 25 mM NaCl, 250 μM TCEP, 5% glycerol. Beads were subsequently washed in 10 column volumes of binding buffer followed by washes with increasing salt concentrations. In all, 10 μl beads were boiled with 2 × SDS-loading buffer after each wash to be analysed by coomassie and western blotting (20 × dilutions of each sample).
Isothermal titration calorimetry
ITC was conducted on an iTC200 Microcalorimeter (GE Healthcare). All proteins were freshly prepared before binding experiments. The sample cell contained 25 μM ΔUblD-Parkin in PBSA and 250 μM TCEP. The syringe contained 500 μM UblD or ubiquitin species. The buffer was used as the reference cell, and titration into buffer of each protein served as controls. In all, 20 injections of 2 μl were delivered with a 2-s addition time and an interval of 200 s. The stirrer speed was set to 1000 r.p.m. Each binding experiment was carried out at 25°C with the samples kept at 4°C before use.
UIM and SH3 effector assays
Indicated quantities of Ubl species (WT and K48A) and E2s (UBE2L3 and UBE2L6) were incubated in Parkin autoubiquitination assays. Custom peptides representing EPS15 WT UIMs (UIM1 -SEEDMIEWAKRESEREEEQR, UIM2 -LNQQEQEDLELAIALSKSEISEA) and mutant UIMs (UIM1m -SEEDMIEWQKRESEREEEQR, UIM2m -LNQQEQEDLELQIQLSKSEISEA) were synthesised by the Peptide Synthesis Laboratory, London Research Institute. Peptide pairs were dissolved in 50 mM Tris–HCl pH 8, 100 mM NaCl, 5% glycerol and a 50 × molar excess was co-incubated with WT Parkin for autoubiquitination assays. SH3 or SH3 mutant domains of endophilin 1A were added at 60 × molar excess in the assay buffer.
NMR experiments
All NMR experiments were collected on a Varian Inova 600 MHz spectrometer equipped with a triple resonance probe using xyz gradients. 1H–15N HSQC spectra were collected using the sensitivity-enhanced method at 25°C (Kay et al, 1992). Multiple samples of 15N-labelled Ubl domain (115 μM) were prepared in 20 mM Tris, 150 mM NaCl, 1 mM DTT in the absence and presence of unlabelled ΔUblD (0, 38, 76, 115 and 150 μM). All spectral parameters used to collect 1H–15N HSQC spectra were identical with the exception of the number of transients, which were increased for samples containing ΔUblD to improve signal to noise of both samples. 1H chemical shifts were referenced directly using a DSS internal standard. Spectra were processed using a 60°-shifted cosine bell weighting function in both 1H and 15N dimensions and baseline corrected using NMRPipe (Deleglio et al, 1995) and visualised using NMRView (Johnson and Blevins, 1994).
Peptide arrays
Peptide arrays representing human full-length Parkin as a series of overlapping peptides (21 residues) were synthesised by the Peptide Synthesis Laboratory, London Research Institute. Recombinant UblD protein was labelled at Cys59 using Fluorescein-5-Maleimide (Thermo Scientific) following the manufacturer's protocol to generate the fluorescein-labelled UblD probe while fluorescein-N-terminal Ubiquitin was commercially available (Boston Biochem). Arrays were activated for interaction assays with 50% v/v ethanol, 10% v/v glacial acetic acid solution (15 min) followed by several washes with array interaction buffer (AI buffer—50 mM Tris pH 8.0, 200 mM NaCl, 250 μM TCEP, 0.05% Tween20). Interaction experiments (50–100 μg of fluorescein UblD/Ub probes in 25 ml AI buffer) were carried out for 2–3 h at 25°C/room temperature and subsequently washed with AI buffer (3–5 × ). Peptide–probe interactions were detected using fluorescence image scanners (Molecular Dynamics STORM 860) and associated imaging software (ImageQuantTL). Experiments were repeated with fresh arrays to confirm positive interactions.
Thioester discharge assays
The experimental setup was adapted from Ozkan et al (2005). Reactions (37°C for 10 min) were setup with 150 nM E1, 2 μM His UBE2L3, 50 μM His Ubiquitin in a buffer containing 50 mM Tris pH 8.0, 2 mM DTT, 5 mM MgCl2 and 5 mM ATP to facilitate formation of His UBE2L3 ∼ His Ub thioester. Subsequently, reactions were arrested with ATPase (1.5 Units Ayprase; Sigma)/EDTA (5 mM) treatment for 10 min at 25°C/room temperature) followed by buffer exchange into 50 mM Tris pH 8.0 using Zeba™ Spin Desalting Columns (Pierce) according to the manufacturer's protocol. Charge and arrest reactions were conducted as master-mixes (55 and 82.5 μl, respectively) and 7.5 μl of arrested samples were subject to thioester discharge (20 min at 25°C/room temperature) with the indicated Parkin E3 species (2.4 μM in 2.5 μl). Reactions were terminated with 10 μl 2 × non-reducing sample buffer (4 M Urea, 150 mM Tris pH 6.8, 5 mM EDTA, 2% SDS, 10% glycerol and bromophenol blue). Samples were further diluted to 40 μl using 50 mM Tris pH 8.0, half of which was subject to reducing conditions (100 μM β-mercaptoethanol). In all, 1/5th of each sample was subject to western blot analysis using anti-His antibody. Blots were developed using ECL Plus Western Blotting Detection Reagents (GE Life Sciences), quantified using CCD camera system (ImageQuant LAS 4000) and associated imaging software (ImageQuantTL). Absolute levels of thioester species were calculated as a percentage of total E2 species detected in the given lane. Experiments were performed at least three times and statistical analysis (one-way ANOVA and Dunnett's multiple comparison post-test) was carried out using GraphPad Prism 5.0c (GraphPad).
Note added in proof
The following paper, recently published (Mol Cell, 10 June), describes an autoinhibition of a RING E3 ligase, cIAP1: Lopez J, John SW, Tenev, T, Rautureau GJ, Hinds MG, Francalanci F, Wilson R, Broemer M, Santoro MM, Day CL, Meier P (2011) CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Mol Cell 42: 569–583.
Supplementary Material
Acknowledgments
We thank Michael Way for extensive comments on the manuscript, Nicola O’Reilly and Dhira Joshi of the Peptide Synthesis Laboratory for the UIM peptides, and Brenda Schulman for the Parkin baculovirus. This work was funded by Cancer Research UK (HW, LB, SJL, AS and VKC) and the Canadian Institute of Health Research (GSS and KRB) and a Bogue Fellowship to VKC.
Author contributions: This project was conceived and designed by VKC and HW. VKC and LB performed the bulk of the experiments, with additional experiments performed by AS, HW, KRB and SJL. VKC, GSS, and HW interpreted the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Biasini E, Fioriti L, Ceglia I, Invernizzi R, Bertoli A, Chiesa R, Forloni G (2004) Proteasome inhibition and aggregation in Parkinson’s disease: a comparative study in untransfected and transfected cells. J Neurochem 88: 545–553 [DOI] [PubMed] [Google Scholar]
- Campanero MR, Flemington EK (1997) Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA 94: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM (2004) S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 304: 1328–1331 [DOI] [PubMed] [Google Scholar]
- Cooper HJ, Heath JK, Jaffray E, Hay RT, Lam TT, Marshall AG (2004) Identification of sites of ubiquitination in proteins: a fourier transform ion cyclotron resonance mass spectrometry approach. Anal Chem 76: 6982–6988 [DOI] [PubMed] [Google Scholar]
- Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, Hirsch E, Rooney T, Fournier A, Brice A (2003) The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet 12: 1427–1437 [DOI] [PubMed] [Google Scholar]
- Corti O, Brice A (2007) Of Parkin and Parkinson’s: light and dark sides of a multifaceted E3 ubiquitin-protein ligase. Drug Discovery Today Dis Mech 4: 121–127 [Google Scholar]
- Dawson TM, Dawson VL (2010) The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord 25: S32–S39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Doss-Pepe EW, Chen L, Madura K (2005) Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J Biol Chem 280: 16619–16624 [DOI] [PubMed] [Google Scholar]
- Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A (2002) Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc Natl Acad Sci USA 99: 14110–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van Bergen En Henegouwen PM, Fon EA (2006) A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol 8: 834–842 [DOI] [PubMed] [Google Scholar]
- Finney N, Walther F, Mantel PY, Stauffer D, Rovelli G, Dev KK (2003) The cellular protein level of parkin is regulated by its ubiquitin-like domain. J Biol Chem 278: 16054–16058 [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O (2006) Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet 15: 2059–2075 [DOI] [PubMed] [Google Scholar]
- Hedrich K, Eskelson C, Wilmot B, Marder K, Harris J, Garrels J, Meija-Santana H, Vieregge P, Jacobs H, Bressman SB, Lang AE, Kann M, Abbruzzese G, Martinelli P, Schwinger E, Ozelius LJ, Pramstaller PP, Klein C, Kramer P (2004) Distribution, type, and origin of Parkin mutations: review and case studies. Mov Disord 19: 1146–1157 [DOI] [PubMed] [Google Scholar]
- Hedrich K, Marder K, Harris J, Kann M, Lynch T, Meija-Santana H, Pramstaller PP, Schwinger E, Bressman SB, Fahn S, Klein C (2002) Evaluation of 50 probands with early-onset Parkinson’s disease for Parkin mutations. Neurology 58: 1239–1246 [DOI] [PubMed] [Google Scholar]
- Henn IH, Gostner JM, Lackner P, Tatzelt J, Winklhofer KF (2005) Pathogenic mutations inactivate parkin by distinct mechanisms. J Neurochem 92: 114–122 [DOI] [PubMed] [Google Scholar]
- Hoenicka J, Vidal L, Morales B, Ampuero I, Jimenez-Jimenez FJ, Berciano J, del Ser T, Jimenez A, Ruiz PG, de Yebenes JG (2002) Molecular findings in familial Parkinson disease in Spain. Arch Neurol 59: 966–970 [DOI] [PubMed] [Google Scholar]
- Hristova VA, Beasley SA, Rylett RJ, Shaw GS (2009) Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J Biol Chem 284: 14978–14986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR (2002) Protein tyrosine kinases: autoregulation and small-molecule inhibition. Curr Opin Struct Biol 12: 735–741 [DOI] [PubMed] [Google Scholar]
- Huynh DP, Scoles DR, Nguyen D, Pulst SM (2003) The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum Mol Genet 12: 2587–2597 [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Takahashi R (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem 275: 35661–35664 [DOI] [PubMed] [Google Scholar]
- Jenner P, Olanow CW (1998) Understanding cell death in Parkinson’s disease. Ann Neurol 44 (3 Suppl 1): S72–S84 [DOI] [PubMed] [Google Scholar]
- Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, Brice A, Seguela P, Fon EA (2007) Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell 18: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA (1994) NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kahns S, Kalai M, Jakobsen LD, Clark BF, Vandenabeele P, Jensen PH (2003) Caspase-1 and caspase-8 cleave and inactivate cellular parkin. J Biol Chem 278: 23376–23380 [DOI] [PubMed] [Google Scholar]
- Kay L, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114: 10663–10665 [Google Scholar]
- Kee Y, Huibregtse JM (2007) Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun 354: 329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM (2010) Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci USA 107: 16691–16696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ (2005) Dopamine covalently modifies and functionally inactivates parkin. Nat Med 11: 1214–1221 [DOI] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25: 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K (2006) Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281: 3204–3209 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM (2008) Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem 105: 1806–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 178: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J (2005) Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA 102: 18890–18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ (2004) Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695: 55–72 [DOI] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, Arends MJ (2010) PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA 107: 15145–15150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Seibler P, Ramirez A, Kock N, Orolicki S, Lohmann K, Klein C (2010) Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet 19: 3124–3137 [DOI] [PubMed] [Google Scholar]
- Safadi SS, Barber KR, Shaw GS (2011) Impact of autosomal recessive Juvenile Parkinson’s disease mutations on the structure and interactions of the parkin ubiquitin-like domain. Biochemistry 50: 2603–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi SS, Shaw GS (2007) A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry 46: 14162–14169 [DOI] [PubMed] [Google Scholar]
- Safadi SS, Shaw GS (2010) Differential interaction of the E3 ligase parkin with the proteasomal subunit S5a and the endocytic protein Eps15. J Biol Chem 285: 1424–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, Kawahara H, Yokosawa H, Hattori N, Mizuno Y, Tanaka K, Kato K (2003) Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep 4: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391: 715–718 [DOI] [PubMed] [Google Scholar]
- Schlehe JS, Lutz AK, Pilsl A, Lammermann K, Grgur K, Henn IH, Tatzelt J, Winklhofer KF (2008) Aberrant folding of pathogenic Parkin mutants: aggregation versus degradation. J Biol Chem 283: 13771–13779 [DOI] [PubMed] [Google Scholar]
- Schlossmacher MG, Frosch MP, Gai WP, Medina M, Sharma N, Forno L, Ochiishi T, Shimura H, Sharon R, Hattori N, Langston JW, Mizuno Y, Hyman BT, Selkoe DJ, Kosik KS (2002) Parkin localizes to the Lewy bodies of Parkinson disease and dementia with Lewy bodies. Am J Pathol 160: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, Koning F, Kloetzel PM, Seeger M (2005) The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol 354: 1021–1027 [DOI] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L (2010) Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet 19: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302–305 [DOI] [PubMed] [Google Scholar]
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37: 735–749 [DOI] [PubMed] [Google Scholar]
- Stavropoulos P, Nagy V, Blobel G, Hoelz A (2008) Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci USA 105: 12236–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EK, Skipper LM (2007) Pathogenic mutations in Parkinson disease. Hum Mutat 28: 641–653 [DOI] [PubMed] [Google Scholar]
- Terreni L, Calabrese E, Calella AM, Forloni G, Mariani C (2001) New mutation (R42P) of the parkin gene in the ubiquitinlike domain associated with parkinsonism. Neurology 56: 463–466 [DOI] [PubMed] [Google Scholar]
- Tomoo K, Mukai Y, In Y, Miyagawa H, Kitamura K, Yamano A, Shindo H, Ishida T (2008) Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin. Biochim Biophys Acta 1784: 1059–1067 [DOI] [PubMed] [Google Scholar]
- Trempe JF, Chen CX, Grenier K, Camacho EM, Kozlov G, McPherson PS, Gehring K, Fon EA (2009) SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol Cell 36: 1034–1047 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Fishman PS, Thakor NV, Oyler GA (2003) Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem 278: 22044–22055 [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B et al. (2004) Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304: 1158–1160 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Lowe J (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67: 67–74 [DOI] [PubMed] [Google Scholar]
- Van Humbeeck C, Waelkens E, Corti O, Brice A, Vandenberghe W (2008) Parkin occurs in a stable, non-covalent, approximately 110-kDa complex in brain. Eur J Neurosci 27: 284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, Ladanyi M, Sander C, Heguy A, Holland EC, Paty PB, Mischel PS, Liau L, Cloughesy TF, Mellinghoff IK, Solit DB et al. (2010) Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet 42: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, de Vries RL, Tocilescu M, Przedborski S (2010) PINK1/Parkin direct mitochondria to autophagy. Autophagy 6: 315–316 [DOI] [PubMed] [Google Scholar]
- Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay SP, Ho MW, Lim TM, Soong TW, Pletnikova O, Troncoso J, Dawson VL, Dawson TM, Lim KL (2005) Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet 14: 3885–3897 [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Gonzalez-de-Chavez F, Wilkes K, O’Farrell C, Farrer MJ (2003) Parkin is not regulated by the unfolded protein response in human neuroblastoma cells. Neurosci Lett 341: 139–142 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD (2007) Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130: 651–662 [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol 3: 939–943 [DOI] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ (2008) Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci USA 105: 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM (2000) Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA 97: 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Menard R, Sulea T (2007) High incidence of ubiquitin-like domains in human ubiquitin-specific proteases. Proteins 69: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.