Abstract
Major histocompatibility complex class I (MHCI) and MHCII proteins differ in structure and sequence. To understand how T cell receptors (TCRs) can use the same set of variable regions to bind both proteins, we have presented the first comparison of a single TCR bound to both MHCI and MHCII ligands. The TCR adopts similar orientations on both ligands with TCR amino acids thought to be evolutionarily conserved for MHC interaction occupying similar positions on the MHCI and MHCII helices. However, the TCR antigen-binding loops use different conformations when interacting with each ligand. Most importantly, we observed alternate TCR core conformations. When bound to MHCI, but not MHCII, Vα disengages from the Jα β-strand, switching Vα’s position relative to Vβ. In several other structures either Vα or Vβ undergoes this same modification. Thus, both TCR V-domains can switch among alternate conformations, perhaps extending their ability to react with different MHC-peptide ligands.
Introduction
Unlike B cells that binds many different types of antigens, T cells bearing αβ antigen receptors (TCRs) usually react only with peptides from foreign antigens bound in the grooves of major histocompatibility complex proteins (MHC). Recent evidence suggests that this bias toward MHC presented antigens is caused by two phenomena: 1) the sequences of TCR variable regions themselves, which appear to have been selected during evolution to have some intrinsic affinity for MHC (Jerne, 1971) and 2) positive and negative selection in the thymus, which allows the maturation of only those thymocytes that bear TCRs that react weakly with self MHC + self peptides (Fink and Bevan, 1978; Zinkernagel et al., 1978). We and others have published data supporting the first idea, showing that in mice and humans the group of TCR variable regions that are related to the mouse Vβ8 or mouse Vα4 families have an intrinsic ability to bind to MHC conferred by 3 amino acids, Y46, Y48 and E54, in the complementarity determining region-2 (CDR2) regions of the Vβ8 relatives and Y29 in the CDR1 loops of the Vα4 relatives (Colf et al., 2007; Dai et al., 2008; Feng et al., 2007; Marrack et al., 2008; Scott-Browne et al., 2009).
Although MHC class I glycoproteins (MHCI) and MHC class II glycoproteins (MHCII) are related in sequence, they have similar but not identical structures. The TCR-exposed surface of MHCI includes two nearly continuous alpha helices, whereas the α-helix of the MHCII α chain is interrupted by a linear stretch of amino acids (Bjorkman et al., 1987; Madden, 1995; Madden et al., 1992). Furthermore, peptides bind to the two MHC classes quite differently. Peptides bind to MHCII in an extended polyproline like mode, sitting low in the binding groove with their ends often extending beyond the ends of the groove. The peptide binding groove of MHCI is closed at both ends. Thus peptides that bind to MHCI are usually shorter, with their termini buried in conserved positions at the ends of the groove and the center of the peptide often bulging up (Engelhard, 1994). These facts, combined with the observation that TCRs rarely cross react between MHCI and MHCII, suggest that TCRs might interact with the two classes of MHC differently. Nevertheless, TCRs use the same set of variable elements (Vαs, Jαs, Vβs, Dβs and Jβs) to react with MHCI and MHCII, although some individual variable elements may be used more often for one class than the other (Sim et al., 1996). Also, the many structures of TCRs bound to MHCI and MHCII show a similar overall orientation of the TCRs on the MHC molecules (Rudolph et al., 2006). Moreover, when these TCRs share a variable element, some of the same CDR1 and CDR2 amino acids are involved in binding to MHCI and MHCII (Burrows et al.; Dai et al., 2008; Feng et al., 2007; Marrack et al., 2008; Rudolph et al., 2006; Scott-Browne et al., 2009).
These observations led us to consider how a single TCR might bind to both MHCI and MHCII ligands. Would the TCR amino acids that we predict to have been built in by evolution to react with MHC be used similarly in the two interactions? Although most T cells with this ability are eliminated by negative selection in the thymus (Huseby et al., 2005; Merkenschlager et al., 1997; Zerrahn et al., 1997), in mice created to have a single MHCII molecule covalently bound to a single peptide, many of these MHC reactive T cells escape negative selection and populate the mature T cell repertoire. These T cells often cross react exuberantly with foreign MHC proteins and T cells that react with both MHCI and MHCII are common in such animals (Huseby et al., 2005; Logunova et al., 2005).
The YAe-62.8 (YAe62) TCR, from a single peptide IAb mouse, has this ability to recognize both MHCI and various alleles of MHCII. We recently reported the structure of the YAe62 TCR bound to the MHCII molecule, IAb, plus a known peptide, p3K (Dai et al., 2008). However the nature of YAe62’s cross reactivity with class I was unknown. Here we identify a peptide that, when bound to the MHCI molecule, Kb, creates a ligand for the YAe62 TCR. We solved the structure of the YAe62 TCR bound to Kb-peptide and compared this structure with that of the same TCR bound to IAb-p3K. The orientation of the TCR on the two ligands is quite similar and the same four Vα and Vβ germline encoded TCR amino acids, previously reported to be conserved for MHCII interaction, are involved in binding to both IAb and Kb. Nevertheless, the YAe62 TCR copes with its two ligands by varying considerably the conformations of its CDR3 amino acids, a strategy that has been described by others for TCRs that react with several ligands of the same class of MHC (Colf et al., 2007; Mazza et al., 2007)
Unexpectedly, the YAe62 TCR uses an additional strategy, not previously noted, to deal with the differences between its two ligands. When bound to the Kb ligand, most of Vα has swiveled away from Vβ leaving its Jα β-strand behind, to mediate the conserved interaction with Vβ, and changing the angle and position of Vα relative to Vβ. An analysis of over 40 published TCR structures shows that this phenomenon has gone unexplored in several other cases involving both TCR Vα and Vβ domains (Archbold et al., 2009; Housset et al., 1997) We hypothesize that this mechanism gives each TCR three potential alternate conformations, thus increasing the diversity of the TCR repertoire and allowing an adjustment of the relationship among the Vα and Vβ CDR loops to accommodate various MHC-peptide ligands.
Results
Identification of a peptide mimotope recognized by YAe62 bound to Kb
YAe62 and 3K-36 are two highly cross-reactive T cells, both produced by immunization with IAb-p3K. Transgenic expression of these TCRs in H-2b mice lacking IAb leads to the development of mature CD8+ T cells, suggesting that, despite their MHCII reactivity, these TCRs can be positively selected on H-2b MHCI molecules and thus might react with an H-2b MHCI presented antigen. For 3K-36, this idea was confirmed by identification of mimotope peptides that could activate CD8+ 3K-36 T cells when presented by Kb (Huseby et al., 2005). In this previous study a peptide was not discovered for YAe62 CD8+ T cells, but they were shown to be activated by MHCII− cells bearing MHCI from H-2k, but not H-2b or H-2q, strongly suggesting a capacity for MHCI-peptide reactivity by YAe62, in this case to an allo-MHCI antigen.
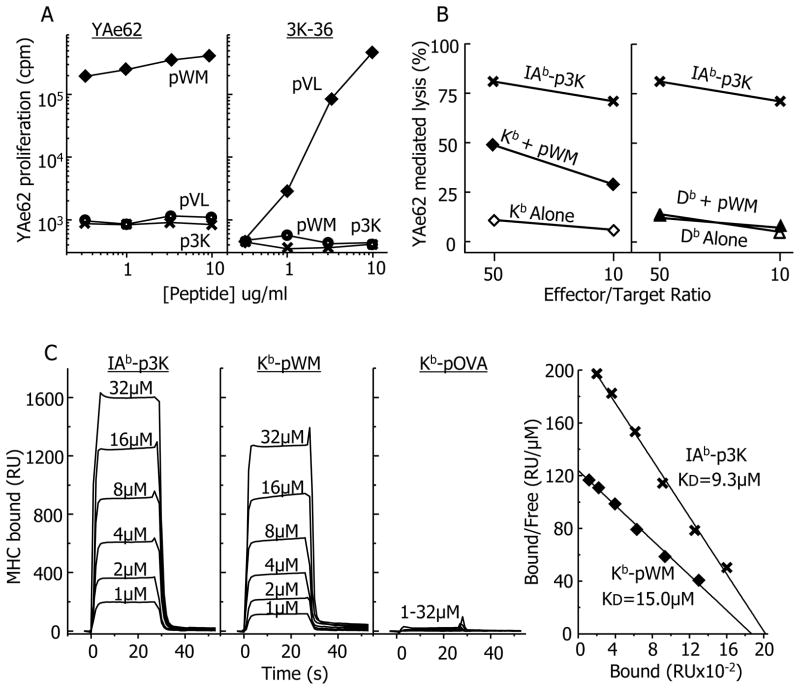
We revisited the question of the MHCI specificity of the YAe62 TCR using positional scanning (Pinilla et al., 1992) to screen a nonamer peptide library for peptides able to activate YAe62 CD8+ T cells from MHCII− mice. After deconvoluting the resultant data, candidate peptides were synthesized and tested for their ability to activate YAe62 CD8+ T cells. Several semi-purified 9-mer peptides with a high degree of sequence homology were found to activate the cells. However when highly purified versions of these peptides were tested, they each failed to activate the T cells. A common contaminating 8-mer peptide, WIYVYRPM (pWM) was identified by mass spectrometry in the semi-purified peptide preparations and was then synthesized de novo. The proliferative responses of CD8+ T cells from Yae62 and 3K-36 TCR transgenic mice to pWM, p3K and pVL (a 3K-36 mimotope peptide), were tested using H-2b, MHCII− presenting spleen cells (Figure 1A). The Yae62 T cells responded strongly to pWM, but not at all to p3K or pVL, whereas the 3K-36 T cells responded to pVL, but not pWM or p3K, confirming the specificity of YAe62 for pWM. To find out which H-2b MHCI molecule presented pWM, we tested activated YAe62-bearing CD8 T cells from YA62 TCR transgenic, MHCII− mice for pWM dependent cytotoxic activity on MHCII− targets expressing either Kb or Db. Cells bearing IAb-p3K were used as positive controls. The results showed that YAe62 recognized pWM presented by Kb, but not Db (Figure 1B).
Figure 1. Identification of a Kb bound mimotope recognized by the YAe62 TCR.
A) The proliferative responses (3H-TdR incorporation) of CD8+ T cells from either YAe62 or 3K-36 TCR transgenic, MHCII− mice (Huseby et al., 2005) to various peptides presented by spleen cells from H-2b, MHCII− mice were measured as described in Extended Experimental Procedures. pVL (VIKWWRRL) is a peptide mimotope specific for the 3K-36 TCR. Similar results were obtained in two other similar experiments. B) The ability of activated CD8+ T cells from YAe62 TCR transgenic mice to lyse 3T3 fibroblasts transduced with either Kb or Db was measured as described in Extended Experimental Procedures. Cells expressing IAb covalently bound to p3K, were used as the positive control. The results of a single experiment are shown. C) Approximately 3000 resonance Units (RU) of biotinylated versions of IAb-p3K, Kb-pMW and Kb-pOVA, as well as an irrelevant MHC molecule, HLA-DR52c, were immobilized in the 4 flow cells of a BIAcore streptavidin biosensor chip. Various concentrations of soluble YAe62 TCR were injected through the 4 flow cells for 30 s. The 3 panels at the left shows the binding of YAe62 TCR to the three ligands (RU) vs. time after correction for the fluid phase surface plasmon resonance signal in the flow cell containing the immobilized control DR52c protein. The net equilibrium RU signals were used to create Scatchard plots for the IAb-p3K and Kb-pWM ligands (right panel). The least squares regression line fit to the data was used to estimate the overall affinity (KD) of the TCR for IAb-p3K and Kb-pMW. Similar results were obtained in two similar experiments.
To study the interaction of the Kb-pWM complex with the YAe62 TCR directly, we produced soluble versions of each in baculovirus infected insect cells (see Extended Experimental Procedures). Briefly, pWM was expressed covalently linked to the N-terminus of beta-2 microglobulin (β2m) via a flexible linker. To improve the stability and homogeneity of the complex, a disulfide bond was introduced by placing cysteines in the linker and in substitution for Tyr84 in the Kb heavy chain (Lybarger et al., 2003). Similarly, as a negative control, Kb was prepared bearing the ovalbumin peptide, SIINFEKL (pOVA). In each case the protein carried a biotinylation tag at the MHC heavy chain C-terminus. As a positive control we used a biotinylated version of soluble IAb-p3K. As described in the Extended Experimental Procedures, the ability of the purified Yae62 TCR to bind the three MHC complexes was assessed by surface plasmon resonance using a BIAcore 2000 instrument (Figure 1C). The TCR bound both IAb-p3K and Kb-pWM with very rapid kinetics (first and seconds panels), but did not bind Kb-pOVA (third panel). The equilibrium binding data were used to calculate the affinity of the two ligands for the TCR (fourth panel). As previously reported, YAe62 had a KD of about 9μM for IAb-p3K (Huseby et al., 2006). It bound less strongly to Kb-pWM, with a KD of about 15μM.
General orientation and footprint of the YAe TCR on IAb –p3K vs Kb-pWM
For crystallography the YAe62 TCR was expressed in E. coliI and refolded and purified as previously described (Dai et al., 2008; Tynan et al., 2007). Kb and β2m covalently bound to pWM were co-expressed in baculovirus infected insect cells as above, but without the biotinylation tag. An equimolar mixture of the two purified proteins was crystallized by the hanging-drop vapor diffusion method (Experimental Procedures). The complex crystallized in two space groups, I222 and P3221. We solved the structure of the complex in both crystals by molecular replacement. Despite the differences in crystal packing, the TCR interaction with Kb-pWM was nearly identical in both initial models. Therefore we solved the higher resolution structure to a final resolution of 2.9Å. The properties of the crystals and details of the data collection, solution and refinement are in the Extended Experimental Procedures and Table S1.
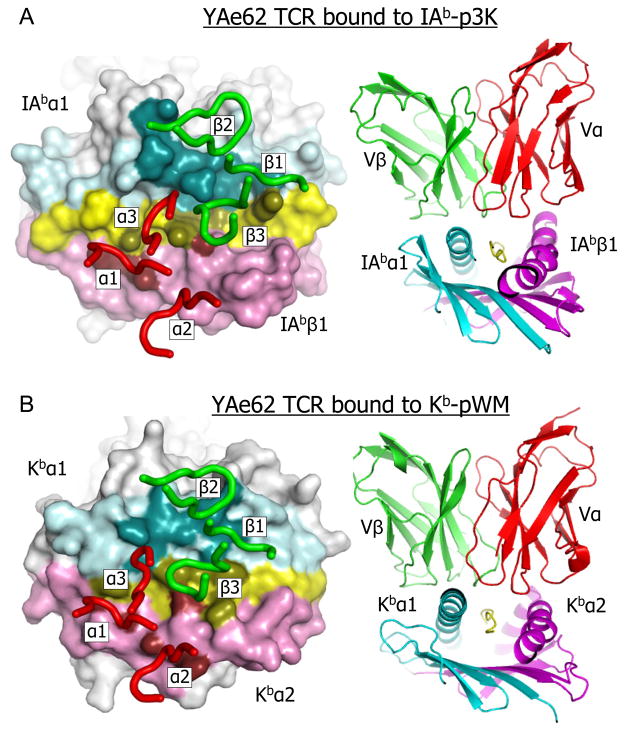
Figure 2 shows the general orientations and footprints of the YAe62 TCR on Kb-pWM and IAb-p3K. When viewed from above, the receptor has the now familiar diagonal orientation on both ligands, but has a slightly shifted position on Kb-pWM compared to IAb-p3K (Figure 2A). However, when viewed from the N-terminal end of the bound peptides the TCR sits somewhat flatter on Kb-pWM than on IAb-p3K (Figure 2B). This results in a larger footprint of the TCR on Kb-pWM (1529Å2) than on IAb-p3K (1178Å2) (Dai et al., 2008)(Figure 2A). A summary of TCR atom to atom contacts with the two ligands is displayed in Table 1 and a complete list of these contacts for the Kb-pWM ligands is included in Table S2. A similar list for the IAb-p3K ligand was previously published (Dai et al., 2008). Despite the larger area of contact, the amino acids of the TCR make many fewer atom-to-atom contacts with the Kb-pWM ligand than with IAb-p3K, perhaps accounting for the lower affinity of the TCR for Kb-pWM. In both cases, the interface between the TCR and the ligand is dominated by van der Waal’s interactions rather than salt bridges or hydrogen bonds.
Figure 2. Orientation and footprint of the YAe62 TCR on its MHCI and MHCII ligands.
A) Left panel- Top view of the water accessible surface of the peptide and the α1 and β1 IAb domains are shown with the following color scheme: helix of the α1 domain, cyan; helix of the β1 domain, magenta; the rest of IAb, white: p3K peptide, yellow. The footprint of the YAe62 TCR on this ligand is shown by coloring MHC-peptide atoms within 4.5 Å of the TCR with darker versions of the same colors,. The YAe62 CDR loops are represented as tubes, Vα (red) and Vβ (green). Right panel – Ribbon representation of the Vα and Vβ domains of the YAe62 TCR bound to the IAb-p3K complex, Vα, red; Vβ, green; IAb α1, cyan; IAb β1, magenta and p3K, yellow. B) As in A, but with the α1 and α2 domain of Kb and pWM instead of the α1 and β1 IAb domains and p3K.
Table 1.
Summary of contacts between the YAe62 TCR and its ligands
| TCR | Atom to Atom Contacts
|
||||||
|---|---|---|---|---|---|---|---|
| MHC Domain | Peptide | MHC Domain | |||||
| CDR | Amino Acid | Abα1 | Kbα1 | p3K | pWM | Abβ1 | Kbα2 |
| α1 | T27 | — | — | 1 | 4 | 1 | 1 |
| G28 | — | — | 1 | 6 | — | — | |
| Y29 | — | — | 5 | — | 29 | 7 | |
|
| |||||||
| α2 | T50 | — | — | — | — | — | 4 |
| T51 | — | — | — | — | 2 | 5 | |
|
| |||||||
| α3 | S93 | — | — | — | 1 | — | — |
| G94 | — | 5 | — | — | — | — | |
| T95 | 8 | 3 | — | — | — | — | |
| Y96 | — | 7 | — | — | — | — | |
|
| |||||||
| β1 | N26 | — | — | 1 | — | — | — |
| N28 | 2 | 19 | 3 | — | — | — | |
| N29 | 5 | — | — | — | — | — | |
|
| |||||||
| β2 | Y46 | 7 | 2 | — | — | — | — |
| Y48 | 53 | 38 | — | — | — | — | |
| G49 | — | 6 | — | — | — | — | |
| A50 | — | 7 | — | — | — | — | |
| T53 | 2 | — | — | — | — | — | |
| E54 | 28 | 1 | — | — | — | — | |
|
| |||||||
| B3 | D93 | — | — | — | 7 | — | — |
| F94 | 40 | — | 3 | 15 | — | 9 | |
| W95 | 48 | 17 | 31 | 7 | 3 | — | |
|
| |||||||
| Total | 193 | 105 | 45 | 40 | 35 | 26 | |
Similar use of conserved CDR1 and CDR2 amino acids in the two structures
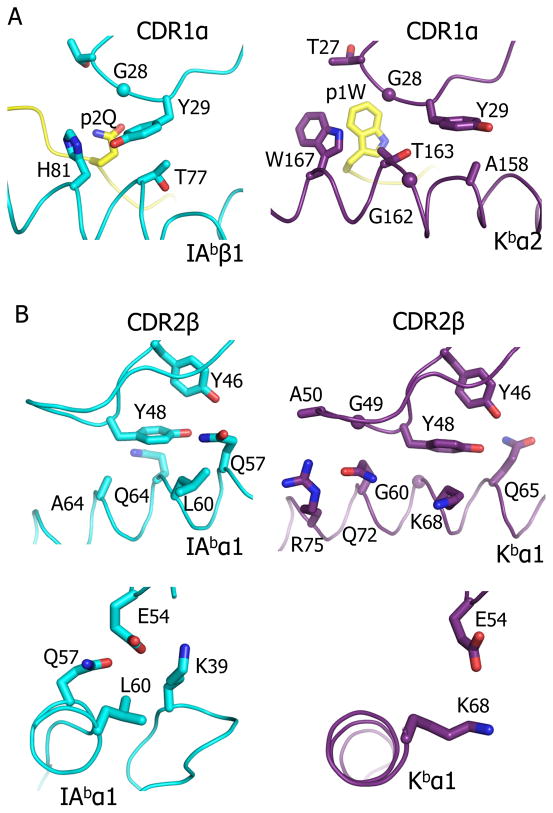
There are both similarities and differences in how the YAe62 CDR1 and CDR2 loops contact the MHCI and MHCII ligands. The tilt of TCR toward the α1-helix of IAb-p3K greatly accentuates ligand interaction with Vβ at the expense of Vα (Table 1 and (Dai et al., 2008) ). In the IAb-p3K complex, CDR1β makes almost no contact, but there is a tight cluster of interaction between Y46, Y48 and E54 of CDR2β and the IAb α1 domain. On the IAb β1-helix, Y29 of CDR1α is the main point of contact. These four amino acids are also involved in the interaction of YAe62 with its Kb-pWM ligand, contacting positions on the MHCI helices which are similar to YAe’s contact positions on MHCII. They have been seen repeatedly contacting similar positions on MHCII molecules in a variety of TCR-MHCII structures involving mouse Vβ8 and Vα4 and related V-regions in mouse and humans and are important for MHCII mediated CD4 T cell thymic selection. Vβ Y46 and Y48 and Vα Y29, when they are present, are also often involved in interactions with MHCI. Thus we and others have suggested that these TCR V region amino acids are evolutionarily conserved for MHC interaction (Dai et al., 2008; Feng et al., 2007; Maynard et al., 2005; Scott-Browne et al., 2009).
For CDR1α, in both the IAb-p3K and Kb-pWM complexes, the aromatic ring of Y29 interacts with a flat orientation on the IAb β1 helix and the Kb α2 helix respectively (Figure 3A). However, to maintain this type of interaction with the Kb helix, Y29 has rotated about its α carbon-β carbon axis about 90º compared to its conformation on the IAb helix, nestling between Kb A158 and T163, as opposed to IAb H81β and T77β. The lack of an amino acid side chain at G162 of Kb facilitates this approach of Y29 to the Kb α-helix.
Figure 3. Conserved amino acids in the YAe62 CDR1α and CDR2β contact both ligands.
The interactions between the YAe62 CDR1α containing αY29 (A), the portion of CDR2β loop containing βY46 and βY48 (B) or β54E (C) with either IAb-p3K (left panels) or Kb-pWM (right panels) are shown. In all panels the TCR loop and the MHC-peptide are represented as α carbon traces with wireframe representations of key amino acid side chains.
In CDR2β, Y48 is a major contributor to both MHCI and MHCII engagement, making numerous contacts to both ligands (Figure 3B, Table 1). Like Vα Y29, the aromatic ring of Vβ Y48 interacts intimately, in a flat orientation, with a similar position on the α1 helix in both structures, facilitated by the lack of an interfering side chain at the adjacent helix position, G69 in the case of Kb and A64 in the case of IAb. A number of previous studies have reported that Vβ Y48 interacts with MHCII in this way, confined to a very precise location, nestled among amino acids, 57, 60, 61 and 64 of the α helix (Feng et al., 2007; Marrack et al., 2008). Vβ Y48 has been observed interacting with a similar location on MHCI in a number of structures involving TCRs related to mouse Vβ8, but in this case it is allowed to roam over a wider area of the exposed α helix on the α1 helix (Figure S1 (Garcia et al., 1998; Miller et al., 2007; Speir et al., 1998; Tynan et al., 2007)).
The position and orientation of Vβ Y46 is almost identical in the two structures, with its aromatic ring at right angles to that of Y48 (an often-seen stable orientation for the interaction of 2 aromatic amino acids) (Figure 3B). In both structures, Y46 makes much less contact with the ligand than does Y48, in each case involving a single MHC amino acid side chain of the a1 helix (Q65 for Kb and Q57 for IAb). One possibility is that the ability of Y46 to orient and support Y48 might be more important than its direct contact with MHC.
With MHCII ligands such as IAb-p3K, CDR2β E54 contacts a conserved Lys on a loop connecting MHCII α1 β strands 3 and 4 (Figure 3C). Since this site does not exist in MHCI ligands, it is not surprising that Vβ E54 is often not seen in contact with MHCI for TCRs bearing Vβs related to mouse Vβ8. However, in the YAe62-Kb-pWM structure there is a single contact between YAe62 E54 and the Kb α1 helix and in two other cases involving a Vβ8 TCR (ref 1LP9 and 1G6R) E54 has been observed in contact with an MHCI α1 helix.
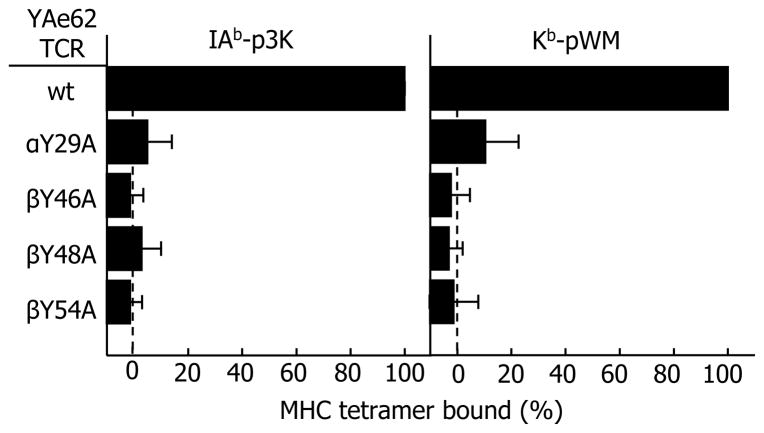
To compare the relative contribution of these four amino acids to the binding of the YAe68 TCR to the two ligands, we introduced the YAe62 TCR, and mutants in which these four amino acids were individually changed to alanine, into a T cell hybridoma lacking any endogenous TCR chains, as previously described (Scott-Browne et al., 2007). The resultant T cell hybridomas were sorted by flow cytometry for roughly equivalent surface expression of their TCRs and co-stained with fluorescent anti-Cβ and tetramers made up of either IAb-p3K or Kb-pWM. Cells were gated on populations with equivalent Cβ staining and the mean fluorescence intensity (MFI) of tetramer staining was calculated. The data are shown in Fig. 4. All four mutations had a dramatic effect on tetramer binding, once again confirming their critical role in MHC interaction. The effects of mutating additional Vβ amino acids to alanine on the binding of the YAe62 TCR to Kb-pWM and IAb-p3K are shown in Figure S2.
Figure 4. Four conserved YAe62 TCR amino acids are essential for recognition of both IAb-p3K and Kb-pWM.
A TCR− T cell hybridoma was transduced with retroviruses encoding either the genes for the wild-type (WT) YAe62 TCR or mutated genes to change individually αY29, βY46, βY48 or βE54 to alanine (Extended Experimental Procedures). The ability of fluorescent tetramers of IAb-p3K or Kb-pWM to bind to the TCRs on these cells was assessed by flowcytometry, co-staining with a Cβ-specific MAb. The results are shown as the mean channel fluorescence obtained with the tetramers on the αβ transduced cells minus that seen on cells transduced with the WT β chain gene only, expressed as the percent of the results with the unmutated TCR. The average +/− SEM of three experiments is shown.
Because of the larger footprint of the YAe62 TCR on Kb-pWM versus IAb-p3K more CDR1 and CDR2 amino acids contribute to the TCR footprint on Kb-pWM than on IAb-p3K despite the overall fewer atom-to-atom contacts (Table 1). For example, in CDR2β, Y48 and E54 are the major amino acids contributing to engagement of IAb-p3K, but for the Kb-pWM ligand CDR2β G49 and A50 also contribute considerably. In CDR2α, only T51 contacts IAb, but both T50 and T51 contact Kb.
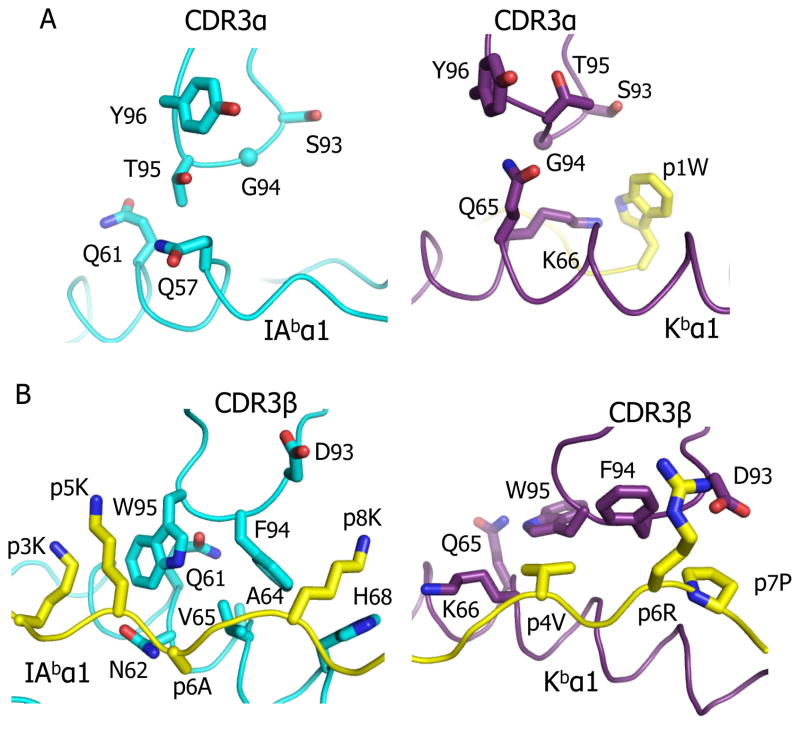
CDR3 loop adjustments to the two ligands
The CDR3s of both Vα and Vβ interact quite differently with IAb-p3K than with Kb-pWM (Fig 5, Table 1, Table S2). In the IAb-p3K structure only T95 in CDR3α makes contact, albeit minimally, with Q58 and Q62 of the IAb β chain, but in the Kb-pWM structure the entire tip of CDR3α (S93, G94, T95 and Y96) makes multiple contacts with both Kb (Q65 and K66) and the p1W of the peptide. (Figure 5A). To accomplish this there also is a considerable conformational change in the CDR3α loop backbone between the two structures. Since, typical of MHC class I bound peptides, the middle of the pWM peptide sits about 2.5Å higher in the peptide binding groove of Kb than p3K does in IAb, it is not surprising that CDR3β has undergone major conformational changes to accommodate this more intimate proximity of the middle of the peptide (Figure 5B). In the IAb-p3K structure, Vβ W95, at the tip of CDR3β, slots between the peptide and the IAb β chain helix, making a major contribution to the overall interface. However, in Kb-pWM, to accommodate the higher position of the peptide, the side chain of W95 swings up and out over the Kb α1 helix making much less overall contact with both the MHC and peptide(Figure 5B). This change in the orientation of CDR3β W95, and also that of CDR3β F94, is permitted by flattening the backbone of CDR3β itself, again illustrating the flexibility with which CDR3 regions adjust to different ligands (Gagnon et al., 2006; Reiser et al., 2003).
Figure 5. Major conformational changes in the CDR3 loops of the YAe62 TCR when bound to IAb-p3K versus Kb-pWM.
The interactions of the αCDR3(A) and βCDR3(B) loops of the YAe62 TCR with IAb-p3K (left) and Kb-pWM (right) are shown. Portions of the CDR3 loops and the MHC/peptide are represented as α carbon traces with wireframe representations of key amino acid side chains.
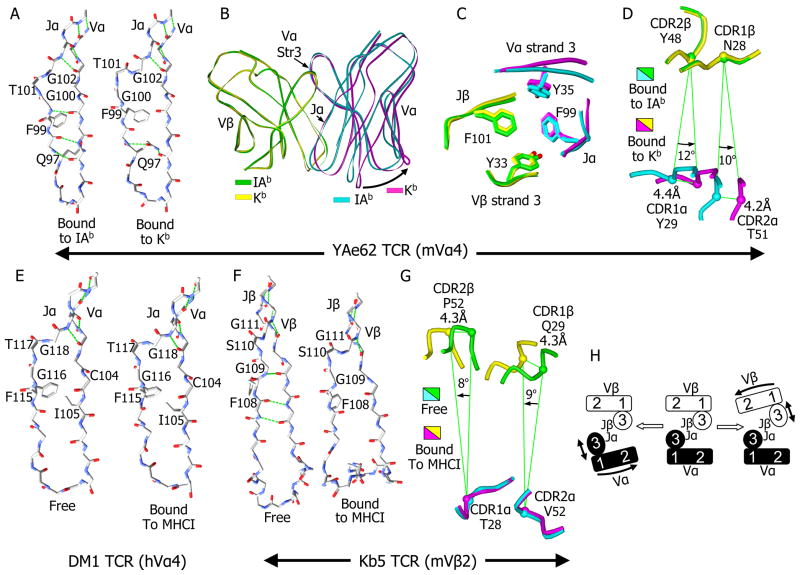
Three alternate conformations of the core TCR structure
The most striking structural change in the YAe62 TCR in binding to Kb-pWM versus IAb-p3K is a disruption in the β-strand core of Vα. When bound to IAb, the core of the YAe62 Vα domain consists two β-strand sheets, typical of immunoglobulin family V domains. One sheet is composed of 4 β-strands (1, 2, 6 and 7) and the other of 5 β-strands (3, 4, 5 and 8 from Vα with 9 from Jα). Also typical of immunoglobulin family V domains, the interaction between the long Vα β-strand 8 and the β-strand of Jα is interrupted at the conserved FGXG Jα motif (Figure 6A, left). However, when bound to Kb-pWM, the lower portion of this interaction is disrupted, breaking three H-bonds, but preserving the interactions between the upper portions of the strands (Figure 6A, right). The side chain of Q97 now swings in to fill the gap, stabilizing the structure by bridging the two β-strands with an H-bond to each strand. This conformational change is clearly seen in the electron density in this region of the TCR (Figure S3A).
Figure 6. Alterations in the relative orientation of Vα to Vβ extend the ability of the TCR to accommodate different MHC ligands.
A) The disruption of the β strand interactions between Jα and Vα of YAe62 when it is bound to Kb-pWM versus IAb-p3K is shown. A wireframe representation of the protein backbone of Jα and the last β strand of Vα, with the side chains for the FGXG conserved motif and αQ97 are shown. Backbone H-bonds, as well as those involving αQ97, are shown in green. For B, C and D the Vα and Vβ domains of the YAe62 TCR bound to IAb-p3K (Vα, cyan; Vβ, green) vs. Kb-pWM (Vα, yellow; Vβ, magenta) were overlaid by Vβ. B) The TCR Vα and Vβ are represented as ribbons. The rotation of Vα in relation to Vβ (arrow) is shown. C) The unchanged positions of the four core aromatic amino acid at the Vα to Vβ interface are shown for the two TCRs. D) A view looking down through the TCR toward the ligands is shown with the CDR1 and CDR2 loops as Cα traces and the positions of the Cα carbon of a central amino acid on each loop labeled. The relative distance and angle of movement of these atoms on the Vα loops relative to the adjacent Vβ atoms is indicated. E) The β strand interactions between Jα and Vα for a representative, DM1, (Archbold et al., 2009), of the three human TCRs bearing Vα4 family members whose structures have been published is shown as in A, either unbound (left) or bound (right) to an MHCI ligand. Side chains for the FGXG conserved motif and for αC104 and αI105 are shown and labeled. F). Same as E but for the mouse Vβ2 bearing TCR, Kb5 (Housset et al., 1997; Reiser et al., 2002). G) Same as D except that the two Kb5 TCRs are overlaid by Vα rather than Vβ. H) A cartoon representation of the relative range in orientation of Vα to Vβ in this and other published TCR structures.
The net result of this change in conformation is that the portion of the Vα domain containing CDR1 and CDR2 swings out away from Vβ (Figure 6B), while not disturbing the portions of the Jα and Vα β-strands 3 (Figure 6B) that stabilize the hydrophobic core of the Vα to Vβ interface that includes F99 of the FGXG motif (Figure 6C). The second glycine (G102) in the FGXG motif is the pivot point of the Vα movement and it is likely that the rotational flexibility of the two glycines in the motif allows Vα to undergo this conformational change.
When the two versions of the TCR are superimposed based on the Vβ domain, and the CDR1 and CDR2 loops are viewed from above down the center of interaction between Vα and Vβ (Figure 6D), one can see that this alternate conformation of the TCR allows the Vα CDR1 and CDR2 loops to rotate about 10–12 degrees in relation to Vβ, changing the relative positions of the Vα and Vβ CDR1 and CDR2 loops in relation to each other by more than 4 Å.
Since the FGXG motif is conserved in nearly all TCR J elements, we examined published TCR-MHC structures to see whether this type of conformational change may have been present, but not explored in the published structures. We superimposed separately the Vα and Vβ domains of more than 40 reported mouse and human TCR structures, involving more than 40 different TCRs (Figure S3B and S3C). In most of these, the J region β-strand has the conventional position in the Vα or Vβ domains. However, we found examples which the J to V interaction undergoes a conformational change that was remarkably similar to that seen in the YAe62 TCR bound to Kb-pWM, one set involving human Vα4 (hTRAV24) and a second involving mouse Vβ2 (mTRBV1).
There are seven structures involving three TCRs (4 for LC13, 2 for DM1 and 1 for KK50) which contain members of the human Vα4 family, but each TCR uses a different Jα element (TRJA 52, 13, 37, respectively). In five cases the TCRs are bound to human MHCI ligands (PDB 3KPR, 3KPS, 1MI5, 3DXA and 2ESV) and in the other two cases the LC13 (PDB 1KGC) and DM1 (PDB 3DX9) TCRs were solved in an unbound form (Archbold et al., 2009; Hoare et al., 2006; Kjer-Nielsen et al., 2002; Kjer-Nielsen et al., 2003; Macdonald et al., 2009). In all seven examples the Vα4 domain has almost the same conformation as that in the YAe62 TCR bound to Kb-pWM (Figure S3B). An example is shown for the bound (Figure 6E, left) and unbound (Figure 6E, right) DM1 TCR. In both cases the Jα β-strand the TCR matches the disrupted conformation seen with the YAe62 bound to Kb-pWM rather than to IAb-p3K. The upper part of the interaction between these strands is intact, but the strands are separated at the second glycine (G118) of the FGXG motif in a manner very similar to that of the YAe62 TCR bound to Kb-pWM (Figure 6A, right).
Since this transformation is seen in the unbound DM1 TCR structure, it seems likely that, in contrast to the bulk of studied TCRs, this altered open conformation is preferred prior to ligand engagement for this TCR. Because these three TCR all use different Jα elements and have very different CDR3α lengths and sequences, we considered whether the preference for this conformation might be inherent in the human Vα4. A unique feature of human Vα4 is the presence of an isoleucine (I105) immediately following the universally conserved cysteine in Vα β-strand 8 (Figure 6E). In nearly all other human and mouse Vα elements, including all of those found in other published structures of MHC-peptide reactive TCRs, this amino acid is an alanine. Its position in all of the human Vα4 structures puts its bulky side chain in juxtaposition to the conserved phenylalanine of the FGXG motif of Jα. This could interfere with the transition to the conventional conformation explaining the predilection of human Vα4 for the more open conformation.
There are five structures involving mouse Vβ2 containing TCRs, three for the Bm3.3 TCR (PDFs 1FO0, 1NAM and 2OL3) and two for the KB5-20 TCR (PDB 1KB5 and 1KJ2) (Housset et al., 1997; Mazza et al., 2007; Reiser et al., 2000; Reiser et al., 2002). The TCRs contain different Jβ elements, TRBJ1-3 and TRBJ2-3, respectively. The Bm3.3 TCR was solved bound to three different MHCI-peptide complexes. In all of these, the Jβ β-strand interacts with Vβ β-strand 8 in a closed conformation, very similar that seen in other Vβ domains (Figure S3C).
However, the exception is the KB5-20 TCR, which was solved bound to a Kb-peptide complex and, in an unbound state, complexed with a monoclonal antibody Fab (Housset et al., 1997; Reiser et al., 2002). In the unbound KB5-20 TCR structure (Figure 6F, left), the Jβ β-strand interaction with Vβ β-strand 8 is nearly identical to that seen in the Bm3.3 structures and in other TCR Vβs. But, as pointed out by the authors who reported these structures, the extra-long CDR3β at the end of these β-strands undergoes a tremendous conformational change upon binding the ligand, collapsing from an extended configuration to a compact one. This change upon binding is accompanied by the separation of the Jβ β-strand and Vβ Vβ-strand 8 that mirrors almost identically the conformational change seen in Vα in the human Vα4 TCRs and in the YAe62 TCR bound to Kb-pWM (Figure 6F, right). The lower part of Jβ separates from Vβ precisely at the second glycine (G111) of the FGXG motif breaking the lower inter-chain H-bonds. Again, the bulk of the Vβ domain containing CDR1 and CDR2 has separated from the β-strand of Jβ which maintains its conserved interaction with Vα. Again, mirroring the case with the YAe62 TCR, when the two versions of the KB5-20 TCR are superimposed based on the Vα domain, and the CDR1 and CDR2 loops are viewed from above down the center of interaction between Vα and Vβ (Figure 6G), one can see that the Vβ CDR1 and CDR2 loops of the ligand bound KB5-20 TCR rotate 8–9 degrees in relation to those of Vα and the relative positions of the Vα and Vβ CDR1 and CDR2 loops move by more than 4 Å. Perhaps because there were so few TCRs for comparison at the time, the authors attributed these structural changes to an unusual situation forced by the collapse of an extra the long CDR3β loop (16 amino acids) upon ligand engagement. However, the Vα CDR3 loop of YAe62 is the shortest of the solved TCRs (8 amino acids) and the human Vα4 containing TCRs assume the open conformation whether or not they are ligated, suggesting that this structural flexibility is inherent in TCR V domains.
Taken together, these structures suggest that in addition to adjustments in the rotomers of individual CDR amino acids and conformational changes in the backbones of CDR loops, a given TCR may switch among at least three distinct alternate conformations depending on whether the VαJα or VβJβ connection is disrupted (Figure 6H). In each case the core JαJβ interaction is not disrupted, but the relation among the CDR1 and CDR2 loops of Vα and Vβ changes. Also, since the disrupted strands support a CDR3 loop, this CDR3 is stretched and changes shape. Whether a fourth conformation involving simultaneous disruption of Vα and Vβ is possible is unknown. However, it might not be possible to preserve the core JαJβ interaction in such a case. Switching among these three alternate conformations can extend the size of the TCR repertoire and may allow the TCR to maintain the conserved points of interaction with MHC with a variety of different MHC alleles, isotypes and classes that differ substantially both in sequence and the spacing between the helices.
Discussion
When the first structures of αβTCRs bound to MHC ligands appeared, the hope was that they would answer the question of why TCRs are confined to MHC ligands. We expected to see easily identifiable conserved interactions of the TCR CDR1 and CDR2 loops with the MHC helices, explaining the evolutionary connection between the TCR and MHC. It has taken 15 years and dozens of TCR-MHC structures to understand that the “rules of engagement” between TCRs and MHC ligands are much more complicated than originally imagined. Their interactions are not conserved in the conventional structural sense, but rather allow for considerably more flexibility, within the confines of certain general rules.
One of these rules has been well-established. In all structures solved so far, the TCR has a diagonal orientation on the MHC (reviewed in (Rudolph et al., 2006). The CDR loops of the TCR usually occupy similar sites in the different structures, with the CDR2 loops interacting particularly with the MHC alpha helices and the very variable CDR3 loops centered on the peptide, while the CDR1 loops often contact both peptide and MHC. This rule applies for both MHCI and MHCII ligands and can be seen in the structure of the YAe62 TCR bound to either IAb-p3K or Kb-pWM. However, within this rule there is considerable latitude in the angle of engagement when viewed from above, in the pitch and yaw when viewed from the side or down the axis of the peptide binding groove (Bjorkman, 1997; Rudolph et al., 2006; Sethi et al.) and, in few extreme cases, involving some autoimmune TCRs, in the position of the TCR along the length of the peptide (Hahn et al., 2005; Li et al., 2005). Accordingly, there are some differences in the angle and yaw of the YAe62 TCR when bound to Kb-pWM compared to IAb-p3K.
With the accumulation of more structures of bound versus free TCRs, individual TCRs bound to different ligands and different TCRs using the same V element, it has also become clear that the general orientation of the TCR on the MHC involves considerable flexibility in the TCR CDR loops, especially the CDR3 loops (Gagnon et al., 2006; Reiser et al., 2003). The loops adjust to different ligands by changing not only the rotamers of their amino acid side chains, but also the conformation of their main chain backbones. This point is well-illustrated in the structures presented here. There are numerous differences in the rotamers of particular CDR amino acids of the YAe62 TCR when engaging its MHCI vs. MHCII ligand. Changes in backbone conformation of the CDR3 loops are particularly extensive.
The semi-conserved orientation of the TCR on MHC has raised the question of whether there are specific germline encoded V region amino acids selected evolutionarily to be linchpin positions for MHC interaction, thus imposing this orientation. The answer to this question has been harder to tease out from the structural data, since it requires comparison of a substantial set of structures involving different MHC ligands and TCRs all of which have the same TCR Vα and/or Vβ element. This requirement has only been met for a handful of V elements, such as those related to mouse Vβ8 and Vα4 discussed here and present in the YAe62 TCR. These V elements contain four amino acids that repeatedly have been observed to interact with the MHCII helices at the same locations (Dai et al., 2008; Feng et al., 2007; Maynard et al., 2005). Those interactions are particularly well-illustrated in the YAe62 complexes with IAb-p3K. We find the same amino acids in similar positions on the Kb-pWM ligand and their mutation dramatically reduced YAe62 binding to both ligands. It is noteworthy that two of these amino acids are tyrosines (αY29 and βY48), whose interaction with the MHC does not require the precise geometry seen in H-bonds, but is dependent rather on the ability of their aromatic ring to create a substantial area of Van der Waals interaction, thus contributing to flexibility in the details of the contact. In both cases the approach of these amino acids to the MHC helix is facilitated by adjacent MHC amino acids lacking an interfering side chain.
Whether the presence of conserved germline encoded MHC interactions will extend to most other V elements will depend on many more new TCR-MHC structures and probably take many years to work out. It could be argued that, given the variability of the TCR angle, pitch and yaw on MHC, these germline encoded interactions will prove to be rare. However, we have seen a very high frequency of highly MHC crossreactive T cells, such as YAe-62, developing in single peptide mice that have limited negative selection (Huseby et al., 2005). We argue that the processes of positive and negative selection in the thymus of normal animals select against these T cells, whose TCRs may display a full set of germline encoded interactions and therefore be likely to be strongly self-reactive. Rather, these processes select for TCRs whose somatically generated CDR3 loops interfere with some, but not all, of the possible germline interactions, steering the TCR specificity toward the peptide while preserving the TCR orientation on MHC via a much lower MHC affinity.
The most unexpected finding in the present study was a substantial disruption of the conventional Jα connection to Vα in the YAe62 TCR when bound to Kb-pWM as compared to IAb-p3K. This type of Vα domain conformational change has not been previously discussed. Nevertheless, our reexamination of previously published TCR structures turned up several examples of a similar altered conformation, one in a Vα and one in a Vβ element (Housset et al., 1997; Mazza et al., 2007; Reiser et al., 2000; Reiser et al., 2002). In each case the J region β strand had separated from the rest of the V element, preserving the critical Vα to Vβ interactions mediated by the two J region β strands. Also in each case, the flex point for this separation is the second glycine in the FGXG motif of the J element, raising the possibility that this motif in TCR J elements may be conserved to function as a swivel point for adjusting the interaction of Vα and Vβ. The result of these conformational changes is to alter the positions of the CDR1 and CDR2 loops of Vα relative to those of Vβ, influencing how these loops approach the MHC helices. This type of adjustment may be used in fine tuning the alignment of the CDR1 and CDR2 loops with particular regions of the MHC helices and as well as in accommodating MHC molecules with different distances between the two helices. Since the β-strands of J and V involved support the CDR3 loop, their repositioning also plays a role in the flexibility of this CDR3.
In the examples so far, this type of conformational change has been seen only in TCRs that interact with MHCI ligands. This may be coincidental, since there are many more TCR-MHCI than TCR-MHCII solved structures. Alternatively, the observation that the positions of the interactions of the amino acids conserved for MHC interaction are more consistent on MHCII ligands than on MHCI ligands may indicate that this additional flexibility in the TCR is more important for proper placement of these amino acids on MHCI ligands. Thus far, all of the examples of this conformational change in Vα involve TCRs constructed with a human Cα. Whether this Cα influences the ability of the Vα to undergo this change cannot be determined from these structures. However, we can say that the human Cα does not automatically induce this change, since there are many TCR structures with human Cα with attached Vα’s in the closed conformation, including that of the YAe62 TCR bound to IAb-p3K.
Unlike B cells that use antigen selected somatic mutation of their rearranged immunoglobulin genes to vastly increase their repertoire and fine tune their specificity for antigen, the T cell repertoire is limited by the set of initial rearrangements of TCR genes. Given that this repertoire is further constrained by the requirement for MHC reactivity and the processes of thymic positive and negative selection, there is a problem how the limited numbers of TCRs in any given animal can cope with the very large number of MHC-peptide combinations with which they may be confronted. This problem appears to have been solved in part by the relatively low affinities needed for TCR interaction with MHC-peptide to trigger response in T cells. In addition, the great flexibility of the CDR regions of TCRs allows a single TCR to bind a number of ligands. Here we have uncovered another phenomenon that allows recognition of more than one ligand by a single TCR, the potential to switch among three alternate conformations by disrupting the J to V interaction of either the Vα or Vβ domain.
Experimental Procedures
Identification of peptide mimotopes
CD8+ YAe62 TCR transgenic spleen cells were incubated with MHC class II-deficient spleen cells pulsed with varying doses of a nonapeptide positional scanning library (N – terminal and C-terminal free, TPI 921) (Pinilla et al., 1992). Proliferative responses were compared to control wells, and ranked and deconvoluted. Mass spectrometry and further examination revealed a stimulatory 8-mer peptide, WIYVYRPM (pWM). Cytotoxicity assays were done using standard techniques (Huseby et al., 1999).
Protein Expression and purification
The DNA encoding Kb (extracellular domains) and β2m covalently attached to pWM or the OVA SIINFEKL peptide (pOVA) were mutated to introduce cysteines into Kb replacing tyrosine 84 and into the linker that attaches β2m to pWM replacing a glycine at position 2 (Lybarger et al., 2003) and were expressed in baculovirus infected insect cells. Soluble YAe62 TCR was expressed in baculovirus as previously described (Dai et al., 2008). For crystallography the Vα and Vβ portions of the YAe62 TCR were produced fused to the extracellular portions of human Cα and Cβ, respectively, in E. coli inclusion bodies (Dai et al., 2008).
Crystal Production and Data Collection
An equimolar mixture of YAe62 TCR and Kb-pWM was crystallized by mixing 0.5 ul of complex solution at a concentration of 15mg/ml with an equal volume of reservoir solution. The complex crystallized in two different space groups.
Structure Determination
The structures of YAe-Kb-pWM complex were determined by molecular replacement using the program Phaser with the YAe62 TCR(PDB 3C60) and Kb-pdEV8(PDB 2CKB) as search models, respectively and manually fitted with program Coot followed by alternating simulated annealing, positional refinement and B factor refinement using the program Phenix.
Structure analysis
Buried molecular surface areas were calculated using GRASP. NCONT in CCP4 was used to analyze the contacts between the TCRs and their ligands.
TCR mutational analysis
The contribution of TCR amino acids to ligand recognition was measured by retroviral transduction of a TCR− hybridoma with wild-type and indicated Ala-substitution mutants of YAe-62.8 Vα and Vβ and measuring the ability of the transductants to respond to ligands and bind tetramers as previously described (Rubtsova et al., 2009; Scott-Browne et al., 2007).
Supplementary Material
HIGHLIGHTS.
A single TCR binds an MHC class I and MHC class II ligand similarly
TCRs use the same key residues in binding different classes of MHC
A single TCR can switch among several core conformations
The structural variability of TCRs increases their repertoire and range of ligands
Acknowledgments
The authors thank Dr Laurent Gapin for thoughtful discussions and the staff of the Advanced Light Source synchrotron facility (BL 8.2.2) for help in the x-ray data collection. This work was supported by USPHS grants AI-18785 and AI-22295. Also we thank the Zuckerman Family/Canyon Ranch and Allen Lapporte for support of the Structural Facility at National Jewish Health.
Footnotes
ACCESSION NUMBER
The coordinates of the YAe62 TCR bound to Kb-pWM have been deposited in the Protein Data Bank under the accession ID 3RGV.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archbold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, Brennan RM, Beddoe T, Wilce MC, Clements CS, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman PJ. MHC restriction in three dimensions: a view of T cell receptor/ligand interactions. Cell. 1997;89:167–170. doi: 10.1016/s0092-8674(00)80195-6. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Burrows SR, Chen Z, Archbold JK, Tynan FE, Beddoe T, Kjer-Nielsen L, Miles JJ, Khanna R, Moss DJ, Liu YC, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci U S A. 107:10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- Fink PJ, Bevan MJ. H-2 antigens of the thymus determine lymphocyte specificity. J Exp Med. 1978;148:766–775. doi: 10.1084/jem.148.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Turner RV, Damirjian M, Wojnarowicz A, Biddison WE, Baker BM. T cell receptor recognition via cooperative conformational plasticity. J Mol Biol. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, Beddoe T, Falco M, Kjer-Nielsen L, Reid HH, et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol. 2006;7:256–264. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
- Housset D, Mazza G, Gregoire C, Piras C, Malissen B, Fontecilla-Camps JC. The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain. EMBO J. 1997;16:4205–4216. doi: 10.1093/emboj/16.14.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- Huseby ES, Ohlen C, Goverman J. Cutting edge: myelin basic protein-specific cytotoxic T cell tolerance is maintained in vivo by a single dominant epitope in H-2k mice. J Immunol. 1999;163:1115–1118. [PubMed] [Google Scholar]
- Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, Kappler JW. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002;10:1521–1532. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunova NN, Viret C, Pobezinsky LA, Miller SA, Kazansky DB, Sundberg JP, Chervonsky AV. Restricted MHC-peptide repertoire predisposes to autoimmunity. J Exp Med. 2005;202:73–84. doi: 10.1084/jem.20050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger L, Yu YY, Miley MJ, Fremont DH, Myers N, Primeau T, Truscott SM, Connolly JM, Hansen TH. Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J Biol Chem. 2003;278:27105–27111. doi: 10.1074/jbc.M303716200. [DOI] [PubMed] [Google Scholar]
- Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MC, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- Madden DR, Gorga JC, Strominger JL, Wiley DC. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Graf D, Lovatt M, Bommhardt U, Zamoyska R, Fisher AG. How many thymocytes audition for selection? J Exp Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PJ, Pazy Y, Conti B, Riddle D, Appella E, Collins EJ. Single MHC mutation eliminates enthalpy associated with T cell receptor binding. J Mol Biol. 2007;373:315–327. doi: 10.1016/j.jmb.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Guimezanes A, Gregoire C, Mosser T, Schmitt-Verhulst AM, Fontecilla-Camps JC, Malissen B, Housset D, Mazza G. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Gregoire C, Darnault C, Mosser T, Guimezanes A, Schmitt-Verhulst AM, Fontecilla-Camps JC, Mazza G, Malissen B, Housset D. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- Rubtsova K, Scott-Browne JP, Crawford F, Dai S, Marrack P, Kappler JW. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci U S A. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi DK, Schubert DA, Anders AK, Heroux A, Bonsor DA, Thomas CP, Sundberg EJ, Pyrdol J, Wucherpfennig KW. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med. doi: 10.1084/jem.20100725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- Speir JA, Garcia KC, Brunmark A, Degano M, Peterson PA, Teyton L, Wilson IA. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 1998;8:553–562. doi: 10.1016/s1074-7613(00)80560-9. [DOI] [PubMed] [Google Scholar]
- Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- Zinkernagel RM, Callahan GN, Althage A, Cooper S, Klein PA, Klein J. On the thymus in the differentiation of “H-2 self-recognition” by T cells: evidence for dual recognition? J Exp Med. 1978;147:882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.