Abstract
It is well accepted that cancer develops following the sequential accumulation of multiple alterations, but how the temporal order of events affects tumor initiation and/or progression remains largely unknown. Here, we describe a mouse model that allows for temporally-distinct cancer mutations. By integrating a Flp-inducible allele of K-rasG12D with established methods for Cre-mediated p53 deletion, we were able to separately control the mutation of these commonly associated cancer genes in vitro and in vivo. We show that delaying p53 deletion relative to K-rasG12D activation reduced tumor burden in mouse model of soft-tissue sarcoma, suggesting that p53 strongly inhibits very early steps of transformation in the muscle. Furthermore, using in vivo RNAi we implicate the p53 target gene p21 as a critical mediator in this process, highlighting cell-cycle arrest as an extremely potent tumor suppressor mechanism.
Keywords: tumor suppression, mouse models, sequential mutagenesis, p53, p21
INTRODUCTION
Tumorigenesis is a multi-step process driven by the accumulation of both genetic and epigenetic alterations in oncogenes and tumor suppressor genes (1). These individual changes occur in a sequential manner and are thought to drive distinct steps in the progression of normal cells to full malignancy (2). Although it is generally believed that the actions of these mutations combine to effect full transformation, it remains unclear how the order of events impacts this process.
While genetically engineered mice have been instrumental in modeling and experimentally validating various attributes of human cancer, most current models that utilize multiple cancer-associated mutations are not designed to perform sequential mutagenesis. As a result, one cannot directly test the importance of mutation timing or order. For example, while the use of multiple alleles controlled by the conditional Cre-LoxP site-specific recombinase (SSR) system allows for the introduction of several cancer relevant mutations, these mutations occur simultaneously at the time of tumor initiation (3). Additional SSR modalities, such as S. cerivisae-derived Flp-Frt, exist but have been used in a more limited fashion in mouse models (4, 5). By combining different SSR systems within the same model one could achieve spatiotemporal control of distinct genetic events provided the recombinases are independently regulated.
Through its ability to respond to various forms of cellular stress by inducing cell cycle arrest or apoptosis, p53 plays a central role in tumor suppression (6). Despite being one of the most thoroughly studied tumor suppressor genes, rather little is known about the precise stage(s) of tumorigenesis at which p53 exerts its functions. In many human cancers, such as those of the lung, colon, and pancreas, p53 alterations have been documented in more advanced stages of tumor development, suggesting that p53 constrains progression of established tumors (7–9). In contrast, the early onset of several cancer types in Li-Fraumeni patients, who inherit a germ-line mutation in p53, argues for a potential role of this tumor suppressor in inhibiting the early stages of transformation in some cell types (10). While these studies establish that p53 mutation correlates with different stages of tumor development, whether these differences underlie specific temporal requirements for p53 loss in distinct tissues remains to be determined.
In addition to mutational data from human tumors, numerous mouse models have documented broad tumor suppressive roles for p53 in numerous contexts (11). Both germline mutant and Cre-LoxP conditional alleles of p53 have been used to promote tumor formation in multiple tissues. However, most of these studies have not been able to pinpoint a specific requirement for p53 mutation as a function of tumor progression. Here we describe a system for sequentially mutating the commonly-associated cancer genes K-ras and p53. By combining a Flp-inducible allele of oncogenic K-ras with an already well-established Cre-regulated p53 deletion allele (12), we were able to separately regulate these mutations, both in cells and in mice. Using this dual SSR strategy, we show a clear effect of p53-mediated tumor suppression in the earliest stages of soft-tissue sarcoma (STS) formation.
MATERIALS AND METHODS
Mouse studies
Information on the construction of the K-rasFSF-G12D targeting construct and mouse strain, as well as details on additional mouse strains used in this study, are found in Supplemental Material. Lung tumors were generated and processed as previously described (16–18), substituting Ad-Flpo (University of Iowa, Gene Transfer Vector Core) for Ad-Cre. Intramuscular viral infections were done as previously shown (20). For GFP marking experiments, needles were dipped in India Ink before injection in order to mark the needle track. Tamoxifen (Sigma) was dissolved in corn oil at 10 mg/ml and injected intraperitoneally every other day for 5 days where applicable. Once masses were visible on the legs, tumors were processed for histology and molecular analysis as described below. For MEF tumor experiments, 1.5 × 10^6 cells were resuspended in 200 ul of PBS and injected subcutaneously. Mice were monitored every few days for tumor formation. Animal studies were approved by Massachusetts Institute of Technology’s (MIT) Committee for Animal Care and conducted in compliance with Animal Welfare Act Regulations and other federal statutes relating to animals and experiments involving animals, and adheres to the principles set forth in the 1996 National Research Council Guide for Care and Use of Laboratory Animals (institutional animal welfare assurance number, A-3125-01).
Cell Culture
Primary MEFs of the indicated genotypes were isolated from E13.5 embryos and propagated in DMEM supplemented with 10% IFS, 5mM glutamine and penicillin/streptomycin. Where applicable 4-hydroxytamoxifen (Sigma) was added to the media at 100nM. Lentiviral infections were performed by directly transferring viral supernatant. For adenovirus infection in vitro Ad-Flpo was added to the media at an MOI of 10.
Genomic PCR analysis
DNA was prepared from MEFs or tumors and subjected to standard PCR analysis. PCR primers are given in Supplemental Material.
Protein extraction and immunoblots
Lysates from MEFs and tumors were prepared and immunoblots were performed as described previously (26). Antibody information is provided in Supplemental Material. Levels of Ras-GTP were determined with the Ras activation kit (Millipore).
Lentiviral vectors and shRNA cloning
The lentivirus containing pgkCre and a U6-shRNA has been described previously (26). The GFP-Cre lentivirus (Ubc-GFP; pgk-Cre) was provided by M. DuPage (Massachusetts Institute of Technology KI, Cambridge, MA). Target sequences for shRNA knockdown were identified and cloned as previously described (26). shRNA sequences are provided in Supplemental Material.
Lentiviral production
Lentivirus was produced as described previously(30).
Histology and Immunohistochemistry
Tissues were fixed in 10% formalin for 6–8 hrs and further processed for histology as previously described (16). For IHC, parraffin-embedded sections were dewaxed, followed by antigen retrieval in 10 mM citrate buffer (pH 6.0) in a pressure cooker. Slides were quenched in 3% hydrogen peroxide and washed in TBST. After blocking in TBST/5% goat serum for 1 hr, the primary antibody (rabbit mAb anti-GFP, #2956- Cell Signaling Technology-CST) was diluted 1:100 in SignalStain Antibody diluent (#8112, CST) and incubated on slides overnight at 4°C. Detection was performed using a biotinylated goat anti-rabbit secondary antibody followed by the Vectastain ABC kit with diaminobenzadine (DAB) (Vector Labs). Slides were counterstained with haematoxylin before coverslipping.
RESULTS
In order to create a model for sequential mutagenesis in vivo, we sought to combine Flp-Frt mediated mutagenesis with already available tools from Cre-LoxP systems. To this end, we first generated a Flp-recombinase inducible allele of oncogenic K-ras (K-rasFSF-G12D), following a very similar strategy used to construct the well-studied Cre-inducible K-rasLSL-G12D allele (13) (Fig 1A and 1B). To characterize the activity of this allele, we first generated mouse embryonic fibroblasts (MEFs) from K-rasFSF-G12D/+ embryos. Introduction of an engineered thermostable version of Flp (Flpe) (14) but not Cre led to removal of the stop cassette, as demonstrated by PCR analysis (Fig 1C). In addition, Flpe expression in K-rasFSF-G12D/+ MEFs induced high levels of active Ras-GTP, along with appropriate downstream signaling events, such as up-regulation of cyclin D1 (Fig 1C).
Figure 1. Construction and initial characterization of K-rasFSF-G12D.
A. Schematic of the targeting strategy for K-rasFSF-G12D, including the (i.) endogenous K-ras locus, (ii.) targeting vector, (iii.) correctly targeted K-rasFSF-G12D locus, and (iv.) recombined K-rasFrt-G12D following Flp expression. Green boxes = exons; thick black bars = probes for Southern blot; star = oncogenic point mutation G12D; B = BamHI; K = KpnI. B. Southern blot of non-targeted (K-ras+/+) and correctly targeted (K-rasFSF-G12D/+) ES cell clones following BamHI/KpnI double digestion and hybridization with an external 5′ probe. C. Characterization of K-rasFSF-G12D/+ MEFs. Top panel: PCR analysis of genomic DNA isolated from K-rasFSF-G12D/+ MEFs infected with lentiviruses expressing Cre (Lenti-Cre) or Flpe (Lenti-Flpe). Bottom panel: Western blot of K-ras+/+ and K-rasFSF-G12D/+ MEFs either uninfected or infected with Lenti-Flpe. D. Representative hematoxylin and eosin (H&E)-stained section of a lung lobe from a K-rasFSF-G12D/+ mouse 16 weeks post Ad-Flpo infection. Scale bar is 500 μm.
We next tested the activity of the K-rasFSF-G12D allele in vivo, which was of particular importance given the evidence that Flpe is much less efficient than Cre in mammalian systems (15). While introduction of adenoviruses or lentiviruses expressing Cre (Ad-Cre or LV-Cre) into the lungs of K-rasLSL-G12D/+ mice results in significant lung tumor formation (16, 17), infection with Ad-Flpe or LV-Flpe failed to generate lung tumors in K-rasFSF-G12D animals (data not shown). However, using a mammalian codon-optimized version of Flp, termed Flpo (15), we were able to initiate numerous lung tumors in K-rasFSF-G12D/+ mice with intratracheal instillation of Ad-Flpo and LV-Flpo (Fig 1D and data not shown). Together, these data indicate that K-rasFSF-G12D is the functional equivalent of the K-rasLSL-G12D allele.
Given the well-known genetic interaction between K-ras and p53 in cellular transformation, we chose to combine mutations in these two genes for initial sequential mutagenesis experiments in MEFs. Previous work had demonstrated that primary MEFs expressing endogenous K-rasG12D have some characteristics of transformation, but they are not fully transformed and require p53 deletion to form tumors in immunocompromised mice (13). With the ability to separate K-ras and p53 mutations in time, we set out to examine the consequences of delayed p53 mutation in K-rasG12D-expressing cells following transplantation into immunocompromised mice.
We generated MEFs containing the K-rasFSF-G12D allele as well as p53flox alleles. To control p53 deletion, we also crossed in the R26CreER-T2 allele, which contains a tamoxifen-inducible allele of Cre driven from the ubiquitously expressed Rosa26 locus (18). Compound mutant MEFs of the following genotypes were used in these experiments: K-rasFSF-G12D/+; R26CreER-T2/+; p53flox/flox (FKCP) and K-rasFSF-G12D/+; R26CreER-T2/+; p53flox/+ (FKC). Infection of these cells with Ad-Flpo led to activation of the K-rasFSF-G12D allele in vitro (designated FK*). FK*CP cells were injected subcutaneously into nu/nu mice and p53 deletion was induced by intraperitoneal (ip) tamoxifen injection at different time points thereafter. As a positive control, we included cells that had been treated one week prior to injection with 4-hydroxytamoxifen (4-OHT) to inactivate p53.
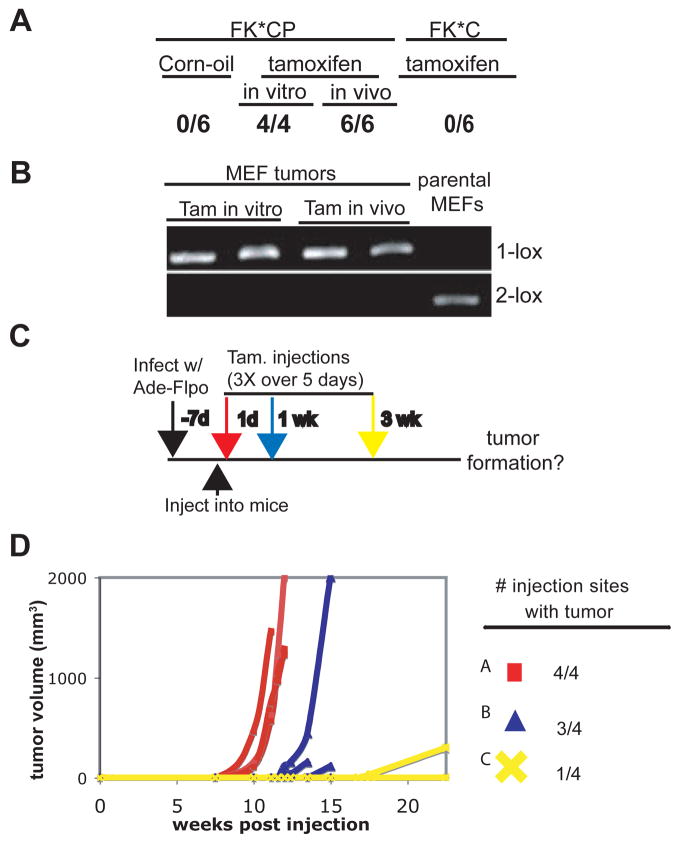
As shown in Figure 2A, when mice injected with FK*CP cells started receiving tamoxifen on the same day of injection, efficient tumor formation was observed, similar to the positive control group. As expected, the resulting tumors showed complete recombination of both p53 alleles (Fig 2B). Importantly, this system showed no leakiness, as mice injected with FK*CP cells and treated with vehicle (corn oil) failed to form tumors (Fig 2A). Additionally, the ability of tamoxifen treatment to promote tumor development depended on complete loss of p53, as shown by the lack of tumorigenecity of FK*C cells following tamoxifen administration (Fig 2A).
Figure 2. Sequential mutagenesis of K-ras and p53 in MEFs.
A. Results of initial transplant experiments, represented as the number of injection sites presenting with tumors following introduction of different MEF lines and various treatment conditions (corn oil or tamoxifen in vivo; 4-OHT in vitro). See text for experimental details. B. Recombination analysis of p53flox alleles in resultant FKCP MEF tumors or the parental MEFs before injection into the animals. C. Schematic of sequential mutagenesis of K-ras and p53. See text for details. D. Growth kinetics and overall incidence of tumors in which p53 was deleted 0 weeks (Group A-red square), 1 week (Group B-blue triangle), or 3 weeks (Group C-yellow X) post subQ injection of the cells into nu/nu mice. Mice were monitored for 23 weeks.
We next investigated the fate of partially transformed FK*CP cells in vivo by addressing how long after injection could tamoxifen treatment still result in tumor formation. FK*CP cells were introduced into different groups of mice that varied in their tamoxifen treatment schedule: beginning 1 day (Group A), 1 week (Group B), or 3 weeks (Group C) following injection of the cells (Fig 2C). Strikingly, while tumor development was efficient in Groups A and B, almost no tumors formed in Group C (Fig 2D). These results suggest that injected FK*CP cells were either removed in a p53-dependent manner or became resistant to p53 loss sometime between one and three weeks following their introduction into the animal. Given the recent evidence that innate immune cells can clear cells undergoing p53-dependent cell cycle arrest/senescence (19), we wondered if FK*CP cells failed to form tumors in Group C because they were removed in this fashion. Therefore, we repeated these experiments using NOD/SCID mice, which are more immunocomprised than the nu/nu strain and deficient in the cell populations implicated in senescent cell clearance. The results of the time-course experiment in NOD/SCID were very similar to those in nu/nu mice (Supplemental Fig 1). These data suggest that innate immune cell-mediated clearance of senescing cells likely does not explain the failure of the partially transformed FK*CP cells to remain responsive to p53 deletion and form tumors following prolonged times in vivo. In summary, although primary MEFs expressing endogenous levels of oncogenic K-ras proliferate continuously in cell culture conditions, transfer into an in vivo environment leads rapidly to engagement of the p53 pathway and irreversible tumor suppression.
Having validated the sequential mutagenesis system using dual SSR technology, we next applied the system to an endogenous tumor model. Previous work had shown that intramuscular infection of Ade-Cre into the limbs of K-rasLSL-G12D/+; p53flox/flox animals resulted in efficient sarcomagenesis (20). However, the presence of just one wild-type allele of p53 completely inhibited tumor formation. Furthermore, careful histological analysis of non-tumor bearing muscles of K-rasLSL-G12D/+; p53flox/+ animals several months after Ade-Cre infection failed to identify any microscopic lesions (data not shown). These results suggest that activation of endogenous K-rasG12D in the muscle leads to rapid p53-dependent tumor suppression.
Despite these initial observations, the exact nature of the tumor suppression and, thus, the fate of K-rasG12D-expressing cells in the p53flox/+ background had not been explored. One possibility was that oncogenic K-ras-positive cells persisted in the muscle, but were limited in their proliferative capacity due to p53-dependent growth arrest. In this case, a secondary mutation in the p53 pathway could potentially unleash the oncogenicity of these latent cells. Another possible scenario was that K-rasG12D activation resulted in a p53-dependent removal or irreversible arrest of the cells, such that subsequent p53 mutations would be ineffective in promoting full transformation of the initially targeted cells.
To distinguish between these two possibilities, we used the sequential mutagenesis strategy. Compound mutant mice of the genotype K-rasFSF-G12D/+; p53flox/flox; R26CreER-T2/CreER-T2 were generated and separated into three groups. All three groups received simultaneous intramuscular infection with Ad-Flpo to activate oncogenic K-ras. They were then segregated based on their tamoxifen treatment schedule and consequent p53 deletion. Tamoxifen was either administered on the same day of Ad-Flpo infection for Group A, or on day 10 post infection (Group B), or on day 21 post infection (Group C) (Fig 3A). If oncogenic K-ras expressing cells persisted and remained fully competent to respond to p53 loss, then all three groups would be expected to form sarcomas efficiently. In contrast, if mutant K-ras cells were removed or rendered refractory to delayed p53 deletion, sarcomagenesis would be inhibited in Groups B and C. In fact, sarcoma formation efficiency varied greatly between the three groups. While all mice in Group A presented with sarcomas, the percentage of effected mice was significantly lower in Group B (56%) and Group C (40%) (Fig 3B). Importantly, tumors from all groups displayed the expected recombination patterns at both K-rasFSF-G12D and p53flox loci (Fig 3D). Of the sarcomas that did form in Group C, some appeared with delayed kinetics compared to Group A (Fig 3C). Taken together, these results suggest that the pool of susceptible K-rasG12D expressing cells in the muscle is significantly diminished over time, either in number or function, by a strong p53 response. Therefore, a mutation in K-ras must be soon followed by disengagement of the p53 pathway for efficient tumorigenesis to occur in the muscle. Alternatively, the inhibition of the p53 pathway would have to occur first in this cell type in order to render the cells sensitive to subsequent oncogene activation.
Figure 3. Delaying p53 loss relative to K-rasG12D activation reduces tumor formation in a mouse model of soft-tissue sarcoma.
A. Experimental outline of sequential mutagenesis strategy, showing the genotype of the compound mutant mice and the treatment regimen. See text for details. B. Bar graph illustrating tumor incidence in mice in which tamoxifen treatment began on day 0 (black bar), day 10 (red bar), or day 21 (green bar) following Ad-Flpo infection. C. Dot plot indicating the kinetics of sarcomagenesis as measured by the time to tumor formation post-p53 loss (tamoxifen treatment). Color scheme is same as in (b) Students T-test was used for statistical significance. D. PCR analysis of K-ras and p53 loci.
The rapid and robust tumor suppression observed in the sequential mutagenesis experiments prompted us to further characterize the activation and function of the p53 pathway in the muscle. The kinetics of the p53 response suggested that this pathway is strongly activated very soon after oncogenic K-ras expression. Previous studies demonstrated that removal of p19Arf, an upstream activator of p53, could substitute for p53 loss in sarcomagenesis (20). Therefore, we monitored p53 pathway activation with the ArfGFP allele, a p19Arf-specific GFP reporter that also functionally inactivates this tumor suppressor (21). Importantly, established sarcomas derived from intramuscular Ad-Cre infection of K-rasLSL-G12D/+; ArfGFP/GFP animals displayed high GFP levels (Fig 4A). To determine if this activation occurs shortly after K-rasG12D expression, we assayed for reporter induction 6 days post Ad-Cre infection in K-rasLSL-G12D/+; ArfGFP/GFP and K-ras+/+; ArfGFP/GFP mice. Interestingly, we observed small areas of intense GFP staining in an oncogenic K-ras-dependent manner (Fig 4A), consistent with the notion that the p19Arf-p53 pathway is rapidly induced in the muscle following oncogenic stress.
Figure 4. Characterization of the p53 response in K-rasG12D-expressing muscle cells.
A. Representative GFP IHC on an established sarcoma (i.) and muscle sections (ii. and iii.) from intramuscular Ad-Cre infection of K-rasLSL-G12D/+; ArfGFP/GFP animals performed either 8 weeks (i.) or 6 days (ii. and iii.) after Ad-Cre infection. The arrow in ii. points to darkly staining cells, shown at higher magnification in iii. Scale bars = 100 μm. B. Kaplan-Meier graph showing sarcoma-free survival in a cohort of K-rasLSL-G12D/+; Rag2−/−; ArfGFP/GFP (red and black lines) or ArfGFP/+ (yellow and green lines) mice infected with lentiviruses expressing Cre and a shRNA targeting either luciferase (black and green lines) or p21 (red and yellow lines). C. Western blot of sarcomas arising from knockdown of either p21 or p19Arf/p16Ink4a in K-rasLSL-G12D/+; ArfGFP/+; Rag2−/− animals. D. GFP IHC on muscle tissue from K-rasLSL-G12D/+(or K-ras+/+) ; p53flox/+; Rag2−/− animals at the indicated times post infection (p.i.) with a Cre-GFP lentivirus. GFP staining is brown, and India Ink, which was used to mark the injection site, stains as black. Numerous infected cells are present in both K-ras+/+ and K-rasLSL-G12D/+ animals, suggesting that there is no significant clearance specific to oncogene-expressing cells. Scale bar = 100 μm.
We next sought to determine the tumor suppressive mechanism downstream of p53. Through its ability to transcriptionally up-regulate a number of different genes under various stress conditions, p53 can induce distinct cell fates, such as apoptosis, cell cycle arrest, or senescence (22). Previously, we showed that deletion of Bak and Bax, two critical mediators of the intrinsic pathway of apoptosis, could not substitute for p53 loss during K-rasG12D-induced sarcomagenesis (20). This suggested that p53 is not functioning through the induction of apoptosis in this setting.
To address if p53 was acting through cell cycle arrest and/or senescence, we focused on the cyclin dependent kinase inhibitor p21, which has been shown to be critical for the G1 cell cycle arrest elicited by p53 in a number of settings (23–25). For these studies, we used our recently reported RNAi-mediated gene silencing method, which involves a lentiviral-based system for in vivo expression of Cre and a shRNA in the context of immunocompromised (Rag2−/−) animals (26). In this system, intramuscular lenti-Cre infection of K-rasLSL-G12D/+; ArfGFP/+ ; Rag2−/− animals failed to generate sarcomas owing to strong p19Arf-p53 pathway activation. However, a shRNA targeting both p19Arf and p16Ink4a could block this tumor suppression and generate sarcomas. To determine if p21 is an important component of p53-dependent tumor suppression in the muscle, we introduced a shRNA targeting p21 into the bifunctional lentiviral vector and infected K-rasLSL-G12D/+; ArfGFP/+ ; Rag2−/− mice. Interestingly, knockdown of p21 was able to promote sarcoma formation (Fig 4B). Molecular analysis confirmed significant knockdown of p21 in the resulting tumors (Fig 4C). In addition, PCR analysis of tumor DNA indicated retention of the wild-type allele of p19Arf, suggesting no additional selective pressure for p53 pathway inactivation during tumor formation (data not shown). Together, these data indicate that p21 is required for the strong tumor suppressor activity of p53 in the muscle following expression of oncogenic K-ras. This suggests that the primary function of p53 in this context is the induction cell cycle arrest, and possibly, senescence.
As the sequential mutagenesis experiments suggested that initially targeted cells lose the capacity for full transformation even after subsequent p53 deletion, this cell cycle arrest must result either in clearance of affected cells and/or be relatively stable and possibly irreversible. To gain further insight into the fate of K-rasG12D-expressing cells, we pursued a cell marking strategy. Administering lentiviruses encoding Cre and GFP allowed us to mark infected cells using immunohistochemistry (IHC) for GFP. As a control, lentiviral infection of K-rasLSL-G12D/+; p53flox/flox; Rag2−/− animals efficiently generated sarcomas that stained for GFP (Supplemental Fig 2). Interestingly, an examination of infected muscles before macroscopic tumor formation revealed regions of GFP+ cells that increased in size over time, most likely corresponding to incipient tumor lesions (Supplemental Fig 2). However, while we could readily detect infected cells in K-rasLSL-G12D/+; p53flox/+; Rag2−/− mice, they never appeared to expand into small lesions, similar to what was seen in K-ras+/+ muscles (Fig 4D). Importantly, many infected cells were present at time points at which p53 deletion was unable to efficiently promote sarcomagenesis in the sequential mutagenesis experiments (Fig 3B and Fig 4D). At present, we have been unable to detect any markers of senescence in these infected cells (data not shown). Although we cannot rule out the possibility that some cells are cleared from the animals, the continued presence of GFP+ cells in these animals might suggest that p53 engages an irreversible cell cycle arrest to potently suppress transformation in muscle cells.
DISCUSSION
The immediate and robust p53-dependent response in muscle cells harboring an oncogenic Ras mutation might partially explain the low frequency of STSs compared to many other malignancies in humans. Interestingly, the incidence of STSs is dramatically elevated in Li-Fraumani patients inheriting a germ-line mutation of p53 (10), indicating that pre-existing deficiencies in p53 function allow for muscle cell transformation, which is otherwise a rare occurrence. Moreover, somatic p53 mutations have been proposed to be relatively early events in other forms of sarcomas (27), further suggesting that muscle cells must overcome an early p53-dependent blockade during tumor initiation. This potent p53-dependent tumor suppression most likely selects for a particular sequence of events in which mutation of p53 occurs very early during spontaneous sarcomagenesis. Because p53 alterations rarely provide an immediate growth advantage in otherwise normal cells, this is not conducive to efficient tumor initiation.
Several recent reports have analyzed p53 function during the early stages of tumorigenesis, although the identity of the cooperating oncogenic event(s) and its precise temporal relationship to p53 manipulation were unknown. In one study, Evans and colleagues demonstrated robust tumor suppression after a small window of p53 activity occurring shortly after irradiation (IR)-induced tumor initiation (28). In a separate study, deleting p53 at different time points after IR revealed that the continued presence of p53 was required to prevent IR-induced tumorigenesis (29). In fact, once p53 was removed, tumor formation occurred with the same kinetics as seen in IR-treated p53−/− animals. Apparently, in this context p53 simply blocked the overt transformation of pre-neoplastic cells in a manner that was completely reversible once p53 was deleted. This is in stark contrast to our observations involving K-rasG12D-expressing muscle cells, where secondary p53 deletion failed to fully recover the tumorigenic capacity of mutated cells. A further understanding of the factors governing p53’s potent tumor suppression in the muscle might guide therapeutic strategies aimed at generating such a strong and irreversible response in more resistant tissues.
Supplementary Material
Acknowledgments
We thank many members of the Jacks lab for helpful advice throughout the course of this project. This work was supported in part y grant 5-U01-CA84306 from the National Institutes of Health, and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. T.J. is a Howard Hughes Medical Institute Investigator and a Daniel K. Ludwig Scholar.
References
- 1.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–58. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 4.Vooijs M, van der Valk M, te Riele H, Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- 5.Awatramani R, Soriano P, Mai JJ, Dymecki S. An Flp indicator mouse expressing alkaline phosphatase from the ROSA26 locus. Nat Genet. 2001;29:257–9. doi: 10.1038/ng1101-257. [DOI] [PubMed] [Google Scholar]
- 6.Zilfou JT, Lowe SW. Tumor Suppressive Functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–22. [PubMed] [Google Scholar]
- 8.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleihues P, Schauble B, zur Hausen A, Esteve J, Ohgaki H. Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol. 1997;150:1–13. [PMC free article] [PubMed] [Google Scholar]
- 11.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9:831–41. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 12.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 13.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–87. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 14.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–62. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 15.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 19.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–7. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 21.Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci USA. 2003;100:15930–5. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 23.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–7. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 24.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 25.Efeyan A, Collado M, Velasco-Miguel S, Serrano M. Genetic dissection of the role of p21Cip1/Waf1 in p53-mediated tumour suppression. Oncogene. 2007;26:1645–49. doi: 10.1038/sj.onc.1209972. [DOI] [PubMed] [Google Scholar]
- 26.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci USA. 2010;107:10184–9. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taubert H, Meye A, Wurl P. Soft tissue sarcomas and p53 mutations. Mol Med. 1998;4:365–72. [PMC free article] [PubMed] [Google Scholar]
- 28.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–7. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 29.Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One. 2009;4:e6654. doi: 10.1371/journal.pone.0006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.