Abstract
Background and Aims
Recent studies indicate that hepatitis C virus (HCV) can modulate the expression of various genes including those involved in interferon signaling, and up-regulation of interferon-stimulated genes by HCV was reported to be strongly associated with treatment outcome. To expand our understanding of the molecular mechanism underlying treatment resistance, we analyzed the direct effects of interferon and/or HCV infection under immunodeficient conditions using cDNA microarray analysis of human hepatocyte chimeric mice.
Methods
Human serum containing HCV genotype 1b was injected into human hepatocyte chimeric mice. IFN-α was administered 8 weeks after inoculation, and 6 hours later human hepatocytes in the mouse livers were collected for microarray analysis.
Results
HCV infection induced a more than 3-fold change in the expression of 181 genes, especially genes related to Organismal Injury and Abnormalities, such as fibrosis or injury of the liver (P = 5.90E-16 ∼ 3.66E-03). IFN administration induced more than 3-fold up-regulation in the expression of 152 genes. Marked induction was observed in the anti-fibrotic chemokines such as CXCL9, suggesting that IFN treatment might lead not only to HCV eradication but also prevention and repair of liver fibrosis. HCV infection appeared to suppress interferon signaling via significant reduction in interferon-induced gene expression in several genes of the IFN signaling pathway, including Mx1, STAT1, and several members of the CXCL and IFI families (P = 6.0E-12). Genes associated with Antimicrobial Response and Inflammatory Response were also significantly repressed (P = 5.22×10−10 ∼ 1.95×10−2).
Conclusions
These results provide molecular insights into possible mechanisms used by HCV to evade innate immune responses, as well as novel therapeutic targets and a potential new indication for interferon therapy.
Introduction
Chronic hepatitis C virus (HCV) infection is one of the most serious global health threats, affecting more than 170 million people worldwide [1]–[3]. Interferon is administered to chronic hepatitis C patients to attempt to eradicate the virus and to prevent the development of advanced liver diseases such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC), with limited success. While the overall eradication rate of HCV has improved since the introduction of pegylated-interferon (PEG-IFN) and ribavirin (RBV) combination therapy, the sustained viral response (SVR) rate of genotype 1b with high viral load still remains only 40–50% [4]–[6]. Viral and host factors, such as HCV RNA titer, viral substitutions in HCV core or NS5A region, age, gender, liver fibrosis, and SNPs in IL-28B locus, are significantly associated with the effects of PEG-IFN and RBV combination therapy [7]–[15], but the precise molecular mechanisms remained unclear.
Recently, some HCV-related structural as well as non-structural proteins have been reported to be associated with host proteins and affect innate immunity or lipid metabolism. RIG-I (retinoic acid inducible gene I) and Mda5 (melanoma differentiation-associated gene 5) are known to activate the type I interferon signaling pathway by interacting with adaptor protein IPS-1/MAVS/VISA/Cardif [16]–[18]. In the presence of HCV infection, the viral non-structural protein NS3/4A, which has serine protease activity, can cleave and inactivate IPS-1 [19]. TLR (Toll like receptor) is a sensor of RNA or DNA and is known to play various roles in viral infection. Abe et al. demonstrated that HCV non-structural protein NS5A inhibits the recruitment of interleukin-1 receptor-associated kinase 1 by interacting with MyD88 and impairs cytokine production in response to TLR ligands [20]. HCV core protein is also known to interact with host proteins. The core protein promotes hepatic steatosis, insulin resistance and hepatocarcinogenesis through activation of host proteins such as PPARα and MAPK [21]–[26]. However, these reports were based on in vitro analysis of cell lines or used human liver tissues in which results were complicated by adaptive immune responses, and it has been difficult to evaluate the direct impact of HCV infection and interferon administration on human hepatocytes.
Mercer and colleagues developed a human hepatocyte chimeric mouse [27] derived from the severely immunocompromised SCID mouse, in which mouse liver cells were extensively replaced with human hepatocytes [27], [28]. This mouse model facilitates continuous HCV infection and makes it possible to analyze the effects of drugs and viral infection on human hepatocytes under immunodeficient conditions [29], [30]. To analyze the putative effects of HCV infection or IFN administration without the adaptive immune response, we constructed an HCV carrier mouse model using the human hepatocyte chimeric mouse and performed cDNA microarray analysis using human hepatocytes dissected from the mouse livers. The results are intended to reflect the direct impacts of HCV infection and IFN administration on human hepatocytes and may help in elucidating HCV immune evasion mechanisms.
Materials and Methods
Human Serum Samples
Serum samples were obtained from HCV carriers after obtaining written informed consent for the donation and evaluation of blood samples. Inocula contained high viral loads of genotype 1b HCV RNA (6.9 log copies/ml). The experimental protocol met the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Hiroshima University Ethical Committee.
Human Hepatocyte Chimeric Mice Experiments
The uPA+/+/SCID+/+ mice and transplantation of human hepatocytes were performed as described previously [28]. All mice were transplanted with hepatocytes from the same donor. Human hepatocyte chimeric mice, in which liver cells were largely (>90%) replaced with human hepatocytes, were used to reduce potential influence by mouse-derived mRNA. The experiments were performed in accordance with the guidelines of the local committee for animal experiments at Hiroshima University.
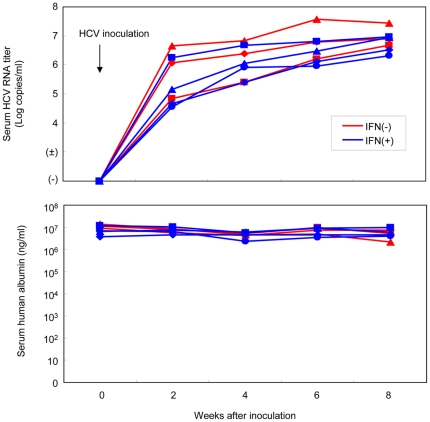
A total of 15 chimeric mice were prepared and assigned to four experimental groups. Group A contained four mice that were neither infected with HCV nor treated with IFN. Group B consisted of four uninfected mice that were administered IFN-α (7,000 IU/g body weight) 6 h before sacrifice. Groups C and D were inoculated via the mouse tail vein with human serum containing 4×105 copies of HCV particles, and Group D was administered INF- α at the same time as Group B. After inoculation, we collected mouse sera every two weeks and analyzed serum HCV RNA levels by real time PCR. All seven mice developed measurable viremia 4 weeks after inoculation. The levels of the virus titer reached over 6 Log10 copies/ml 8 weeks after inoculation (Figure 1). Conversely, serum human albumin levels remained more than 2×106 ng/ml in each mouse during 6 weeks after inoculation (Figure 1). Eight weeks after inoculation, when serum HCV RNA levels had plateaued, IFN-α (7,000 IU/g body weight) was administered to the four mice in Group D as well as the four uninfected mice in Group B. Six hours after IFN administration all 15 mice were sacrificed. Infection, extraction of serum samples, and sacrifice were performed under ether anesthesia as described previously [29]–[31]. Human albumin levels in mouse serum were measured with a Human Albumin enzyme-linked immunosorbent assay (ELISA) Quantitation kit (Bethyl Laboratories Inc., Montgomery, TX) according to the instructions provided by the manufacturer. Serum samples obtained from mice were aliquoted and stored in liquid nitrogen until use.
Figure 1. Change in HCV titers and human albumin levels in mouse serum.
HCV RNA titers (upper panel) and human albumin levels (lower panel) in chimeric mouse sera after inoculation are shown. The horizontal axis indicates weeks after inoculation. Mouse sera were collected every two weeks after inoculation, and serum HCV RNA and human albumin levels were measured. Results were similar for all mice.
Analysis of HCV markers
For quantitative analysis of HCV RNA, 10 µl samples of mouse serum were used. Total RNA was extracted using Sepa Gene RV-R (Sanko Junyaku Co., Ltd., Tokyo, Japan) and dissolved with 8.8 µL of RNase free water and reverse transcribed (RT). RT reactions were performed with 20 µl of the reaction mixtures, containing random primer (Takara Bio Inc., Shiga, Japan), RT buffer and M-MLV reverse transcriptase (ReverTra Ace, TOYOBO Co., Osaka, Japan) according to the instructions provided by the manufacturer. After the RT reaction, HCV RNA was quantified by real-time PCR using the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplification was performed as described previously [29], [30]. The lower detection limit of this assay is 300 copies. For detection of small amounts of HCV RNA, we also performed nested PCR. Amplification conditions were as described previously [29], [30].
Dissection of mouse livers and total RNA extraction from human heptocytes in the mouse livers
All 15 chimeric mice were sacrificed by anesthesia with diethyl ether. Human hepatocytes were finely dissected from mouse livers, submerged in RNA later® solution (Applied Biosystems), and stored in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Mini Kit according to the manufacturer protocol (Qiagen Inc., Valencia, CA). RNA quality was assessed using ultraviolet absorption at 260 nm/280 nm (NanoDrop Technologies, Wilmington, DE) and agarose gel electrophoresis. Microarray analysis was performed using the Affymetrix GeneChip Human Gene U133Plus2.0 Array, which interrogates 38,500 genes across 54,675 distinct probes (Affymetrix, Santa Clara, CA). The Affymetrix GeneChip Whole Transcript Sense Target Labeling Assay Manual Version 4 was used for complementary DNA (cDNA) generation, hybridization, and array processing. Briefly, 300 ng of total RNA underwent first-strand and second-strand cDNA synthesis. Complementary RNA was generated and used to produce sense-strand cDNA, which was fragmented and end-labeled with biotin. Biotin-labeled cDNA was hybridized to the Human Gene 1.0 ST Array for 16 hours at 45°C using the GeneChip Hybridization Oven 640 (Affymetrix). Washing and staining with streptavidin-phycoerythrin was performed using the GeneChip Fluidics Station 450, and images were acquired using the Affymetrix Scanner 3000 (Affymetrix).
Microarray Data Analysis and Hierarchical Clustering
Fluorescence intensities captured by the Affymetrix GeneChip Scanner were converted to numerical values using the Affymetrix GeneChip Operating Software, were log2 transformed, and were standardized using quantile normalization with the Robust Multiarray Analysis (RMA) algorithm [32], [33]; this method normalizes the distribution of probe intensities for all the gene arrays in a given set.
Obtained gene expression profiles were analyzed using GeneSpring GX 10.0.2 software (Tomy Digital Biology, Tokyo, Japan). Expression ratios were calculated and normalized per chip to the 50th percentile and finally normalized per gene to medians. We worked on a pre-screened list of 32,885 probes obtained after filtering the data for outliers, negative and positive controls, and on the quality flag Cy3 signals being “well above background.” To pass this last flag, Cy3 net signals needed to be positive and significant, with g(r)BGSubSignal greater than 2.6 g(r) BG_SD. To determine if there were genes differentially expressed among samples, we performed two Welch's t-tests (P<0.01) on this prescreened list of genes: one without correction and one with Benjamini and Hochberg's correction. Complete linkage hierarchical clustering analysis was applied using Euclidean distance, and differentially expressed genes were annotated using the information from the Gene Ontology Consortium. Global molecular networks and comparisons of canonical pathways were generated using Ingenuity™ Pathway Analysis 8.6 (Ingenuity™ Systems, CA, USA).
Real time PCR for analyzing the mRNA expression in the human hepatocytes
Total RNA was extracted from the implanted human hepatocytes in the mouse livers using RNeasy Mini Kit (Qiagen) and reverse-transcribed using ReverTra Ace (TOYOBO, Osaka, Japan) with random primer in accordance with the instructions supplied by the manufacturer. The selected cDNA were quantified by real-time PCR using the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA), and the expression of GAPDH served as a control. Amplification was performed in a 25 µl reaction mixture containing 12.5 µl SYBR Green PCR Master Mix (Applied Biosystems), 5 pmol of forward primer, 5 pmol of reverse primer, and 1 µl of cDNA solution. After incubation for 2 min at 50°C, the sample was denatured for 10 min at 95°C, followed by a PCR cycling program consisting of 40 cycles of 15 s at 95°C, 30 s at 55°C, and 60 s at 60°C.
Statistical analysis
Differences between groups were examined for statistical significance using the Student's t- test.
Results
Change of gene expression with HCV infection
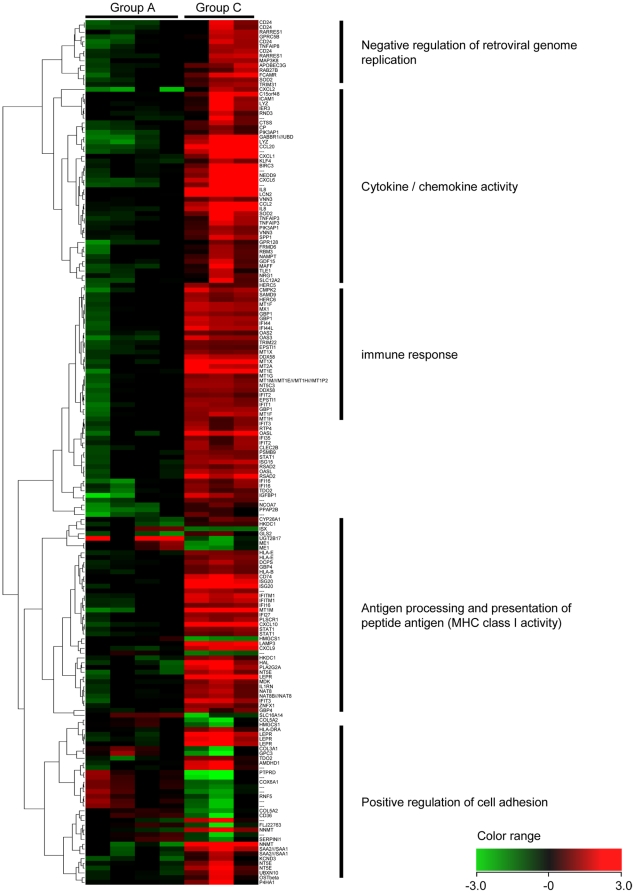
To analyze the effect of HCV infection on gene expression in human hepatocytes, we compared the gene expression profiles between Group A (without HCV infection) and Group C (with HCV infection). Among the 2,519 genes that remained significant after screening by Welch's t-test, more than 3.0-fold expression changes between groups were observed in 181 genes. 157 of these 181 genes were up-regulated following HCV infection, and the other 24 were down-regulated. Cluster analysis of the 181 genes is shown in Figure 2, and the top 20 up-/down-regulated genes by HCV infection are listed in Tables 1 and 2, respectively.
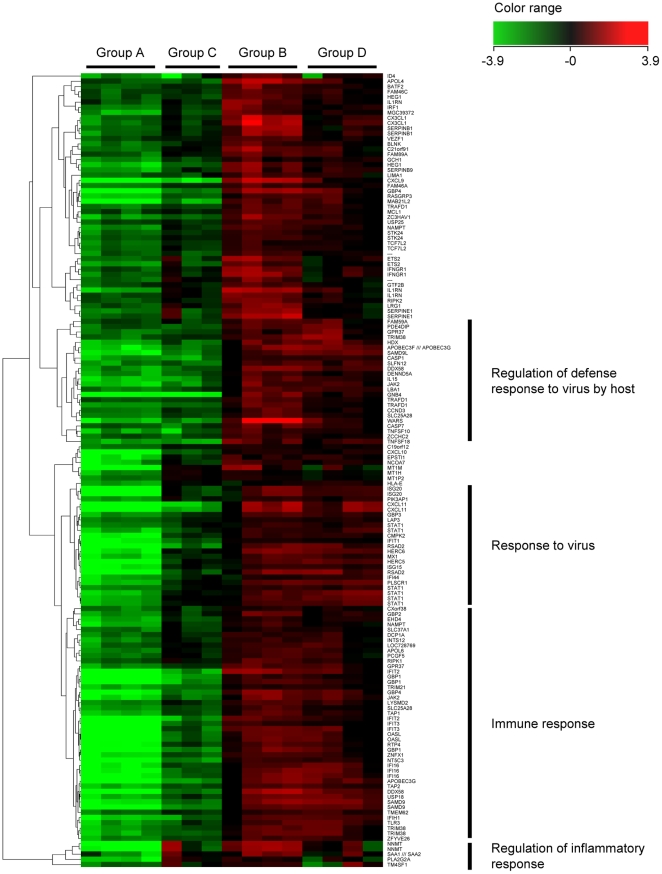
Figure 2. Hierarchical clustering analysis of 181 genes associated with HCV infection.
To analyze the influence of HCV infection on human hepatocytes, clustering analysis on gene expression was performed between Group A (without HCV infection; 4 columns on the left side) and Group C (with HCV infection; 3 columns on the right side). 157 genes were up-regulated following HCV infection, including interferon-stimulated genes (ISGs) such as MX1 and genes in the CXCL and IFI families, and 24 genes were down-regulated, including ME1 and HMGCS1.
Table 1. The top 20 genes up-regulated with HCV infection.
| Probe set | Unigene code | Gene symbol | Fold change | P value |
| 202237_at | Hs.503911 | NNMT | 33.16 | 1.66E-03 |
| 205476_at | Hs.75498 | CCL20 | 30.23 | 1.59E-04 |
| 202859_x_at | Hs.551925 | IL8 | 30.16 | 4.42E-04 |
| 206336_at | Hs.164021 | CXCL6 | 25.52 | 1.86E-03 |
| 217546_at | Hs.647370 | MT1M | 24.69 | 2.46E-04 |
| 212531_at | Hs.204238 | LCN2 | 24.17 | 9.19E-04 |
| 209894_at | Hs.705413 | LEPR | 23.77 | 5.83E-04 |
| 204533_at | Hs.632586 | CXCL10 | 23.61 | 1.47E-05 |
| 213797_at | Hs.17518 | RSAD2 | 20.43 | 7.31E-05 |
| 204439_at | Hs.715563 | IFI44L | 17.92 | 9.73E-04 |
| 213975_s_at | Hs.706744 | LYZ | 15.22 | 1.10E-03 |
| 206643_at | Hs.190783 | HAL | 14.88 | 3.98E-03 |
| 216598_s_at | Hs.303649 | CCL2 | 14.76 | 6.99E-03 |
| 235229_at | Hs.332649 | 13.93 | 6.22E-04 | |
| 205890_s_at | Hs.714406 | GABBR1///UBD | 13.67 | 1.46E-03 |
| 33304_at | Hs.459265 | ISG20 | 13.58 | 5.61E-05 |
| 205569_at | Hs.518448 | LAMP3 | 10.96 | 3.58E-05 |
| 204470_at | Hs.789 | CXCL1 | 10.90 | 9.06E-03 |
| 208607_s_at | Hs.632144 | SAA1///SAA2 | 10.40 | 4.56E-03 |
| 205302_at | Hs.642938 | IGFBP1 | 9.55 | 1.10E-02 |
Table 2. The top 20 genes down-regulated with HCV infection.
| Probe set | Unigene code | Gene symbol | Fold change | P value |
| 207245_at | Hs.575083 | UGT2B17 | 20.04 | 2.15E-02 |
| 214043_at | Hs.446083 | PTPRD | 6.81 | 2.57E-02 |
| 214416_at | Hs.702961 | 6.36 | 3.51E-02 | |
| 209220_at | Hs.713537 | GPC3 | 5.40 | 4.40E-02 |
| 238029_s_at | Hs.504317 | SLC16A14 | 4.90 | 1.16E-02 |
| 231594_at | 4.59 | 1.87E-02 | ||
| 1556824_at | Hs.702604 | 4.40 | 2.89E-02 | |
| 232707_at | Hs.567637 | ISX | 4.30 | 6.52E-03 |
| 205822_s_at | Hs.397729 | HMGCS1 | 4.23 | 1.95E-04 |
| 204058_at | Hs.21160 | ME1 | 4.06 | 2.15E-02 |
| 1555084_at | 3.95 | 4.10E-02 | ||
| 215076_s_at | Hs.443625 | COL3A1 | 3.92 | 2.99E-02 |
| 209555_s_at | Hs.120949 | CD36 | 3.91 | 4.49E-02 |
| 221729_at | Hs.445827 | COL5A2 | 3.89 | 2.60E-02 |
| 217676_at | Hs.696837 | 3.86 | 8.73E-03 | |
| 233604_at | Hs.280892 | FLJ22763 | 3.82 | 2.44E-02 |
| 1563298_at | Hs.352254 | 3.64 | 1.97E-02 | |
| 224344_at | Hs.497118 | COX6A1 | 3.34 | 1.68E-02 |
| 237031_at | Hs.146276 | 3.22 | 4.62E-04 | |
| 216018_at | Hs.534342 | RNF5 | 3.19 | 3.13E-02 |
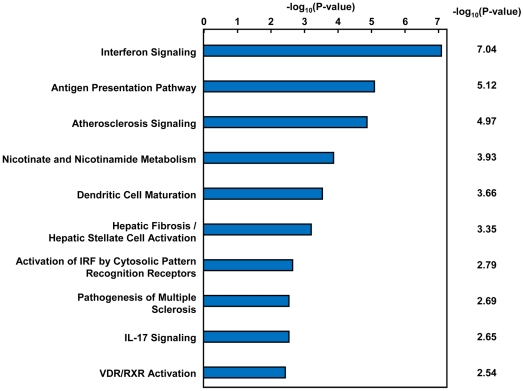
It is well known that chronic HCV infection triggers multiple biological responses. To analyze biological significance and regulatory pathways involved in the changes observed, we performed network analysis with the 181 genes using Ingenuity™ Pathway Analysis (IPA). As shown in Table 3, most of the 181 genes (e.g. CXCL9, CXCL10, IFIT3 and Mx1, which are well known interferon-stimulated genes (ISGs)) belonged to categories such as Organismal Injury and Abnormalities, Inflammatory Response, and Cell-To-Cell Signaling and Interaction. Through canonical pathway analysis of the 181 genes using Ingenuity Pathways Analysis, 10 canonical pathways significantly affected by HCV infection were identified, with interferon signaling as the most significant (Figure 3). These results indicate that the intra-hepatic innate immune response was strongly activated by HCV infection in human hepatocytes.
Table 3. The effect of HCV infection on biological functions by category.
| Category | P value | Up-regulated genes in network | Down-regulated genes in network | ||
| Number of genes | Representative genes | Number of genes | Representative genes | ||
| Organismal Injury and Abnormalities | 5.90E-16–3.66E-03 | 27 | CXCL1, CXCL6, CXCL9, CXCL10, IFIT1, IFIT3, MX1, etc. | 1 | SERPINI1 |
| Cancer | 1.81E-13–5.73E-03 | 54 | BIRC3, CXCL9, CXCL10, GBP1, IFIT3, IGFBP1, ISG20, MAP3K8, etc. | 4 | CD36, COL3A1, GPC3, RNF5 |
| Inflammatory Response | 9.31E-13–5.89E-03 | 39 | APOBEC3G, CCL2, CXCL9, CXCL10, IL8, MX1, STAT1, TRIM22, etc | 2 | CD36, COL3A1 |
| Cell-To-Cell Signaling and Interaction | 4.95E-10–4.99E-03 | 30 | CCL2, CD74, CXCL1, CXCL2, CXCL9, ICAM1, IL8, NRG1, STAT1, etc. | 3 | CD36, SERPINI1, GPC3 |
| Hematological System Development and Function | 4.95E-10–5.95E-03 | 36 | CCL2, CCL20, CXCL9, CXCL10, IL8, IL1RN, TNFAIP3, etc | 1 | CD36 |
| Immune Cell Trafficking | 4.95E-10–5.73E-03 | 26 | CCL2, CCL20, CTSS, CXCL6, CXCL9, CXCL10, MDK, NEDD9, etc. | 1 | CD36 |
| Infection Mechanism | 5.03E-10–3.66E-03 | 16 | CCL2, CXCL9, CXCL10, DDX58, IFIT1, IL8, ISG20, MX1, RSAD2, STAT1, etc. | 0 | |
| Infectious Disease | 5.03E-10–5.46E-03 | 26 | APOBEC3G, CXCL9, CXCL10, DDX58, MT1X, STAT1, TNFAIP3, etc | 2 | CD36, HMGCS1 |
| Reproductive System Disease | 6.43E-10–1.37E-03 | 42 | CCL2, CXCL1, CXCL2, IFIT1, IGFBP1, KLF4, MAP3K8, NEDD9, SPP1, etc. | 1 | RNF5 |
| Cellular Movement | 6.64E-10–5.91E-03 | 31 | IGFBP1, IL8, KLF4, MDK, NEDD9, NRG1, RARRES1, SOD2, TNFAIP8, etc | 2 | CD36, RNF5 |
Figure 3. The effects of HCV infection on canonical pathways.
To analyze the effects of HCV infection on canonical pathways, pathway analysis was performed using the 181 genes identified to be significantly up- or down-regulated following HCV infection. The IFN signaling pathway was the most significantly affected by HCV infection. Statistical analysis was performed using Fisher's exact test.
Change of gene expression with interferon treatment
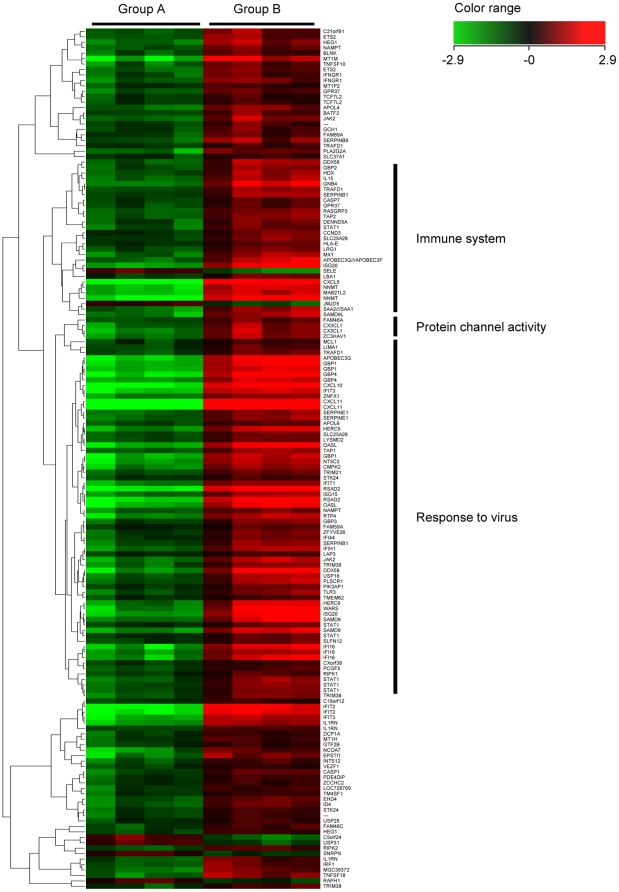
To analyze the direct effects of IFN in human hepatocytes, we compared gene expression profiles between Group A (without IFN treatment) and Group B (with IFN treatment). Out of the 218 genes that remained significant after screening by Welch's t-tests and Benjamini-Hochberg correction for multiple testing, 158 had a greater than 3.0-fold change between groups. 152 of the 158 genes were up-regulated following IFN administration, and the other 6 were down-regulated. Cluster analysis of the 158 selected genes is shown in Figure 4. The top 35 up-regulated genes (>10.0-fold changes), which include many well-known ISGs (e.g., members of the CXCL and IFI families), and the 6 down-regulated genes are listed in Tables 4 and 5, respectively.
Figure 4. Hierarchical clustering analysis of 158 genes associated with IFN treatment.
To analyze the effects of IFN in human hepatocytes, clustering analysis was performed between Group A (without IFN treatment; 4 columns on the left side) and Group B (with IFN treatment; 4 columns on the right side). 152 genes were up-regulated, and 6 genes were down-regulated following IFN treatment. Several well-known interferon-stimulated genes (ISGs), including CXCL9, Mx1, ISG20 and OASL, were among the up-regulated genes.
Table 4. The top 35 genes up-regulated with IFN treatment.
| Probe set | Unigene code | Gene symbol | Fold change | P values |
| 211122_s_at | Hs.632592 | CXCL11 | 482.47 | 1.30E-06 |
| 203915_at | Hs.77367 | CXCL9 | 216.26 | 1.35E-07 |
| 242625_at | Hs.17518 | RSAD2 | 101.24 | 1.26E-05 |
| 202237_at | Hs.503911 | NNMT | 86.80 | 6.52E-06 |
| 217502_at | Hs.437609 | IFIT2 | 75.05 | 1.73E-06 |
| 204533_at | Hs.632586 | CXCL10 | 67.43 | 2.72E-07 |
| 217546_at | Hs.647370 | MT1M | 46.69 | 1.12E-04 |
| 235175_at | Hs.409925 | GBP4 | 44.94 | 1.03E-06 |
| 204205_at | Hs.660143 | APOBEC3G | 43.39 | 5.55E-06 |
| 204747_at | Hs.714337 | IFIT3 | 32.73 | 1.17E-06 |
| 218943_s_at | Hs.190622 | DDX58 | 32.19 | 1.44E-04 |
| 33304_at | Hs.459265 | ISG20 | 31.97 | 3.94E-05 |
| 202269_x_at | Hs.62661 | GBP1 | 31.73 | 5.30E-06 |
| 210797_s_at | Hs.118633 | OASL | 31.59 | 9.21E-06 |
| 200629_at | Hs.497599 | WARS | 29.65 | 2.28E-04 |
| 206332_s_at | Hs.380250 | IFI16 | 26.37 | 3.80E-05 |
| 210302_s_at | Hs.584852 | MAB21L2 | 24.31 | 5.31E-07 |
| 228531_at | Hs.65641 | SAMD9 | 18.65 | 1.45E-05 |
| 223298_s_at | Hs.487933 | NT5C3 | 17.48 | 6.65E-06 |
| 219863_at | Hs.26663 | HERC5 | 17.02 | 2.29E-05 |
| 225710_at | Hs.173030 | GNB4 | 16.98 | 3.30E-05 |
| 219684_at | Hs.43388 | RTP4 | 16.38 | 2.55E-05 |
| 212657_s_at | Hs.81134 | IL1RN | 15.18 | 1.33E-06 |
| 219352_at | Hs.529317 | HERC6 | 14.86 | 1.31E-04 |
| 226702_at | Hs.7155 | CMPK2 | 12.50 | 1.86E-05 |
| 205842_s_at | Hs.656213 | JAK2 | 12.49 | 6.16E-05 |
| 230036_at | Hs.489118 | SAMD9L | 11.98 | 7.84E-05 |
| 214995_s_at | Hs.660143 | APOBEC3F///APOBEC3G | 11.62 | 1.58E-04 |
| 823_at | Hs.531668 | CX3CL1 | 11.15 | 1.07E-04 |
| 203153_at | Hs.20315 | IFIT1 | 10.84 | 5.93E-06 |
| 225076_s_at | Hs.371794 | ZNFX1 | 10.40 | 1.38E-06 |
| 213069_at | Hs.477420 | HEG1 | 10.37 | 3.52E-05 |
| 205483_s_at | Hs.458485 | ISG15 | 10.34 | 1.29E-05 |
| 235276_at | Hs.546467 | EPSTI1 | 10.21 | 2.10E-04 |
| 219209_at | Hs.163173 | IFIH1 | 10.05 | 4.06E-05 |
Table 5. The top 6 genes down-regulated with IFN treatment.
| Probe set | Unigene code | Gene symbol | Fold change | P value |
| 206211_at | SELE | 5.83 | 6.11E-05 | |
| 224875_at | C5orf24 | 5.46 | 1.11E-04 | |
| 227256_at | Hs.183817 | USP31 | 3.94 | 7.27E-05 |
| 220070_at | Hs.145717 | JMJD5 | 3.87 | 5.04E-05 |
| 1552482_at | Hs.471162 | RAPH1 | 3.31 | 1.73E-04 |
| 226587_at | Hs.592473 | SNRPN | 3.17 | 6.13E-05 |
The effect of HCV infection on IFN response
To analyze the effect of HCV infection on IFN response, we focused on the 152 genes that were up-regulated following IFN administration and compared gene expression ratios between Groups A and B (gene expression changes by IFN without HCV infection) and between Groups C and D (gene expression changes by IFN with HCV infection). In 69.7% (106/152) of the IFN-induced genes, IFN responsiveness was significantly reduced following HCV infection (Figure 5). The top 20 genes are shown in Table 6. Although viral titers differed among mice, we found no correlation between IFN responsiveness and HCV RNA titer. We performed pathway analysis to identify significant associations with canonical pathways, and the top 5 associated pathways are shown in Table 7. IFN responsiveness was significantly reduced following HCV infection in several canonical pathways, and the IFN signaling pathway, in particular, was strongly associated. To verify the effects of HCV infection and/or IFN treatment on gene expression, signal intensities of genes involved in the IFN and JAK-STAT signaling pathways were analyzed. As shown in Figure 6A, among 28 representative genes in the IFN signaling pathway, signal intensities of 22 genes could be analyzed through cDNA microarray analysis. In all genes except IFNAR1, expression was up-regulated following HCV infection, whereas IFN responsiveness was suppressed as a result of HCV infection (Figure 6B). 16 out of 22 genes in the JAK-STAT signal pathway could be analyzed via cDNA microarray analysis (Figure 6C), and 12 of the 16 genes were up-regulated following HCV infection, whereas IFN responsiveness was suppressed in 9 genes (Figure 6D).
Figure 5. Hierarchical clustering analysis of 152 genes associated with IFN administration with or without HCV infection.
To analyze the effect of HCV infection on IFN response, gene expression ratios between Groups A and B (gene expression changes by IFN without HCV infection) and those between Groups C and D (gene expression changes by IFN with HCV infection) were compared in the 152 IFN-induced genes. 69.7% of the selected genes showed reduced IFN responsiveness following HCV infection.
Table 6. The top 20 genes in which IFN-induced up-regulation is inhibited following HCV infection.
| Probe Set ID | Gene symbol | Fold change | P value | |
| HCV infection (-) | HCV infection (+) | |||
| 235175_at | GBP4 | 44.94 | 5.50 | 2.93E-07 |
| 231577_s_at | GBP1 | 24.60 | 4.89 | 6.15E-07 |
| 218943_s_at | DDX58 | 32.19 | 5.56 | 1.26E-05 |
| 226702_at | CMPK2 | 12.50 | 2.13 | 2.35E-05 |
| 225973_at | TAP2 | 7.07 | 2.84 | 3.31E-05 |
| 229450_at | IFIT3 | 20.66 | 2.74 | 6.37E-05 |
| 217739_s_at | NAMPT | 5.91 | 1.95 | 6.51E-05 |
| 213797_at | RSAD2 | 69.70 | 4.50 | 7.01E-05 |
| 210797_s_at | OASL | 31.59 | 3.53 | 1.02E-04 |
| 218508_at | DCP1A | 3.56 | 1.65 | 1.36E-04 |
| 228531_at | SAMD9 | 18.65 | 5.87 | 1.45E-04 |
| 204804_at | TRIM21 | 5.37 | 2.69 | 1.47E-04 |
| 219209_at | IFIH1 | 10.05 | 4.69 | 1.67E-04 |
| 219684_at | RTP4 | 16.38 | 2.55 | 1.98E-04 |
| 239186_at | MGC39372 | 7.61 | 2.57 | 2.27E-04 |
| 219211_at | USP18 | 8.72 | 3.74 | 2.40E-04 |
| 225076_s_at | ZNFX1 | 10.40 | 2.91 | 2.91E-04 |
| 204698_at | ISG20 | 31.29 | 3.33 | 3.00E-04 |
| 223192_at | SLC25A28 | 5.05 | 2.20 | 3.25E-04 |
| 228439_at | BATF2 | 4.59 | 1.83 | 3.28E-04 |
Table 7. The top 5 canonical pathways associated with the 106 genes in which IFN response is suppressed by HCV infection.
| Category | P value | Ratio | Associated genes |
| Interferon Signaling | 4.41E-11 | 7/30 genes | IFIT1, IFIT3, IFNGR1, IRF1, MX1, STAT1, TAP1 |
| Type I Diabetes Mellitus Signaling | 1.08E-04 | 5/119 genes | HLA-E, IFNGR1, IRF1, RIPK1, STAT1 |
| Antigen Presentation Pathway | 5.29E-04 | 3/39 genes | HLA-E, TAP1, TAP2 |
| Primary Immunodeficiency Signaling | 1.64E-03 | 3/63 genes | BLNK, TAP1, TAP2 |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 2.95E-03 | 3/73 genes | ISG15, RIPK1, STAT1 |
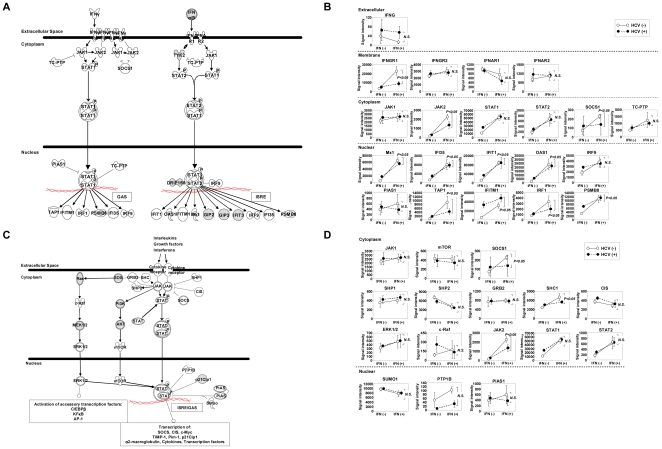
Figure 6. Changes in expression of genes in the IFN and JAK-STAT signaling pathways under HCV infection and/or IFN administration.
A) An overview of the IFN signaling pathway consisting of 26 representative genes is shown. Genes illustrated as gray shapes were not included in this study. B) Relative expression levels of genes with/without HCV infection and/or IFN administration were plotted (closed dots: with HCV infection; open dots: without HCV infection) using microarray data. The slopes of the dashed and solid lines represent IFN responsiveness with and without HCV infection, respectively. In 21 of the 22 examined genes in the IFN signaling pathway, signal intensities increased and IFN responsiveness was repressed following HCV infection. Student's t-test was used for statistical analysis. C) An overview of the JAK-STAT signaling pathway consisting of 22 representative gene products is shown. Genes illustrated as gray shapes were not included in this study. D) Relative expression levels of genes with/without HCV infection and/or IFN administration were plotted using microarray data (closed dots: with HCV infection; open dots: without HCV infection). Signal intensities increased following HCV infection in 16 of 22 genes in the JAK-STAT signaling pathway, and IFN response was suppressed in 9 genes. Statistical analysis was performed using Student's t-test.
On the other hand, only 33 genes (21.7%), including several ISGs, such as GBP1, GBP4 and IFIT3, remained responsive to IFN in the presence of HCV and were expressed more than 3.0-fold higher in Group D compared to Group C (Table 8). Pathway analysis indicated that these 33 genes were significantly associated with Antimicrobial Response and Inflammatory Response (P = 5.22×10−10 ∼ 1.95×10−2). Changes in mRNA expression for 29 down-regulated genes, including ISG20, WARS, Mx1, CXCL10, IFNGR1 and IFITM1 were verified by real time PCR (data not shown).
Table 8. The 33 genes that remained more than 3-fold up-regulated following IFN treatment in HCV-infected mice.
| ID | Symbol | Location | Type(s) |
| 214995_s_at | APOBEC3F | unknown | enzyme |
| 204205_at | APOBEC3G | Nucleus | enzyme |
| 206011_at | CASP1 | Cytoplasm | peptidase |
| 204533_at | CXCL10 | Extracellular Space | cytokine |
| 210163_at | CXCL11 | Extracellular Space | cytokine |
| 203915_at | CXCL9 | Extracellular Space | cytokine |
| 218943_s_at | DDX58 | Cytoplasm | enzyme |
| 231577_s_at | GBP1 (includes EG:2633) | Cytoplasm | enzyme |
| 235175_at | GBP4 (includes EG:115361) | Cytoplasm | enzyme |
| 225710_at | GNB4 | Plasma Membrane | enzyme |
| 1553646_at | HDX | unknown | other |
| 213069_at | HEG1 | unknown | other |
| 206332_s_at | IFI16 | Nucleus | transcription regulator |
| 219209_at | IFIH1 | Nucleus | enzyme |
| 217502_at | IFIT2 | unknown | other |
| 204747_at | IFIT3 | Cytoplasm | other |
| 205992_s_at | IL15 | Extracellular Space | cytokine |
| 204698_at | ISG20 | Nucleus | enzyme |
| 205841_at | JAK2 | Cytoplasm | kinase |
| 210302_s_at | MAB21L2 | unknown | other |
| 223298_s_at | NT5C3 | Cytoplasm | phosphatase |
| 205660_at | OASL | unknown | enzyme |
| 205801_s_at | RASGRP3 | Cytoplasm | other |
| 242625_at | RSAD2 | unknown | enzyme |
| 228531_at | SAMD9 | unknown | other |
| 230036_at | SAMD9L | unknown | other |
| 219885_at | SLFN12 | Nucleus | enzyme |
| 206271_at | TLR3 | Plasma Membrane | transmembrane receptor |
| 214329_x_at | TNFSF10 | Extracellular Space | cytokine |
| 221371_at | TNFSF18 | Extracellular Space | cytokine |
| 203610_s_at | TRIM38 | unknown | other |
| 219211_at | USP18 | Cytoplasm | peptidase |
| 200629_at | WARS | Cytoplasm | enzyme |
Discussion
We previously developed a human hepatocyte chimeric mouse model that can be chronically infected with hepatitis B and C viruses [29]–[31]. This mouse model has enabled us to analyze the effect of viral infection and the response to medication under immunodeficient conditions. Microarray analyses using the human hepatocyte chimeric mouse model with HCV infection have recently been reported, and HCV infection was found to affect expression of genes related to innate antiviral immune response, lipid metabolism and apoptosis via ER stress [34], [35]. Whereas these reports were concerned especially with host specific responses to HCV infection, no studies addressing viral modulation of the IFN response have been reported, even though such studies might be important for understanding viral evasion mechanisms in response to IFN therapy and for improving therapy effectiveness for chronic hepatitis C. Therefore, in this study we performed cDNA microarray analysis using a human hepatocyte chimeric mouse model and obtained gene expression profiles to investigate direct influences of HCV infection on IFN responses in human hepatocytes.
First, we evaluated host response to HCV infection in human hepatocytes by comparing profiles between groups A (without HCV infection) and C (with HCV infection). 181 genes were significantly up- or down-regulated following HCV infection. Canonical pathway analysis revealed that genes involved in IFN signaling were the most strongly up-regulated following HCV infection (Figure 3). These findings are mostly consistent with previous studies [36], [37]. On the other hand, while no genes involved in lipid metabolism showed any significant induction by HCV infection in this study, Walters et al. reported that HCV-infected chimeric mice exhibited host-specific induction in the expression of lipid metabolism genes [35]. However, we used hepatocytes from a single donor, whereas Walters et al. used hepatocytes from multiple donors, so our results are not necessarily inconsistent with their findings that HCV infection causes induction of lipid metabolism genes in a host-specific manner.
Although several cDNA microarray analyses have also been performed using human liver tissues obtained after hepatic resection, the largest difference between human and chimeric mouse livers is the presence or absence of human lymphocytes. According to the previous report using human liver tissues, genes involved in the innate immune response, as well as cell cycle, growth and communication, were up-regulated by HCV infection [38]. In the present study using SCID-derived mice, genes involved in immune response (e.g. OAS2, Mx1, IFI27 and IFI44L), cell cycle and growth (e.g. HERC5) and cell communication (e.g. HLA-B) were similarly up-regulated by HCV infection. However, Apolipoprotein L, Cold autoinflammatory syndrome 1, CD97 antigen, and HLA-DQ, which are mainly expressed in lymphocytes, were not observed to be up-regulated by HCV infection in the chimeric mice. These results demonstrate that the chimeric mouse model accurately reflects intracellular responses to HCV infection without the lymphocytic immune response.
To verify the microarray results, expression data were compared with previously published microarray data on the GEO website (http://www.ncbi.nlm.nih.gov/geo/). Previously published microarray data showed up-regulation of IGFBP7, IFI27, HLA-B, and CD74 in HCV-infected liver tissues compared to non-infected liver tissues (fold changes were 2.1, 2.2, 2.1 and 2.3, respectively) [39]. Likewise, we found that IFI27, HLA-B, and CD74 were up-regulated following HCV infection (fold changes were 3.6, 3.3, and 6.6, respectively). These three genes are associated with MHC class I activity, suggesting that intra-cellular immunity in human hepatocytes was activated following HCV infection both in human subjects and in chimeric mouse livers. Metallothionein 1G (MT1G) expression was also found to be up-regulated by HCV infection in both the current and published studies [39], [40]. Although metallothionein isoforms are associated with collagen deposition [41], members of the metallothionein family may be up-regulated and induce liver fibrosis in response to HCV infection.
In this study, genes associated with Organismal Injury and Abnormalities were found to be up-regulated in response HCV infection (Table 3), and some genes in this category, such as CXCL9, CXCL10 and IFIT3, maintained high IFN responsiveness under HCV infection (Table 8). These results suggest that protective responses to fibrosis or hepatic injury were activated at the start of HCV infection and remained activated until complete eradication of HCV from hepatocytes was achieved.
Secondly, we compared gene expression profiles between groups A (without IFN treatment) and B (with IFN treatment) to evaluate IFN response without HCV infection. IFN-α stimulates the intracellular IFN-signaling cascade after binding to the IFN-α receptor and mediates the transcriptional activation of IFN-stimulated genes [42]–[47]. More than 3.0-fold up-regulation was observed 6hrs after IFN treatment in 152 genes. Known ISGs such as those in the CXCL family (CXCL9, CXCL10 and CXCL11), the IFIT family (IFIT2 and IFIT3) and the APOBEC family (APOBEC3G) were included among the top 20 genes up-regulated following IFN treatment (Table 4). The APOBEC family is well known to have anti-viral effects by inducing genomic hypermutation in human immunodeficiency virus and hepatitis B virus [48]–[57]. APOBEC3G expression has been reported to be elevated in patients infected with HCV [58], although it is not clear whether APOBEC3G can block HCV replication. On the other hand, CXCL9 and IFIT3 were reported to relate to liver fibrosis in chronic hepatitis C patients. Serum CXCL9 concentrations correlated with the levels of fibrosis in chronic hepatitis C patients, and CXCL9 has been shown to exert anti-fibrotic effects in vitro and in vivo [59]. IFIT3 expression is also reportedly up-regulated in the transition from mild to moderate fibrosis [60]. The results of this study suggest that IFN treatment might lead not only to HCV eradication but also help to prevent and repair liver fibrosis by inducing these key molecules.
We focused on the 152 genes up-regulated (> 3.0 fold) as a result of IFN administration and evaluated the effect of HCV infection on IFN response among these genes. As shown in Table 8, although several ISGs still showed high response to IFN treatment in the presence of HCV infection, 7 genes in the IFN Signaling pathway became unresponsive (Table 6). Reduction in IFN responsiveness was also observed for STAT1 (4.27 fold in the absence of HCV to 2.29 fold in the presence of HCV, P = 4.04×10−4), as well as 5 of 7 genes downstream of STAT1 (IFIT1, IFIT3, IRF1, MX1, and TAP1). As shown in Figure 3, IFN signaling was activated in the presence of HCV, and the expression of STAT1 was more than 3.0 fold up-regulated by HCV infection (data not shown). STAT1 expression was highest in mice with both HCV infection and IFN treatment, but downstream genes such as MX1, IFIT1 and IFIT3 showed reduced IFN response. Sarasin-Filipowicz et al. reported that IFN-induced STAT1 phosphorylation was stronger in rapid responders than in non-rapid responders [61]. Reduced induction of genes downstream of STAT1 by IFN under HCV infection might reflect reduced phosphorylation of STAT1, although we did not quantify STAT1 phosphorylation in this study.
Recently, an IL-28B genetic polymorphism strongly associated with response to IFN-α plus ribavirin combination therapy [12], as well as with hepatic ISG expression [62], was identified. Further studies using chimeric mice transplanted with hepatocytes carrying different genotypes of candidate genes such as IL-28B will be important in order to elucidate possible mechanisms underlying host-specific responses.
In conclusion, we performed cDNA microarray analysis using HCV-infected human hepatocyte chimeric mice, which allowed us to analyze the direct effects of IFN treatment and HCV infection without the confounding effects of the lymphocytic immunological response. These results might provide molecular insights into possible mechanisms used by HCV to evade IFN-induced immune responses, as well as suggest novel therapeutic targets and a potential new indication for interferon therapy. Further analysis of the genes identified in our study would be worthwhile in order to improve efficacy of the therapy for chronic hepatitis C.
Acknowledgments
This study was carried out at the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University and the Analysis Center of Life Science, Hiroshima University. The authors thank Rie Akiyama and Ruri Mikami for their excellent technical assistance, and Aya Furukawa for clerical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Ministry of Education, Sports, Culture and Technology and Ministry of Health, Labor and Welfare (Grants-in-Aid for scientific research and development). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, et al. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 2.Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 3.Lee SH, Kim YK, Kim CS, Seol SK, Kim J, et al. E2 of hepatitis C virus inhibits apoptosis. J Immunol. 2005;175:8226–8235. doi: 10.4049/jimmunol.175.12.8226. [DOI] [PubMed] [Google Scholar]
- 4.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 5.Hoofnagle JH, Ghany MG, Kleiner DE, Doo E, Heller T, et al. Maintenance therapy with ribavirin in patients with chronic hepatitis C who fail to respond to combination therapy with interferon alfa and ribavirin. Hepatology. 2003;38:66–74. doi: 10.1053/jhep.2003.50258. [DOI] [PubMed] [Google Scholar]
- 6.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 7.Abbate I, Lo Iacono O, Di Stefano R, Cappiello G, Girardi E, et al. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J Hepatol. 2004;40:831–836. doi: 10.1016/j.jhep.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, et al. Prediction of response to pegylated interferon and ribavirin in hepatitis C by polymorphisms in the viral core protein and very early dynamics of viremia. Intervirology. 2007;50:361–368. doi: 10.1159/000107707. [DOI] [PubMed] [Google Scholar]
- 9.Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–380. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 10.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 12.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura S, Tsuge M, Hatakeyama T, Abe H, Imamura M, et al. Amino acid substitutions in core and NS5A regions of the HCV genome can predict virological decrease with pegylated interferon plus ribavirin therapy. Antivir Ther. 2010;15:1087–1097. doi: 10.3851/IMP1674. [DOI] [PubMed] [Google Scholar]
- 14.Sezaki H, Suzuki F, Kawamura Y, Yatsuji H, Hosaka T, et al. Dig Dis Sci; 2008. Poor Response to Pegylated Interferon and Ribavirin in Older Women Infected with Hepatitis C Virus of Genotype 1b in High Viral Loads. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 17.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe T, Kaname Y, Hamamoto I, Tsuda Y, Wen X, et al. Hepatitis C virus nonstructural protein 5A modulates the toll-like receptor-MyD88-dependent signaling pathway in macrophage cell lines. J Virol. 2007;81:8953–8966. doi: 10.1128/JVI.00649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriishi K, Mochizuki R, Moriya K, Miyamoto H, Mori Y, et al. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2007;104:1661–1666. doi: 10.1073/pnas.0607312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriishi K, Okabayashi T, Nakai K, Moriya K, Koike K, et al. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 24.Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, et al. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365–4370. [PubMed] [Google Scholar]
- 25.Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, et al. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78(Pt 7):1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, et al. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 28.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraga N, Imamura M, Tsuge M, Noguchi C, Takahashi S, et al. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis C virus and its susceptibility to interferon. FEBS Lett. 2007;581:1983–1987. doi: 10.1016/j.febslet.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Kimura T, Imamura M, Hiraga N, Hatakeyama T, Miki D, et al. Establishment of an infectious genotype 1b hepatitis C virus clone in human hepatocyte chimeric mice. J Gen Virol. 2008;89:2108–2113. doi: 10.1099/vir.0.83658-0. [DOI] [PubMed] [Google Scholar]
- 31.Tsuge M, Hiraga N, Takaishi H, Noguchi C, Oga H, et al. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus. Hepatology. 2005;42:1046–1054. doi: 10.1002/hep.20892. [DOI] [PubMed] [Google Scholar]
- 32.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 34.Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, et al. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5:e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters KA, Joyce MA, Thompson JC, Smith MW, Yeh MM, et al. Host-specific response to HCV infection in the chimeric SCID-beige/Alb-uPA mouse model: role of the innate antiviral immune response. PLoS Pathog. 2006;2:e59. doi: 10.1371/journal.ppat.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanford RE, Guerra B, Lee H, Chavez D, Brasky KM, et al. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 38.Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, et al. Different signaling pathways in the livers of patients with chronic hepatitis B or chronic hepatitis C. Hepatology. 2006;44:1122–1138. doi: 10.1002/hep.21383. [DOI] [PubMed] [Google Scholar]
- 39.Caillot F, Derambure C, Bioulac-Sage P, Francois A, Scotte M, et al. Transient and etiology-related transcription regulation in cirrhosis prior to hepatocellular carcinoma occurrence. World J Gastroenterol. 2009;15:300–309. doi: 10.3748/wjg.15.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caillot F, Hiron M, Goria O, Gueudin M, Francois A, et al. Novel serum markers of fibrosis progression for the follow-up of hepatitis C virus-infected patients. Am J Pathol. 2009;175:46–53. doi: 10.2353/ajpath.2009.080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh PP, Li JJ, Yip GW, Lo SL, Guo CH, et al. Modulation of metallothionein isoforms is associated with collagen deposition in proliferating keloid fibroblasts in vitro. Exp Dermatol. 2010;19:987–993. doi: 10.1111/j.1600-0625.2010.01124.x. [DOI] [PubMed] [Google Scholar]
- 42.Brassard DL, Grace MJ, Bordens RW. Interferon-alpha as an immunotherapeutic protein. J Leukoc Biol. 2002;71:565–581. [PubMed] [Google Scholar]
- 43.Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 44.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 45.Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp Hematol. 1999;27:1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 46.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staeheli P, Pitossi F, Pavlovic J. Mx proteins: GTPases with antiviral activity. Trends Cell Biol. 1993;3:268–272. doi: 10.1016/0962-8924(93)90055-6. [DOI] [PubMed] [Google Scholar]
- 48.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 49.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 50.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 51.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 52.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 53.Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, et al. Dual effect of APOBEC3G on Hepatitis B virus. J Gen Virol. 2007;88:432–440. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- 54.Noguchi C, Ishino H, Tsuge M, Fujimoto Y, Imamura M, et al. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–633. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- 55.Rosler C, Kock J, Kann M, Malim MH, Blum HE, et al. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 56.Suspene R, Sommer P, Henry M, Ferris S, Guetard D, et al. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komohara Y, Yano H, Shichijo S, Shimotohno K, Itoh K, et al. High expression of APOBEC3G in patients infected with hepatitis C virus. J Mol Histol. 2006;37:327–332. doi: 10.1007/s10735-006-9059-0. [DOI] [PubMed] [Google Scholar]
- 59.Wasmuth HE, Lammert F, Zaldivar MM, Weiskirchen R, Hellerbrand C, et al. Antifibrotic effects of CXCL9 and its receptor CXCR3 in livers of mice and humans. Gastroenterology. 2009;137:309–319, 319 e301-303. doi: 10.1053/j.gastro.2009.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asselah T, Bieche I, Laurendeau I, Paradis V, Vidaud D, et al. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology. 2005;129:2064–2075. doi: 10.1053/j.gastro.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 61.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]