Abstract
EnvZ and OmpR constitute the bacterial two-component signal transduction system known to mediate osmotic stress response in a number of Gram-negative bacteria. In an effort to understand the mechanism through which Shewanella oneidensis senses and responds to environmental osmolarity changes, structure of the ompR-envZ operon was determined with Northern blotting assay and roles of the EnvZ/OmpR two-component system in response to various stresses were investigated with mutational analysis, quantitative reverse transcriptase PCR (qRT-PCR), and phenotype microarrays. Results from the mutational analysis and qRT-PCR suggested that the EnvZ/OmpR system contributed to osmotic stress response of S. oneidensis and very likely engaged a similar strategy employed by E. coli, which involved reciprocal regulation of two major porin coding genes. Additionally, the ompR-envZ system was also found related to cell motility. We further showed that the ompR-envZ dependent regulation of porin genes and motility resided almost completely on ompR and only partially on envZ, indicating additional mechanisms for OmpR phosphorylation. In contrast to E. coli lacking ompR-envZ, however, growth of S. oneidensis did not show a significant dependence on ompR-envZ even under osmotic stress. Further analysis with phenotype microarrays revealed that the S. oneidensis strains lacking a complete ompR-envZ system displayed hypersensitivities to a number of agents, especially in alkaline environment. Taken together, our results suggest that the function of the ompR-envZ system in S. oneidensis, although still connected with osmoregulation, has diverged considerably from that of E. coli. Additional mechanism must exist to support growth of S. oneidensis under osmotic stress.
Introduction
Osmotic stress caused by changes of environmental osmotic strength is among the environmental stresses of great physiological relevance to microbes [1]. To cope with osmotic stress, bacteria have developed a number of strategies for effective adaptation. These strategies and their underlying mechanisms have been extensively reviewed [2]–[3]. In Escherichia coli, it is known that the EnvZ/OmpR two-component system plays a central role in mediating signal transduction in response to osmotic stress [4]–[6]. EnvZ is a transmembrane histidine kinase that monitors environmental osmolarity. At high osmolarity, EnvZ autophosphorylates and transfers the phosphoryl group to the response regulator OmpR, leading to formation of phosphorylated OmpR (OmpR-P). OmpR-P then binds to the promoter regions of outer membrane porin genes ompF and ompC and differentially modulates their expression according to the cellular OmpR-P level [7]. EnvZ also acts as a phosphatase that dephosphorylates OmpR-P once the osmotic stress fades away. Thus, environmental osmolarity affects the porin composition by the sum of EnvZ kinase and phosphatase activities in vivo [8]. In recent years, the regulatory scope of the EnvZ/OmpR system has been extended to genes related to virulence in Shigella flexneri, fatty acid receptor, peptide permease, and flagella in E. coli, as well as acid shock and stationary-phase acid tolerance response in Salmonella [9]–[15]. Deletion of EnvZ/OmpR was reported to have an impact on expression of more than 100 genes in E. coli, causing drastic changes in cell growth and important cell functions such as metabolism and motility [16].
Shewanella oneidensis MR-1, a facultative anaerobic γ-proteobacterium, is widely distributed in nature [17]. For more than a decade, this bacterium has received intensive studies owing to its remarkably diverse respiratory capacities and the potential for environmental remediation [18]. Despite the grand effort to characterize the transcriptomic responses triggered by various environmental stresses [19]–[22], little is known about how this bacterium senses these insults and modulates gene expression in response. Even though it is believed to posses more than 130 two-component proteins for signal transduction and gene regulation [18], to our knowledge only one pair of the known two-component systems, namely the Arc system have been characterized in S. oneidensis to date [23]–[25]. The Arc system in S. oneidensis differs substantially from that in E. coli, in terms of the structure (no full length ArcB homologue), regulon, and physiological function [23]–[25].
In this study, we chose to investigate the EnvZ/OmpR two-component system in S. oneidensis to address whether this system 1) is of general importance to signal perception and transduction post osmolarity changes and beyond, and 2) has deviated structurally and functionally from its counterpart of E. coli, like the Arc system did. Our results suggest that, although the ompR/envZ null mutant did not show any significant growth defect under conditions tested, these proteins indeed are involved in regulating the expression of porin genes in response to osmolarity changes. Moreover, the EnvZ/OmpR system appears to be important in bacterial tolerance to alkaline environments and affects cell motility. We further showed that the ompR-envZ dependent regulation of porin genes and motility resided almost completely on ompR and only partially on envZ, indicating additional mechanisms for ompR phosphorylation.
Results
Determination of the structure of the ompR-envZ operon
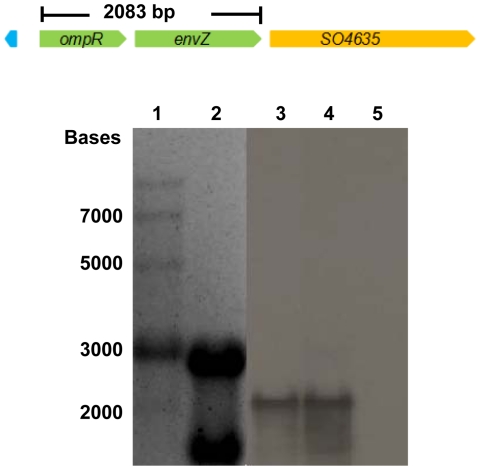
The EnvZ/OmpR system is known to be one of the major regulatory systems controlling the expression of outer membrane porins in response to osmolarity in many Gram-negative bacteria [4], [5]. To determine roles of the EnvZ/OmpR system in S. oneidensis, we first determined the structure of the ompR-envZ operon. Annotation of the genome sequence for S. oneidensis revealed the presence of both ompR and envZ homologues. Comparison of the deduced amino acid sequence showed that S. oneidensis OmpR (242 a. a.) and EnvZ (439 a. a.) share a high degree of identity to their homologues in E. coli (79% and 43%, respectively) and V. cholerae (241 a. a. 78% and 439 a. a. 42%, respectively), indicating potential for similar biological functions [26]. Although the coding sequences of ompR and envZ were arranged sequentially as in E. coli, appearance of SO4635, a methyl-accepting chemotaxis protein coding gene, immediately following envZ in the same orientation (Fig. 1) raised the possibility that ompR, envZ and SO4635 constitute a distinct type of three-gene operon (as opposed to the usual ompR-envZ two-gene operon) in S. oneidensis. To determine whether SO4635 is included in the operon, a Northern blotting assay was performed with RNAs from mid-exponential phase cells. In the analysis, approximately 300 bp internal fragments for each ORF were synthesized by PCR with primers listed in Table S1. As shown in Fig. 1, a∼2.3 kb transcript was detected with probes for either ompR or envZ whereas no such transcript was found with the probe for SO4635. Transcript for SO4635 alone is about 2 kb long and was not detected at all with the corresponding probe. Therefore, we conclude that ompR and envZ are co-transcribed as a single operon, while so4635 is transcribed separately. The function of SO4635 was not pursuit further.
Figure 1. Northern blot assay of the ompR-envZ operon.
Total RNA used in the analysis was from mid-exponential growing MR-1 cells. Organization of ompR-envZ and adjacent genes is shown. Lanes: 1, RNA ladder; 2, total RNA control (same amount of RNA stained with ethidium bromide); 3, RNA hybridized with probe specific for ompR; 4, RNA hybridized with probe specific for envZ; 5, RNA hybridized with probe specific for so4635.
Growth of S. oneidensis under osmotic stress
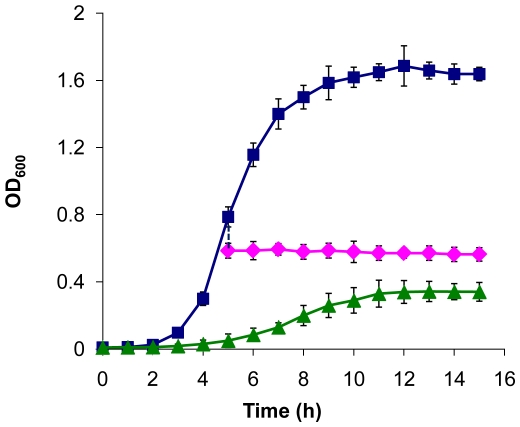
To probe the effect of osmotic stress on S. oneidensis cells, we examined growth of MR-1 in LB medium with 20% of sucrose at 30°C under aerobic conditions. Our results showed that the effect of 20% sucrose on growth of MR-1 was significant (Fig. 2). Even though MR-1 cells were able to survive and grow under osmotic stress, final cell densities are much lower for cultures grown under osmotic stress compared to those under normal conditions, with a maximum cell density of 0.38 (OD600). To determine if the final densities represented an irreversible cessation of growth, stressed cells were centrifuged and resuspended in LB (with no sucrose added) to 0.38 of OD600. Growth resumed shortly and reached a final densities of 1.8 (optical value of final density under normal conditions) of OD600 (data not shown). However, when 20% sucrose (final concentration) was introduced into the cultures of MR-1 at their late exponential phases (OD600≈0.8) to impose an osmotic shock, cells stopped growing immediately and the growth never resumed in this sucrose containing medium. Further examination revealed that if the osmotic shock was introduced before the culture reached OD600 of 0.38, cell growth can resume in the 20%-sucrose-containing medium, but only to a final OD600 of 0.38, indicating that 0.38 of OD600 was the maximum cell density permissible by 20% sucrose. Overall, results presented here suggest that the osmotic stress by 20% sucrose prevents S. oneidensis cells from growing to high cell densities but such an inhibition is temporary and reversible.
Figure 2. Growth of S. oneidensis strains at 30°C in LB medium under normal or osmotic stress conditions.
Under normal condition (blue square), osmotic stress condition (20% sucrose) (green triangle), or grown in LB and then transferred to the osmotic shock condition (purple diamond). The growth curves are the average curves for at least three replicate samples and the error bars represent standard error of the mean (SEM).
Construction and characterization of mutants devoid of the EnvZ/OmpR system
To assess the impact of the EnvZ/OmpR system on growth of S. oneidensis, the ompR, envZ, single and ompR/envZ double deletion mutants, designated as HG4633 (ΔompR), HG4634 (ΔenvZ), and HG4633/4 (ΔompRΔenvZ), were created. A mutagenesis system for constructing deletion mutants in S. oneidensis MR-1 has previously been developed and successfully utilized [23], [27]. The deletion was confirmed by PCR and DNA sequencing (data not shown).
Physiological role of the EnvZ/OmpR system in S. oneidensis was first assessed by growing our mutant strains under either normal or stressed (imposed by 20% sucrose) condition. Strikingly, compared to the parental strain, neither the S. oneidensis ΔompRΔenvZ double mutant nor either of the two single mutants ΔompR and ΔenvZ exhibited any noticeable differences in growth, in terms of generation time and maximum cell density (data not shown) under either tested conditions. In contrast, deletion of EnvZ/OmpR was reported to significantly slow down the growth of E. coli in LB [16]. These results imply that EnvZ/OmpR system may have diverged considerably between S. oneidensis and E. coli in terms of their physiological roles.
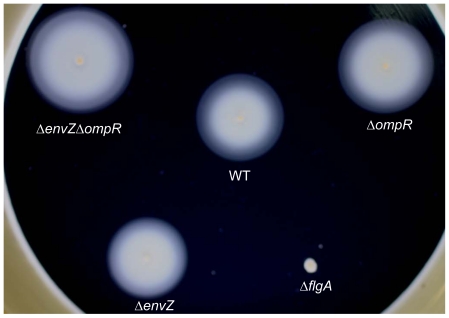
Motility of these mutants was also assayed, as significant down-regulation of the chemotaxis/motility-related genes was previously observed in salt stressed S. oneidensis [22], and EnvZ/OmpR was reported to regulate genes involved in flagella synthesis in E. coli [16]. Interestingly, our results showed that although motility of ΔenvZ was the same as its parental strain, ΔompR and the double mutant displayed increased motility (Fig. 3). To ensure that the observed phenotype was due to mutation per se, we cloned the operon of envZ-ompR along with its promoter into pBBR1MCS-2 and introduced the resultant construct into either the ompR or the double mutation strain. Swimming motility of the plasmid-borne ompR reduced to the level indistinguishable from their parental strains (data not shown). These results suggest that the OmpR may play a role in regulating motility-related genes.
Figure 3. Motility of the ΔenvZ, ΔompR, ΔenvZΔompR mutants.
Both swimming and swarming motility assays were performed. The parental wild type and an aflagellated strain were included as controls. Experiments were performed five times and results were statistically consistent (p<0.05). Only swimming results were shown as swimming and swarming are in agreement with each other.
The ompR-envZ operon is induced under osmotic stress
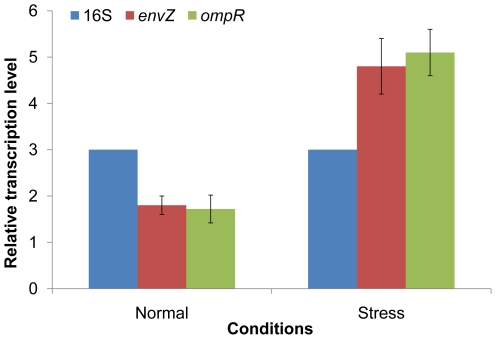
Apparently, both ompR and envZ are transcribed in the exponentially growing wild-type cells as their mRNAs allowed determination of the operon structure. However, lack of apparent growth phenotypes of the three S. oneidensis mutants under either normal or stressed conditions raised a question about whether the system was osmo-responsive. To test this, we performed qRT-PCR to examine the amount of the ompR-envZ transcripts in cells under normal and stressed conditions. Transcripts of ompR and envZ were virtually the same in wild-type under the same conditions, in agreement with the observation that they are on a single polycistronic mRNA. However, compared to the untreated control, transcription of these two genes was induced approximately 2.8-fold in cells exposed to 20% sucrose (Fig. 4), suggesting potential involvement of ompR-envZ in response to osmotic stress.
Figure 4. Transcription of envZ and ompR in S. oneidensis under normal and stressed conditions.
The relative transcription levels of envZ and ompR were presented as fold increases with respect to expression of 16S rRNA gene. The results were means plus SEM from three independent experiments.
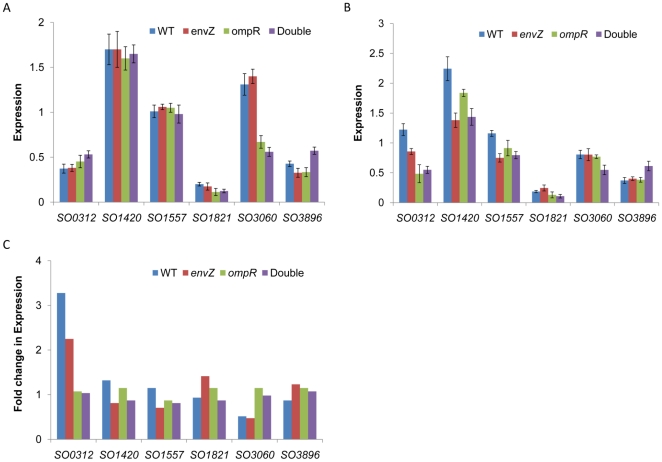
SO0312 and SO3060 are likely homologues of E. coli ompC and ompF respectively
The best known osmotic stress response in Gram-negative bacteria is the alteration of membrane porin composition mediated by EnvZ/OmpR system. Hence one of the main goals of this study was to determine the homologues of E. coli porin coding genes ompC and ompF in S. oneidensis. Although OmpC and OmpF share substantial similarity in sequence and three-dimensional structure, their expression is regulated reciprocally by the EnvZ/OmpR two-component system upon changes in environmental osmolarity [28]. Elevated osmolarity leads to preferential expression of ompC and repression of ompF, whereas low osmolarity favors the opposite. S. oneidensis has 6 putative outer-membrane porin-coding genes: SO0312, SO1420, SO1557, SO1821, SO3060, and SO3896. All of them are similar in size (321 – 398 a. a.) and significantly homologous to both ompC and ompF of E. coli.
To determine their involvement in osmotic stress response, expression changes of these genes in wild-type and the three mutant strains (ΔompR, ΔenvZ, and ΔompRΔenvZ) under normal and stressed conditions were compared (Fig. 5). Expression of these genes under normal conditions except for SO3060 was indistinguishable across all tested strains although their absolute transcript levels varied substantially in each strain (Fig. 5A). The fact that SO3060 expressed at levels significantly lower in ompR and double mutant strains than those in wild-type and ΔenvZ, suggest that the gene may be under control of OmpR. Under stressed conditions, SO0312 displayed a responsive expression pattern while the rest 5 genes were not affected (Fig. 5B). We then compared expression of these genes under the two tested conditions as presented in Fig. 5C. Results from the wild-type revealed that transcription of SO0312 and SO3060 was drastically affected by the stress imposed by 20% sucrose while the other four genes were hardly responsive. The high osmolarity induced SO0312 but repressed SO3060 in wild-type strain, suggesting that SO0312 and SO3060 may be the counterparts of E. coli ompC and ompF, respectively, and S. oneidensis very likely employed the same reciprocal transcriptional regulation scheme to modulate membrane porin composition in response to osmotic stress. Strikingly, the same trend was observed in the ΔenvZ mutant, implicating that the two-component system may still be able to function, at least partially, in the absence of the sensory kinase. In contrast, such response was not found in either ΔompR or the double mutant. These findings indicate existence of EnvZ-independent mechanism for OmpR phosphorylation. Intriguingly, it is know in E. coli that ArcB, histidine kinase of the ArcBA two component system can phosphorylate and cross regulate OmpR [29]. To test whether the same mechanism is responsible for OmpR phosphorylation in the absence of EnvZ in MR-1, we examined OmpC expression in mutants devoid of ArcS or both ArcS and EnvZ with qRT-PCR. Additionally, we also employed bacterial two-hybrid system to test interaction of HptA with OmpR. ArcS and HptA are proteins found to function as ArcB in MR-1, with each resembling part of E. coli ArcB protein [23]-[25]. Unfortunately, neither was ArcS found to affect OmpC expression in response to osmo-stress, nor was HptA detected to interact with OmpR (data not shown), suggesting that the S. oneidensis counterpart to E. coli ArcB may not be able to cross-phosphorylate OmpR.
Figure 5. Transcription of six outer membrane porin genes in S. oneidensis strains under normal and stressed conditions.
All data were normalized to expression of 16S rRNA gene. The absolute expression levels of these genes under normal and stressed conditions were presented in (A) and (B), respectively. The relative transcription levels of these genes were presented as fold changes in each strain between two tested conditions in (C). The results were means plus SEM from three independent experiments.
Functional analysis of the EnvZ/OmpR system
The results presented above indicated that the EnvZ/OmpR system probably still contributes to osmotic stress response (very likely by modulating membrane porin composition) and affect cell motility. However, under the conditions tested, lack of EnvZ/OmpR did not cause a pleiotropic effect as observed in E. coli [16]. To evaluate impact of the EnvZ/OmpR system on metabolism of S. oneidensis under a broader set of conditions, capacity of the wild-type, ΔompR, ΔenvZ, and ΔompRΔenvZ strains to metabolize 95 different carbon sources was tested using PM1 phenotype microarrays from Biolog (information about carbon sources at http://www.biolog.com/pdf/PM1-PM10.pdf). All these strains displayed positive reaction with carbon sources A03 (N-acetyl-D-glucosamine), B09 (D, L-Lactic acid), C05 (tween 20), D05 (tween 40), D12 (uridine), E05 (tween 80), E11 (2′-deoxyadenosine), E12 (adenosine), F12 (inosine), G10 (methylpyruvate), and H08 (pyruvic acid) (Fig. 6A). However, no difference was observed between profiles obtained from the wild-type and any of the mutants. In contrast, the same phenotype microarrays revealed that the E. coli EnvZ/OmpR mutants displayed increased consumption of D-glucose, D-fructose, mannitol, and N-acetyl-D-glucosamine [30]. Collectively, these results indicate that that the EnvZ/OmpR system is unlikely to be involved in regulation of carbon metabolism in S. oneidensis.
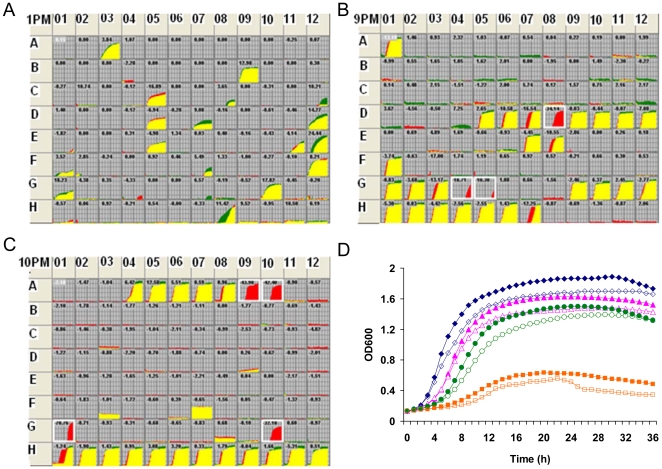
Figure 6. Involvement of the EnvZ-OmpR system in metabolism and stress response.
(A, B, C) The envZ/ompR mutation strain was subjected to PM analysis (3 PMs). The PM kinetic profile shown by comparing the ompR-envZ mutant (green) and its wild-type parental strain (red). Red indicates a stronger response by MR-1, and green indicates a stronger response by the mutant; when the two strains have equivalent metabolism or equivalent growth responses in a well, the red and green kinetic graphs overlap and are yellow. A box around a growth curves indicates a significant difference in response. (D). Independent growth studies in LB with various concentrations of sodium sulfate. 2%, diamond; 3%, triangle; 4%, cycle; 5%, square. In all cases, MR-1, closed; ompR-envZ-, open. Experiments were performed three times independently and consistent results were obtained. For clarity, error bars (SEM, < 5%) were omitted in the figure.
We then examined whether the EnvZ/OmpR system has an impact on bacterial response to various stresses using PM9 and PM10 phenotype microarrays. Lack of significant difference in profiles between the three mutants used (data not shown) indicates that both the sensor (EnvZ) and the regulator (OmpR) appear to be essential to the functionality of the two-component system in the context of the PM analysis. To be concise, we used the phenotype microarray results of ΔompRΔenvZ strain to represent those of all mutant strains unless otherwise noted. As shown in Fig. 6B and C, the mutant was notably hypersensitive to 5% sodium sulfate (PM09, D08), 200 mM sodium phosphate at pH 7.0 (PM09, G04), 20 mM sodium benzoate at pH 5.2 (PM09, G05), pH 8.5 (PM10, A09), pH 9 (PM10, A10), pH 9.5 + anthranilic acid (PM10, G01), and pH 9.5 + tryptamine (PM10, G10). In contrast to E. coli ompR-envZ mutants, the S. oneidensis ompR-envZ mutants were not hypersensitive to ethylene glycol. It is worth noting that most of these hypersensitivities were observed in alkaline environment, suggesting that the two-component system may be involved in bacterial response to alkaline stresses. Interestingly, in Salmonella enteric the system plays a significant role in the acid stress response [31].
To confirm the phenotypes revealed by PM analysis, hypersensitivity of the mutation strains to 5% sodium sulfate was assessed by an independent growth study. All the wild-type and mutant strains were investigated in LB supplemented with sodium sulfate at concentrations of 2, 3, 4, and 5% (Fig. 6D). Consistent with the PM analysis, 5% sodium sulfate was found to inhibit growth of the bacteria, with a more significant impact on the mutant strain. Interestingly, in the growth study, the mutant displayed hypersensitivity to all tested concentrations of sodium sulfate, although to much lesser extent at lower concentrations compared to 5%. The fact that PM analysis failed to reveal these milder effects could probably be attributed to the limited sensitivity of the colorimetric mechanism employed by the approach. Nonetheless, our results suggest that even though PM analysis may not be suitable to pick up significant subtle differences, it provides a valuable tool to examine over thousands of cellular phenotypes in a high-throughput fashion and provide meaningful multi-dimensional biochemical profiles of organisms. These profiles alone, or combined with transcriptomes and proteomes, will greatly facilitate the future researches in all directions.
Discussion
Two-component signal transduction systems composed of a sensory histidine kinase and a response regulator are broadly utilized by bacteria for effective acclimation to environmental changes. Given the number of studies aiming to dissect stress responses of S. oneidensis performed, it is surprising that the Arc two-component system is the only one being scrutinized [23]–[25]. Additionally, the Arc system is so atypical that it does not only replace the full length ArcB with two proteins but also differs substantially in physiological function from its E. coli counterpart. Thus, investigation into other two-component systems of S. oneidensis seems demanding.
Among the most extensively studied, EnvZ/OmpR is known to govern the response to osmotic changes by modulating expression of porin coding genes ompC and ompF and thereby outer membrane permeability [32]. The objective of present work was to investigate whether the EnvZ/OmpR system is conserved structurally and functionally in S. oneidensis. The Northern blotting assay revealed that genes coding for EnvZ and OmpR constitutes an ompR-envZ operon which was transcribed as a single polycistronic mRNA. Transcription of this operon was found to increase ∼2.8 fold under osmotic stress, consistent with the system being involved in osmotic stress response. In two-component systems, differences at the transcriptional level have been largely overlooked as the phosphorylation states of the response regulator was widely accepted as the determining factor for functionality [33]–[34]. Our results indicated that increased transcription of ompR-envZ might also be a strategy that S. oneidensis cells adopt to tackle the osmotic stress problem. An exemplary strategy of such kind was employed by the two-component system NRI-NRII: members of the NRI regulon are known to require different amount of the response regulator for activation or repression, so different response can be triggered at different expression level of NRI-NRII [35]. By this line of reasoning, the increment observed in envZ/ompR transcription could also be perceived as hinting an OmpR regulon of a considerable size.
In addition, we presented evidence that, among the 6 putative porin coding genes found in the S. oneidensis genome, SO0312 and SO3060 were very likely counterparts of E. coli ompC and ompF, respectively. We showed that expression of these two genes responded reciprocally to osmotic stresses in an OmpR-dependent manner, while only partial dependency was observed for EnvZ. In addition, motility was also found to increase in the absence of OmpR but not EnvZ. These results on one hand suggest that EnvZ/OmpR is still linked to osmotic stress response, and indicate the presence of additional mechanisms responsible for OmpR phosphorylation on the other hand. Phosphorylation of OmpR in the absence of EnvZ can be attributed to cross-regulation among two-component systems [36]. Although ArcB was known to regulate OmpR in E. coli [29], such effect was not observed in our study. It is not surprising however, given that no apparent orthologue of ArcB exits in MR-1 and its role is played by two proteins together. Moreover, MR-1 possesses a much larger arsenal of two-component systems compared to E. coli [18]; it is poised to employ a more sophisticated cross-talking signal transduction network. In addition to non-cognitive histidine kinases, even non-enzymatic phosphorylation agents such as acetyl phosphate, a global signal that feeds into various two-component systems [37], could also be responsible for OmpR phosphorylation. This may be particularly worth noting because acetyl phosphate was involved with the EnvZ/OmpR system as a phosphoryl group donor and its level correlated with the level of phosphorylated OmpR in E. coli [38]–[39]. Although phosphorylation of OmpR by agents other than EnvZ is not expected to be as efficient as EnvZ, dephosphorylation of OmpR is also greatly reduced in ΔenvZ due to the lack of phosphatase activity of EnvZ. Hence we postulate that such EnvZ-independent phosphorylation could still support OmpR-P accumulation to physiologically relevant levels to regulate genes such as ompC and ompF in the absence of EnvZ.
In contrast to the significant growth impairment reported for a E. coli strain devoid of the entire system [16], none of the three deletion mutants (ΔompR, ΔenvZ, ΔompRΔenvZ) of S. oneidensis displayed noticeable growth defect compared to the parental wild type strain even under the osmotic stress (by 20% sucrose). This apparent robustness of growth to loss of EnvZ/OmpR could potentially be due to additional mechanisms coping with osmotic changes. For example, RcsB-RcsC was reported to respond to osmolarity in Salmonella typhi [40], and OmpR-independent mechanism influencing ompC and ompF expression was also suggested to exist in E. coli [41]. Given the presence of shewanellae in a large spectrum of environmental conditions that covering a broad osmolarity range [42]–[48], it is not surprising if organisms in this genus have developed sophisticated multi-tiered defense mechanism to deal with osmotic changes. Moreover, as far as the data from Phenotype Microarrays were considered, the ΔompRΔenvZ mutants of E. coli and S. oneidensis exhibited drastic differences [27], suggesting that the function of EnvZ/OmpR has diverged significantly between the two organisms. In particular, the apparent importance of EnvZ/OmpR under alkaline conditions in S. oneidensis indicates that this system has shifted focus to a different set of environmental insults and acquired new roles.
In conclusion, we show in this work that the EnvZ/OmpR system in S. oneidensis, despite a few similarities, departs significantly from that system in E. coli. We speculate that these findings reflect a general trend of functional divergence of two-component systems in S. oneidensis compared to the Gram-negative model bacterium E. coli. Although two-component systems are ubiquitous signal transduction modules to bacteria, the functions of specific two-component systems however, can be tailored in different organism in a natural-habitat-specific manner.
Methods
Bacterial strains, plasmids, and culture conditions
A list of all bacterial strains and plasmids used in this study is given in Table 1. S. oneidensis and E. coli strains were grown in Luria-Bertani (LB, Difco, Detroit, MI) medium at 30 and 37°C, respectively. When needed, the growth medium was supplemented with antibiotics at the following concentrations: ampicillin at 50 µg/ml, and gentamycin at 15 µg/ml. The suicide vector pDS3.0 has been described elsewhere [27].
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
| E. coli strain | ||
| WM3064 | Donor strain for conjugation; ΔdapA | [49] |
| S. oneidensis strains | ||
| MR-1 | Wild-type | Lab stock |
| HG4633 | ompR in-frame mutant derived from MR-1; ΔompR | This study |
| HG4634 | envZ in-frame mutant derived from MR-1; ΔenvZ | This study |
| HG4633-4 | ompR-envZ in-frame double mutant derived from MR-1; ΔompRΔenvZ | This study |
| HG3256 | flgA in-frame mutant derived from MR-1; ΔflgA, aflagelated | [50] |
| Plasmids | ||
| pDS3.0 | Apr, Gmr, derivative from suicide vector pCVD442 | [27] |
| pDS3-OMPR | pDS3.0 containing the PCR fragment for deleting ompR | This study |
| pDS3-ENVZ | pDS3.0 containing the PCR fragment for deleting envZ | This study |
| pDS3-OED | pDS3.0 containing the PCR fragment for deleting ompR and envZ | This study |
| pBBR1MCS-2 | Broad host Kmr vector used for complementation | [51] |
| pBBR-OED | pBBR1MCS-2 containing the envZ-ompR operon with its promoter | This study |
Construction of S. oneidensis in-frame deletion strains
The mutagenesis method for both single and double in-frame deletion mutations were the same except that different sets of primers were used to generate insertion fragments. Primers used in this study were listed in Table S1 of the supporting information. Below the method used for construction of the ompR in-frame deletion mutation strain HG4633 (ΔompR) was briefly described.
Two fragments flanking ompR were amplified by PCR with primers SO4633-5-F and SO4633-5-R, primers SO4633-3-F and SO4633-3-R respectively, and joined together by the second round PCR with primers SO4633-5-F and SO4633-3-R. The resulting fragment was digested with SacI (New England Biolabs, Beverly, MA) and then ligated into SacI site of pDS3.0 treated with the shrimp alkaline phosphatase (Roche Diagnostics, Mannheim, Germany), resulting in the plasmid pDS3-OMPR. E. coli WM3064 cells containing pDS3-OMPR were used for conjugal transfer of pDS3-OMPR to S. oneidensis MR-1 as described previously [27]. In-frame deletion mutations were screened by colony PCR amplification and verified by DNA sequencing of the PCR-amplified DNA fragment containing the mutated region. For strains HG4634 (ΔenvZ) and HG4633/4 (ΔompRΔenvZ), primers used were: SO4634-5-F and SO4634-5-R, SO4634-3-F and SO4634-3-R; SO4633-5-F and SO4633-5-R, SO4634-3-F and SO4634-3-R, respectively.
Growth of wild-type and mutant strains on normal and osmotic stress conditions
A single colony of each S. oneidensis strain was used to inoculate 5 ml of LB in 50 ml plastic tubes and grown overnight at 30°C (optimal growth temperature) on a rotary platform (200 rpm). This culture was then used to inoculate 30 ml of LB medium with or without 20% sucrose pre-warmed to 30°C in a 250 ml shake flask at an OD600 of 0.01 and the flask was shaken on a rotary platform (250 rpm) at 30°C. For the osmotic shock, cells were grown to exponential phase and diluted with prewarmed LB supplemented with 50% sucrose to the final sucrose concentration of 20%. For RNA work, cells before and 10 min after addition of sucrose were pelleted by centrifugation for 30 seconds, frozen immediately in liquid nitrogen, and then stored at −80°C. Three individual cultures were grown in parallel as biological replicates. Growth was measured every 30 min for normal condition and every 4 h for stress condition. Three individual cultures for each strain were grown simultaneously. Growth was measured every 30 min and 4 h under normal and stress conditions, respectively.
Swarming and swimming motility assay and mutation complementation
A fresh colony of tested strains was grown to an OD600 of 0.8 in LB media. The cultures (1 µl) were spotted onto a swarming LB plate (0.5% agar) or stabbed into a swimming LB plate (0.2% agar). All plates were incubated at the room temperature for 48 h. For complementation, DNA fragments containing envZ and ompR as well as their promoter were generated by PCR amplification with MR-1 genomic DNA as the template using primers SO4633/4-COM-F and SO4633/4-COM-R, respectively as listed in Table S1. These fragments were digested with SacI and ligated to SacI-digested pBBR1MCS-2 to form pBBR-OED, which was electroporated into WM3064. Introduction of pBBR-OED into all mutants constructed in this study was done by conjugation, and kanamycin-resistant colonies were selected. The presence of pBBR-OED in the mutants was confirmed by plasmid purification and restriction enzyme digestion.
Northern blot hybridization
In order to determine the genes that constitute the ompR operon, Northern blotting was performed with total RNA extracted from exponentially growing oneidensis MR-1 cells (OD600≈0.4). RNA was extracted using Trizol (Invitrogen) and RNeasy kit (Qiagen) as described previously [19]. All chemicals used in this experiment were obtained from Roche Diagnostics (Roche Diagnostics, Indianapolis, IN) and every step was performed according to the manufacturer's instructions unless otherwise indicated. Digoxigenin (DIG)-labeled DNA probes were generated using the PCR DIG Probe synthesis kit. The primers used to generate probes of ca. 300 bp for ompR, envZ and so4635 were listed in Table S1. 15 µg (per lane) of the same total RNA used in RT-PCR was electrophoresed on a 1.2% agarose gel in 1x MOPS containing 2% formaldehyde. Gels were run for 5 h at 55 V in the cold. Lanes corresponding to the molecular weight markers (RNA ladder, New England Biolabs, Beverly, MA) and control RNA sample were cut out, stained with ethidium bromide and photographed under UV light. Transfer of RNA on the rest of the gel to nylon membranes was done by upward capillary blotting with 20x SSC. The RNA was then bound to the nylon membrane using Stratalinker 1800 at 120 mJ for 1 min. After prehybridization, hybridization was performed overnight at 50°C using approximate 4 ng of labeled probe per ml of DIG hybridization buffer. Washing of the membrane and detection of specific transcripts on the blots were carried out using washing and blocking reagents and the DIG luminescent detection kit. Signals were visualized by exposure to X-ray film (Kodak, Rochester, NY). The size of the target mRNA was estimated from the sizes of 16S and 23S rRNA, and the RNA ladder detected by ethidium bromide staining before membrane transfer.
Quantitative RT-PCR (qRT-PCR)
RNA templates for the analysis were extracted from the wild-type and mutant cells collected 10 minute after 20% sucrose was introduced. Primers were designed using Omiga software (Oxford Molecular Ltd., San Diego, CA) and were synthesized by Applied Biosystems (Table S1). PCR products amplified from these ORFs were single-band fragments of 99–101 bp in length, as confirmed by agarose gel electrophoresis. A 100-bp fragment of the arcA gene, which was amplified by PCR with genomic DNA as the template, was used to construct the standard curve. The reaction was performed with 50 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C and monitored in an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA). The transcriptional levels of target genes were normalized against the expression of 16S rRNA gene as internal control. The expression of each gene was determined from three replicates on a single qRT-PCR experiment.
Bacterial two-hybrid assay
The BacterioMatch II Two-Hybrid system was used to investigate protein-protein interaction between HptA and OmpR in vivo in E. coli cells according to manufacturer's instructions. Briefly, the genes htpA and envZ (without the first 114 bases encoding the first membrane-spanning region), were cloned into plasmid pBT separately, and ompR gene was cloned into plasmid pTRG. After verification by sequencing, the resultant plasmids were co-transformed into BacterioMatch II Validation Reporter Competent Cells on M9 salt agar plates containing 25 mg/ml chloramphenicol and 12.5 mg/ml tetracycline with or without 3-amino-1,2,4-triazole (3-AT). And pBT-LGF2, pTRG-Gal11P, empty pBT and pTRG were used as positive and negative controls. The plates were incubated at 37°C for 24 h and then moved to room temperature in a dark location (to preserve the tetracycline) for an additional 16 h. The strength of interaction typically correlated with the ratio of colonies obtained on the selective plated compared to on the non-selective plates. The positive interactions were confirmed by streaking colonies on plates containing both 3-AT and streptomycin (12.5 mg/ml).
Phenotype Microarrays
Phenotype microarray (PM) plates (Biolog Inc., Hayward, California) were used to examine the involvement of the EnvZ/OmpR two-component system in carbon metabolism and various stresses according to the manufacturer's instruction. The altered phenotypes of the ΔompR, ΔenvZ, and ΔompRΔenvZ mutants were assessed by comparing to their parental strain MR-1. The mutant strains were recorded as a green trace and MR-1 was recorded as a red trace in OmniLog Incubator-Reader. Color-coded kinetic graphs could then be overlaid by the OmniLog PM bioinformatic software, and differences were visualized and quantified according to the manufacturer's instruction. To confirm the PM analysis results, growth of the wild-type and mutation strains in LB supplemented with selected substances identified by PM analysis was investigated.
Supporting Information
Primers used in this study.
(PDF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was financially supported by Major Program of Science and Technology Department of Zhejiang (2009C12061), by the Fundamental Research Funds for the Central Universities, and by Major State Basic Research Development Program (973 Program: 2010CB833803) to HG; and by Zhejiang R&D program (2009R50G2010001) and Department of education of Zhejiang Province (Z200909658) to JY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Csonka LN, Epstein W. Osmoregulation. In: Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium cellular and molecular biology. Washington DC: ASM Press; 1996. pp. 1210–1224. [Google Scholar]
- 2.Bremer E, Kramer R. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria. In: Gisela Storz RH-A, editor. Bacterial stress responses. Washington, D.C.: American Society for Microbiology; 2000. pp. 79–97. [Google Scholar]
- 3.Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 4.Cai SJ, Inouye M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Cai SJ, Inouye M. Interaction of EnvZ, a sensory histidine kinase, with phosphorylated OmpR, the cognate response regulator. Mol Microbiol. 2002;46:1283–1294. doi: 10.1046/j.1365-2958.2002.03240.x. [DOI] [PubMed] [Google Scholar]
- 8.Mattison K, Kenney LJ. Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem. 2002;277:11143–11148. doi: 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- 9.Shin S, Park C. Modulation of flagellar expression in Escherichiacoli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Morales O, Fernandez-Mora M, Hernandez-Lucas I, Vazquez A, Puente JL, et al. Salmonella enterica serovar Typhimurium ompS1 and ompS2 mutants are attenuated for virulence in mice. Infect Immun. 2006;74:1398–1402. doi: 10.1128/IAI.74.2.1398-1402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang IS, Audia JP, Park YK, Foster JW. Autoinduction of the OmpR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol Microbiol. 2002;44:1235–1250. doi: 10.1046/j.1365-2958.2002.02937.x. [DOI] [PubMed] [Google Scholar]
- 12.Bang IS, Kim BH, Foster JW, Park YK. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardini ML, Fontaine A, Sansonetti PJ. The two-component regulatory system OmpR-EnvZ controls the virulence of Shigella flexneri. J Bacteriol. 1990;172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh EB, Siino DF, Igo MM. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J Bacteriol. 2004;186:4019–4024. doi: 10.1128/JB.186.12.4019-4024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills SD, Ruschkowski SR, Stein MA, Finlay BB. Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of Salmonella-induced filaments. Infect Immun. 1998;66:1806–1811. doi: 10.1128/iai.66.4.1806-1811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, et al. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 17.Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int J Syst Bacteriol. 1999;49:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 18.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 19.Gao HC, Wang Y, Liu XD, Yan TF, Wu LY, et al. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao HC, Yang ZMK, Wu LY, Thompson DK, Zhou JZ. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol. 2006;188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaphart AB, Thompson DK, Huang K, Alm E, Wan XF, et al. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J Bacteriol. 2006;188:1633–1642. doi: 10.1128/JB.188.4.1633-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YQ, Gao WM, Wang Y, Wu LY, Liu XD, et al. Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J Bacteriol. 2005;187:2501–2507. doi: 10.1128/JB.187.7.2501-2507.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao HC, Wang XH, Yang ZK, Palzkill T, Zhou JZ. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gralnick JA, Brown CT, Newman DK. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56:1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 25.Lassak J, Henche AL, Binnenkade L, Thormann KM. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl Environ Microbiol. 2010;76:3263–3274. doi: 10.1128/AEM.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao WM, Liu YQ, Giometti CS, Tollaksen SL, Khare T, et al. Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genomics. 2006;7:76. doi: 10.1186/1471-2164-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno T, Kato M, Jo YL, Mizushima S. Interaction of OmpR, a positive regulator, with the osmoregulated ompC and ompF genes of Escherichia coli - studies with wild-type and mutant OmpR proteins. J Biol Chem. 1988;263:1008–1012. [PubMed] [Google Scholar]
- 29.Matsubara M, Kitaoka SI, Takeda SI, Mizuno T. Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes cells. 2000;5:555–569. doi: 10.1046/j.1365-2443.2000.00347.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhou L, Lei XH, Bochner BR, Wanner BL. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol. 2003;185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rychlik I, Barrow PA. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev. 2005;29:1021–1040. doi: 10.1016/j.femsre.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Aiba H, Mizuno T. Phosphorylation of a bacterial activator protein, OmpR, by a protein-kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett. 1990;261:19–22. doi: 10.1016/0014-5793(90)80626-t. [DOI] [PubMed] [Google Scholar]
- 33.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 34.Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 36.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genetics. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69 doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsubara M, Mizuno T. EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci Biotechn Bioch. 1999;63:408–414. doi: 10.1271/bbb.63.408. [DOI] [PubMed] [Google Scholar]
- 39.Heyde M, Laloi P, Portalier R. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J Bacteriol. 2000;182:198–202. doi: 10.1128/jb.182.1.198-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arricau N, Hermant D, Waxin H, Ecobichon C, Duffey PS, et al. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol. 1998;29:835–850. doi: 10.1046/j.1365-2958.1998.00976.x. [DOI] [PubMed] [Google Scholar]
- 41.Pratt LA, Hsing WH, Gibson KE, Silhavy TJ. From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 42.Semple KM, Westlake DWS. Characterization of iron-reducing Alteromonas-putrefaciens strains from oil-field fluids. Can J Microbiol. 1987;33:366–371. [Google Scholar]
- 43.Brettar I, Hofle MG. Nitrous-oxide producing heterotrophic bacteria from the water column of the central baltic - abundance and molecular-identification. Mar Ecol Prog Ser. 1993;94:253–265. [Google Scholar]
- 44.Bowman JP, McCammon SA, Nichols DS, Skerratt JH, Rea SM, et al. Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5 omega 3) and grow anaerobically by dissimilatory Fe(III) reduction. Int J Syst Bacteriol. 1997;47:1040–1047. doi: 10.1099/00207713-47-4-1040. [DOI] [PubMed] [Google Scholar]
- 45.Yamada M, Nakasone K, Tamegai H, Kato C, Usami R, et al. Pressure regulation of soluble cytochromes c in a deep-sea piezophilic bacterium, Shewanella violacea. J Bacteriol. 2000;182:2945–2952. doi: 10.1128/jb.182.10.2945-2952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Obraztova A, Stewart N, Popa R, Fredrickson JK, et al. Shewanella loihica sp. nov., isolated from iron-rich microbial mats in the Pacific Ocean. Int J Syst Evol Microbiol. 2006;56:1911–1916. doi: 10.1099/ijs.0.64354-0. [DOI] [PubMed] [Google Scholar]
- 47.Skerratt JH, Bowman JP, Nichols PD. Shewanella olleyana sp nov., a marine species isolated from a temperate estuary which produces high levels of polyunsaturated fatty acids. Int J Syst Evol Microbiol. 2002;52:2101–2106. doi: 10.1099/00207713-52-6-2101. [DOI] [PubMed] [Google Scholar]
- 48.Pagani L, Lang A, Vedovelli C, Moling O, Rimenti G, et al. Soft tissue infection and bacteremia caused by Shewanella putrefaciens. J Clin Microbiol. 2003;41:2240–2241. doi: 10.1128/JCM.41.5.2240-2241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang Y, Gao H, Chen J, Dong Y, Wu L, et al. Pellicle formation in Shewanella oneidensis. BMC Microbiol. 2010;10:291. doi: 10.1186/1471-2180-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
(PDF)