Abstract
Background
Evolutionary theories of aging propose that longevity evolves as a competition between reproduction and somatic maintenance for a finite pool of resources. Reproduction is thought to shorten lifespan by depleting resources from processes promoting somatic maintenance. Maternal yolk production, vitellogenesis, represents a significant maternal cost for reproduction and is suppressed under genetic and environmental conditions that extend lifespan. However, little is known about the pathways regulating vitellogenesis in response to prolongevity cues.
Results
In order to identify mechanisms that suppress vitellogenesis under prolongevity conditions, we studied factors regulating vitellogenesis in C. elegans nematodes. In C. elegans, vitellogenesis is depressed in the absence of insulin-like signaling (IIS). We found that the C. elegans daf-2/IIS pathway regulates vitellogenesis through two mechanisms. vit-2 transcript levels in daf-2 mutants were indirectly regulated through a germline-dependent signal, and could be rescued by introduction of daf-2(+) sperm. However, yolk protein (YP) levels in daf-2 mutants were also regulated by germline-independent posttranscriptional mechanisms.
Conclusions
C. elegans vitellogenesis is regulated transcriptionally and posttranscriptionally in response to environmental and reproductive cues. The daf-2 pathway suppressed vitellogenesis through transcriptional mechanisms reflecting reproductive phenotypes, as well as distinct posttranscriptional mechanisms. This study reveals that pleiotropic effects of IIS pathway mutations can converge on a common downstream target, vitellogenesis, as a mechanism to modulate longevity.
Background
According to evolutionary theories of aging, lifespan evolves as a trade-off between the metabolic costs of somatic maintenance with those of reproduction. Reproductive processes such as egg production and progeny rearing are energy-intense and drain resources away from processes that promote somatic maintenance. Studies have provided evidence for a trade-off between reproduction and survival. For example, experimentally increased egg production in wild seabirds is associated with lower rates of postmigratory return to breeding grounds [1,2]. One mechanism by which organisms can regulate the relative burdens of reproduction and somatic maintenance in response to environmental conditions is through phenotypic plasticity of life-history traits, such as growth and reproduction [3]. Plasticity in life-history traits can affect lifespan directly, such as to delay reproduction until environmental conditions improve, or indirectly, as a consequence of elevated stress resistance.
One highly plastic reproductive process is vitellogenesis, the process of maternal yolk production that provides the major nutrient source for developing embryos. Vitellogenin genes are expressed in adult females within tissues specialized for yolk production, such as the insect fat body and avian liver. Vitellogenesis has been well-studied in birds and insects and is hormonally regulated in response to environmental conditions. In insects, vitellogenesis is induced by the coordinate action of 20-hydroxyecdysone and juvenile hormone (JH). Insect JH is regulated, in turn, by insulin-like signaling (IIS). Eggs in Drosophila insulin receptor mutants fail to become vitellogenic and this phenotype can be rescued by methoprene, a JH analog [4,5]. In Aedes aegypti mosquitoes, vitellogenin gene expression is synergistically enhanced by ecdysone and insulin signaling, via the nutrient sensing protein, TOR [6].
IIS is also required for vitellogenesis in C. elegans nematodes [7-9]. In C. elegans, IIS is mediated by the insulin receptor-like protein (IR), DAF-2 which transduces signals via AGE-1/PI3K to antagonize the activity of DAF-16, a FoxO transcription factor [10-13]. Under conditions that reduce DAF-2/IR signaling, levels of vit gene transcripts and yolk proteins (YP) are both reduced [7,14]. In addition to suppressing vitellogenesis, defective IIS prolongs adult lifespan in C. elegans and Drosophila [15]. The mechanism by which IIS modulates longevity is likely to be multifactorial and complex [16]. Vitellogenesis appears to be one target of the IIS prolongevity mechanism, possibly for the purpose of reallocating resources to somatic maintenance. Consistently, RNAi knockdown of vitellogenin gene expression could extend lifespan of wildtype adults [7]. Fecundity has been linked to longevity in daf-2 mutants. Sixteen daf-2 alleles were associated with reduced brood size at nonpermissive temperatures [17]. However, certain alleles, termed class 1, had brood sizes ranging from 85-100% that of wildtype, while class 2 alleles had brood sizes ranging from 60-93% of wildtype. In insects, IIS regulates vitellogenesis hormonally through JH. However, it is not known whether C. elegans IIS also regulates vitellogenesis through hormonal effectors or by direct action in the intestine, the site of vitellogenin production.
To address this question, we identified factors modulating C. elegans vitellogenesis and then examined whether these factors were modified by IIS or were IIS-independent. The C. elegans genome contains six vitellogenin (vit) genes. The genes, vit-1, -2, -3, -4 and -5, all contribute to the pool of YP170, the major yolk protein. vit-6 encodes a protein that is processed into YP115 and YP88. Our analyses show that IIS stimulates vitellogenin gene transcription through a sperm-dependent signal, implying the existence of germline-dependent hormonal regulators of vitellogenesis in C. elegans. However, yolk protein levels correlated poorly with vit-2 mRNA and were regulated by IIS independently of sperm and nutrients. Based on these findings, we propose that vit genes may be transcribed in excess and titrated to reflect reproductive needs. Furthermore, IIS deficits appear to suppress vitellogenesis convergently through germline-dependent transcriptional mechanisms and a separate posttranscriptional mechanism.
Results
Differential regulation of vit-2 transcription and translation
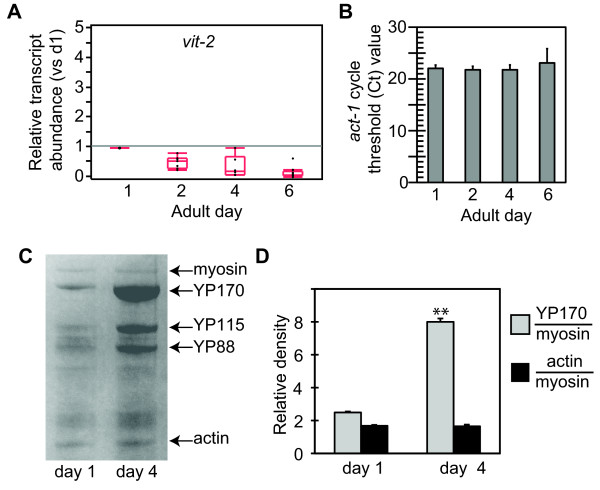
To measure vitellogenesis in C. elegans adult hermaphrodites, we examined vit mRNA levels and yolk protein (YP) levels. First, quantitative RT-PCR was used to determine levels of the vit transcripts contributing to the pool of YP170, the major C. elegans yolk protein. We developed primer sets that specifically recognized vit-2 or vit-5, but not vit-1, -3 or -4, which have very similar sequence. For simplicity, the term "vit" may be used to refer to vit-2/5. Samples were collected at four time points across the adult reproductive period, between adult days 1 to 6. In wildtype worms, the abundance of vit-2 and -5 transcripts declined progressively from adult days 1 to 6, resulting in approximately 7-fold reduction over this period (p ≤ 0.0001, ANOVA) (Figure 1A, Table 1). In comparison, absolute levels of act-1 mRNA did not decline over this period, indicating that transcription was not globally reduced and these changes were relatively specific to vit genes (p = 0.49, ANOVA) (Figure 1B).
Figure 1.
Regulation of vit-2 mRNA and yolk protein levels in young adult wildtype C. elegans hermaphrodites. A) Quantitation of vit-2 mRNA in wildtype adult hermaphrodites, relative to act-1, normalized to day 1 levels. Levels of vit-2 mRNA decline progressively during the first 6 days of adulthood in wildtype hermaphrodites. mRNA levels were measured in independent trials using pools of 30 hermaphrodites at indicated adult ages. Results were normalized to mRNA levels in day 1 adult wildtype hermaphrodites, relative to act-1. Results were statistically analyzed and are presented as quantile plots. Black dots indicate results from individual trials. The median value is indicated by a horizontal line within the quantile box plot. The 25th and 75th quantiles are represented by the upper and lower ends of the boxplot, respectively. Whisker lines extending from the box designate the outer-most data point that falls within 1.5 × the range between the 25th and 75th percentiles. Summary statistics are presented in Table 1. B) Absolute values of act-1 mRNA as determined using the cycle threshold (Ct). Levels act-1 mRNA remained constant between adult days 1-6 in wildtype hermaphrodites (p = 0.49, ANOVA). Data are 5 independent measurements at each age. C) Analysis of yolk protein (YP) abundance by SDS-PAGE of total lysates from wildtype hermaphrodites stained with Coomassie blue. Arrows indicate YP170, YP115 and YP88, and myosin and actin which provided standards for YP quantitation. YP levels relative to myosin were elevated in day 4 hermaphrodites compared to day 1, although actin levels remained constant. Gel results are quantified in D). Error bars, standard deviation; ** p < 0.01, T-test.
Table 1.
Vitellogenin transcriptional regulation in daf-2 pathway mutants
| Genotype | Day | vit-2 # | vit-5 # | sod-3 # | Trials |
|---|---|---|---|---|---|
| Wildtype | 1 | 1.0 | 1.0 | 1.0 | 17 |
| Wildtype | 2 | 0.54(0.20) | 0.75 (0.30) | 0.90 (0.31) | 7 (6^) |
| Wildtype | 4 | 0.39(0.35) | 0.61 (0.59) | 0.77 (0.54) | 6 (5^) |
| Wildtype | 6 | 0.19(0.16) | 0.23(0.25) | 1.1 (0.96) | 13 (12^) |
| daf-2 (e1368) | 1 | 1.9 (1.4) | 1.5 (1.0) | 9.9 (6.9)* | 6 |
| daf-2 (e1368) | 2 | 0.74 (0.10) | 0.55 (1.1) | 5.7 (6.2) | 3 |
| daf-2 (e1368) | 4 | 0.095 | 0.086 | 2.53 | 1 |
| daf-2 (e1368) | 6 | 0.10(0.08) | 0.10(0.10) | 4.5 (1.2)* | 3 |
| daf-2 (e1370) | 1 | 1.6 (1.3) | 0.86 (0.95) | 20.4 (11.5)* | 8 |
| daf-2 (e1370) | 2 | 0.64 (0.48) | 0.16 (0.14)* | 21.1 (4.9)* | 3 |
| daf-2 (e1370) | 4 | 0.13(0.14) | 0.04(0.05) | 15.4 (14.5) | 3 |
| daf-2 (e1370) | 6 | 0.05(0.03)* | 0.03(0.03)* | 21.7 (16.1)* | 6 |
|
daf-16 (mgDf50); daf-2(e1370) |
1 | 2.1 (1.8) | 1.8 (1.8) | 1.05 (0.27) | 6 (5^) |
|
daf-16 (mgDf50); daf-2(e1370) |
2 | 0.29 (1.0) | 0.40 (0.19) | 0.56 (0.30) | 2 |
|
daf-16 (mgDf50); daf-2(e1370) |
4 | 0.29 (0.43) | 0.47 (0.61) | 0.24 (0.10) | 3 |
|
daf-16 (mgDf50); daf-2(e1370) |
6 | 0.28 (0.29) | 0.35 (0.32) | 0.34 (0.21)* | 2 |
# mRNA levels relative to day 1 wildtype (SD).
Bold, p < 0.05 vs day 1, within genotype, T-test.
^ Trials in which sod-3 was measured.
* p < 0.05 vs wildtype, same day, T-test.
To determine the concordance between vit message and protein levels during this period, we also examined yolk proteins directly in wildtype hermaphrodites. Yolk proteins (YP) are the most abundant proteins in the worm and are easily detected in Coomassie-stained crude lysates after polyacrylamide gel electrophoresis [18]. We confirmed the identities of bands corresponding to YP170, YP115, YP88, actin (42 kDa) and myosin (200 kDa) by mass spectrometry. We quantified the levels of each protein by densitometry and calculated abundance relative to myosin, as an internal standard, as well as an independent loading standard run on the same gel. In contrast to the progressive decline observed for vit-2 and -5 transcripts, YP levels increased in abundance nearly 4-fold by adult day 4 (p ≤ 0.0001, T-test) (Figure 1C,D). Actin levels did not change over this period. Furthermore, the absolute levels of myosin relative to a loading standard were constant, showing there was no overall change in protein levels. Rather, the increased level of YP170 reflected specific increases in this pool.
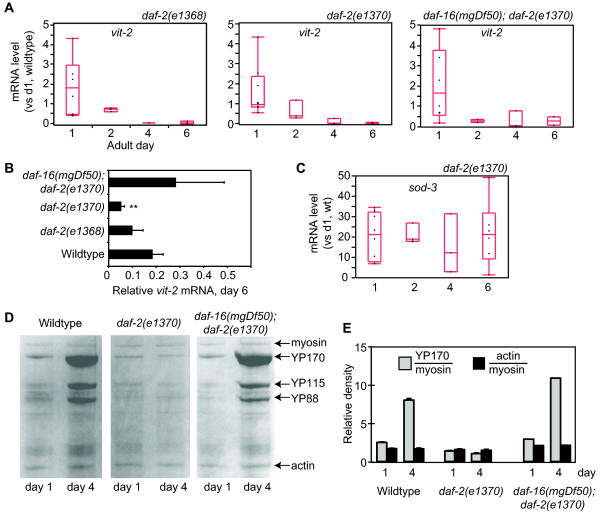
We next examined vit mRNA levels in daf-2 mutant hermaphrodites. Most daf-2 mutations prolong adult lifespan but have pleitropic effects on other adult processes. The class 1 daf-2(e1368) mutation extends adult lifespan, but does not strongly affect other adult traits, such as fertility, while the class 2 daf-2(e1370) mutation is more pleitropic and negatively affects fertility [17]. We observed that vit-2 and -5 mRNA levels were similar to wildtype in both daf-2 mutants on days 1, 2 and 4, but were strongly reduced in daf-2(e1370) hermaphrodites on adult day 6 (Figure 2, Table 1). Reduced vit-2 transcript abundance in daf-2 hermaphrodites was dependent on daf-16, which encodes a FOXO transcription factor that is the major target of signaling by the DAF-2/insulin receptor, and vit mRNAs were present at normal levels in day 6 daf-16(mgDf50); daf-2(e1370) hermaphrodites (Figure 2A,B, Table 1) [12,19,20]. We investigated whether sod-3, a direct target of DAF-16, also changed progressively between adult days 1 and 6 [21-23]. Unlike vit-2, sod-3 mRNA did not undergo a progressive change, but was maintained at high levels in daf-2(e1370) mutants across this period (p = 0.92, ANOVA) (Figure 2C, Table 1). This suggests that vit-2/-5 repression in daf-2(e1370) adults occurs through a different mechanism than sod-3 activation. In contrast to the progressive decline we observed for vit transcripts in daf-2 hermaphrodites, YP levels were depressed in daf-2(e1370) hermaphrodites for the entire reproductive period (2-fold in day 1 adults vs wildtype, p ≤ 0.001; 8-fold in day 4 adults vs. wildtype, p ≤ 0.0001) (Figure 2D,E). YP suppression in daf-2(e1370) hermaphrodites was also daf-16-dependent. Although YP levels were consistently lower in daf-2(e1370) adults, actin was present at normal levels (Figure 2E).
Figure 2.
Suppression of vitellogenesis in daf-2(e1370) hermaphrodites. A) vit-2 mRNA levels relative to day 1 wildtype hermaphrodites in daf-2(e1368), daf-2(e1370) and daf-16(mgDf50); daf-2(e1370) adult hermaphrodites. (Left) vit-2 mRNA levels were similar to wildtype in daf-2(e1368). (Center) In daf-2(1370) hermaphrodites, vit-2 mRNA levels were similar to wildtype on days 1-4, but were significantly lower on day 6 (p = 0.001, T-test). (Right) The reduction of vit-2 mRNA in daf-2(e1370) was daf-16-dependent and was suppressed in daf-16(mgDf50); daf-2(e1370) hermaphrodites. See Table 1 for statistical summary. B) Direct comparison of vit-2 mRNA levels in day 6 adults of indicated genotype. Error bars, SEM; ** p < 0.001, T-test vs. wildtype day 6. C) sod-3 mRNA levels in daf-2(e1370) hermaphrodites. There was no statistically significant change between days 1-6 (p = 0.78, ANOVA). D) Coomassie-stained SDS-PAGE analysis of total lysates for measuring YP levels. E) Quantification of YP170 and actin abundance relative to myosin, as indicated. In day 1 daf-2(e1370) hermaphrodites, YP170 levels were reduced compared to wildtype and daf-16(mgDf50); daf-2(e1370) hermaphrodites (p < 0.01, T-test). In addition, YP170 increased in day 4 wildtype and daf-16(mgDf50); daf-2(e1370) hermaphrodites, but remained low in daf-2(e1370) hermaphrodites (p < 0.01, e1370 vs wildtype, day 4). There were no significant differences in actin levels, relative to myosin, between strains.
Sperm stimulate vit mRNA levels in hermaphrodites
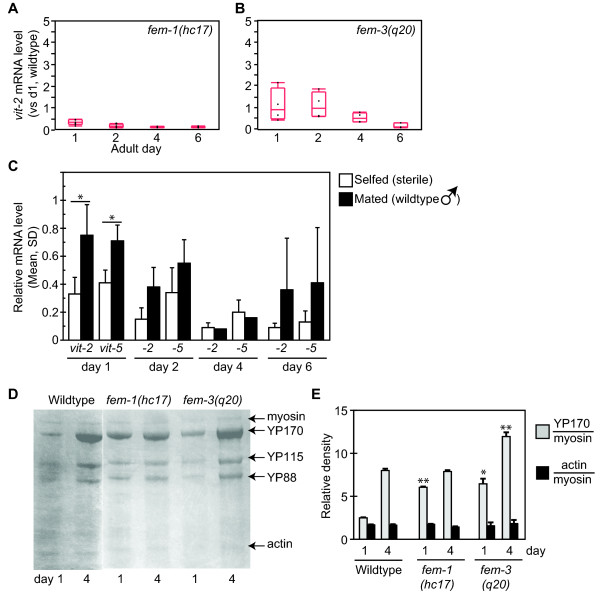
We next considered whether the progressive decline in vit mRNA levels could reflect a physiological change that occurred between adult days 1 and 6. This period encompasses the fertile period, when C. elegans hermaphrodites lay a brood of approximately 200 eggs. Sperm abundance limits fertility in wildtype hermaphrodites, and cessation of egg production reflects sperm depletion [24]. To investigate the relationship between reproduction and vit expression, we examined vit transcripts and YP levels in mutants with reproductive defects. In fem-1(hc17) hermaphrodites, which fail to produce sperm [25], vit-2 and vit-5 mRNA were significantly reduced on adult days 1 and 2 compared to wildtype (Figure 3A, Table 2). These transcripts remained suppressed at days 4 and 6, but were not statistically significantly different than in the wildtype at these ages. In contrast, vit-2 and -5 mRNA levels were normal in fem-3(q20gf) animals, which develop a masculinized germline producing primarily sperm (Figure 3B, Table 2) [26]. The levels of sod-3 mRNA were roughly normal in both strains, indicating that the fem-1 and fem-3 mutations did not have pleitropic effects on daf-2/daf-16 pathway activity (Table 2). Thus, vit transcriptional suppression may be correlated with deficits in sperm, but not oocyte, production.
Figure 3.
Regulation of vitellogenesis in sterile mutants. A, B) vit-2 mRNA levels in sperm-deficient fem-1(hc17) (A) and oocyte-deficient fem-3(q20gf) (B) hermaphrodites, relative to day 1 wildtype hermaphrodites. See Table 2 for statistical summary. C) vit-2 and -5 mRNA levels in fem-1(hc17) hermaphrodites of indicated adult age who were maintained in the absence (unfilled) or presence (filled) of wildtype males (*p < 0.05 mated versus unmated; T-test). D, E) YP levels in sterile fem-1(hc17) and fem-3(q20gf) adults on days 1 and 4. *p < 0.05, **p < 0.01 versus wildtype hermaphrodites of the same age, T-test.
Table 2.
Levels of vit-2, vit-5 and sod-3 mRNA in sterile strains
| Genotype | Day | vit-2 # | vit-5 # | sod-3 # |
|---|---|---|---|---|
| fem-1(hc17) | 1 | 0.33 (0.12, 7)* | 0.041 (0.09, 7)* | 0.93 (0.1, 6) |
| fem-1(hc17) | 2 | 0.15 (0.08, 4)* | 0.34 (0.18, 4)* | 3.0 (3.7, 2) |
| fem-1(hc17) | 4 | 0.092 (0.03, 3) | 0.20 (0.09, 3) | 1.2 (1.4, 2) |
| fem-1(hc17) | 6 | 0.094 (0.03, 7) | 0.13 (0.08, 7) | 0.63 (0.5, 6) |
| fem-3(q20) | 1 | 1.1 (0.76, 4) | 1.3 (1.1, 4) | 0.98 (1.2, 4) |
| fem-3(q20) | 2 | 1.1 (0.60, 4) | 1.1 (0.54, 4) | 4.4 (6.8, 4) |
| fem-3(q20) | 4 | 0.53 (0.22, 4) | 0.43 (0.18, 4) | 1.69 (1.2, 4) |
| fem-3(q20) | 6 | 0.16 (0.12, 3) | 0.05 (0.04, 3)* | 2.72 (2.31, 3) |
| fem-1(hc17) × wildtype males | 1 | 0.75 (0.22, 4)^ | 0.71 (0.23, 4) | 0.53 (0.10, 4) |
| fem-1(hc17) × wildtype males | 2 | 0.38 (0.14, 2) | 0.55 (0.17, 2) | 0.12 (0.15, 2) |
| fem-1(hc17) × wildtype males | 4 | 0.082 (1 trial) | 0.16 (1 trial) | 0.24 (1 trial) |
| fem-1(hc17) × wildtype males | 6 | 0.357 (0.37, 3) | 0.41 (0.4, 3) | 0.20 (0.12, 3) |
# Average mRNA level (SD, trials)
* p < 0.05 vs. wildtype, same day, T-test.
^ p < 0.05, vs. fem-1(hc17) unmated, T-test.
To test this hypothesis, we examined whether wildtype sperm could stimulate vit-2 mRNA levels in sperm-deficient fem-1(hc17) hermaphrodites by measuring mRNA after allowing them to mate with wildtype males. We confirmed that mating rescued fem-1(hc17) sterility by the appearance of fertile heterozygous progeny (not shown). Sterile fem-1 hermaphrodites co-cultured with wildtype males exhibited a broad range of vit-2 mRNA levels which, on average, were elevated compared to unmated fem-1 hermaphrodites (Figure 3C, Table 2). On adult day 1, we observed a statistically significant elevation in vit-2 mRNA levels from mating (p = 0.02; T-test). Mating had no effect on sod-3 mRNA (p = 0.81). The progressive decline in vit-2 mRNA between adult days 1 and 6 was also abrogated by coculture with wildtype males (vit-2 mRNA days 1-6: p < 0.001 selfed, p = 0.17 mated; ANOVA). These results demonstrate that vit transcriptional suppression in sterile fem-1(hc17) hermaphrodites could be rescued by the introduction of wildtype sperm from mating.
To assess the impact of reduced vit transcripts on overall yolk protein production, we examined YP170 levels in sterile fem-1(hc17) adult hermaphrodites. In comparison to wildtype adults, YP levels relative to myosin were normal or elevated in fem-1(hc17) adults (Figure 3D,E). Yolk protein levels were also elevated in germline-masculinized fem-3(q20gf) hermaphrodites. This indicates that the sperm-dependent stimulation in vit-2/5 mRNA levels is likely to be dispensable for YP production under normal conditions.
Deficits in IIS suppress vitellogenesis through two separable effects
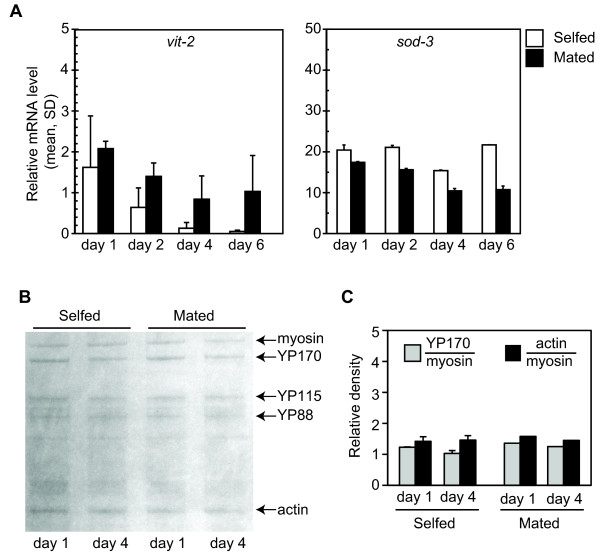
Our analysis showed that vit-2/-5 transcriptional suppression in daf-2(e1370) hermaphrodites followed the same temporal pattern as in wildtype hermaphrodites. Since vit-2 mRNA was stimulated by sperm in fem-1(hc17) hermaphrodites, we tested whether wildtype sperm could also enhance vit-2 mRNA levels in daf-2(e1370) hermaphrodites. When daf-2(e1370) hermaphrodites were cocultured with wildtype males, vit-2 mRNA levels were stabilized during days 1-6, compared to self-fertilized hermaphrodites (p = 0.016 in day 1-6 unmated hermaphrodites; p = 0.1 in day 1-6 mated hermaphrodites; ANOVA)(Figure 4A). Similar results were obtained for vit-5 mRNA (not shown). Mating had no significant effect on sod-3 message levels in daf-2(e1370) hermaphrodites, consistent with regulation of these targets through distinct mechanisms (p = 0.17 in day 1-6 unmated hermaphrodites; p = 0.88 in day 1-6 mated hermaphrodites; ANOVA). As noted for fem-1, we detected a broad range of vit transcript levels in mated daf-2(e1370) hermaphrodites, such that relative differences in vit transcripts of mated compared to selfed daf-2(e1370) hermaphrodites were not statistically significant at any single timepoint. This variation is likely to reflect differences in mating efficiency between trials [24]. We confirmed that mating occurred by the presence of heterozygous male progeny. Mating did not significantly alter progeny production nor have adverse effects for the mated hermaphrodites, such as death from internal hatching of embryos (not shown). Although mating stimulated levels of vit-2 transcripts in daf-2(e1370) hermaphrodites, there was no effect on YP (Figure 4B,C). In both mated and unmated daf-2(e1370) hermaphrodites, YP170 levels were equivalently reduced in comparison to wildtype.
Figure 4.
Vitellogenesis in daf-2(e1370) hermaphrodites mated with wildtype males. A) vit-2 and sod-3 mRNA levels, relative to wildtype day 1, in daf-2(e1370) hermaphrodites maintained at 25°C in the absence (unfilled bars) or presence (filled bars) of wildtype males. The levels of vit-2 mRNA declined between days 1-6 of adulthood in unmated (self-fertile) hermaphrodites (p = 0.016, ANOVA). This decline was abrogated in daf-2(e1370) hermaphrodites cocultured with wildtype males (p = 0.1, ANOVA). No significant change was detected in sod-3 mRNA levels in unmated or mated hermaphrodites (unmated, p = 0.17; mated, p = 0.88; ANOVA). Similar results were obtained for vit-5 mRNA. Unmated, 8 replicates were analyzed for day 1, 3 replicates for days 2 and 4, and 6 replicates for day 6; mated, 3 replicates were analyzed for each age. B) YP levels visualized by Coomassie after SDS-PAGE in daf-2(e1370) hermaphrodites maintained as for (A). C) Densitometric quantitation of YP170 and actin (control), relative to myosin, in (B). Although mating with wildtype males could stabilize vit-2 mRNA, there was no stimulation of YP levels.
Cis-acting requirements for vitellogenin repression in daf-2(e1370) adults
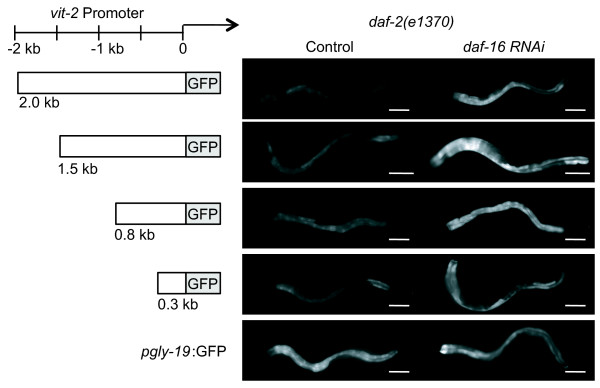
To further characterize the basis for YP suppression in daf-2(e1370) hermaphrodites, we tested whether a GFP reporter expressed from the vit-2 promoter could recapitulate daf-2 regulation of endogenous YP. Using an pvit-2:gfp reporter containing 2 kb of upstream promoter sequence (pvit-2(2.0):gfp), we compared GFP fluorescence in daf-2(e1370) day 1 adult hermaphrodites grown on daf-16 RNAi or control conditions. The pvit-2:gfp reporter was a transgene maintained as a stably-transmitted extrachromosomal array. If the pvit-2:gfp reporter were a target for daf-2 suppression of vitellogenesis, then we expected daf-16 RNAi to block daf-16 activity and allow increased GFP expression in a daf-2(e1370) background. Consistently, the level of GFP fluorescence was higher in daf-2(e1370) hermaphrodites treated with daf-16 RNAi than untreated animals (68%, 37%, 24% and 35% increase by daf-16 RNAi in 4 independent lines) (Figure 5, Table 3). As a negative control, we found that daf-16 RNAi did not affect GFP expression from the intestinal gly-19 promoter [27], which is independent of daf-2-pathway regulation (-3% and -28% change in pgly-19:GFP fluorescence after daf-16 RNAi in 2 independent lines).
Figure 5.
Expression of pvit-2:gfp transcriptional reporters in daf-2(e1370) hermaphrodites in the presence or absence of daf-16 activity. Left, schematic illustration of vit-2 promoter fragments used to express gfp. As a control, GFP expression was examined from the pgly-19:gfp transcriptional reporter, containing the 1910 bp gly-19 promoter [27]. Right, representative images of GFP fluorescence in daf-2(e1370) day 1 adult hermaphrodites maintained on control bacteria (L4440) or daf-16 dsRNA-expressing bacteria to induce daf-16 RNAi. In all experiments, animals were raised at 15°C to the L4-adult molt and then transferred to 25°C, the nonpermissive condition for certain daf-2(e1370) phenotypes. GFP expression from all vit-2 promoters was increased under daf-16 RNAi conditions, indicating that daf-16 activity suppressed expression from each vit-2 promoter fragment. GFP expression from the gly-19 promoter was unaffected by daf-16 RNAi. Bars, 100 microns. Images were collected with identical acquisition settings. Statistical summary presented in Table 3.
Table 3.
Effect of daf-1 6 RNAi on pvit-2:gfp expression in daf-2(e1370) adult hermaphrodites
| Reporter | Line 1^ | Line 2^ | Line 3^ | Line 4^ |
|---|---|---|---|---|
| pvit-2(2.0):gfp | 68%(0, 1) | 37%(0.1, 5) | 30%(0.2, 5) | 35%(0, 1) |
| pvit-2(1.5):gfp | 47%(0.04, 2) | |||
| pvit-2(0.8):gfp | 26% (0.2, 5)* | 48%(0.2, 2) | ||
| pvit-2(0.3):gfp | 30%(0, 1) | 40%(0, 1) | 24%(0, 1) | |
| pgly-19:gfp | -3% (0, 1) | -28%(0, 1) | ||
^ Relative GFP fluorescence (SD, trials). Data show % change (SD, trials) of average maximum GFP fluorescence from daf-16 RNAi versus control in daf-2(e1370) hermaphrodites. Positive values indicate increased pvit-2:GFP fluorescence associated with reduced daf-16 activity from RNAi. Bold indicates GFP intensity was significantly different in daf-16 RNAi-treated animals in all trials (p < 0.05, T-test).
* GFP fluorescence intensity increased statistically significantly in 3/5 trials (p ≤ 0.05, T-test). In the remaining 2 trials, % GFP intensity change on daf-16 RNA was +5% (p = 0.3) and +6% (p = 0.6)
Using GFP reporters expressed from subfragments of the 2-kb vit-2 promoter, we identified a cis-regulatory region responsive to daf-16 activity. Deletions of 0.5, 1.2 and 1.7 kb upstream of the vit-2 translational start sequence did not disrupt daf-16-dependent repression of pvit-2:GFP expression in daf-2(e1370) hermaphrodites (Figure 5 Table 3). Thus, daf-16 regulation of YP requires only a short 300-bp vit-2 promoter, but not additional upstream sequences. The 300-bp vit-2 promoter contains binding sites for three transcription factors that direct vitellogenenin expression in the adult hermaphrodite intestine: the ELT-2 GATA factor, the male-specific transcriptional repressor MAB-3, and an unidentified factor which binds to VPE2 repeat sequences in the vit-2 promoter [28-30]. DAF-16 recognizes its transcriptional targets through a binding site with consensus GTAAAc/tA [22]. Although the vit-2 promoter contains this sequence within the 5-kb upstream of the translational start [7], we did not detect any matches within the 300-bp promoter fragment sufficient for daf-16-mediated repression.
We also examined gfp mRNA levels in animals containing an integrated pvit-2(2.0):gfp transgene. As for the endogenous vit-2 locus, gfp mRNA was not detectable in wildtype or daf-2(e1370) adults immediately after the L4-adult molt (day 0), but increased dramatically by adult day 1 (Table 4). Levels of gfp mRNA were similar in day 1 wildtype and daf-2(e1370) adults (p = 0.771, T-test, 3 trials). This observation suggests that, like endogenous vit-2, the pvit-2:gfp transgene was not subject to transcriptional suppression at this time point. This is consistent with the conclusion that daf-16 activity in daf-2(e1370) adults acts to repress GFP levels at a posttranscriptional step.
Table 4.
Levels of pvit-2:gfp mRNA in wildtype and daf-2(e1370) adults
| Wildtype | daf-2(e1370) | |||
|---|---|---|---|---|
| Adult day | vit-2^ | gfp^ | vit-2^ | gfp^ |
| 0 | 0.007 | 0.001 | 0.008 | 0.001 |
| 1 | 1.00 (0) | 1.00 (0) | 2.60 (0.566)# | 1.10 (0.503)∞ |
Day 0, 1 trial; day 1, 3 trials.
^ Mean mRNA level (SD); mRNA level relative to act-1, normalized to wildtype day 1 hermaphrodites.
# p = 0.04, vs. vit-2 mRNA in wildtype day 1, T-test.
∞ p = 0.8, vs pvit-2:gfp mRNA in wildtype day 1, T-test.
Discussion
This study's goal was to characterize the effect of daf-2 pathway activity on vitellogenesis in C. elegans. Our findings corroborate previous reports showing that daf-16 activity suppresses vitellogenesis at both the transcriptional and translational levels [7-9,14]. However, our temporal analysis of this suppression do not support a simple model where DAF-16 acts as a direct transcriptional repressor of vit genes. First, YP levels were reduced in a daf-16-dependent manner in young adults, although vit mRNA levels were normal or elevated at this stage. Second, wildtype sperm stimulated vit-2 mRNA non-autonomously in daf-2 hermaphrodites, but failed to increase YP levels. Third, we observed daf-16-dependent suppression of pvit-2(0.3 kb):gfp expression, although this promoter fragment lacks DBE-like DAF-16 binding sites, with the caveat that DAF-16 might bind to non-DBE sites in the vit-2 promoter [31]. Finally, we note that vit genes were not represented among 103 direct DAF-16 targets identified by chromatin immunoprecipitation [23].
Since these data do not seemingly support the simple model that DAF-16 functions as a transcriptional repressor at the vit loci, we propose an alternative model whereby daf-16 represses vitellogenesis through two parallel pathways that independently regulate vit transcription and translation (Figure 6). At the transcriptional level, increased daf-16 activity in daf-2(e1370) adults was associated with a progressive decline of vit mRNA which mirrored a similar progressive decline of vit mRNA in wildtype adults. We also found that vit mRNA levels were low in a feminized mutant unable to produce sperm, but were normal in a masculinized mutant that produces sperm but not oocytes. Together, these observations suggest a role for sperm in stimulating vit mRNA levels in early adulthood. The daf-2 pathway promotes germcell proliferation and daf-2(e1370) hermaphrodites have low fertility at 25°C [17,32,33]. Because the introduction of wildtype sperm could stimulate vit mRNA levels in daf-2(e1370) hermaphrodites, we theorize that daf-2(e1370) hermaphrodites are deficient in a sperm-derived signal that simulates vit-2/-5 gene transcription.
Figure 6.
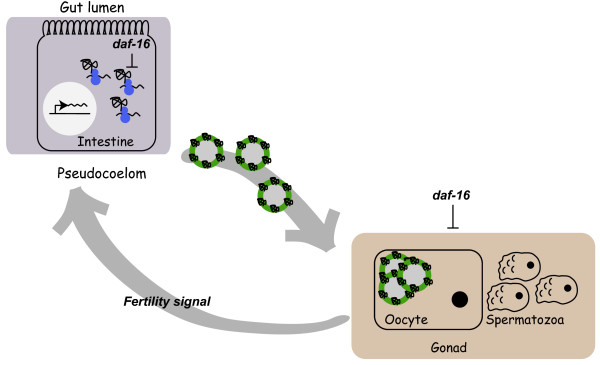
Model of factors regulating C. elegans vitellogenesis. In intestinal cells of adult hermaphrodites (left), vit genes are transcribed and translated into yolk proteins (YP), which function as ApoB-100-like lipid transport proteins for mobilization of nutrients from the intestine to the oocytes. After synthesis in the intestine, YP are exported into the pseudocoelomic space and taken up by oocytes in the gonad (right). daf-16 activity appears to affect vitellogenesis through two separable pathways. In intestinal cells, daf-16 activity represses YP production at a posttranscriptional level. In addition, increased daf-16 activity in daf-2(e1370) hermaphrodites suppresses a germline-dependent signal that stimulates vit-2 mRNA production. The daf-16-dependent regulation of germline-dependent vit-2 mRNA stimulation may reflect daf-2 and daf-16 regulation of germline proliferation [22].
In contrast to the progressive decline of vit mRNA in daf-2(e1370) hermaphrodites, yolk protein production was suppressed constitutively throughout adulthood. This observation suggests a shift of translation regulation. Consistently, GFP expression from the pvit-2(2.0):gfp transcriptional reporter was suppressed in a daf-16-dependent manner, although gfp mRNA levels were comparable in wildtype and daf-2(e1370) hermaphrodites. When under stress, cells respond by upregulating translation of stress-resistance proteins, such as heat-shock proteins and chaperones. At the same time, cells repress translation of non-essential proteins to conserve energy supplies [34]. One interpretation of our results is that daf-2(e1370) adults undergo a similar translational switch to repress YP synthesis in early adulthood. Genetic results indicate that such a shift in translational regulation should be regulated by one or more DAF-16 target genes. It is not currently known if specific DAF-16 targets may regulate translation. However, MS analysis of 1685 proteins in daf-2(e1370) and wildtype adults identified 86 proteins with differential abundance, only 35 of which had been previously identified as DAF-16 target genes [14]. While the remaining 51 proteins might be novel DAF-16 targets, as proposed [14], it is also possible that these might represent additional secondary targets of a daf-16-dependent shift in translational regulation. We note that global translational reprogramming in response to thermal stress (35°C, 2 hours) occurs normally in daf-2(e1370) adults [35]. Thus, daf-2/daf-16 pathway activity may regulate translational reprogramming at 25°C, a relatively nonstressful condition, but not under severe stress, such as at 35°C.
We were surprised to observe significant discordance between vit transcript and YP levels under the conditions we examined in the course of this study. In wildtype adults, YP170 increased during adulthood although vit-2 and -5 mRNA decreased. Furthermore, YP170 was reduced in daf-2(e1370) day 1 adults which contained normal levels of vit-2 and -5 mRNA. We acknowledge the caveat that other vit loci (vit-1, -3 and -4), which contribute to the YP170 pool, may have increased expression to make up for the decline in vit-2 and -5. However, other studies have observed these 5 loci to be expressed at similar levels [8] (S.K. Rao & C.A.Wolkow, unpublished results). In addition, alternative regulation of these loci could not account for the discrepancy between vit mRNA and YP170 in day 1 daf-2(e1370) adults.
Discrepancies between mRNA and protein levels have been observed under other conditions. Studies in yeast have examined the correlation between transcript and protein abundance on a large scale. With the exception of highly expressed genes, these studies report little correlation between relative levels of mRNA and protein, concluding that transcript levels may be inadequate reflections of the true cellular composition [36]. One factor that could contribute to discordance between vit transcript and YP levels is the potential accumulation of YP within embryos in the hermaphrodite uterus or the pseudocoelomic body cavity. Thus, older hermaphrodites containing more eggs in the uterus might also contain more yolk in bulk lysates. However, this explanation does not account for our finding that fem-3(q20) hermaphrodites, which fail to make oocytes, contained normal levels of YP170. In this strain, YP are likely to accumulate in the pseudocoelom. Rather, we hypothesize that vit genes are transcribed at high levels on the first day of adulthood, in anticipation of reproductive needs. Then, as reproduction proceeds over the next 3-5 days, vit mRNA levels are titrated downward in response to actual reproductive demand. Providing excess vit transcripts in early adulthood may ensure adequate YP synthesis capacity for maximum reproductive rate.
Conclusions
Vitellogenins have been repeatedly identified as targets repressed by daf-16 activity and downregulated under prolongevity conditions [7-9,14]. Furthermore, vitellogenin downregulation by RNAi was itself sufficient to extend adult C. elegans lifespan [7]. These findings indicate that longevity and somatic maintenance in daf-2 mutants is likely to reflect combinatorial effects of protective enzyme induction and reproductive suppression. Our studies show that daf-2 pathway deficits suppress vitellogenesis through two separable mechanisms. This reveals new insight that longevity assurance in daf-2 mutants can result from convergent regulation of vitellogenesis by pleitropic phenotypes in these mutants.
Methods
Strains and growth conditions
The strains used in this study were N2 Bristol [wildtype], DR1572 [daf-2(e1368)], CB1370 [daf-2(e1370)], CY312 [daf-16(mgDf50); daf-2(e1370)], BA17 [fem-1(hc17)], JK816 [fem-3(q20)], CY625 [daf-2(e1370); bvIs7 (pvit-2:gfp; gcy-7:gfp)]. Strains were obtained from Caenoharbditis Genetics Center at the University of Minnesota. All strains were maintained at 15°C on nematode growth medium (NGM) agar plates seeded with OP50 E. coli strain following standard protocols [37]. For mating assays, animals were cultured from embryos at 15°C for 4-5 days to the young adult (day 0) stage. Mating assays were carried out using at a ratio of 1 hermaphrodite to 2 males and then shifted to 25°C. 30 hermaphrodites were harvested from mating plates on days 1, 2, 4 and 6 for Q-PCR analysis. For nutrient deprivation experiments, young adult animals (day 0) raised at 15°C were shifted to 25°C for 24 hours (day 1). Day 1 animals were kept on NGM plates with OP50 bacteria (fed) or transferred to plates without food for 6 hours (fasted). Worms were then harvested and processed for Q-PCR analysis or protein gel analysis.
Quantitative real time RT-PCR
Worms were allowed to lay eggs on NGM agar plates and grown at 15°C for 4-5 days to obtain synchronized populations. Young adult (YA) worms were then shifted to 25°C and harvested on day 1 (fertile adult), day 2, day 4 and day 6 (post-reproductive adult) by picking 30 worms into M9 Buffer. Total RNA was isolated using Absolutely RNA Miniprep Kit (Stratagene, La Jolla, CA). cDNA was synthesized using Stratascript First-Strand Synthesis System (Stratagene). Triplicate 25 μl quantitative real time RT-PCR (qPCR) reactions were set-up in 96-well plates using 2 × SYBRGreen master mix (Applied Biosystems, Foster City, CA), and reactions were run on MJ Research Opticon thermal cycler (BioRad, Hercules, CA). Data was analyzed using the Ct method and relative mRNA levels is reported as the mRNA abundance of each gene relative to the mRNA abundance of the control gene, act-1 (β-actin). The following qPCR primers were used: vit-2, 5'-GACACCGAGCTCATCCGCCCA and 5'-TTCCTTCTCTCCATTGACCT; vit-5, 5'-GGCAATTTGTTAAGCCACAA and 5'-CCTCCTTTGGTCCAGAAACCT; sod-3, 5'-CCAACCAGCGCTGAAATTCAATGG and 5'-GGAACCGAAGTCGCGCTTAATAGT; act-1, 5'-CCAGGAATTGCTGATCGTATGCAGAA and 5'-TGGAGAGGGGAAGCGAGGATAGA; gfp, 5' CTGGAGTTGTCCCAATTCTTG and 5'- AAGCATTGAACACCATAACAGAAA. Results of individual trials were statistically analysed using JMP 7.0 (SAS, Cary, NC) to obtain p-values of significance using one-way ANOVA and T-test.
Protein analysis
Worms were allowed to lay eggs on NGM agar plates and grown at 15°C for 4-5 days to obtain synchronized populations. Young adult (YA) worms were then shifted to 25°C and harvested on adult days 1 and 4 by picking 100 worms into M9 Buffer, diluted in 2 × Laemmli Sample Buffer (Bio-Rad) containing beta-mercaptoethanol and incubated in 70°C water bath for 15 min. The worm mixture was vortexed every 5 min during the incubation and spun for 5 min to remove insoluble precipitates prior to loading on a NuPAGE 4-12% Bis-Tris gel (Invitrogen). Gels were stained and destained following standard protocols. Protein bands were quantified using ImageJ (NIH Image).
For protein identification, Coomassie-stained protein bands were excised, micro-dissected and destained with 50% acetonitrile (ACN) with 50 mM ammonium bicarbonate. Proteins were reduced with 10 mM DTT for 30 minutes at room temperature (RT) and alkylated with 10 mg/mL iodoacetamide (Sigma) for 20 min at RT. After alkylation, gel pieces were treated with trypsin (Sequencing grade, Promega) in 50 mM ammonium bicarbonate overnight at 37°C. The resultant peptides were extracted from the gel with 0.1% TFA in water and then 0.1% TFA in 70% ACN for 10 minutes, collected in eppendorf tubes and dried using a vacuum drier (Savant, USA). Peptide samples were desalted with ZipTip (Millipore), dried again, and stored at -80°C for LC-MS/MS analysis.
LC-MS/MS analyses were performed using an LXQ linear ion trap mass spectrometer coupled to a Surveyor LC system (Thermo Fisher Scientific). Peptide samples were loaded onto pre-equilibrated analytical columns using a pressure-loader. The capillary-tip-columns were affixed to the MS ion-source system and reverse-phase chromatographic separations were carried out using the following gradient setting; 100% A (0.1% formic acid in water) for 5 minutes; 0-50% B (0.1% formic acid in ACN) for 60 minutes; 50-70% B for 5 minutes; 70% B for 5 minutes. Pump flow rates were controlled to deliver 200 nL/min to the analytical column, which was achieved by splitting pump flow at a 1:1000 ratio. LXQ settings were as follows: spray voltage, 1.6 kV; 1 microscan for MS scans at maximum inject time 10 ms with mass range 400-1400 m/z, 3 microscans for MS/MS at maximum inject time100 ms with automatic mass range. The LXQ was operated in a data-dependent mode, that is, one MS scan for precursor ions followed by four data-dependent MS/MS scans for precursor ions above a threshold ion count of 500 with normalized collision energy value of 35%.
The MS/MS data were analyzed with Mascot software v2.3.0 (Matrix Sciences). MS/MS peak lists were generated using lcq_dta.exe from the BioworksBrowser v3.3.1 SP1 (Thermo Fisher Scientific) with the following options applied: grouping tolerance, 0; intermediate scans, 0; minimal scans per group, 0; precursor charge state analysis, auto. The generated lists were searched against Sprot C. elegans database with the following criteria: enzyme, trypsin (KR/P); missed cleavage sites, 3; peptide tolerance, 1.0 amu; fragment ions tolerance, 0.5 amu; variable modifications, carbamidomethylation (+57Da) and methionine oxidation (+16Da); decoy database, on. To ensure the false discovery rate (FDR) below 1%, the search results were further filtered with various significant p-values.
pvit-2:gfp reporter analysis
The pvit-2:gfp reporter was constructed using a 2-kb fragment upstream of the vit-2 presumptive ATG start site, which was PCR amplified with primers containing unique BamHI and KpnI restriction sites and ligated to pPD95.75 vector (gift of A. Fire, Stanford University). Animals were transformed by standard microinjection-mediated transformation using pvit-2:gfp plasmid DNA and gcy-7:gfp coinjection marker at a final concentration of 25 ng/μL and 100 ng/μL, respectively. For vit-2 promoter deletion analysis, the 2-kb vit-2 promoter was digested and re-ligated to remove 5' regions of the promoter and generate smaller fragments. Animals were photographed under epi-fluorescence using a 10 × objective on a Nikon E800 with a Hamamatsu Orca ER camera using Openlab software (Improvision). Exposure times were constant for each trial, although exposure times varied between some trials. For GFP quantification, ImageJ (NIH) was used to determine the maximum pixel intensity for a region-of-interest encompassing the intestine. Individual GFP intensity values were measured for an average of 14 animals/trial (range, 3-23 animals) and the values for daf-16 RNAi versus L4440 were compared for each trial of each transgene by T-test (Excel). Percentage changes in GFP fluorescence were calculated from the average maximum GFP intensity measured in each experiment after normalization for exposure time, if necessary.
RNA-interference by feeding
RNA-interference (RNAi) knockdown of daf-16 in C. elegans was achieved by feeding animals E. coli strain HT115 expressing daf-16 dsRNA or contained a control plasmid (L4440) [38]. To obtain synchronous populations, 5-10 adult wildtype or daf-2(e1370) adults carrying pvit-2:gfp transgenic arrays were allowed to lay eggs for 4-6 hrs on NGM agar plates supplemented with ampicillin (100 μg/mL) and IPTG (1 mM). Animals were exposed to daf-16 RNAi from hatching, and grown at 15°C for 4-5 days to YA stage. Animals were then shifted to 25°C and images were captured on days 1, 2, and 4 for GFP fluorescence analysis. GFP fluorescence intensity was measured using ImageJ.
Authors' contributions
ASD, WBI, MAW and CAW designed the experiments. ASD conducted the experiments, except WBI constructed the pvit-2:gfp plasmids and strains and performed GFP expression analysis in Figure 5. SP and SM conducted LC/MS/MS protein identification. ASD and CAW wrote the manuscript
All authors read and approved the manuscript.
Contributor Information
Ana S DePina, Email: adepina1@jhmi.edu.
Wendy B Iser, Email: iserwe@grc.nia.nih.gov.
Sung-Soo Park, Email: parkss@mail.nih.gov.
Stuart Maudsley, Email: maudsleyst@grc.nia.nih.gov.
Mark A Wilson, Email: wilsonma@grc.nia.nih.gov.
Catherine A Wolkow, Email: wolkowca@gmail.com.
Acknowledgements
This work was carried out with funds from the Intramural Research Program of the National Institute on Aging, NIH. We are grateful to members of the NIA Laboratory of Neuroscience and Baltimore/DC area worm club for helpful comments.
References
- Visser ME, Lessells CM. The costs of egg production and incubation in great tits (Parus major) Proc Biol Sci. 2001;268:1271–7. doi: 10.1098/rspb.2001.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager RG, Monaghan P, Houston DC. The cost of egg production: increased egg production reduces future fitness in gulls. J Avian Biol. 2001;32:159–166. doi: 10.1034/j.1600-048X.2001.320209.x. [DOI] [Google Scholar]
- Caswell H. Phenotypic plasticity in life-history traits: Demographic effects and evolutionary consequences. Amer Zool. 1983;23:35–46. [Google Scholar]
- Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37:1317–26. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, Riddle DL. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–15. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–21. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–9. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–20. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–3. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–94. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–55. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189–96. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–71. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–93. [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–34. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–7. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–21. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–35. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–19. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. 2007;303:434–47. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris M, Spieth J, Madej C, Lea K, Blumenthal T. Analysis of the VPE sequences in the Caenorhabditis elegans vit-2 promoter with extrachromosomal tandem array-containing transgenic strains. Mol Cell Biol. 1994;14:484–91. doi: 10.1128/mcb.14.1.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302:627–45. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–81. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–84. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. pp. 671–80. [DOI] [PMC free article] [PubMed]
- Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-beta and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–37. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–72. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]