Abstract
Aims
We compared the applicability of catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) and FISH to enumerate prokaryotic populations in ultra-oligotrophic alpine ground waters and bottled mineral water
Methods and Results
The fluorescent oligonucleotide probes EUB338 and the EUB338mix (EUB338/EUB338-II/EUB338-III) were used to enumerate Bacteria and probes EURY806 and CREN537 for Euryarchaea and Crenarchaea, respectively. Improved detection of Planctomycetales by probe EUB338-II was tested using a different permeabilization step (proteinase K instead of lysozyme). Total detection efficiency of cells in spring water (normalized to DAPI counts) of four different alpine karst aquifers was on average 83% for CARD-FISH and only 15% for FISH. Applying CARD-FISH on bottled natural mineral waters resulted in an average total hybridization efficiency of 89%, with 78% (range 77% - 96%) Bacteria and 11% (range 3% - 22%) of cells identified as Archaea.
Conclusions
CARD-FISH appears to result in substantially higher recovery efficiency than the conventional FISH approach and hence, is more suitable for the enumeration of specific prokaryotic groups in ground- and drinking water.
Significance and Impact of the study
This study represents the first evaluation of CARD-FISH on ultra-oligotrophic ground- and drinking water. Results are relevant for basic research and drinking water distributors. Archaea can comprise a significant fraction of the prokaryotic community in bottled mineral water.
Keywords: Archaea, Bacteria, CARD-FISH, FISH, drinking water, oligotrophic ground- and mineral water
INTRODUCTION
Well and spring water from oligotrophic ground water habitats are of great importance for public water supply in many areas throughout the world. Thus, understanding the biogeochemistry and microbiology of these aquatic resources is of general interest as it has an impact on the ability to distribute and store this water (i.e., “biostability”) in water supply systems. Only limited information exists on the occurrence and significance of prokaryotic populations, both, in the aquifer itself as well as in the abstracted water (Ghiorse and Wilson 1988, Gounot 1994, Griebler, et al. 1999). Studying the microbiology and ecology of these oligotrophic to ultra-oligotrophic systems provides also knowledge on basic ecological questions such as adaptation and survival strategies of microorganisms under nutrient-poor conditions. However, a basic requirement to study these microorganisms in such systems is the availability of methods which allow for sensitive, specific and quantitative detection of prokaryotic cells in a reduced metabolic state.
During the last decade, the fluorescence in situ hybridization (FISH) method ascended toward an indispensable tool to enumerate specific prokaryotic populations in marine and freshwater habitats (Behrens, et al. 2003, Chen, et al. 2004, DeLong, et al. 1989, Sekar, et al. 2004, Yeates, et al. 2003). Although FISH has gained widespread acceptance, there are some technical and conceptual problems that can lead to a great variability in the detection of target cells, especially under oligotrophic conditions (Bouvier and del Giorgio 2003, Oda, et al. 2000, Pernthaler, et al. 2002b). Frequently only a low fraction of all planktonic cells (<50%) can be visualized by FISH in fresh water systems (Sekar, et al. 2003) as well as in coastal surface waters (Pernthaler, et al. 2002a). Considerable efforts have been made to overcome some of these problems (Amann, et al. 1990, Behrens, et al. 2003, DeLong, et al. 1999, Fuchs, et al. 2000, Ouverney and Fuhrman 1997, Pernthaler, et al. 2002b). Probably the most significant improvement was the development of the catalyzed reporter deposition FISH (CARD-FISH) (Pernthaler, et al. 2002a, Schönhuber, et al. 1997), facilitating the detection of cells with low ribosomal content. The critical step in the CARD-FISH approach is the diffusion of large molecules, in this case the horse radish peroxidase (HRP)-labelled probe, into whole cells embedded in an agarose matrix. A directed permeabilization of prokaryotic cell walls prior to the hybridization step is therefore of crucial importance to enable the penetration of the probe (Pernthaler, et al. 2002a).
Another challenge when using FISH-based protocols is the fact that only a small fraction of the occurring prokaryotes are taxonomically characterized. Consequently, all rRNA-targeting probes have to be continuously reevaluated and modified as new sequence data is reported (Daims, et al. 1999). For example, the probe EUB338 was used to enumerate Bacteria, however, studies indicate that this probe is insufficient for detecting all bacterial groups (Hugenholtz, et al. 2001). Therefore two supplementary probes, EUB338-II (targeting Planctomycetales) and EUB338-III (targeting Verrucomicrobia), were designed for a more complete detection of this domain (Daims, et al. 1999). Unfortunately, there are few studies to date which evaluated the effectiveness of the probe mix (EUB I to III) when performing CARD-FISH in freshwater environments. In addition, there are only very limited data about the applicability of FISH-based protocols to detect members of the domain Archaea in fresh water systems (Bouvier and del Giorgio 2003).
The aim of this study was to evaluate the applicability of CARD-FISH to enumerate specific prokaryotic populations in ultra- oligotrophic ground and drinking water from various environments. As a first step, the CARD-FISH protocol was adapted (i.e. optimization of permeabilization and amplification time) and evaluated (i.e. cell loss determination, effect of different enzymatic treatments on Planctomycetales permeabilization) for prokaryotic cells from oligotrophic freshwater habitats. The adapted protocol was then used to compare the CARD-FISH procedure with the conventional FISH method at spring water samples from four different alpine karst springs covering a gradient from extremely low to high surface-influenced karst habitats (Farnleitner et al, 2005). A higher mean detection efficiency of CARD-FISH compared to FISH was obtained by applying a EUB338mix (EUB338, EUB338-II, EUB-III) and an improved protocol for archaeal cell detection (Teira, et al. 2004). Finally CARD-FISH in bottled drinking water samples, originating from various European groundwater habitats, demonstrated its general applicability for water samples from oligotrophic and ultra-oligotrophic environments.
MATERIALS AND METHODS
Study sites and sample preparation
The selected ground waters from four different alpine springs are located in the Northern Calcareous Alps in Austria (Farnleitner et al, 2005). DKAS 1 and HKAS 3 show a discharge regime with very low variations and an average water storage capacity of > 20 years. In contrast the discharge response after precipitation is very quick at LKAS 2 and SKAS 9, and the average water retention time is in the range of only a few years. Total prokaryotic abundance in the springs is in the range of about 107 to 108 cells L−1, reflecting their ultra-oligotrophic conditions (Farnleitner, et al. 2005) Because of varying substrate supply and surface runoff conditions, cell counts from aquifers with high water retention times (i.e. DKAS 1 and HKAS 3) are typically lower than in more dynamic aquifers (i.e. LKAS 2 and SKAS 9) (Farnleitner, et al. 2005).
Samples were collected from January 2004 to July 2004 at three to four weeks intervals, directly at the spring outlet. Duplicate samples of 60 ml were taken and immediately fixed with paraformaldehyde (final concentration 2%) at 4°C in the dark for 14–18 hours. Subsequently the samples were filtered through polycarbonate filters (0.2-μm pore-size; 25mm diameter; type GTTP; Millipore Corp. Bedford, MA), supported by cellulose acetate filters (0.45-μm pore-size; Millipore), to ensure an equal distribution of the cells on the filter surface, washed twice with 5ml of 0.2μm filtered Milli-Q water, air dried and stored at −20°C. In addition, the abundance of major prokaryotic groups was examined in 12 different brands of mineral waters, differing in their geological origin and bottled in different European countries. Their brands were made anonymous and they were randomly designated from 1 to 12. Bottles of mineral water were sampled aseptically. After mixing the water in the bottle, duplicate samples of 90 ml were taken from each bottle immediately after opening it and treated as described.
FISH
Filters from spring water samples were cut into halves. One was kept at −20°C for CARD-FISH the other was used for FISH and further cut into three parts for hybridization with different probes. The FISH procedure is based on the standard protocol (Glöckner, et al. 1999, Manz, et al. 1993) using 5ng/μl of the respective probe. All probes were purchased from Thermoelectron (Germany). Oligonucleotide probes for fluorescence in situ hybridization were 5`monolabeled with the indocarbocyanine dye Cy3. One section of the filter was hybridized with the EUB338mix containing probes EUB338 (5`-GCTGCCTCCCGTAGGAGT-3`; targeting Bacteria), EUB338-II (5`-GCAGCCACCCGTAGGTGT-3`; targeting Planctomycetales), EUB338-III (5`-GCTGCCACCCGTAGGTGT-3`; targeting Verrucomicrobia) (Daims, et al. 1999) simultaneously, one section with probe EUB338 only and one section with probe NON338 as a control for non-specific binding (Amann, et al. 1995). After incubation in a prewarmed washing solution (Glöckner, et al. 1999), filter sections were dipped in 80% ethanol, dried on Whatman 3M paper (Whatman Ltd., Austria) and placed on a glass slide. Subsequently they were mounted with a drop of DAPI mix (5.5 parts Citifluor [Citifluor, Ltd.], 1 part Vectashield [Vector Laboratories, Inc.], 0.5 parts PBS with DAPI at a final concentration of 1 μg/ml) for counterstaining the cells. At the first two sampling dates additional filter sections were hybridized with Cy3-monolabeled Archaeal probes (EURY806, 5`-CACAGCGTTTACACCTAG-3` targeting Euryarchaea; CREN537, 5`-TGACCACTTGAGGTGCTG-3`; targeting Crenarchaea) (Teira, et al. 2004). The probes target the same cells as the polynucleotide probes used in previous studies (DeLong, et al. 1999, Karner, et al. 2001). Positive controls were performed with prefiltered (0.2μm) spring water samples (2 samples from each DKAS 1 and LKAS 2) spiked with E.coli (strain K12 (DSM4509 harvested at late exponential phase, purchased from DSMZ)) at two different final concentrations (103 cells ml−1 and 105 cells ml−1). After immediate fixation as described above they were hybridized with the EUB338mix.
CARD-FISH
The remaining filter halves were dipped in low-gelling-point agarose (0.1% [wt/vol] Biozym, USA; in Milli-Q water) dried upside down on a glass petri dish at 37°C and dehydrated in 96% (vol/vol) ethanol (Pernthaler, et al. 2002a). Subsequently they were cut into four to five sections for hybridization with different HRP-labelled oligonucleotide probes. CARD-FISH was based on the protocol for marine habitats (Teira, et al. 2004) using the same permeabilization, hybridization and washing conditions, however, with some modifications as subsequently described. Cell wall permeabilization was done by incubation of the respective filter sections in either lysozyme (10 mg/ml; Sigma) for Bacteria (probes EUB338, EUB338-III, NON338) or proteinase K- (0.2μl/ml; 1.844U/mg, 10.9mg/ml, Fluka) solution (0.05 M EDTA, 0.1 M Tris-HCl [pH 8]) for Archaea (EURY806, CREN537) and EUB338-II at 37°C for 1 h. Filters were washed three times with Milli-Q water and subsequently incubated in 0.01M HCl for 20min at room temperature to inhibit potentially present intracellular peroxidase and residual proteinase K. After two more washing steps in Milli-Q water, filters were dehydrated with 96% ethanol and dried at room temperature. Filter sections were hybridized at 35°C (Schönhuber, et al. 1997) for 10 to 12 h with the EUB338mix, EUB338, EURY806, CREN537 and NON338, respectively. Three hundred μl of hybridization buffer (0.9 M NaCl, 20mM Tris-HCl [pH 7.5], 10% [wt/vol] dextran sulphate, 0.02% [wt/vol] sodium dodecyl sulphate (SDS), 1% blocking reagent [Boehringer Mannheim, Mannheim, Germany], and 55% [vol/vol] formamide [for EUB338, EUB338-II, EUB338-III and NON338] or 20% [vol/vol] formamide [for EURY806 and CREN 537]) were transferred into a 0.7-ml reaction vial. The respective HRP-probe was added to a final concentration of 0.28ng/μl (0.05 μM). After hybridization the filter sections were transferred into 50ml of prewarmed washing buffer (5 mM EDTA [pH 8], 20 mM Tris-HCl [pH 7.4 to 7.6], 0.01% [wt/vol] SDS) containing 13 mM NaCl (for EUB338, EUB338-II, EUB338-III and NON338) and 145 mM NaCl (for EURY806 and CREN 537) at 37°C for 10 min. Thereafter all sections were transferred in phosphate-buffer saline (PBS) (145 mM NaCl, 1.4 mM NaH2PO4, 8 mM Na2HPO4 [pH 7.6]) amended with 0.05% Triton X-100 (PBS-T) at room temperature for 15min. After removal of excess buffer, the filter sections were put into a 1.5 ml reaction vial containing 493 μl of amplification buffer (10% [wt/vol] dextran sulphate, 2 M NaCl, 0.1% [wt/vol] blocking reagent, and 0.0015% H2O2 [freshly prepared] in PBS) and 5 μl of tyramide-Alexa488 (1mg/ml; Molecular Probes, Europe) and incubated at 37°C for 45min. Finally filter sections were washed at room temperature in PBS-T (15 min) and Milli-Q, dehydrated in 96% ethanol and air dried. They were put on a slide, counterstained with DAPI-mix and stored at −20°C until further processing. At three sampling dates additional sections were hybridized with EUB338 only to determine the fraction of bacteria targeted with the probes EUB-II and EUB-III by comparison with the EUB338mix. Different times for permeabilization, hybridization and amplification were tested to optimize the procedure for fresh water environment. At the first two sampling dates additional filter sections from spring water samples were stained with DAPI to determine cell losses during the CARD-FISH procedure. In addition a test series was carried out to examine differences in the hybridization signal of probe EUB338-II when cells were either permeabilized with lysozyme or proteinase K. Additional control experiments were performed without any probe to test for remaining intrinsic peroxidase activity. Filters from mineral water samples were hybridized with the EUB338mix and a mix of the used archaea probes (EURY806 and CREN537).
Image acquisition and analysis
The stained filter sections were examined on a Leitz DMRB microscope equipped with a HBO 50-W Hg lamp, a 100x Plan Apochromat oil objective and appropriate filter sets for DAPI, Cy3 and Alexa488 (Chroma Tech. Corp., Rockingham,USA). First DAPI-stained cells were counted in one randomly selected microscopic field, followed by the determination of the fraction of Cy3- (FISH) or Alexa488-(CARD-FISH) stained cells in the same field. At least 800 to 900 DAPI-stained cells were counted per filter piece. Images were taken with a KAPPA CF15/4MCC camera system.
Statistical Analysis
In order to obtain a standardized indication of variability to compare the FISH- and the CARD-FISH data set, the parametric coefficient of variation (%CV) was used (100 × SD divided by the arithmetic mean). In addition to the comparison of the percentage of total hybridization for FISH and CARD-FISH results, CVs for the respective EUB338mix data sets were calculated. All further statistical comparisons were performed in SPSS version 13.0 using the nonparametric Wilcoxon test with related samples and Spearman rank correlation.
RESULTS
FISH in oligotrophic ground water
The average total prokaryotic abundance of all four springs stained by DAPI was 2.7× 104 cells ml−1 ranging from a minimum of 2.2 × 104 in February to a maximum of 3.4 × 104 in January (Tab.1). In all samples the fraction of hybridized cells was higher after FISH with the EUB338mix (on average 5% higher) than with the EUB338 probe alone (p < 0.01, n = 24; nonparametric Wilcoxon test, related samples). Only very few cells were hybridized with the NON338-probe (mean 0.5%). After subtracting the counts from the negative control the mean percentage of DAPI-stained cells visualized with the EUB338mix was 15% ± 6% (± standard error; n = 56) and signal intensity of hybridized cells was rather poor. Positive controls proved that the low detection rate with FISH was not caused by any technical problem. E.coli cells in spiked samples (0.2μm filtered spring water) could be hybridized (91-95% of DAPI-stainable E.coli cells) and gave bright fluorescence signals. No cells could be visualized after FISH with the archaeal oligonucleotide probes CREN537 and EURY806 (data not shown).
TABLE 1.
Average detection rates by FISH for DKAS 1, LKAS 2, HKAS 3 and SKAS 9 (n = 56; 7 sampling dates, four springs, two replicates per sample).
| Date | Total cell counts [104cells ml−1] |
Fraction % of total cells detected with probe* |
% of total hybridization§ |
Range min - max |
|||
|---|---|---|---|---|---|---|---|
| EUB338 | EUBmix† | ΔEUB‡ | NON338 | ||||
| 07.01.04 | 3.4 | 21 | 25 | 4 | 1.6 | 23 | 8 – 43 |
| 02.02.04 | 2.9 | 13 | 22 | 9 | 0.0 | 22 | 10 – 34 |
| 25.02.04 | 2.2 | n.a.** | 11 | n.a. | 0.4 | 11 | 0 – 32 |
| 23.03.04 | 2.7 | 12 | 16 | 4 | 0.2 | 16 | 0 – 47 |
| 19.04.04 | 2.4 | n.a. | 19 | n.a. | 0.6 | 19 | 0 – 43 |
| 17.05.04 | 3.0 | 6 | 9 | 3 | 0.7 | 9 | 0 – 23 |
| 14.06.04 | 2.3 | n.a. | 9 | n.a. | 0.2 | 8 | 0 – 27 |
|
| |||||||
| Avg. †† | 2.7 | 13 | 16 | 5 | 0.5 | 15 | 0 – 47 |
Percent detection of DAPI-stained cells. Between 800 and 900 DAPI-stained cells were counted per filter piece. Average values based on 8 analysis.
Hybridization with a mixture of probes EUB338, EUB338 II and EUB338 III.
Difference between fraction (%) EUB338 and EUBmix.
Percentage of total hybridization of probes (EUBmix) normalized to total DAPI counts. Numbers have been corrected by substracting NON338 counts.
not analysed
total average
CARD-FISH in oligotrophic ground water
The described CARD-FISH protocol resulted in high fractions of hybridized prokaryotic cells and intense fluorescent labelling of the cells in water samples from all four springs (Tab.2). The average numbers of DAPI-stainable cells (2.7 × 104 cells ml−1) were not significantly different from the results of the corresponding DAPI counter-stains during CARD-FISH analysis (p = 0.66, n = 16; nonparametric Wilcoxon test, related samples). Therefore no loss of DAPI-stainable cells during the CARD-FISH procedure could be detected (data not shown). The mean percentage of cells hybridized with the EUB338 probe alone was 66% ± 8% (± standard error; n = 24), increasing on average by 9% when the EUB338mix was applied (Tab. 2). The fraction of hybridized cells with the EUB338mix was 75% ± 7%. Permeabilization with proteinase K resulted in an archaeal contribution of 6% to 12% of DAPI-stainable cells with the two specific archaeal probes. The mean percentage of prokaryotic cells identified as Euryarchaea was 6% ± 2%, the fraction identified as Crenarchaea was 3% ± 1%. After subtracting counts from the negative control (~0.6%) the total DAPI-stained cells detected with all five oligonucleotide probes averaged 83% (± 6%). After lysozyme treatment 1% of the total cells could be visualized with probe EUB338-II in DKAS 1 and 3% in LKAS 2. Proteinase K treatment augmented these fractions to 2% in DKAS 1 and 6% in LKAS 2 (p < 0.01, n = 16, nonparametric Wilcoxon test, related samples). As expected, controls without probe, to test for remaining intrinsic peroxidase activity, did not give any fluorescent signal.
TABLE 2.
Average detection rates by CARD-FISH for DKAS 1, LKAS 2, HKAS 3 and SKAS 9 (n = 56; 7 sampling dates, four springs, two replicates per sample).
| Date | Total cell counts [104cells ml−1] |
Fraction % of total cells detected with probe* |
% of total hybridization§ |
Range min - max |
|||||
|---|---|---|---|---|---|---|---|---|---|
| EUB338 | EUB-mix† | ΔEUB‡ | EURY806 | CREN537 | NON338 | ||||
| 07.01.04 | 3.4 | 59 | 74 | 15 | 6 | 4 | 1 | 82 | 74 – 96 |
| 02.02.04 | 2.4 | n.a** | 73 | n.a. | 7 | 1 | 0.6 | 80 | 55 - 94 |
| 25.02.04 | 2.1 | n.a. | 62 | n.a. | 10 | 2 | 0.3 | 74 | 64 - 84 |
| 23.03.04 | 2.6 | 63 | 74 | 11 | 4 | 5 | 0.5 | 82 | 67 - 96 |
| 19.04.04 | 3.1 | n.a. | 79 | n.a. | 3 | 3 | 0.7 | 85 | 83 - 88 |
| 17.05.04 | 3.0 | 75 | 80 | 6 | 4 | 3 | 0.4 | 86 | 81 - 94 |
| 14.06.04 | 2.5 | n.a. | 82 | n.a. | 5 | 4 | 0.5 | 91 | 82 - 99 |
|
| |||||||||
| Avg. †† | 2.7 | 66 | 75 | 10 | 6 | 3 | 0.6 | 83 | 55 - 99 |
Percent detection of DAPI-stained cells. Between 800 and 900 DAPI-stained cells were counted per filter piece. Average values based on 8 analysis.
Hybridization with a mixture of probes EUB338, EUB338 II and EUB338 III.
Difference between fraction (%) EUB338 and EUBmix.
Percentage of total hybridization of probes (EUBmix, EURY806, CREN537) normalized to total DAPI counts. Numbers have been corrected by substracting NON338 counts.
not analysed
total average
Comparison between FISH and CARD-FISH
The CARD-FISH protocol was not only superior in terms of the intensity of fluorescence labelling but also in regard to mean detection efficiency (15% for FISH versus 83% for CARD-FISH) (Fig.1). Total recovery efficiency of prokaryotic cells using CARD-FISH ranged from 68% for SKAS 9 in winter to 99% for HKAS 3 in spring and was significantly higher than the maximum value reached with the FISH protocol (34%) throughout the entire investigation period (Fig.2). Nevertheless, the recovery efficiency using CARD-FISH decreased in all four springs during the winter months and steadily increased from end of February until June (Spearman correlation (independent data sets); r = 0.61, p < 0.01, n = 40). Generally, a higher recovery efficiency was obtained for DKAS 1 and HKAS 3, the two springs representing the hydrogeological more stable spring type than for LKAS 2 and SKAS 9 (Fig. 3). None of these trends could be discerned with FISH results, where the highest recovery was found in DKAS 1 during the winter period and the lowest value (7%) was also in DKAS 1 in May. Furthermore, variability in the recovery efficiency for FISH, indicated by the parametric coefficient of variation, was rather high (CV 47.5%, n=28) compared to CARD-FISH (CV 11.5%, n=56). Variability within the data sets obtained with the EUB338mix showed similar results (FISH, CV 47.5%, n=56; CARD-FISH, CV 14.3%, n=56). One drawback when using CARD-FISH, however, is that cell shapes can hardly be obtained. Image analysis after FISH allows for determination of cell morphologies or sizes (Alfreider, et al. 1996). In contrast to that even the morphological differentiation between rods and cocci is sometimes ambiguous after CARD-FISH hybridization, especially in environments where the majority of cells are rather small, because the cells are getting bloated during the amplification step.
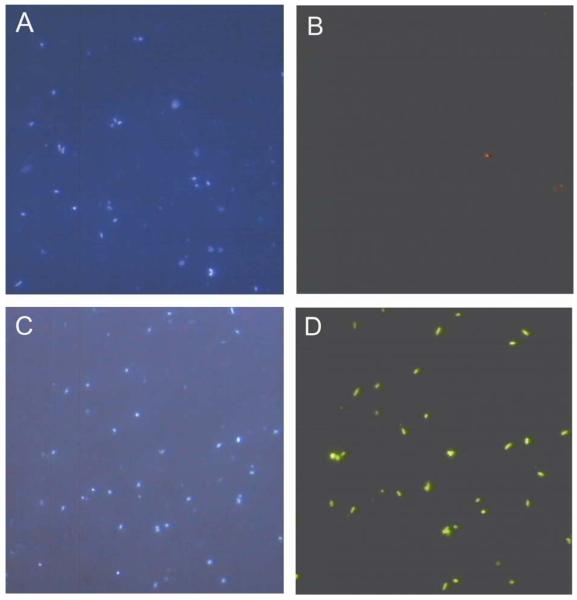
Fig.1.
Photomicrographs of DAPI-stained hybridized cells from DKAS 1. Pictures (A) and (B) depict corresponding DAPI-stained vs. FISH-labelled (Cy3 - probes) cells. Pictures (C) and (D) depict corresponding DAPI-stained vs. CARD-FISH-labelled (Alexa488-labeled tyramide) cells. Both samples were hybridized with the EUB338mix.
Fig.2.
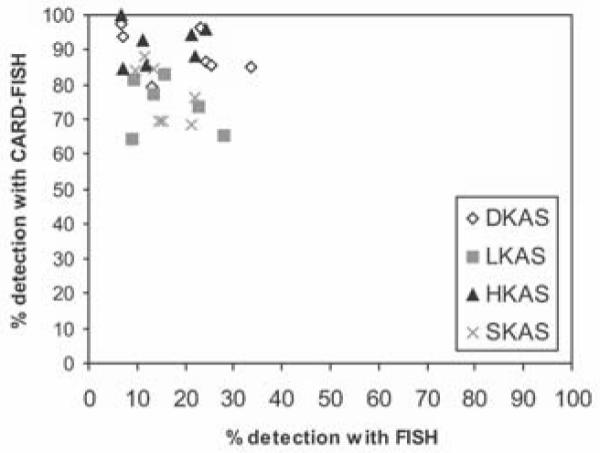
Comparison of total detection efficiency (% of all probe-hybridized cells normalized to total DAPI-stained cells) of the FISH and the CARD-FISH approach. Both methods were applied to prokaryotic cells immobilized on the same filter (probes, EUB338, EUB338-II, EUB338-III, EURY806, CREN537).
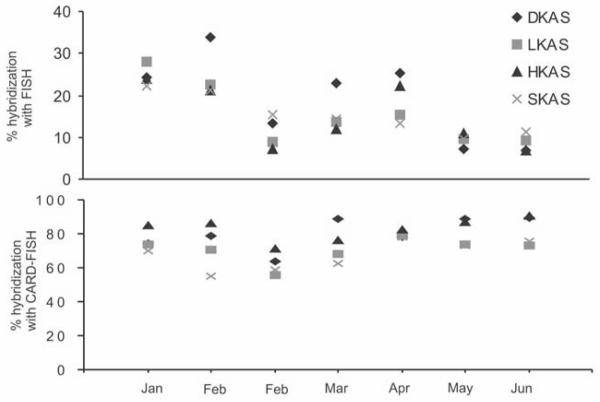
Fig.3.
Percentage of prokaryotes detected by the different probes used to cover the bacterial and archaeal populations from the four springs over a 6 months periode using (A) the FISH protocol and (B) the CARD-FISH protocol. Different scales were used at the y-axis. DKAS 1 and HKAS 3 represent systems with higher average water residence time whereas LKAS 2 and SKAS 9 represent spring types vulnerable to immediate surface influence.
CARD-FISH analysis of mineral water
The CARD-FISH method was applied on samples from 7 carbonated and 5 non-carbonated mineral waters originating from geologically different aquifers (Tab.3). Total counts of DAPI-stained cells ranged between 4 × 103 and 1.1 × 105 cells ml−1. The mean percentage of prokaryotic populations hybridized with all five oligonucleotides probes was 89% ± 5%, of which 78% were Bacteria (stained with the EUB338mix) and 11%, (range 1% to 22%) were Archaea (stained by the ARCHmix) (Tab. 4). Negative controls ranged between 0% and 0.4% (data not shown) and were not subtracted from total hybridization results.
TABLE 3.
Chemical characterization of different mineral waters*
| Type | Cations [mg/l] |
Anions [mg/l] |
TDS† [mg/l] |
||||||
|---|---|---|---|---|---|---|---|---|---|
| K+ | Na+ | Mg2+ | Ca2+ | Cl− | SO42− | NO3− | HCO3− | ||
| 1 | 5.4 | 52.1 | 6.1 | 8.1 | 16 | 1.5 | n.a. | 164 | 192 |
| 2 | 5 | 115 | 9.2 | 45 | 109 | 50 | 2 | n.a. | 36 |
| 3 | 2.9 | 14.2 | 1.7 | 29.5 | 8.3 | 31.4 | 5.8 | 78 | 186 |
| 4 | 6.2 | 11.2 | 8 | 11.5 | 13.5 | 8.1 | 6.3 | 71 | 130 |
| 5 | n.a. ‡ | 7.3 | 19.9 | 9.1 | n.a. | 105 | n.a. | 258 | 403 |
| 6 | 0.5 | 3 | 1.3 | 4.5 | 5 | 4 | 1.9 | 15 | 33 |
| 7 | 1 | 5 | 24 | 78 | 4.5 | 10 | 3.8 | 357 | 309 |
| 8 | 1.8 | 11.4 | 43.3 | 110 | 21 | 229 | n.a. | 255 | 691 |
| 9 | 2 | 13.9 | 65.6 | 146.4 | 8 | 298.6 | 0.5 | 421 | 1000 |
| 10 | 4.1 | 22.3 | 50.1 | 123.6 | 15.4 | 286 | 1 | 298 | 801 |
| 11 | 3.3 | 28.3 | 17.5 | 87.1 | 3 | 12.8 | n.a. | 414 | 610 |
| 12 | n.a. | 11.4 | 9.9 | 27 | 12.7 | 17.2 | n.a. | 107 | n.a. |
parameters were copied from bottles
total dissolved solids
not available
TABLE 4.
Average detection rates by CARD-FISH for different mineral waters (n = 24; twelve bottles, in duplicate). Samples 1-7 represent carbonated mineral water, samples 8-12 represent non-carbonated mineral water.
| Type | Country | Total cell counts [104cells ml−1] |
Fraction % of total cells detected with probes* |
% of total hybridization§ |
Range min - max |
|
|---|---|---|---|---|---|---|
| EUBmix† | ARCHmix‡ | |||||
| 1 | Spain | 2.0 | 96 | 3 | 99 | 98 – 99 |
| 2 | Iceland | 0.4 | 80 | 8 | 89 | 85 – 92 |
| 3 | Western-Austria | 8.3 | 85 | 7 | 93 | 89 – 96 |
| 4 | Germany | 6.0 | 77 | 15 | 92 | 85 – 99 |
| 5 | Switzerland | 2.9 | 82 | 6 | 87 | 83 – 91 |
| 6 | Belgium | 10.5 | 80 | 11 | 91 | 87 – 95 |
| 7 | France | 3.4 | 80 | 13 | 92 | 85 – 99 |
| 8 | Central Austria | 0.8 | 73 | 20 | 93 | 90 – 95 |
| 9 | Eastern Austria | 0.4 | 68 | 18 | 86 | 84 – 87 |
| 10 | Western Austria | 0.4 | 76 | 6 | 82 | 78 – 85 |
| 11 | Central Austria | 7.7 | 66 | 22 | 88 | 85 – 99 |
| 12 | Spain | 1.8 | 79 | 1 | 80 | 75 – 86 |
|
| ||||||
| Avg. ** | 3.7 | 78 | 11 | 89 | 75 – 99 | |
Percent detection of DAPI-stained cells. Between 800 and 900 DAPI-stained cells were counted per filter piece. Average values based on 2 analysis.
Hybridization with a mixture of probes EUB338, EUB338 II and EUB338 III.
Hybridization with a mixture of probes EURY806 and CREN 537.
Percentage of total hybridization of probes (EUBmix, ARCHmix) normalized to total DAPI counts.
total average
DISCUSSION
CARD-FISH versus FISH in oligotrophic freshwater environments
The presented data demonstrate that CARD-FISH is an effective tool to overcome significant problems occurring when using FISH in oligotrophic and ultra-oligotrophic ground water environments (e.g., no detection of cells with low ribosomal content). Although FISH was successfully applied in various habitats (Franks, et al. 1998, Glöckner, et al. 1999, Nielsen, et al. 2002), the method is limited in certain environments e.g. (Bouvier and del Giorgio 2003, Kenzaka, et al. 1998, Pernthaler, et al. 2002a, Pernthaler, et al. 2002b). Hybridization assays carried out in ice, snow and drinking waters gave an estimated mean recovery efficiency for Bacteria lower than 40% (Bouvier and del Giorgio 2003). Our study demonstrates that FISH detection rates in ultra-oligotrophic systems can be as low as 8 to 23 % of DAPI stainable cells. The CARD-FISH method is hardly affected by a low number of ribosomes, although hybridization results might still be influenced by modification (Kalpaxis, et al. 1998) or degradation (Davis, et al. 1986) of target sites, providing an possible explanation for the decrease of detection efficiency observed with CARD-FISH during February and March (Fig. 3). It can be speculated that these modifications, besides a low ribosomal content, could be responsible for the high variability of total hybridization results with FISH (Fig. 3), also found in other studies where slow growing communities prevail (Bouvier and del Giorgio 2003). Another reason for the observed decrease in the recovery efficiency using CARD-FISH during the winter months could be alterations in bacterial or archaeal cell walls induced by stress conditions (Gounot and Russell 1999) or increased carbon limitation (Nystrom, et al. 1992), rendering the cell wall more resistant to the permeabilization step and prevent the probe from entering the cell (Wagner, et al. 1998). As this study identified organisms on a low taxonomic resolution level, it remains unknown whether the lower recovery efficiency during the winter months is caused by a less efficient hybridization of the entire community or individual groups (Farnleitner, et al. 2005). Although CARD-FISH results showed a fraction of at least 6 % belonging to the domain Archaea (Tab. 2) in these karst water aquifers, identification with FISH was neither possible for Euryarchaea nor for Crenarchaea (Tab. 1). Crenarchaeota comprised 3% of the total archaeal community. This group appears to be ubiquitously present in aquatic habitates and soil and has been reported in a wide variety of temperate and cold environments (Sjöling and Cowan 2003). The majority of the archaeal community was identified as Euryarchaeota (6%). The recovery efficiency using CARD-FISH likely represents the upper level of active prokaryotes since DAPI not only stains viable cells, but also detects dead or ghost cells lacking ribosomes and hence, are not detectable by CARD-FISH (Zweifel and Hagström 1995).
Using the EUB338mix
The importance of an optimized permeabilization step becomes evident when using the EUB338mix. Probe EUB338-II was designed for the order Planctomycetales (Schlesner 1994, Ward, et al. 1995). Planctomycetales exhibit some unique characters, most importantly their cell wall is similar to that of Archaea (Kandler and König 1998, Neef, et al. 1998). This validates our results indicating better permeabilization of Planctomycetales when cell walls with proteinase K than with lysozyme. Some studies describe Planctomycetes attached to particulate matter or cell clusters and suggest a possible role in the degradation of more recalcitrant compounds (Neef, et al. 1998). This could be advantageous in alpine karst ecosystems and one reason for the high detection rate in LKAS 2 (6%) than in DKAS 1 (2%), as LKAS 2 is a more dynamic spring type and likely contains more particulate matter (Farnleitner, et al. 2005).
CARD-FISH in natural mineral water
Although consumption of natural mineral water is constantly increasing, there are only very few studies on prokaryotic community composition in this environment. Most approaches are based on isolation and subsequent identification (Defives, et al. 1999, Dewettinck, et al. 2001, Leclerc and Moreau 2002) and/or subsequent molecular typing (Nichols, et al. 2003, Villari, et al. 2003) and are therefore suffering from well-recognized cultivation biases. Evaluation of new methods for microbiological assessments of the microbial community in mineral water are not only interesting due to the unique habitat characteristics, but are an industry-driven need (Ramalho, et al. 2001). The CARD-FISH approach we used, applying the EUB338mix and ARCHmix, gain reliable and reproducible results. Even with total cell numbers as low as 3.6 × 103 cells ml−1, total hybridization rates were still higher than 80% (Tab.4). Our data strongly indicates the presence of an archaeal community in all ground-mineral waters analyzed, comprising between 1 and 20 % of the total prokaryotic abundance. Recently, changes in bacterial community composition in mineral water after bottling were reported using FISH (Loy, et al. 2005). Although only 63% of all cells present were detected after the initial growth phase (about one week after bottling), unreliability of FISH in oligotrophic systems became especially evident during the first days after bottling. The number of FISH-detectable cells was only up on a maximum level of 3% when total cell counts remained constant at 2 × 103 cells ml−1. This highlights the (in)sensitivity of the FISH method when prokaryotic activity is low (Jones, et al. 1999, Leclerc and Moreau 2002) such as in bottled mineral water. The low detection rate is in accordance with earlier findings that report hybridization of 5% of total counts with a bacteria-specific probe (Leclerc and Moreau 2002). As bottles in our study were not obtained directly from the producer, the microbial community was analysed after an unknown time span after bottling and thus, after the initial growth-phase, when total cell counts were already at a stable level (Defives, et al. 1999, Dewettinck, et al. 2001). Although the applicability of CARD-FISH for a time course analysis during the first weeks after bottling, was not tested, the rather small variations in prokaryotic abundance among the different brands of mineral water indicate that this method is as an adequate tool for future investigations. It would be important to further examine not only community shifts after the bottling-process but also the autochthonous prokaryotic community of the source habitat and vulnerability indicators with more specific probes using the CARD-FISH approach. A possible combination of CARD-FISH with microautoradiogaphy (MAR) (Teira, et al. 2004) would not only provide reliable data on the abundance of particular populations but also on the physiological state of the community. In conclusion, the applicability of FISH and CARD-FISH in ultra-oligotrophic freshwater systems was compared. We found severe limitations for the FISH approach in environments where the prokaryotic community is likely to be in a starvation-survival state. Total hybridization rates with CARD-FISH were at the upper end of what might be reached with this method. CARD-FISH was found to be a useful and straightforward tool to obtain enfolding insights into the prokaryotic community composition including the archaeal fraction in ultra-oligotrophic aquatic environments like ground- and drinking water.
ACKNOWLEDGEMENTS
This study was funded by the FWF project P18247-B06 granted to A.H.F. Special thanks also to Hermann Kain for crucial technical assistance.
REFERENCES
- Alfreider A, Pernthaler J, Amann R, Sattler B, Glockner FO, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–70. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Fuchs B, Mueller F, Amann R. Is the in situ accessibility of the 16S rRNA of E.coli for Cy3-labeled oligonucleotide probes predictid by a three-dimensional structure model of the 30S ribosomal subunite? Appl Environ Microbiol. 2003;69:4935–4941. doi: 10.1128/AEM.69.8.4935-4941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Rühland C, Inácio J, Huber H, Fonseca Á, Spencer-Martins I, Fuchs BM, Amann R. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea and Eucarya to Cy3-laeled oligonucleotide probes. Appl Environ Microbiol. 2003;69:1748–1758. doi: 10.1128/AEM.69.3.1748-1758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier T, del Giorgio PA. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): A quantitative review of published reports. FEMS Microbiol Rev. 2003;44:3–15. doi: 10.1016/S0168-6496(02)00461-0. [DOI] [PubMed] [Google Scholar]
- Chen H, Ponniah G, Salonen N, Blum P. Culture-independent analysis of fecal enterobacteria in environmental samples by single-cell mRNA profiling. Appl Environ Microbiol. 2004;70:4432–4439. doi: 10.1128/AEM.70.8.4432-4439.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. System. Appl. Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Davis BD, Luger SM, Tai PC. Role of ribosome degradation in the death of starved Escherichia Coli cells. Journal of Bacteriology. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defives C, Guyard S, Oularé MM, Mary P, Hornez JP. Total counts, culturable and viable, and non-culturable microflora of a French mineral water: a case study. Journal of Applied Microbiology. 1999;86:1033–1038. doi: 10.1046/j.1365-2672.1999.00794.x. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Taylor LT, Marsh TL, Preston CM. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–63. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Dewettinck T, Hulsbosch W, Van Hege K, Top EM, Verstraete W. Molecular fingerprinting of bacterial populations in groundwater and bottled mineral drinking water. Appl Microbiol Biotechnol. 2001;57:412–418. doi: 10.1007/s002530100797. [DOI] [PubMed] [Google Scholar]
- Farnleitner AH, Wilhartitz I, Kirschner AKT, Stadler H, Burtscher MM, Hornek R, Szewzyk U, Herndl G, Mach R. Bacterial dynamics in spring water of two contrasting alpine karst aquifers indicates stable autochthonous microbial endokarst communities and morphological adaptations to hydrological conditions. Environmental Microbiology. 2005;7:1248–1259. doi: 10.1111/j.1462-2920.2005.00810.x. [DOI] [PubMed] [Google Scholar]
- Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–45. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Glockner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–7. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiorse WC, Wilson JT. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–72. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–6. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounot AM. Microbial oxidation and reduction of manganese: consequences in groundwater and applications. FEMS Microbiol Rev. 1994;14:339–49. doi: 10.1111/j.1574-6976.1994.tb00108.x. [DOI] [PubMed] [Google Scholar]
- Griebler C, Mindl B, Danielopol DL. Biofilme in Grundwasser-Ökosystemen. Schriftenreihe des Österreichischen Wasser- und Abwallwirtschaftsverbandes. 1999;127:23–51. [Google Scholar]
- Hugenholtz P, Tyson GW, Blackall LL. Design anf evaluation of 16S rRNA-targeted oligonucleotide probes for fluorscence in situ hybridization. Methods in Molecular Biology. 2001;179:29–42. doi: 10.1385/1-59259-238-4:029. [DOI] [PubMed] [Google Scholar]
- Jones CR, Chamberlain AHL, Adams MR. An investigation of the presence of ultramicrocells in natural mineral water. Letters in Applied Microbiology. 1999;28:275–279. doi: 10.1046/j.1365-2672.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Kalpaxis DL, Karahalios P, Papapetropoulou M. Changes in ribosomal activity of Escherichia coli cells during prolonged culture in sea salts medium. J Bacteriol. 1998;180:3114–9. doi: 10.1128/jb.180.12.3114-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, König H. Cell wall polymers in Archaea (Archaebacteria) CMLS. Cell.Mol.Life Sci. 1998;54:305–308. doi: 10.1007/s000180050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Kenzaka T, Yamaguchi N, Tani K, Nasu M. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology. 1998;144:2085–2093. doi: 10.1099/00221287-144-8-2085. [DOI] [PubMed] [Google Scholar]
- Leclerc H, Moreau A. Microbiological safety of natural mineral water. FEMS Microbiol Rev. 2002;26:207–222. doi: 10.1111/j.1574-6976.2002.tb00611.x. [DOI] [PubMed] [Google Scholar]
- Loy A, Beisker W, Meier H. Diversity of Bacteria growing in natural mineral water after bottling. Appl Environ Microbiol. 2005;71:3624–3632. doi: 10.1128/AEM.71.7.3624-3632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W, Szewzyk U, Ericsson P, Amann R, Schleifer KH, Stenstrom TA. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–8. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef A, Amann R, Schlesner H, Schleifer KH. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- Nichols RAB, Campbell BM, Smith HV. Identification of Cryptosporidium spp. oocysts in United Kingdom noncarbonated natural drinking waters and drinking waters by using a modified nested PCR-restriction fragment length polymorphism assay. Appl Environ Microbiol. 2003;69:4183–4189. doi: 10.1128/AEM.69.7.4183-4189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JL, Juretschko S, Wagner M, Nielsen PH. Abundance and phylogenetic affiliation of iron reducers in activated sludge as assessed by fluorescence in situ hybridization and microautoradiography. Appl Environ Microbiol. 2002;68:4629–4636. doi: 10.1128/AEM.68.9.4629-4636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T, Olsson RM, Kjelleberg S. Survival, stress resistance, and alterations in protein expression in the marine vibrio sp. strain S14 during starvation for different individual nutrients. Appl Environ Microbiol. 1992;58:55–65. doi: 10.1128/aem.58.1.55-65.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Slagman SJ, Meijer WG, Forney LJ, Gottschal JC. Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. Fems Microbiology Ecology. 2000;32:205–213. doi: 10.1111/j.1574-6941.2000.tb00713.x. [DOI] [PubMed] [Google Scholar]
- Ouverney CC, Fuhrman JA. Increase in fluorescence intensity of 16S rRNA in situ hybridization in natural samples treated with chloramphenicol. Appl. Environ. Microbiol. 1997;63:2735–2740. doi: 10.1128/aem.63.7.2735-2740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002a;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R. Comparison of fluorescenly labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol. 2002b;68:661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho R, Cunha J, Teixeira P, Gibbs PA. Improved methods for the enumeration of heterotrophic bacteria in bottled mineral waters. J Microbiol Methods. 2001;44:97–103. doi: 10.1016/s0167-7012(00)00237-2. [DOI] [PubMed] [Google Scholar]
- Schlesner H. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Plamctomycetales from various aquatic habitats using dilute media. System. Appl. Microbiol. 1994;17:135–145. [Google Scholar]
- Schönhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–73. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R, Fuchs B, Amann R, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Applied and Environmental Microbiology. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöling S, Cowan DA. High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles. 2003;7:275–282. doi: 10.1007/s00792-003-0321-z. [DOI] [PubMed] [Google Scholar]
- Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villari P, Crispino M, Montuori P, Boccia S. Molecular typing of Aeromonas isolates in natural mineral water. Appl Environ Microbiol. 2003;69:697–701. doi: 10.1128/AEM.69.1.697-701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Schmid M, Juretschko S, Trebesius K, Bubert A, Goebel W, Schleifer KH. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol Letters. 1998;160:159–168. doi: 10.1111/j.1574-6968.1998.tb12906.x. [DOI] [PubMed] [Google Scholar]
- Ward N, Rainey FA, Stackebrandt E, Schlesner H. Unraveling the extent of diversity within the order Planctomycetales. Appl Environ Microbiol. 1995;61:2270–5. doi: 10.1128/aem.61.6.2270-2275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates C, Saunders AM, Crocetti GR, Blackall LL. Limitations of the widely used GAM42a and BET42a probes targeting bacteria in the Gammaproteobacteria radiation. Microbiology. 2003;149:1239–1247. doi: 10.1099/mic.0.26112-0. [DOI] [PubMed] [Google Scholar]
- Zweifel UL, Hagström A. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Applied and Environmental Microbiology. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]