Abstract
A large range of debilitating medical conditions1 are linked to protein misfolding, which may compete with productive folding particularly in proteins containing multiple domains2. With 75% of the eukaryotic proteome consisting of multidomain proteins, how is inter-domain misfolding avoided? It has been proposed that maintaining low sequence identity between covalently linked domains is a mechanism to avoid misfolding3. Here we use single-molecule Förster Resonance Energy Transfer (FRET) experiments4,5 to detect and quantify rare misfolding events in tandem Ig domains from the I-band of titin under native conditions. About 5.5% of molecules with identical domains misfold during refolding in vitro and form a surprisingly stable state with an unfolding half time of several days. Tandem arrays of immunoglobulin-like (Ig-like) domains in humans exhibit significantly lower sequence identity between neighbouring domains than between non-adjacent domains3. In particular, the sequence identity of neighbouring domains has been found to be preferentially below 40%3. Interestingly we observe no misfolding for a tandem of naturally neighbouring domains with low sequence identity (24%), whereas misfolding occurs between domains which are 42% identical. Coarse-grained molecular simulations predict the formation of domain-swapped structures, which are in excellent agreement with the observed transfer efficiency of the misfolded species. We infer that the interactions underlying misfolding are very specific and result in a sequence-specific domain swapping mechanism. Diversifying the sequence between neighbouring domains appears to be a successful evolutionary strategy to avoid misfolding in multidomain proteins.
Multidomain proteins comprise covalently-linked, frequently similar domains, resulting in high effective local protein concentration. It is therefore probable that these proteins have evolved to avoid inter-domain misfolding in vivo. While co-translational, “domain-by-domain” folding is assumed to be important for avoiding misfolding6, many proteins that are long-lived or subject to tensile forces will fold and unfold numerous times during their lifetime and may thus be particularly vulnerable to misfolding. The giant muscle protein titin, for example, undergoes reversible domain unfolding which may play a role in muscle elasticity7.
Single-molecule techniques are ideal for detecting rare events8 such as misfolding in native conditions. Indeed, the first evidence for misfolding of adjacent domains in long tandem arrays of the well-characterised 27th domain from the I-band of titin, I27, was obtained using single-molecule atomic force microscopy (AFM)9. An alternative approach is single-molecule FRET4,5, whose great sensitivity allows the detection of very small populations. FRET enables the mapping of intramolecular distances by means of the distance-dependent efficiency of excitation energy transfer between a donor and acceptor fluorophore attached to specific positions of the protein10 (for details, see Supplementary Fig. 1).
We hypothesised that denaturation of tandem constructs of titin domains with guanidinium chloride (GdmCl), followed by rapid refolding into native conditions, might allow formation of misfolded species, which should be detectable using single-molecule FRET11. We labelled a tandem construct of I27 (I27-I27) in the A-strand of domain 1 (E3C) and the G-strand of domain 2 (N83C) (Fig. 1a) with a donor (Alexa Fluor 488) and an acceptor (Alexa Fluor 594) fluorophore, attached via cysteine residues engineered on the protein surface. For a misfolded domain to be formed by strands from domains 1 and 2, we predicted that these two strands must be adjacent for this domain to have the mechanical properties observed in the previous AFM experiments9. The correctly folded tandem would then have low transfer efficiency, while a misfolded state would have high transfer efficiency. A monomer of I27 was labelled in the corresponding positions to provide a model for the misfolded state (Fig. 1b). Labelling was found to have little effect on the stability of I27 (Supplementary Fig. 2), and doubly-labelled proteins that had not previously been unfolded in GdmCl (‘never-unfolded’) show single, correctly folded populations with transfer efficiencies (E) of 0.37 (±0.01) and 0.93 (±0.01) for the tandem I27-I27 and monomer I27, respectively (Fig. 2 a & b).
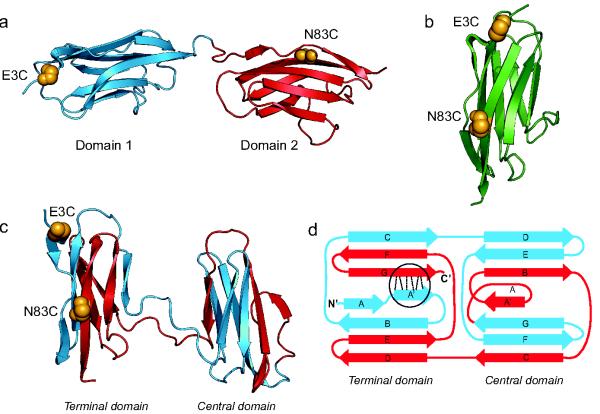
Figure 1.
Structures of Native and Misfolded I27 Constructs. (a) Natively folded I27-I27 tandem repeat with labelling positions highlighted (golden spheres). (b) Native I27 crystal structure (1tit.pdb) with labelling positions corresponding to those expected for the misfolded state of I27-I27. (c) One of the domain-swapped misfolded state structures formed in Gō-model simulations. (d) Schematic of this misfolded state topology: hydrogen bonds that are perpendicular to the direction of applied force in AFM mechanical unfolding are shown by dashed lines (circled). Four other misfolded state topologies were populated in the simulations (Supplementary Fig. 5b). We note that we cannot distinguish between such topologies from the results presented here.
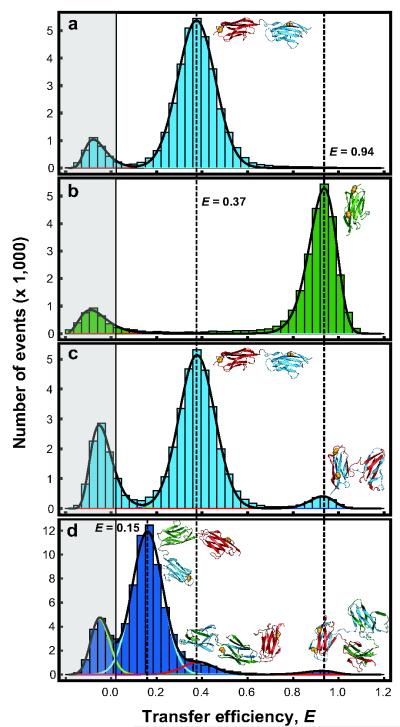
Figure 2.
Transfer Efficiency Histograms of Doubly-Labelled I27 Constructs. (a) ‘Never-unfolded’ I27-I27. (b) ‘Never-unfolded’ monomeric I27. (c) Refolded I27-I27. (d) Refolded I27-I27-I27; fits of individual populations shown as coloured lines for clarity. Histograms are fitted with normal or log-normal distributions. The peak in the grey shaded area consists of events from molecules without an active acceptor fluorophore28. Note that in these experiments a short four-amino acid linker (Arg-Ser-Glu-Leu) is included between the domains in the I27-I27 tandem to allow direct comparison with previous AFM and aggregation experiments3,9. ‘Never-unfolded’ I27-I27-I27 is shown in Supplementary Fig. 6.
We conducted refolding experiments by diluting unfolded I27-I27 into refolding buffer. The resulting transfer efficiency histograms now exhibited two populations (Fig. 2c): one corresponding to the correctly folded native state (E = 0.37), and one with precisely the same transfer efficiency as the analogously labelled monomer (E = 0.93). This observation reveals that the A-strand of the first domain and the G-strand of the second domain are arranged as in the monomer. A quantitative analysis (Supplementary Table 1) showed that 5.5(±0.2)% of the molecules are found in the misfolded form.
Based on the results of the AFM studies9 we had supposed that the misfolded species consisted of a single strand-swapped titin domain with the remaining sequence unstructured, and thus with an unfolding time similar to that of a native domain (τ ≈ 34 minutes12). We therefore investigated the unfolding kinetics of the misfolded state13 (Fig. 3a). At high GdmCl concentrations the decay in the number of high-transfer efficiency events (E > 0.8), corresponding to the misfolded state, is fitted well by a single exponential (Fig. 3b) with rate constants slightly higher than the unfolding rate constant for I27wt determined in ensemble measurements12 (Fig. 3c). We can also estimate the unfolding rate constant of the correctly folded species, which agrees well with the ensemble data (Fig. 3c) (see Supplementary Fig. 3 and Supplementary Information). In the absence of denaturant, however, the misfolded species was surprisingly long lived, converting to the correctly folded form only on a time scale of days (Supplementary Fig. 4). The formation of the misfolded structure is thus under kinetic, rather than thermodynamic, control. Its remarkable kinetic stability clearly distinguishes the misfolded species described here from short-lived, partially folded intermediates14-16 sometimes termed “misfolded” because they contain some non-native interactions17.
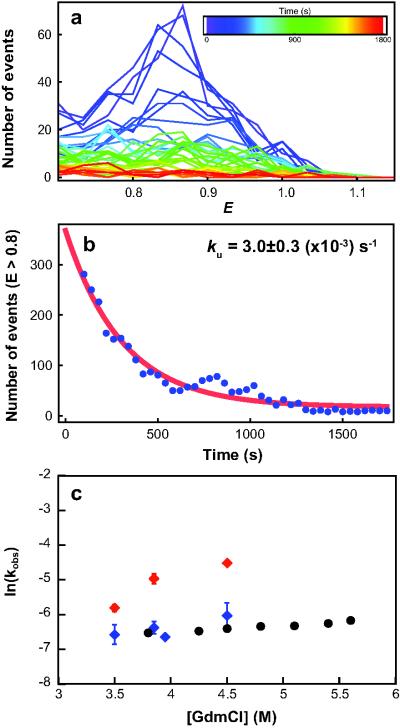
Figure 3.
Unfolding Kinetics. (a) Evolution of transfer efficiency histograms over time (E ≥ 0.7) from single-molecule double-jump experiments in which refolded/misfolded I27-I27 (doubly-labelled) was unfolded in 3.5 M GdmCl. Histograms were constructed for a moving window of 120s that was shifted by 30s for each increment (inset, colour key). (b) The number of events with E > 0.8 for each histogram in (a) was summed and the resulting kinetics fitted with a single exponential decay. The rate constants are unaffected by different window sizes or the use of non-overlapping windows. (c) Unfolding rate constants for I27wt monomer (black) (ensemble data from12)*, and for the misfolded and natively folded states of I27-I27 from single-molecule measurements (red and blue, respectively). The error bars represent the standard error of the fit (see Methods). Note that for some data points the error bars are smaller than the symbols. *I27 domains have the same unfolding rate constants in tandem repeat proteins as in isolated domains.
An explanation for the slow unfolding under native conditions is suggested by folding simulations of I27-I27 with a Gō-like model18. In these simulations, only native interactions are attractive, and interactions between a given pair of residues are considered equal, independently of whether they are in the same or different domains. While most trajectories result in two correctly folded domains, misfolded species with two fully-folded, strand-swapped domains are occasionally formed. Five different strand-swapped topologies were observed (Fig. 1c,d and Supplementary Fig. 5). Such an extensively misfolded structure explains its persistence; correct folding cannot occur while either misfolded domain remains folded. Since refolding rate constants are much higher than unfolding rate constants under native conditions, the simultaneous unfolding of both domains is very unlikely, and conversion to the native state is extremely slow19.
We can test the validity of our model further by investigating the refolding of a three-domain tandem of I27 with the FRET labels in domain 1 (E3C) and domain 3 (N83C). If domain-swapped structures were to be formed, we would expect to see two misfolded populations: one with the FRET efficiency of the monomer (misfolding between domains 1 and 3) and another population with the efficiency of the I27-I27 tandem (misfolding between domains 1 and 2 or 2 and 3). This is precisely what we observe (Fig. 2 d). The proportion of monomer-like (high-FRET) species in the trimeric tandem is significantly lower than before (2.8(±0.6)%); this is likely to reflect the lower probability of association between domains that are more distant in sequence. The population of the misfolded species with dimer-like FRET efficiency (9(±2)%) was instead almost twice as high as in the two-domain tandem (5.5(±0.2)%); this is probably due to the two alternative possibilities to misfold in an analogous way to the dimeric tandem (domain 1 with 2 and domain 2 with 3). Simulations with the Gō-like model also predict domain-swapped structures with monomer- and dimer-like FRET efficiencies, with relative populations similar to experiment (Supplementary Fig. 6).
Much work has been dedicated to investigating the sequence specificity of protein aggregation20,21, including the hypothesis that there is selective pressure to prevent oligimerisation by a domain-swapping mechanism22,23. Misfolding is often considered to precede aggregation, suggesting that sequence-specific behaviour observed in aggregation also applies to misfolding. Our single-molecule FRET experiments now allow us to test this hypothesis directly by investigating mixed tandem constructs, I27-I28 and I27-I32 (Fig. 4b). Indeed I27-I28, natural neighbours in titin with only 24% sequence identity, did not yield any detectable population of high-FRET misfolded species upon refolding (Fig. 4c,d). However, misfolding is seen upon refolding of I27-I32 (sequence identity 42%) (Fig. 4e,f), to the same extent as I27-I27 (Supplementary Table 1). This misfolded species also unfolds with the same rate constant as that of I27-I27 (Supplementary Fig. 7). The I27-I32 misfold is consistent with previous experiments showing chimeric domains of I27 and I32 to be stable24,25. This result strongly supports the idea that protein misfolding is sequence-specific. In proteins where sequence identity between neighbouring domains is high, the topology may prevent formation of stable misfolded species19.
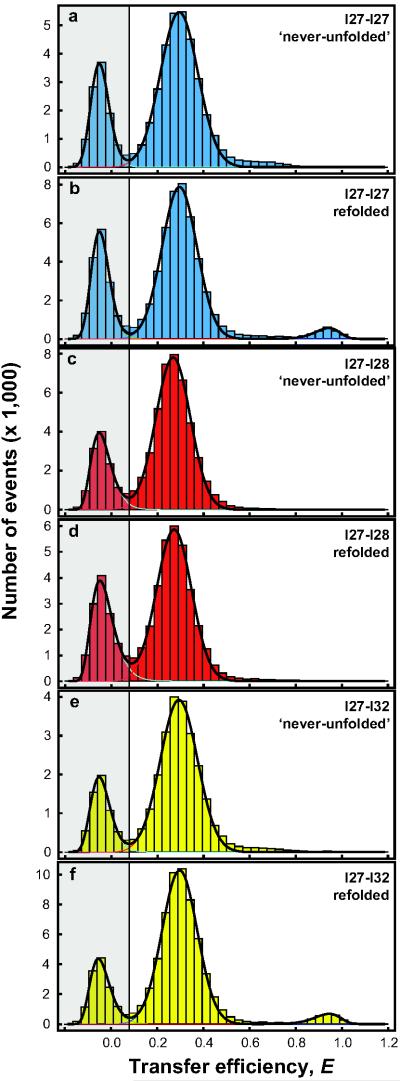
Figure 4.
Transfer Efficiency Histograms of Tandem Constructs with Identical and Non-identical Domains. (a) and (b) I27-I27 ‘never-unfolded’ control and refolded, respectively. (c) and (d) I27-I28 ‘never-unfolded’ control and refolded, respectively. (e) and (f) I27-I32 ‘never-unfolded’ control and refolded, respectively. In order to mimic the natural protein, there was no linker added between the domains in these experiments. Note that the frequency of misfolding was the same for I27-I27 with and without the linker, 5.5(±0.2)% and 5.7(±0.5)%, respectively (compare Fig. 2c with Fig. 4b and Supplementary Table 1). Addition of the four amino acid linker also made no difference to the results for I27-I28 (Supplementary Fig. 9).
Misfolding in our experiments is more frequent than had been observed in AFM experiments9, suggesting that the tethering in those experiments reduces misfolding; this might be advantageous for titin domains in vivo. Unfolding of the misfolded species observed with AFM showed them to have the same mechanical resistance as correctly folded I27, but twice the chain length is released upon unfolding. Our results are entirely compatible with this finding. While the misfolded species has two folded domains, and is thus stable in folding conditions, only the terminal domain would experience shearing of the H-bonds between the parallel A’ and G strands (Fig. 1d circled) perpendicular to the direction of the applied force, in the same way as the correctly folded I2726, resulting in the same mechanical stability. Since force is not applied to the A and G strands in the central domain, this domain is likely to unfold at low force, together with the terminal domain. This hypothesis is supported by simulations (Supplementary Fig. 8).
In summary, our results suggest that diversifying the sequence composition between neighbouring domains is an effective evolutionary strategy to ensure efficient folding in multidomain proteins and avoid the formation of stable misfolded species. This adds a significant piece of the puzzle in understanding the problems encountered during the crucial evolutionary transition from single to multi-domain proteins.
Methods Summary
For details of protein production, ensemble equilibrium measurements and labelling see Methods. Single-molecule experiments, instrumentation, data reduction and analysis are detailed in Methods. The resulting relative populations from all experiments and analysis techniques are summarised in Supplementary Table 1. Folding simulations using a Gō-like model were run using the CHARMM code27 as described in Methods. For details of mechanical unfolding simulations see Methods.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust (grant number 064417), the Swiss National Science Foundation (to B.S.), and the Swiss National Center of Competence in Research for Structural Biology (to B.S.). M.B.B was supported by a UK Medical Research Council studentship. A.B. is supported by a Marie Curie Intra-European Fellowship. R.B. is supported by a Royal Society University Research Fellowship. J.C. is a Wellcome Trust Senior Research Fellow. We thank H. Hofmann, A. Soranno and A. Hoffmann for helpful discussions and valuable contributions to data analysis.
Methods
Protein expression and labelling
Cysteine residues were introduced by site-directed mutagenesis. E3C in domain 1 (always I27) and N83C in domain 2 if I27 or I32, and K83C for I28 (with the numbering relative to a single domain), for the two domain constructs and E3C in domain 1 and N83C in domain 3 for the three domain construct of I27. DNA sequencing confirmed the mutagenesis.
I27 monomer and the I27-I27, I27-I27-I27, I27-I28, and I27-I32 tandems, with the engineered surface cysteines, were expressed as described previously29,30. Labelling was carried out using Alexa Fluor 488 (donor) and Alexa Fluor 594 (acceptor) maleimides (Invitrogen) according to the manufacturer’s procedures. Both dyes were mixed simultaneously with reduced protein in equimolar ratios and incubated at 4°C for ~10 hours. Un-reacted dye was removed by gel filtration and the differently labelled variants were separated by ion-exchange chromatography (MonoQ 5/50 GL; GE Healthcare Biosciences AB, Uppsala, Sweden). I27 has two intrinsic cysteines that were not removed as they are buried in the native state and all labelling was carried out on folded protein in native conditions.
Ensemble measurements
Equilibrium measurements were performed for the doubly labelled I27 monomer to check the effect of labelling (Supplementary Fig. 2). Experiments were performed in GdmCl on a Cary Eclipse fluorimeter (Varian Inc., CA, USA) monitoring intrinsic tryptophan fluorescence as described previously29, but with lower protein concentrations (0.05-0.5 μM) and the addition of 0.001% Tween 20.
Single-molecule instrumentation
Observations of single-molecule fluorescence were made using a custom-built confocal microscope equipped with a continuous-wave 488 nm solid-state laser (FCD488-010, JDSU, Milpitas, CA, USA) and an Olympus UplanApo 60x/1.20W objective. After a dichroic mirror that separates excitation and emission light (500DCXR, Chroma Technology, Rockingham, VT), fluorescence emission passed through a 100 μm pinhole and was split by a second dichroic mirror (585DCXR, Chroma Technology) into donor and acceptor fluorescence. Donor fluorescence then passed a filter (ET525/50M, Chroma Technology) before being focused onto a single-photon avalanche diode (MPD 100ct, Micro Photon Devices, Bolzano, Italy) while acceptor fluorescence passed a filter (QT 650/100) before being focused onto a single-photon avalanche diode (SPCM-AQR-13, PerkinElmer Optoelectronics, Vaudreuil, QC, Canada). The arrival time of every photon was recorded with a two-channel time-correlated single-photon counting module (PicoHarp300, PicoQuant, Berlin, Germany). All measurements were performed with a laser power of 100 μW, measured at the back aperture of the objective (beam waist: 8 mm).
Single-molecule equilibrium measurements
All experiments were performed at protein concentrations between 0.5 and 25 pM, in the same final solution conditions: PBS; 0.001% Tween 20; 140 mM β-mercaptoethanol; 20 mM cysteamine hydrochloride (single-molecule buffer). Tween 20 (Pierce) was used to prevent surface adhesion of the proteins31, while the photoprotective agents β-mercaptoethanol (Sigma) and cysteamine hydrochloride (Sigma) were employed to minimise chromophore damage32. ‘Never-unfolded’ experiments were conducted by mixing protein in PBS (0.01% Tween, 10 mM β-mercaptoethanol) 1:99 with 0.04 M GdmCl in single-molecule buffer (to mimic the final conditions in the refolding experiments). Refolding experiments were performed by mixing protein unfolded in 4.4M GdmCl (PBS, 0.01% Tween, 10 mM β-mercaptoethanol) 1:100 with single-molecule buffer, to a final GdmCl concentration of 0.04 M. 8 to 10 hour measurements were made for all constructs with one or more repeats. The absence of aggregates was ensured in all experimental conditions as previously described33. Fluorescence bursts were identified by combining successive photons separated by 150 μs or less, and events comprised of 35 or more photons were kept for analysis. Transfer efficiencies were corrected for quantum yields, cross-talk, and direct excitation as described previously34,35.
Populations of correctly folded and misfolded molecules in the transfer efficiency histograms were quantified using two methods. Where possible, peaks were fitted using a Gaussian distribution (for populations where 0.1 ≤ E ≤ 0.8) or a log-normal distribution (for populations where E ≥ 0.8), and the resulting fits integrated. Transfer efficiencies quoted in the main text denote the average E obtained from these fits with standard deviations calculated from multiple experiments. The populations of misfolded species were determined relative to the sum of natively folded and misfolded populations. Alternatively, ranges of transfer efficiencies were chosen for each population, the corresponding number of bursts summed, and the relative populations of misfolded species computed. Only the latter method was amenable for the ‘never-unfolded’ control measurements and all experiments involving I27-I28 tandems as no misfolded population was observed. The resulting relative populations from all experiments and analysis techniques are summarised in Supplementary Table 1.
Single-molecule kinetic measurements
Unfolding of the misfolded species was achieved by mixing a previously refolded protein sample (prepared as for refolded experiments above, but with higher protein concentration) with GdmCl in single-molecule buffer. A moving-window analysis13 was applied to the time-resolved photon trajectory with a window size (Δt) of 120s. Transfer efficiency histograms were calculated from the bursts in that time window, and the window was shifted by Δt/3 to form each successive time-point (Fig. 4a). A time t = ts+Δt/2, where ts is the start time of the window, was assigned to each histogram, and the number of events with E ≥ 0.8 in each histogram as a function of time was fitted to a single exponential (Fig. 4b). The resulting rate constants were robust for different window sizes and non-overlapping windows. Standard errors were taken from the covariance matrix of the fit (weighted by the average inverse variance of the residuals of the data points with respect to an unweighted fit).
The analysis of the unfolding of the native state from the same type of experiments is detailed in Supplementary Information and Supplementary Fig. 5.
Simulations
A coarse-grained Gō-like model was generated based on the structure of I27 (1tit.pdb36) using a standard procedure37. Briefly, all bond lengths are fixed by constraints, harmonic terms are used for the angles, a knowledge based potential for the torsion angles, and non-bonded interactions are treated with a Gō-like potential in which only interactions formed between residues in the folded protein are attractive (with relative strengths given by the Miyazawa-Jernigan matrix), all others being repulsive. Two or three identical I27 sequences were linked by four residue repulsive linkers to treat the two and three domain tandems, respectively. Interactions between residues i and j in different domains were treated exactly like the interactions between those residues in the same domain, and interactions with the linker were repulsive. A simulation temperature was chosen such that the folding barrier was at least 3 kBT, by using the free energy barrier projected onto the fraction of native contacts as a lower bound. Folding simulations were run, starting from fully extended configurations, using Langevin dynamics with a friction of 0.1 ps−1 and a time step of 10 fs. The final structures were clustered using a simple leader-follower algorithm with a cut-off of 0.15 nm.
Mechanical unfolding simulations were performed in which a force of 150 pN was applied to the ends of the I27-I27 tandem, starting from structures belonging to either the folded cluster or one of the misfolded clusters, and monitoring unfolding by the end-end distance. The CHARMM molecular simulation package was used for all calculations27.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information: Supporting information is provided with this document.
References
- 1.Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Annu Rev Genomics Hum Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- 2.Jaenicke R, Seckler R. Protein misassembly in vitro. Adv. Protein Chem. 1997;50:1–59. doi: 10.1016/s0065-3233(08)60318-6. [DOI] [PubMed] [Google Scholar]
- 3.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 4.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 5.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorov AN, Baldwin TO. Cotranslational protein folding. J. Biol. Chem. 1997;272:32715–32718. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- 7.Li H, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 8.Borgia A, Williams PM, Clarke J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 2008;77:101–125. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 9.Oberhauser AF, Marszalek PE, Carrion-Vasquez M, Fernandez JM. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999;6:1025–1028. doi: 10.1038/14907. [DOI] [PubMed] [Google Scholar]
- 10.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 11.Gambin Y, et al. Direct single-molecule observation of a protein living in two opposed native structures. Proc. Natl. Acad. Sci. USA. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler SB, Clarke J. Mapping the folding pathway of an immunoglobulin domain: structural detail from phi value analysis and movement of the transition state. Structure. 2001;9:355–366. doi: 10.1016/s0969-2126(01)00596-2. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann H, et al. Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc. Natl. Acad. Sci. USA. 2010;107:11793–11798. doi: 10.1073/pnas.1002356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivarsson Y, Travaglini-Allocatelli C, Brunori M, Gianni S. Folding and misfolding in a naturally occurring circularly permuted PDZ domain. J Biol Chem. 2008;283:8954–8960. doi: 10.1074/jbc.M707424200. [DOI] [PubMed] [Google Scholar]
- 15.Gianni S, et al. Structural characterization of a misfolded intermediate populated during the folding process of a PDZ domain. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1956. [DOI] [PubMed] [Google Scholar]
- 16.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329:1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 17.Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: misfolding on a path to the native state. Nat Struct Biol. 2002;9:209–216. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 18.Yang S, et al. Domain swapping is a consequence of minimal frustration. Proc. Natl. Acad. Sci. USA. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora P, Hammes GG, Oas TG. Folding mechanism of a multiple independently-folding domain protein: double B domain of protein A. Biochemistry. 2006;45:12312–12324. doi: 10.1021/bi060923s. [DOI] [PubMed] [Google Scholar]
- 20.Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49:117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 21.Straub JE, Thirumalai D. Principles governing oligomer formation in amyloidogenic peptides. Curr Opin Struct Biol. 2010;20:187–195. doi: 10.1016/j.sbi.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Mitraki A. Protein aggregation from inclusion bodies to amyloid and biomaterials. Adv Protein Chem Struct Biol. 2010;79:89–125. doi: 10.1016/S1876-1623(10)79003-9. [DOI] [PubMed] [Google Scholar]
- 24.Borgia A, Steward A, Clarke J. An effective strategy for the design of proteins with enhanced mechanical stability. Angew. Chem. Int. Ed. Engl. 2008;47:6900–6903. doi: 10.1002/anie.200801761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balamurali MM, et al. Recombination of protein fragments: a promising approach toward engineering proteins with novel nanomechanical properties. Protein Sci. 2008;17:1815–1826. doi: 10.1110/ps.036376.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 28.Dahan M, et al. Ratiometric measurement and identification of single diffusing molecules. Chemical Physics. 1999;247:85–106. [Google Scholar]
References
- 29.Scott KA, Steward A, Fowler SB, Clarke J. Titin; a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 2002;315:819–829. doi: 10.1006/jmbi.2001.5260. [DOI] [PubMed] [Google Scholar]
- 30.Steward A, Toca-Herrera JL, Clarke J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 2002;11:2179–2183. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 32.Nettels D, et al. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. USA. 2009 doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hillger F, et al. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew. Chem. Int. Ed. Engl. 2008;47:6184–6188. doi: 10.1002/anie.200800298. [DOI] [PubMed] [Google Scholar]
- 34.Schuler B. Single-molecule fluorescence spectroscopy of protein folding. Chemphyschem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann A, et al. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Improta S, Politou AS, Pastore A. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure. 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 37.Karanicolas J, Brooks CL., 3rd. Improved Go-like models demonstrate the robustness of protein folding mechanisms towards non-native interactions. J. Mol. Biol. 2003;334:309–325. doi: 10.1016/j.jmb.2003.09.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.