Abstract
The gene for the atypical Notch ligand Delta-like homologue 1 (Dlk1) encodes membrane-bound and secreted isoforms functioning in multiple developmental processes in vitro and in vivo. Dlk1, a member of a cluster of imprinted genes, is expressed from the paternally-inherited chromosome1,2. Here we show that mice deficient in Dlk1 exhibit defects in postnatal neurogenesis within the subventricular zone (SVZ), a developmental continuum resulting in depletion of mature neurons in the olfactory bulb. We show that DLK1 is a factor secreted by niche-astrocytes, while its membrane-bound isoform is present in neural stem cells (NSCs) being required for the inductive effect of secreted DLK1 on self-renewal. Surprisingly, we find a requirement for Dlk1 expressed from both maternal and paternally inherited chromosomes. Selective absence of Dlk1 imprinting in both NSCs and niche astrocytes is associated with postnatal acquisition of DNA methylation at the germ line-derived imprinting control region (IG-DMR). The results emphasize molecular relationships between NSCs and niche-astrocytes identifying a signalling system coded by a single gene functioning co-ordinately in both cell types. The modulation of genomic imprinting in a stem cell environment adds a new level of epigenetic regulation to the establishment and maintenance of the niche raising wider questions about the adaptability, function, and evolution of imprinting within specific developmental contexts.
The mammalian adult brain is generally post-mitotic, but reservoirs of NSCs bearing features of astroglial cells in the hippocampus and SVZ support lifelong neurogenesis3,4.. The SVZ is a very active germinal niche in which production of neurons occurs via a transit-amplifying progenitor population (TAP)4, giving rise to migrating neuroblasts that integrate into the olfactory bulb (OB) circuitry5. The specialized microenvironment containing niche-astrocytes regulates long-term maintenance of NSCs ensuring continual neurogenesis3,4,6. An emerging hypothesis is that NSCs and niche-astrocytes are established within the postnatal radial glia/astrocytic lineage7, however, the potential lineage relationships and cell-cell interactions between them are not completely understood6,7,8.
Dlk1 encodes a transmembrane protein belonging to the Notch/Delta/Serrate family of signalling molecules with key roles in differentiation9-11. Dlk1 is widely expressed during embryonic development12-14 and is dosage-sensitive with over-expression causing phenotypes ranging from prenatal lethality to defects in postnatal energy homeostasis15. Few tissues retain Dlk1 expression postnatally and deletion experiments reveal in vivo functions in adipogenesis and haematopoiesis10,11,16. We detected DLK1 in neurogenic areas of the prenatal telencephalon and in the postnatal SVZ with a peak at day 7 (P7). In the adult brain, Dlk1 expression is mainly restricted to neurons of several areas including the ventral tegmental area14, the septum and the ventral striatum but the protein is still detected in specific cell types of the mature SVZ, in particular NSCs (GFAP+/SOX2+/Nestin+) and niche-astrocytes (GFAP+/SOX2−/S100β−). Differentiated parenchymal astrocytes (GFAP+/S100β+), βIII-tubulin+ neuroblasts and IB4+ ependymocytes, are not DLK1+ (Supplementary Fig. 1a-j).
To test for potential roles for DLK1 in neurogenesis, we analysed brain germinal regions in embryos and postnatal mice harbouring a targeted mutation in the Dlk1 gene11. Although we observed expression of DLK1 both in progenitor cells and in differentiating neurons in E12.5 and E14.5 ganglionic eminences, we did not observe any differences in progenitor cell activity or in neurogenesis in the mantle between Dlk1 wild-type and mutant embryos (Supplementary Fig. 2a-c), indicating that DLK1 is dispensable during embryonic neurogenesis. In contrast, we observed increased radial glia/NSC activity in the developing SVZ at P7, indicated by higher numbers of GFAP+/Ki67+ cells resulting in increased doublecortin (DCX)+ neuroblasts (Fig. 1a,b; Supplementary Fig. 3a), suggesting failure to maintain the slower dividing stem cell pool in the postnatal SVZ.
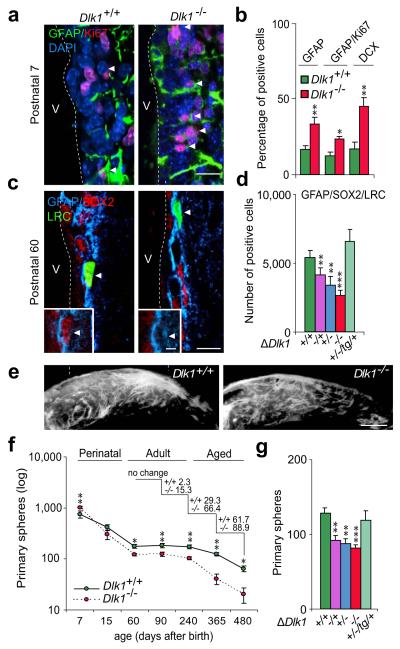
Figure 1. DLK1 regulates postnatal neurogenesis.
(a) Immunohistochemistry for GFAP (green) and Ki67 (red) within the postnatal 7 (P7) SVZ of wild-type and Dlk1−/− mice. (b) Percentage of cell-types in the P7 SVZ from wild-type and Dlk1−/− mice. (c) Immunohistochemistry for GFAP (blue), SOX2 (red) and BrdU (green) within the adult SVZ of wild-type and Dlk1−/− mice. (d) Number of GFAP+/SOX2+/BrdU-label retaining cells (LRC)+ in the SVZ of mice with deletions of maternally-inherited (Dlk1−/+), paternally-inherited (Dlk1+/−), or both alleles (Dlk1−/−). Light-green bar represents paternal transmission mutant mice in a Dlk1 transgenic background (Dlk1+/−/Dlk1tg/+). (e) Whole-mount for DCX+ migrating neuroblasts within the SVZ. (f) Primary spheres from wild-type and Dlk1 mutant SVZs at different developmental stages. (g) Primary spheres from adult SVZ of different Dlk1 mutants. *p<0.05, **p<0.01, ***p<0.001. Error bars, s.e.m. of five experiments (n=4 cultures per genotype). V, ventricle. Scale bars; in a, c, 20 μm (inset 10 μm).
P7 is a transition time-point between development of the embryonic germinal layer and the mature SVZ17 so we next evaluated adult mice. P60 wild-type and Dlk1 mutant mice were injected with 5-bromo-2-deoxyuridine (BrdU), and sacrificed one month later to mark BrdU label-retaining cells (LRCs), i.e. relatively quiescent NSCs and cells that abandon the cell cycle shortly after labelling, such as terminally differentiated cells within the SVZ and newly-born OB neurons18. The numbers of BrdU+/GFAP+/SOX2+ or GFAP+/Nestin+ cells were significantly reduced in Dlk1−/− mice (Fig. 1c,d; Supplementary Fig. 3b,c). However, the percentage of GFAP+/Ki67+ cells was similar between genotypes (Supplementary Fig. 3d,e), indicating a change in NSC number but not their cycling parameters. Fewer NSCs resulted in fewer MASH1+ TAP and DCX+ neuroblasts (Supplementary Fig. 3c, e-f) resulting in a less densely populated rostral migratory stream (Fig. 1e). Moreover, the numbers of post-mitotic calretinin (CR)+ and tyrosine-hydroxylase (TH)+ newly-born BrdU+ neurons in the granular and periglomerular layers of the mutant OB were significantly reduced (Supplementary Fig. 1g).
It has been demonstrated that disruption of quiescence at early postnatal periods leads to loss of stem cell potential and depletion of the NSC pool later in life19,20. To test whether DLK1 was indeed required for NSC maintenance we evaluated the size of the stem cell pool over time by determining primary neurosphere yield at different ages. A transient higher yield in primary neurospheres from P7 SVZ mutant tissue was followed by an increase in their progressive decline as the animals aged (Fig. 1g) suggesting that postnatal expression of DLK1 is required for life-long maintenance of the NSC pool.
To evaluate dosage effects of the Dlk1 gene, mice carrying the mutation on the maternally (Dlk1−/+) or the paternally inherited allele (Dlk1+/−) were analyzed. Interestingly, numbers of GFAP+/SOX2+/BrdU+ LRCs and of newly-generated OB neurons were reduced in both Dlk1 heterozygotes (Fig. 1d; Supplementary Fig. 3g). Consistent with the defects observed in vivo, SVZs from Dlk1−/+, Dlk1+/− and Dlk1−/− all yielded significantly fewer primary neurospheres than wild-type tissue though brain-size was normal (Fig. 1f; Supplementary Fig. 3h,i). We confirmed that these phenotypes were specifically due to a reduction in the available levels of DLK1, by generating paternal transmission Dlk1 mutants also hemizygous for a Dlk1 expressing transgene (Dlk1−+/−;Dlk1tg/+)15. The number of GFAP+/SOX2+/BrdU+ LRCs in vivo and in vitro neurospheres obtained from double mutant SVZs was not significantly different to their wild-type littermates indicating mutant rescue (Fig. 1d,g).
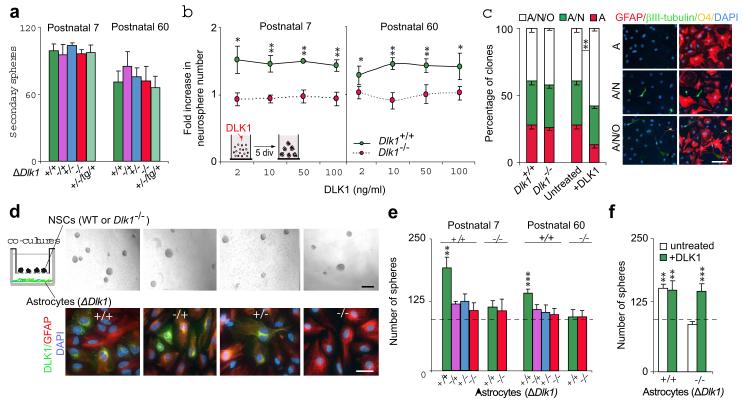
Despite being reduced in number, mutant neurospheres displayed normal clonogenic capacity upon passage (Fig. 2a), suggesting that DLK1 acts as a postnatal niche-secreted factor in vivo. Treatment of P7 and P60 cultures with recombinant mouse DLK1 resulted in 40-50% more neurospheres (Fig. 2b) and significantly higher numbers of secondary spheres were formed from primary neurospheres which had been grown (pre-treated) in DLK1-supplemented medium (Supplementary Fig. 4a). These increases were not due to DLK1 promoting NSC survival (Supplementary Fig. 4b), suggesting that addition of DLK1 specifically increases self-renewing symmetrical divisions. Moreover, multipotentiality in clonal differentiation assays is increased by exogenous DLK1 (Fig. 2c), further supporting a role for DLK1 as a niche factor. In order to further evaluate this, we co-cultured astrocytes acutely isolated from the SVZs of wild-type, Dlk1−/+, Dlk1+/− and Dlk1−/− P7 and P60 mice8 with wild-type NSCs using transwell inserts (Fig. 2d). Wild-type niche-astrocytes induced a significant increase in neurosphere formation which was abrogated when niche-astrocytes were derived from Dlk1 mutants (Fig. 2d,e). The reduction in neurosphere number in Dlk1−/− astrocyte-conditioned medium was rescued by the exogenous addition of recombinant DLK1 (Fig. 2f), an indication that DLK1 secreted by SVZ niche-astrocytes, probably in combination with other niche factors, regulates NSC self-renewal.
Figure 2. DLK1 is secreted by postnatal SVZ niche-astrocytes.
(a) Secondary spheres formed from wild-type and Dlk1−/− primary neurospheres at P7 and P60. (b) Primary spheres from Dlk1+/+ and Dlk1−/− SVZ, after DLK1 treatment. (c) Quantification of unipotent (astrocytes), bipotent (astrocytes/neurons) and tripotent (astrocytes/ neurons/oligondendrocytes) clones derived from Dlk1+/+ and Dlk1−/− neurospheres grown in the presence or absence of DLK1 (left panel). Immunocytochemistry for GFAP (red), βIII-tubulin (green) and O4 (yellow) in differentiated neurospheres (right panel). (d) Primary neurospheres co-cultured with wild-type or Dlk1 mutant niche-astrocytes (upper panels). Immunocytochemistry for GFAP (red) and DLK1 (green) in niche-astrocyte cultures (lower panels). (e) Quantification of P7 and P60 co-culture experiments shown in d. (f) Primary spheres co-cultured with Dlk1+/+ or Dlk1−/− astrocytes and treated with DLK1. Dashed-lines indicate non co-cultured spheres. *p<0.05, **p<0.01, ***p<0.001. Error bars, s.e.m of triplicate cultures (3-6 samples per genotype). Scale bars: in c, 50 μm; in d (upper panel, 100 μm; lower panel, 30 μm).
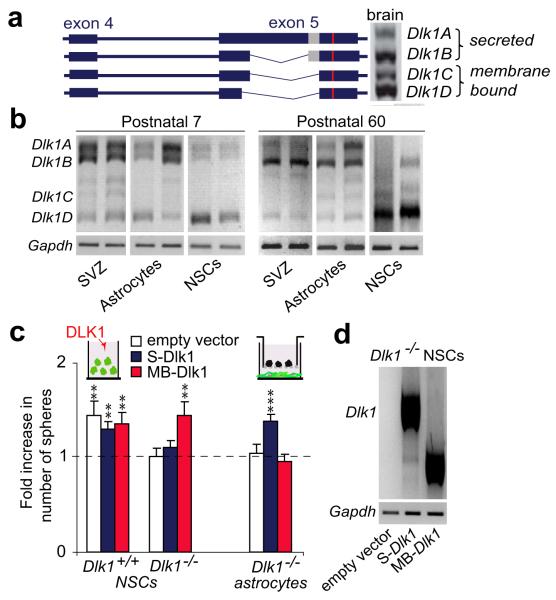
Alternatively spliced transcripts of Dlk1 (Fig. 3a) encoding protein isoforms that are either membrane-tethered or proteolytically-cleaved and secreted, have been described21. Secreted isoforms DLK1A and DLK1B contain a juxtamembrane motif for cleavage by extracellular proteases, absent from membrane-bound DLK1C and DLK1D (Fig. 3a). Secreted isoforms represent the predominant type in acutely isolated niche-astrocytes. Membrane-bound isoforms are preferentially expressed by P7 and P60 NSCs (Fig. 3b). Interestingly, exogenously added DLK1 or co-culture with niche-astrocytes did not increase neurosphere formation in Dlk1−/− NSC cultures (Fig. 2b,e) indicating that the membrane-tethered form of DLK1 in NSCs contributes to the response to soluble DLK1.
Figure 3. NSCs require membrane-bound DLK1 to respond to niche-secreted DLK1.
(a) Dlk1 transcripts in whole brain. Proteolytic cleavage domain (grey box) is shown. (b) Semi-quantitative PCR of Dlk1 isoforms in SVZ, NSCs and niche-astrocytes at postnatal 7 and 60. (c) Neurospheres generated from Dlk1−/− or Dlk1+/+ cultures nucleofected with pIRES-GFP, pIRES-GFP-S-Dlk1 (secreted isoform) or pIRES-GFP-MB-Dlk1 (membrane-bound isoform) after DLK1 treatment or from wild-type NSCs co-cultured with transduced Dlk1−/− astrocytes. (d) Semi-quantitative PCR of Dlk1 isoforms in nucleofected NSCs. **p<0.01, ***p<0.001. Error bars: s.e.m; triplicate cultures (n=6 animals per genotype).
To confirm whether membrane-bound DLK1 is required for the response to soluble DLK1, GFP-tagged vectors expressing membrane (MB-Dlk1) or secreted (S-Dlk1) isoforms were nucleofected into Dlk1+/+ and Dlk1−/− NSCs, and neurosphere formation in response to exogenous DLK1 was determined. Dlk1−/− NSCs regained the response to exogenous DLK1 only when expressing the membrane-bound isoform (Fig. 3c; Supplementary Fig. 4c,d). Furthermore, expression of S-Dlk1 but not MB-Dlk1 in Dlk1−/− niche-astrocytes rescued wild-type NSC response in co-cultures (Fig. 3c), indicating that membrane-bound DLK1 in NSCs is stimulated by secreted DLK1. Interestingly, increased expression of Dlk1 above wild-type levels does not influence NSCs response (Fig. 3c; Supplementary Fig. 4e). It is worth noting that responses elicited by Jagged122 and PEDF23 are unperturbed in Dlk1 mutant NSCs. Moreover, no change was observed in Notch activity after addition of recombinant DLK1 to wild-type NSCs, suggesting a Notch-independent role for DLK1 (Supplementary Fig. 5a-d).
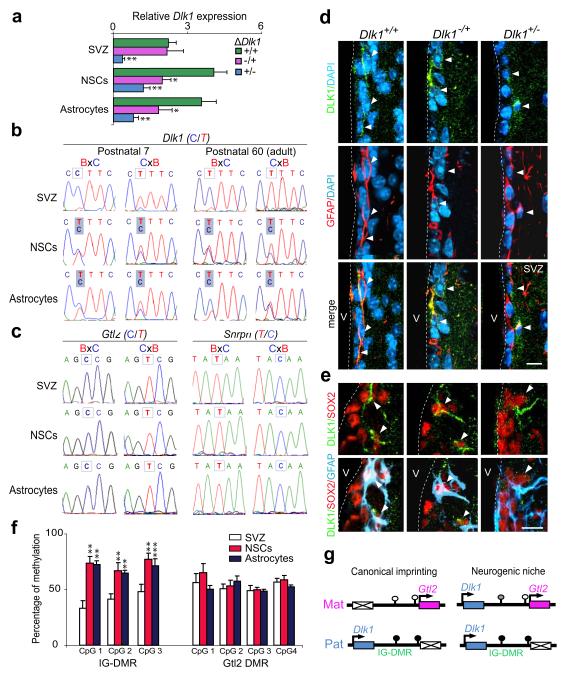
Dlk1, canonically expressed from the paternally-inherited chromosome, belongs to the Dlk1-Dio3 imprinted gene cluster on mouse chromosome 12. Northern-blots of adult brain confirmed Dlk1 transcription from the paternal allele (Supplementary Fig. 6a). Moreover, we found low levels of expression of maternal Dlk1 in paternal heterozygotes supporting canonical expression from the paternal allele in septal neurons and ventral striatum (Supplementary Fig. 6b,c). Nonetheless, we had observed similar neurogenic phenotypes in Dlk1−/+, Dlk1+/− and Dlk1−/− mutants, and niche-astrocytes from Dlk1+/− expressed reduced but detectable levels of DLK1 protein (Fig. 1d,g; Fig. 2d), suggesting a requirement for Dlk1 from the maternally-inherited allele in postnatal neurogenesis. Consistent with the neurogenesis phenotypes, expression was reduced in Dlk1−/+, Dlk1+/− and Dlk1−/− vs. Dlk1+/+ NSCs and niche-astrocytes, indicating activity from both parental alleles in the neurogenic population (Fig. 4a). We therefore assayed Dlk1 imprinting in wild-type postnatal and adult F1 hybrid offspring from reciprocal crosses of Mus musculus domesticus (C57BL6J) and Mus musculus castaneus (Cast/Ei) parents (Supplementary Fig. 6d). SVZ tissue from reciprocal hybrids showed the expected paternal expression of Dlk1 whereas neurospheres and niche-astrocytes showed biallelic expression of both membrane-bound and secreted isoforms (Fig. 4b; Supplementary Fig.6e). Importantly, other imprinted genes such as the adjacent Gtl2 gene, and Snrpn on chromosome 7C, maintained imprinting in NSCs and niche-astrocytes (Fig. 4c). To determine if biallelic expression was reflected in DLK1 protein levels in vivo, immunostaining in combination with GFAP and SOX2 was performed on maternal and paternal heterozygotes. This confirmed expression of DLK1 from both parental chromosomes in the NSCs of the neurogenic zone (Fig. 4d,e). In contrast, postmitotic neurons in non-neurogenic regions showed clear imprinting of Dlk1 (Supplementary Fig. 6b,c). Dlk1 was canonically imprinted in NSCs or astrocytes derived from E14.5 embryonic and new-born SVZ, becoming biallelic by P7 (Supplementary Fig. 6f). These data demonstrate specific and selective absence of Dlk1 imprinting in the NSC and niche-astrocyte populations commencing at postnatal stages and continuing to adulthood. This indicates that the mechanism conferring postnatal biallelic expression can override the imprint specifically and selectively at Dlk1, and that this regulation is required for normal neurogenesis.
Figure 4. NSCs and niche-astrocytes selectively lose Dlk1 imprinting postnatally.
(a) Quantitative PCR of Dlk1 expression in SVZ, NSCs and niche-astrocytes derived from Dlk1+/+, Dlk1−/+ and Dlk1+/− mice. (b) Dlk1 allele-specific expression of postnatal 7 and 60 SVZ, NSCs, and niche-astrocytes derived from reciprocal F1 hybrid offspring from Mus musculus domesticus (BL6) and Mus musculus castaneus (Cast). (c) Gtl2 and Snrpn allele-specific expression. (d) Immunohistochemistry for GFAP (red) and DLK1 (green) in the SVZ of Dlk1+/+, Dlk1−/+ and Dlk+/− mice. (e) Immunohistochemistry for GFAP (blue), SOX2 (red), and DLK1 (green) in the SVZ of Dlk1 mutant mice. Arrowheads indicate positive cells. (f) Methylation at the IG-DMR and Gtl2 promoter DMR. (g) Schematic representation of the Dlk1-Gtl2 domain in the neurogenic niche. Open and closed circles represent unmethylated and hypermethylated CpGs respectively. *p<0.05, **p<0.01. Error bars: s.e.m; n=10 per group and three bisulfite conversions. Scale bars: in c, 10 μm; in d, 7 μm.
Dlk1-Dio3 domain imprinting is controlled by the intergenic differentially methylated region (IG-DMR) located between Dlk1 and the adjacent non-coding RNA gene Gtl22,24,25. This germline-derived mark, characterised by hypomethylation on the maternal chromosome30, is required for post-fertilization acquisition of differential methylation at the secondary Gtl2-DMR at its promoter. Paternal IG-DMR methylation is required for expression of Dlk1 and repression of Gtl2 on the paternally-inherited chromosome26. We measured methylation at these DMRs and found no change at the Gtl2-DMR, consistent with retention of its imprinting. In contrast, hypermethylation at the IG-DMR was observed in NSCs and niche-astrocytes indicating that absence of Dlk1 imprinting is associated with gain of methylation at the germ-line DMR postnatally (Fig. 4f,g; Supplementary Fig. 7a,b).
In conclusion, our data support a role for DLK1 in a neurogenic continuum initiating at the early postnatal period that is maintained over the lifetime of the aging animal (Supplementary Fig. 8). Compromised neurogenesis appears to be driven by an early increase, followed by depletion of the stem cell pool. In the postnatal SVZ, niche-astrocyte-derived soluble DLK1, signals through a membrane-bound form of DLK1 in NSCs to regulate stem cell number with a continued requirement for DLK1 in their maintenance with age. Importantly, our findings indicate that NSCs and niche-astrocytes are distinguished early by expression of DLK1 membrane and secreted isoforms respectively and that differential processing of a single gene confers distinct functional properties to these two cell types. Also noteworthy is the epigenetically regulated selective absence of imprinting resulting in a biallelic dosage of Dlk1 required for normal neurogenesis. Biallelic expression of imprinted genes in specific cell types may constitute an important regulatory event in their developmental programme. The possibility that loss or gain of genomic imprinting might be used as a dynamic developmental mechanism to control gene dosage has wider implications for our understanding of the function and evolution of imprinting, and implies that the epigenetic mechanisms controlling the process in somatic lineages may be adaptable to the environmental niche in which they are acting.
Methods summary
Dlk1 mutant and transgenic mice were maintained as described11,15. For immunohistochemistry, vibratome sections from 4% paraformaldehyde-fixed brains were used. Neurosphere and primary astrocytes cultures were established as described6,8. DLK1 recombinant protein (ENZO, Life Sciences) was added to medium at the time of seeding. For co-culture experiments, a transwell insert (Millipore) was used and dissociated NSCs were seeded in the upper compartment. Neurospheres were nucleofected using a Mouse NSC Nucleofector Kit (Amaxa Biosystems, Germany) as previously described23. Constructs used were pIRES-GFP empty vector (Invitrogen), pIRES-GFP-S-Dlk1 (S-Dlk1) or pIRES-GFP-MB-Dlk1 (MB-Dlk1). Cells were seeded 48h after nucleofection for neurosphere assessment and immunocytochemistry as previously described18. Antibodies used are noted in Supplemental Methods. For qPCR using SYBR Green, RNA was isolated with Trizol (Invitrogen) and reverse-transcribed using SuperScript II RT (Invitrogen). Imprinting assays were based on PCR amplification and sequencing of samples derived from reciprocal F1 hybrid offspring of Mus musculus domesticus (C57BL/6) and Mus musculus castaneus (Cast/Ei). Bisulfite mutagenesis-based cytosine methylation analysis was as described previously24.
Methods
Animals and in vivo manipulations
Generation and genotyping of Dlk1 mutant and transgenic mice have been described previously11,15. Except for hybrid analyses, experiments were done with mice on a C57BL6 genetic background and their corresponding wild-type littermates. Housing of mice and all experiments were carried out in accordance with UK Government Home Office licensing procedures.
Tissue preparation and Immunohistochemistry
BrdU administration regimes have been previously detailed18. Two-to-four month-old mice were injected intraperitoneally with 50 mg BrdU/kg of body weight every 2 hours for 12 consecutive hours (7 injections in total). Animals were deeply anesthetized and transcardially perfused 30 days after the injections with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer pH 7.4 (PB) and their brains were vibratome-sectioned at 40 μm. The sections were blocked in 10% FBS (vol/vol, Invitrogen) and 0.1% Triton X-100 (vol/vol) in PBS (0.9% NaCl in PB) for 1 h and incubated overnight. For embryonic studies pregnant females were injected intraperitoneally with a single injection of 100 mg BrdU/kg body weight and embryos (E12.5 and E14.5) harvested one hour later. Embryonic and postnatal brains were dissected in PBS and fixed for 2h in 4% PFA at 4°C. Fixed samples were cryoprotected overnight in 20% sucrose/PBS at 4°C and mounted in OCT Compound (VWR) and sectioned coronally (20 μm) with a cryostat (CM3050S, Leica). For immunohistochemistry, cryostat sections were washed in PBS, and blocked at room temperature for 1 h in PBST (PBS, 0.1% Triton X-100) supplemented with 10% goat serum (Vector Laboratories). Primary antibodies and dilutions used were: mouse anti-BrdU (1:250; Dako), Ki67 (1:200; Novocastra), βIII-tubulin (1:250; Covance), Nestin (1:500, Chemicon) or Mash1 (1:100; Beckton Dickinson), goat anti-Sox2 (1:100; R&D Systems) or Doublecortin (DCX; 1:300; Santa Cruz), rabbit anti-GFAP (1:500; Dako), S100β (1:100, Dako), Tyrosine Hydroxylase (TH; 1:2000; Sigma), Calretinin (CR; 1:4000; Swant) or pH3 (1:500; Upstate) and rat anti-DLK1 (1:100; ENZO Life Sciences). For ependymal cell detection in vivo, sections were labeled with fluorescein isothiocyanate (FITC)-conjugated lectin IB4 (Sigma; 1:1,000). For BrdU detection sections were pre-incubated in 2 N HCl for 30 min at 37 °C and neutralized in 0.1 M sodium borate (pH 8.5). Secondary antibodies Alexa 488-conjugated donkey anti-mouse, Alexa 488-conjugated anti-rat, Alexa 647-conjugated donkey anti-rabbit (Molecular Probes), and CY3-conjugated donkey anti-goat (Jackson ImmunoResearch Laboratories) were used at 1:500 for 1 h. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI; 1 mg/ml). For DLK1 staining Dlk1−/− SVZ sections or cultures were used to confirm antibody specificity. Inmunofluorescence was analyzed using a Leica Multispectral confocal microscope (Leica). For SVZ whole-mounts we used protocols established previously27. Briefly the lateral walls of the lateral ventricle were dissected out and the resulting whole mounts were fixed for 1.5 hr in 4% paraformaldehyde and washed overnight at 4°C in PBS. For DCX staining, whole mounts were washed 3 times in PBS containing 0.5% Triton X-100 for 15 min each, blocked for 2 hr in blocking solution (10% FBS (vol/vol) and 0.5% Triton X-100 (vol/vol) in PBS (0.9% NaCl in PB), incubated for 72 hr at 4°C in goat anti-DCX (1:500 dilution; Santa Cruz). Secondary antibody Alexa 488-conjugated anti-goat (Jackson ImmunoResearch Laboratories) was used at 1:500. The stained walls were mounted with Fluorsave (Calbiochem) between two coverslips.
Neural Stem Cell and primary astrocyte cultures
Methods for NSC culture and self-renewal assessment, as well as BrdU immunocytochemistry in neurosphere cultures, have been previously described in detail18. Single cells from SVZ dissociates were seeded at very low density (2.5 cell/μl) in neurosphere growth medium with EGF and FGF which was supplemented with recombinant DLK1 (mouse):Fc (human) protein (2-100 ng/ml; produced in HEK 293 cells; ENZO Life Sciences), Jagged1 (1 μg/ml; Calbiochem), or PEDF (50 ng/ml; Bioproducts MD), and analyzed for primary neurosphere formation after 5 days in vitro. For self-renewal assays primary spheres formed in any condition were treated with Accutase (0.5 mM; Sigma) for 10 min, mechanically dissociated to a single-cell suspension and re-plated in growth medium containing EGF and FGF. Cell apoptosis and viability in single cells and incipient neurospheres were determined at 24 and 72 hours after plating, as previously described23. Multipotency capacity was analyzed by seeding individual passage 2-3 neurospheres of similar sizes in Matrigel-coated 96-well plates for 7 days in vitro (2% fetal bovine serum, vol/vol) before fixation in 4% PFA. No less than 50 clones were analyzed for each condition and the experiment as was previously described23. Distribution of unipotent (A, Astrocytes GFAP+), bipotent (A/N, GFAP+ Astrocytes and βIII-tubulin+ Neurons) and tripotent (A/N/O, GFAP+ Astrocytes, βIII-tubulin+ Neurons and O4+ Oligondendrocytes) clones was determined. To generate primary astrocyte cultures we used established methods as described previously by the Gage laboratory and others6,28,29. Importantly, no sub-culturing was made to the astrocyte cultures that were used for co-culture experiments. In brief, isolated adult SVZs were transferred to EBSS containing 1.0 mg/ml papain (Worthington DBA), 0.2 mg/ml L-cystein (Sigma), and 0.2 mg/ml EDTA (Sigma) and incubated in this mixture for 30 min at 37 °C. Tissue was then rinsed in DMEM/F12 medium (1:1 v/v; Invitrogen) and carefully triturated with a fire-polished Pasteur pipette to a single cell suspension. Isolated cells were collected by centrifugation, resuspended in astrocyte medium (DMEM/F12 medium containing 2 mM L-glutamine, 0.6% glucose, 9.6 g/ml putrescine, 6.3 ng/ml progesterone, 5.2 ng/ml sodium selenite, 0.025 mg/ml insulin, 0.1 mg/ml transferring and supplemented with 10% fetal bovine serum). Single cell suspensions from the SVZ were seeded onto Matrigel (1x; Becton Dickinson) treated wells in astrocyte medium and cultured in 37 °C incubator with 5% CO2 until astrocytes reached confluence (approx 6 days). The medium was changed every 2 days. The flasks were then shaken at 100 rotations per min for 3 hours at room temperature to shake off proliferating cells, oligodendrocyte progenitors and neurons. For co-culture experiments, the astrocyte culture medium was changed to neurosphere growth medium with mitogens and 2 h afterwards, a transwell insert (0.4-μm pore; 12-mm diameter; Millipore) was placed above and dissociated NSCs were seeded in the upper compartment. The astrocyte purity of the cultures was established by immunostaining against GFAP. Primary antibodies and dilutions used in vitro were: mouse anti-BrdU (1:500; Dako), O4 (1:2, Developmental Studies Hybridoma Bank) or βIII-tubulin (1:300; Covance), rabbit anti-GFAP (1:700; Dako), NICD (1:200, Abcam) or Caspase 3 (1:300, Cell Signaling), and rat anti-DLK1 (1:100; ENZO Life Sciences). Secondary antibodies Alexa 488-conjugated donkey anti-mouse, Alexa 488-conjugated anti-rat, Alexa 647-conjugated donkey anti-rabbit (Molecular Probes), and CY3-conjugated donkey anti-goat (Jackson ImmunoResearch Laboratories) were used at 1:500 for 1 h. DAPI (1 mg/ml) was used for counterstaining.
Cell transduction and luciferase assays
Constructs representing the predominant forms of Dlk1 generated by alternate splicing and proteolysis were generated previously21. We subcloned them in a pIRESh-GFPII vector (Invitrogen). NSCs grown for 2 days (passage 4–6) were nucleofected with 2–4 μg pIRESh-GFPII empty vector, pIRESh-GFPII-S-Dlk1 (S-Dlk1; secreted form, contains sequences from full length Dlk1A up to the juxtamembrane cleavage domain (amino acid 303) or pIRESh-GFPII-MB-Dlk1 (MB-Dlk1; membrane-bound form, lacks coding sequence for the juxtamembrane motif, amino acids 230-303, that is the substrate for cleavage and so represents Dlk1C) using a Mouse NSC Nucleofector Kit (Amaxa Biosystems, Germany) as described by the manufacturer. Cells were dissociated 2 days after nucleofection and seeded at low-density (2.5 cells per μl) in 96-well plates for neurosphere assessment. In reporter assays with a firefly luciferase-based construct we electroporated 2-4 μg of DNA from the construct 4xwtCBF1-luc30 and 50 ng of a Renilla luciferase construct as internal control. After 24–36 h, transduced cells (passage 4–6) were dissociated, plated in the presence or absence of DLK1 (ENZO; 50 ng/ml) or PEDF (100 ng/ml) as a positive control, and cultured for 24 h before being harvested for analysis. Efficiency was around 80% in all cases. Luciferase activity was measured in cell lysates using the Dual Luciferase Assay System (Promega) and promoter activity was defined as the ratio between the firefly and Renilla luciferase activities.
Expression Studies
RNAs were extracted with Trizol (Invitrogen), following manufacturer’s guidelines. For Northern blots mRNA was isolated from 75 μg of total RNA using Dynabeads Oligo (dT)25 kit (Invitrogen) following the supplied protocol. We used probes as described previously for Dlk1 and Gapdh2. For quantitative PCR (qPCR) 1 μg of total RNA was DNase-treated with RQ1 RNase-free DNase (Promega) following the manufacturer’s guidelines. All cDNA was synthesized using random primers and SuperScript II RT reverse transcriptase (Invitrogen), following standard procedures. Quantitative PCR was used to measure expression levels of Dlk1 normalized to Gapdh and was performed on a DNA Engine 2 Opticon detection system (Bio-Rad) using SYBR Green I as a double-strand DNA-specific binding dye. Thermocycling was performed in a final volume of 20 μl containing 2 μl of cDNA sample (diluted 1:5), 20 pmol of each primer (Dlk1-F, 5′-GAAAGGACTGCCAGCACAAG-3′; Dlk1-R, 3′-CACAGAAGTTGCCTGAGAAGC-5′; Gapdh-F, 5′-CCATCACCATCTTCCAGGAG-3′, Gapdh-R, 5′-GCATGGACTGTGGTCATGAG-3′), 2 mM MgCl2, 0.2 mM dNTP mixture, 1X Taq reaction buffer, 0.5 U HotStart Taq DNA polymerase, and 0.5 μl of a 1:3000 dilution of SYBER Green I. Semi-quantitative PCR (sqPCR) was performed to determine the different Dlk1 isoforms, and levels of transcripts were quantified by densitometric analysis of PCR bands in ethidium bromide-stained electrophoresis gels and normalized to Gapdh levels obtained at 20 cycles. Primers used were: Dlk1Splice-F, 5′-CTGCACACCTGGGTTCTCTG-3′; DlkSplice-R, 3′-TCCTCATCACCAGCCTCCTT-5′. In situ hybridization was conducted on PFA-fixed 40 μm vibratome adult brain sections according to procedures previously described15.
Imprinting assays
All imprinting assays were based on PCR amplification followed by direct sequencing to analyze parental specific expression of the genes. PCR reactions were purified with the QiAquick kit (Qiagen) prior to sequencing. We used reciprocal F1 hybrid offspring Mus musculus domesticus (strain C57BL/6, abbreviated BL6) and Mus musculus castaneus (strain Cast/Ei, abbreviated Cast) subspecies in which we had identified a single-nucleotide polymorphism between the two subspecies. The sequences of murine Dlk1 were obtained from GenBank (Accession number: NM010052). The Dlk1 polymorphism, located at exon 6 nucleotide 721, is a ‘T’ in BL6 mice and a ‘C’ in Cast mice. The Dlk1 imprinting assay used the following primers to amplify a fragment between exons 4 and 5: Dlk1-F, 5′-ACCCCCTGCGCCAACAATGG-3′ and Dlk1-R, 3′-GGGGTGAAGGGCCTGGGGAGT-5′, with thermal cycler conditions 94°C 1 min, 60°C 1 min, 72°C 1 min for 30 cycles. Primer Dlk1seq, 5′-AGAAGAAGAACCTCCTGTTGCA-3′, was used for direct reverse PCR fragment sequencing. The sequences of murine Gtl2 were obtained from GenBank (Accession number: Y13832). The Gtl2 polymorphism, located at exon 8, nucleotide 26, is a ‘G’ in BL6 mice and an ‘A’ in Cast mice. The Gtl2 imprinting assay used the following primers to amplify a fragment between exons 6 and 8: Gtl2-F, 5′-CTTGCTGGCCCTGGAGAT-3′and Gtl2-R, 3′-AACGTGTTGTGCGTGAAGTC-5′ with thermal cycler conditions 94°C 1 min, 59°C 1 min, 72°C 1 min for 33 cycles. Primer Gtl2-R was used for direct sequencing. The sequences of murine Snrpn were obtained from GenBank (Accession number: NM013670). The Snrpn polymorphism, located at exon 10 nucleotide 1270 is a ‘C’ in BL6 mice and a ‘T’ in Cast mice. The Snrpn imprinting assay used the following primers to amplify a fragment between exons 9 and 10: Snrpn-F, 5′-CATTATGGCTCCTCCACCTG-3′ and Snrpn-R, 3′-GTACCTGCAAGCTTTTTGACCC-5′, with thermal cycler conditions 94°C 1 min, 61°C 1 min, 72°C 1 min for 30 cycles. Primer Snrpn-F was used for direct sequencing. Genomic DNA sequences traces from BL6 and Cast SVZ, were used to identify strain-specific polymorphisms. (Supplementary Fig. 6d).
Combined bisulfite restriction analysis (COBRA) and pyrosequencing
DNA methylation level was quantified using COBRA and pyrosequencing. The method for bisulfite-based cytosine methylation analysis was adapted from Takada et al., 200224. DNA was denaturated by adding NaOH to a final concentration of 0.4 N. After 15 min incubation at 50°C, DNA was treated with 5 M sodium bisulfite/100 mM hydroxyquinone and incubated 18 h at 50°C. DNA was then desalted with Qiaquick PCR purification Kit (Qiagen) according manufacturer and desulphonated by adding NaOH to a final concentration of 0.3 N and incubating it 15 min at 37°C. DNA was finally neutralized and precipitated by adding 10 μg pf glycogen, 3 M of ammonium acetate and 200 μl of absolute ethanol and by incubating it overnight at −20°C. To generate the product for each target two rounds of PCR were done with fully or semi-nested primer pairs. Primers set for Gtl2 DMR (intron 1) were CT-F1 (5′-TGG TTT GGG GGT AGT TTT TTA TTG TAG-3′) and CT-R1 (5′-AAA AAA TAC AAA TAA ATT AAT TAA CAA ATC ACA AA -3′) for the first PCR. For the second round of PCR the following primers were used: CT-F2 (5′-ATT TTT AAA GAT GGT TGA TGT GGG TTT-3′) and CT-R1. Primers set for Gtl2 DMR (promoter) were GPFO (5′-TTT TAT TTA TAA TTA GGG TTT ATG TAG GGA AA-3′) and GPR3 (5′-TCT TCA TTT TTA CAC ATC CTT TCA AA-3′) for the first PCR and GPF2 (5′-GTA ATT TGT TAT AGA TTG GGG GGT TT-3′) and GPR4 (5′-CTT TCA AAC AAA AAA TAA CTA ACC ACT CAC CAA-3′) for the second round PCR. Primers for the IG-DMR first round PCR were IGOF (5′-GTG TTA AGG TAT ATT ATG TTA GTG TTA GG-3′) and IGOIR (5′-TAC AAC CCT TCC CTC ACT CCA AAA ATT-3′) and for the second round PCR IGIF (5′-ATA TTA TGT TAG TGT TAG GAA GGA TTG TG-3′) and IGOIR. First and second PCRs were run in 25 μl in the presence of 2U of HotStar Taq polymerase (Qiagen), 1X manufacture’s buffer, 3 mM MgCl2, 400 μM dNTPs and 1μM primers. PCR conditions were 10 cycles at 95°C for 40 secs, 53°C.for 40 secs and 72°C for 1 min followed by 30 cycles of 95°C for 30 secs, 53°C.for 30 secs and 72°C for 1 min. The PCR products were digested with appropriate restriction enzymes (RE) for combined bisulfite restriction analysis, representing each enzyme a representative methylation site. For IG-DMR NruI, Sau3AI and MluI were used; for Gtl2 DMR HincII, HpyCH4IV and for Gtl2 intron 1, HhaI and HpyCH4IV. MseI was used in all reaction as a bisulfite conversion control. The entire process was repeated three times and produced similar results. For pyrosequencing analysis a biotin-labeled primer was used to purify the final PCR product using Sepharose beads. The PCR product was bound to Streptavidin Sepharose High Performance (Amersham Biosciences), purified, washed, denatured with 0.2 mol/L NaOH, and washed again. Pyrosequencing primer (0.3 μM) was then annealed to the purified single-stranded PCR product and pyrosequencing was performed using the PyroMark Q96MD pyrosequencing system (Qiagen, Germany). Primers used: IG-DMR-F: 5′-GTGGTTTGTTATGGGTAAGTT-3′ and IG-DMR-R: 5′-CCCTTCCCTCACTCCAAAAATT-3′.
Statistical analyses
All statistical tests were performed using the GraphPad Prism Software version 4.00 for Windows, GraphPad Software, San Diego California USA (www.graphpad.com). The significance of the difference between groups was evaluated by analysis of variance followed by a two-tailed Student’s t-test in all experiments. When comparisons were performed with relative values (normalized values and percentages), data were first normalized by using an arcsen transformation. Treatment experiments were analyzed by Paired-T Test. Data is presented as the mean ± standard error of the mean (s.e.m) and the number of experiments performed with independent cultures/animals (n) is indicated in the legends.
METHODS REFERENCES
- 27.Mirzadeh Z, et al. The subventricular zone en-face: wholemount staining and ependymal flow. J Vis Exp. 2010;39 doi: 10.3791/1938. pii: 1938. doi: 10.3791/1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lie DC, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437(7063):1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 29.García-Marqués J, De Carlos JA, Greer CA, López-Mascaraque L. Different astroglia permissivity controls the migration of olfactory bulb interneuron precursors. Glia. 2010;58(2):218–230. doi: 10.1002/glia.20918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh JJ, et al. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16(3):952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary Material
Acknowledgments
We are grateful to Prof J Herbert and Prof EB Keverne for sharing expertise during the course of this work. We thank members of the Ferguson-Smith lab for helpful discussions and Bowen Sun, Dionne Gray, Scott Curran, Isabel Gutteridge, Rebecca Rancourt and Xavier d’Anglemont de Tassigny for technical assistance. The work was funded by grants from the MRC, and Wellcome Trust to AFS and grants from MICINN (SAF program), MSC (TerCel and Ciberned), and Generalitat Valenciana (Prometeo) to IF. SF is a recipient of a University of Cambridge Herchel-Smith Fellowship.
References
- 1.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14(16):1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 2.Takada S, et al. Delta-like and Gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10(18):1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- 3.Riquelme PA, Drapeau E, Doetsch F. Brain micro-ecologies: neural stem cell niches in the adult mammalian brain. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):123–137. doi: 10.1098/rstb.2006.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma DK, Bonaguidi MA, Ming GL, Song H. Adult neural stem cells in the mammalian central nervous system. Cell Res. 2009;19(6):672–682. doi: 10.1038/cr.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lledo PM, Merkle FT, Alvarez-Buylla Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31(8):392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417(6884):39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 7.Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6(11):1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 8.Kornyei Z, et al. Humoral and contact interactions in astroglia/stem cell co-cultures in the course of glia-induced neurogenesis. Glia. 2005;49(3):430–444. doi: 10.1002/glia.20123. [DOI] [PubMed] [Google Scholar]
- 9.Moon YS, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22(15):5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdallah BM, et al. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19(5):841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- 11.Raghunandan R, et al. Dlk1 influences differentiation and function of B lymphocytes. Stem Cells Dev. 2008;17(3):495–507. doi: 10.1089/scd.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis E, et al. Ectopic expression of DLK1 protein in skeletal muscle of padumnal heterozygotes causes the callipyge phenotype. Curr Biol. 2004;14(20):1858–1862. doi: 10.1016/j.cub.2004.09.079. [DOI] [PubMed] [Google Scholar]
- 13.Christophersen NS, et al. Midbrain expression of Delta-like 1 homologue is regulated by GDNF and is associated with dopaminergic differentiation. Exp Neurol. 2007;204(2):791–801. doi: 10.1016/j.expneurol.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Bauer M, et al. Delta-like 1 participates in the specification of ventral midbrain progenitor derived dopaminergic neurons. J Neurochem. 2008;104(4):1101–1115. doi: 10.1111/j.1471-4159.2007.05037.x. [DOI] [PubMed] [Google Scholar]
- 15.da Rocha ST, et al. Gene dosage effects of the imprinted delta-like homologue 1 (Dlk1/Pref1) in development: implications for the evolution of imprinting. PLoS Genet. 2009;5(2):e1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Forman SJ, Bhatia R. Expression of DLK1 in hematopoietic cells results in inhibition of differentiation and proliferation. Oncogene. 2005;24(27):4472–4476. doi: 10.1038/sj.onc.1208637. [DOI] [PubMed] [Google Scholar]
- 17.Tramontin AD, García-Verdugo JM, Lim DA, Alvarez-Buylla A. Postnatal development of radial glia and the ventricular zone (VZ): a continuum of the neural stem cell compartment. Cereb Cortex. 2003;13(6):580–587. doi: 10.1093/cercor/13.6.580. [DOI] [PubMed] [Google Scholar]
- 18.Ferron SR, et al. A combined ex/in vivo assay to detect effects of exogenously added factors in neural stem cells. Nat Protoc. 2007;2(4):849–859. doi: 10.1038/nprot.2007.104. [DOI] [PubMed] [Google Scholar]
- 19.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19(6):756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray SJ, Takada S, Harrison E, Shen SC, Ferguson-Smith AC. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol. 2008;8:11. doi: 10.1186/1471-213X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyfeler Y, et al. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. Embo J. 2005;24(19):3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreu-Agullo C, Morante-Redolat JM, Delgado AC, Farinas I. Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat Neurosci. 2009;12(12):1514–1523. doi: 10.1038/nn.2437. [DOI] [PubMed] [Google Scholar]
- 24.Takada S, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet. 2002;11(1):77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35(1):97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 26.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.