Abstract
Purpose
Utilizing cell-based approaches to identify genetic markers predictive of patients’ risk for poor response prior to chemotherapy.
Experimental Design
We performed genome-wide association studies (GWASs) to identify SNPs associated with cellular sensitivity to carboplatin through their effects on mRNA expression using International HapMap lymphoblastoid cell lines (LCLs) and replicated them in additional LCLs. SNPs passing both stages of the cell-based study were tested for association with progression free survival (PFS) in patients. Phase-1 validation was based on 377 ovarian cancer patients receiving at least 4-cycle of carboplatin and paclitaxel from the Australian Ovarian Cancer Study (AOCS). Positive associations were then assessed in the phase-2 validation analysis of 1,326 patients from the Ovarian Cancer Association Consortium and The Cancer Genome Atlas.
Results
In the initial GWAS, 342 SNPs were associated with carboplatin-induced cytotoxicity, of which 18 unique SNPs were retained after assessing their association with gene expression. One SNP (rs1649942) was replicated in an independent LCL set (p-valueBonferroni adjusted=9×10−3). It was found to be significantly associated with decreased PFS in phase-1 AOCS patients (Pper-allele=2×10−2), with a stronger effect in the subset of women with optimally debulked tumours (Pper-allele =4×10−3). rs1649942 was also associated with poorer overall survival in women with optimally debulked tumours (Pper-allele=9×10−3). However, this SNP was not significant in the phase-2 validation with patients from numerous cohorts.
Conclusion
This study demonstrates the potential of cell-based, genome-wide approaches to identify germ-line predictors of treatment outcome and highlights the need for extensive validation in patients to assess their clinical effect.
Keywords: ovarian cancer, pharmacogenomics, carboplatin, genome-wide study, HapMap CEU
Introduction
Ovarian cancer is the fifth leading cause of cancer mortality among women (1). Treatment of advanced disease consists of a platinum agent (usually carboplatin) and a taxane (usually paclitaxel) following cytoreductive surgery (2). Despite the high initial response rate to this chemotherapy, a proportion of cancers are intrinsically resistant to therapy (3), and susceptibility to side-effects is variable, with some patients developing severe carboplatin-induced myelosuppression (4). Clinically useful predictors that identify individuals most likely to benefit from carboplatin, or for that matter most chemotherapy, are lacking. Hence, identifying patients prior to treatment who are less likely to benefit from or most likely to experience adverse events from chemotherapeutic agents is essential.
The most relevant system for pharmacogenomic discovery in oncology are humans; however, executing pharmacogenomic clinical trials with enough power to detect true genetic signals in the presence of multiple confounding factors such as concomitant medications, dosage, and diet is extremely costly and difficult. Therefore, cell-based models evaluating gene expression, genetic polymorphisms and/or other biomarkers have been developed to help predict chemotherapy-induced response and toxicity (5). One such model utilizes International HapMap lymphoblastoid cell lines (LCLs) that have extensive and publicly available genotypic information, enabling genome-wide association studies (GWAS) that identify, in an unbiased fashion, genotype-phenotype relationships (5). The advantage to using LCLs in pharmacogenomics discovery is that they can be grown under identical conditions, allowing the phenotype to be tested in a well-controlled, isolated system without many of the confounders found in vivo. Most importantly, HapMap LCLs have publicly available genotypic data and are now part of the 1000 Genomes Project (6). Utilizing International HapMap LCLs, we developed a genome-wide model (referred to as the “triangle model”) that integrates genotype, gene expression and in vitro cytotoxicity data, to identify genetic polymorphisms associated with cellular sensitivity to chemotherapeutics (7–10). The successful clinical validation of these cell-based model findings was recently reported in a small head and neck cancer trial (11).
The goal of the present study was to use this cell model to discover genetic variants associated with cellular sensitivity to carboplatin that could be tested in a large cohort of clinical samples from patients treated with carboplatin. We hypothesized that genetic variants identified in our cell-based model would have utility in identifying patients treated with carboplatin with different clinical outcomes.
Materials and Methods
Genome-wide approach to identify genetic polymorphisms that are associated with carboplatin sensitivity
EBV-transformed LCLs derived from thirty Centre d’ Etude du Polymorphisme Humain (CEPH) trios from Utah residents with ancestry from Northern and Western Europe (HAPMAPPT01, CEU) along with 52 unrelated CEPH LCLs (8) were purchased from the Coriell Institute for Medical Research (Camden, NJ). Cell growth inhibition was evaluated at concentrations of 0, 10, 20, 40 and 80 µM of carboplatin for 72h and reported previously (11, 12). The concentration required to inhibit 50% cellular growth (IC50) was determined for each LCL through curve fitting and used as an indicator of carboplatin sensitivity.
The genome-wide approach that incorporates genome-wide SNPs, gene expression and carboplatin IC50 to identify genetic predictors of platinum sensitivity was described previously (7–9, 11). Briefly, SNP genotypes from the CEU population were downloaded from the International HapMap database (http://www.HapMap.org, Release 22). 2,286,186 SNPs with MAF > 5% and no Mendelian inheritance transmission errors in the CEU trios were used. IC50 values were log2 transformed to obtain approximate normally distributed phenotypes. The quantitative transmission disequilibrium test (QTDT) was performed to identify any genotype-cytotoxicity associations using the QTDT software (http://www.sph.umich.edu/csg/abecasis/QTDT) (13) with sex as a covariate. P ≤ 1×10−4 was used to select SNPs to carry forward in the analysis. The LD patterns at selected SNPs within each population were evaluated using Haploview version 3.32 (http://www.broad.mit.edu/mpg/haploview/). To detect evidence of recent positive selection in the genomic regions of interest, we employed the Haplotter online tool (http://haplotter.uchicago.edu/) developed by the Pritchard group to compute the integrated haplotype score (iHS) (14). The iHS quantifies the amount of extended haplotype homozygosity at a locus along the ancestral allele background relative to the derived allele background. Since iHS is standardized with mean 0 and variance 1, a positive iHS score greater than 2 means that haplotypes on the ancestral allele background are longer compared to the derived allele background.
Baseline gene expression was evaluated in 87 CEU LCLs using the Affymetrix GeneChip® Human Exon 1.0 ST array (Exon Array), as previously described (10). The gene expression data described in this study has been deposited into GEO (GenBank Accession No: GSE7761). Genes were evaluated as “transcript clusters”, each of which refers to a cluster of one or more exons covering a genic region. Transcript cluster expression summarizes all exonic transcriptional evidence for a known or putative gene. SNPs derived from the genotype-IC50 association analysis in CEU were tested for their association with gene expression using the QTDT test with gender as a covariate as described previously (9). A Bonferroni correction using the number of expressed genes tested (pc<5×10−2) was used to adjust for multiple testing.
To examine the relationship between gene expression and sensitivity to carboplatin, a general linear model was constructed with log2-transformed carboplatin IC50 as the dependent variable and transformed gene expression level together with an indicator for gender as the independent variables (9). If a SNP was significantly associated with carboplatin IC50 and the same SNP was significantly associated with gene expression, then the above approach was used to test whether gene expression significantly predicted IC50. Transcript cluster expression, with gender as a covariate, was tested as a predictor of carboplatin sensitivity in the CEU population. P < 5×10−2 was considered statistically significant. A target gene in this analysis was defined as one whose expression was associated with one or more SNP genotypes, and whose expression significantly correlated with carboplatin IC50.
Genotype carboplatin sensitivity analysis on replication sample set
NCI and NHGRI have jointly published a set of guidelines for designing a replication study following GWAS (15). Our study sought to adhere closely to these guidelines. A set of 52 unrelated LCLs that are part of the same genetic ancestry (CEPH/Utah) but are not part of HAPMAPPT01 LCLs that were used in the discovery set was evaluated for carboplatin sensitivity. Phenotypic data was log2 transformed prior to association analysis as was done for the discovery set. Eighteen SNPs identified in the discovery step (after removing redundant SNPs with linkage disequilibrium (LD)>0.8) were genotyped through Sequenom MassARRAY iPLEX platform. Genotype-phenotype association evaluation was conducted using linear regression. Additive genetic effects were assumed for these associations. Bonferroni adjusted p<5×10−2 was considered statistically significant.
Validation of selected SNPs in clinical samples
SNPs found to be significantly associated with carboplatin sensitivity in vitro were evaluated in invasive ovarian cancer patients receiving primary chemotherapy treatment of a minimum of 4 cycles of paclitaxel (175 or 135 mg/m2) and carboplatin (area under the curve, 5 or 6) at 3 weekly intervals, using a two-phase validation approach. In phase 1 we analyzed genotype data from 377 non-Hispanic white patients from the Australian Ovarian Cancer Study (AOCS), both overall and according to debulking status (optimally debulked patients ≤1 cm residual disease, and suboptimally debulked patients >1cm residual disease). In phase 2 we further evaluated positive findings in 1,326 non-Hispanic White invasive ovarian cancer patients of all histologies and morphologies receiving primary chemotherapy treatment of minimum of 4 cycles of paclitaxel and carboplatin known, or presumed, to have had the standard dosages as given to the patients in phase 1. Phase 2 patients were derived from six studies participating in the Ovarian Cancer Association Consortium (OCAC), plus additional AOCS patients not included in phase 1 validation analysis, and The Cancer Genome Atlas (TCGA); ethnicity was self-reported. Details of study design and patient ascertainment have been previously described elsewhere (16, 17) and are summarized in Supplementary Table 1. Clinical characteristics of patients participating in phase 1 and 2 validation studies are summarized in Supplementary Table 2. In AOCS, BEL, LAX and RPX, progression-free survival (PFS) was defined as the time interval between the date of histologic diagnosis and the first confirmed sign of disease recurrence, or progression, based on Response Evaluation Criteria in Solid Tumors (RECIST) criteria, modified for ovarian cancer as defined by the Gynecologic Cancer InterGroup (GCIG), and previously described (18, 19). For SCO, progression was defined as 1) a 25% or greater increase in the size of at least one bidimensionally or unidimensionally measurable lesion, 2) a clear worsening from previous assessment of any evaluable disease (note that worsening of existing non-evaluable disease did not constitute progression), 3) the reappearance of any lesion that had disappeared, with the exception of ascitic or pleural fluid that was drained and recurred within 3 months of drainage, or 4) the appearance of any new lesion and/or site. For MAL, progression was determined by ultrasound or defined by CA125 values (increase in CA125 to > 35 U/ML from a CA125 value lower than 35 U/ML after the primary treatment). For MAY, progression was defined as radiographic evidence of recurrence or initiation of second-line therapy. No consistent definition of progression was used in TCGA. All studies participating in phase 2 validation were compared at baseline to assess differences in median PFS across sites (Supplementary Figure 1). Overall survival was the interval between the date of diagnosis and death from any cause. All studies have received approval from their respective human research ethics committees, and all OCAC participants provided written informed consent. Details of TCGA can be found at http://cancergenome.nih.gov/.
DNA extraction, genotyping methods and quality assurance for all samples available for genotyping have been previously described (18). Genotype data for TCGA patients was downloaded through the TCGA data portal and assessed for ancestral outliers; patients of European descent were included in phase 2 analyses. SNPs found to be significantly associated with carboplatin sensitivity were further evaluated by inferring the missing genotypes in TCGA samples with the reference of the 1000 Genomes Data (1000G 2010-06 release, CEU). We performed the standard two-stage imputation in MACH 1.0 (20). Good imputation quality was attained by applying the following quality controls: imputed R2>0.3; MAF>1%; and p-value from Hardy-Weinberg Equilibrium test <1 × 10−6.
The primary test for association was the relationship between SNP genotypes and progression-free survival (PFS) and overall survival (OS). The Kaplan-Meier (KM) product limit method was used to estimate and plot the PFS and OS probabilities. Cox proportional hazards models were used to obtain hazard ratios (HR) and 95% confidence intervals (CI) adjusted for the effects of FIGO stage and residual disease (nil, ≤1 cm, >1 cm and ≤ 2 cm, >2 cm). Assuming log additive effects, the risks associated with each additional minor allele were estimated by fitting the number of rare alleles carried as a continuous covariate. Risks associated with heterozygosity and homozygosity for the minor allele of SNPs associated with outcome were also estimated. To account for differences in coding of residual disease and tumor characteristics across different studies in phase 2 analyses, estimates were additionally adjusted for tumor histology and grade, and residual disease was fitted as a dichotomous covariate (nil vs. any). Also, to allow for variation in time from diagnosis to study entry across phase 2 studies, PFS and OS data were left-truncated, with time at risk starting on date of diagnosis, and time under observation beginning at the time of study entry. Additionally, OS data were right censored at 5 years post-diagnosis in order to reduce the number of deaths unrelated to ovarian cancer. Summary per-allele estimates for PFS and OS for all phase 2 validation studies were obtained using a weighted meta-analysis of site-specific loge hazard ratios. All tests for association were two-tailed, statistical significance was assessed at the conventional level of P<5×10−2, and analyses were performed in STATA SE v. 11 (Stata Corp.) and the R project for Statistical Computing.
Target gene expression evaluation in NCI-60 datasets
To explore the potential mechanism of action for our identified and validated genetic variation(s), we examined the degree of correlation between the target gene expression and cellular (using both LCLs and NCI60 tumor cells) susceptibility to carboplatin. We downloaded the NCI-60 microarray expression and GI50 datasets (released in March, 2007) from the DTP/NCI Molecular Target Databases (21, 22). These datasets are comprised of gene expression data on untreated NCI-60 cell lines using different microarray platforms along with GI50 data. Linear regression was performed between the expression of a gene of interest and log10 carboplatin GI50 in all 60 tumor cell lines as well as in a subset of ovarian cancer cell lines (n=7). p<5×10−2 was considered statistically significant.
Results
Phenotyping the discovery and replication samples
Cellular growth inhibition was evaluated in 87 HapMap Phase II CEU LCLs (discovery) and an independent set of 52 CEPH LCLs (replication). The average log2-transformed carboplatin IC50 values were not significantly different between these two sets of samples (4.49 and 4.60 µM for discovery and replication samples, respectively; p=0.46). The discovery set results have been described previously (12).
Genome-wide approach to identify genetic polymorphisms that are associated with carboplatin sensitivity
The overview of study workflow and number of findings from each analysis is shown in Table 1. We identified a total of 342 SNPs that were strongly associated with carboplatin IC50 phenotype at p ≤ 10−4in the LCL discovery samples (11). All 342 SNPs are listed in Supplementary Table 3. A binomial test showed a significant difference between our findings and random expectation (p < 10−5), suggesting that our association test results are not likely to be an artifact.
Table 1.
Overview of study workflow and number of findings from each analysis
| Preclinical Cell-Based Model | Clinical Studies | |||||
|---|---|---|---|---|---|---|
| Discovery | Replication | Clinical evaluation | ||||
| Approach | GWAS between 2 ×106 SNPs and carboplatin-IC50 in HapMap Phase I/II CEU LCLs | Association between 342 SNPs and 13,314 transcript clusters (genes) expression | Linear correlation between experession of 29 genes and carboplatin-IC50 | Association between 17 unique SNPs and carboplatin IC50 in 52 non-HapMap Phase I/II CEPH LCLs | Association between 1 SNP and PFS and OS in 377 AOCS patient samples | Association between 1 SNP and PFS in 1326 OCAC and TCGA patient samples |
| P-value cut-off | P ≤ 10−4 | P < 3.7 × 10−6 | P < 5×10−2 | P < 5×10−2 | P < 5×10−2 | P < 5×10−2 |
| Findings | 342 SNPs | 31 SNPs | 26 genes (also associated with 18 unique SNPs | 1 SNP | 1 SNP | --- |
SNPs found to be associated with carboplatin sensitivity (n = 342) were further evaluated for their functional relevance using 13,314 transcript clusters (representing 10,830 genes) expression. These analyses narrowed our candidate SNP list to 31 SNPs (Bonferroni corrected based on number of transcript clusters Pc < 5×10−2) associated with the expression of 29 different genes. After removing SNPs in high LD (r2 ≥ 0.8), our SNP list was further reduced to 18 unique signals. The final analysis examined the correlation between the expression of these “target genes” (defined as genes whose expression was associated with carboplatin sensitivity related SNPs) and carboplatin IC50 phenotype using a general linear model (7). The expression of 24 transcript clusters (representing 26 genes) was correlated to carboplatin IC50 at p<5×10−2. The p values were used not to assess significance but as a tool to filter SNPs that show at least moderate evidence of being functional by being correlated to genes whose expression relates to drug sensitivity (11).
Evaluation of SNPs in a replication set
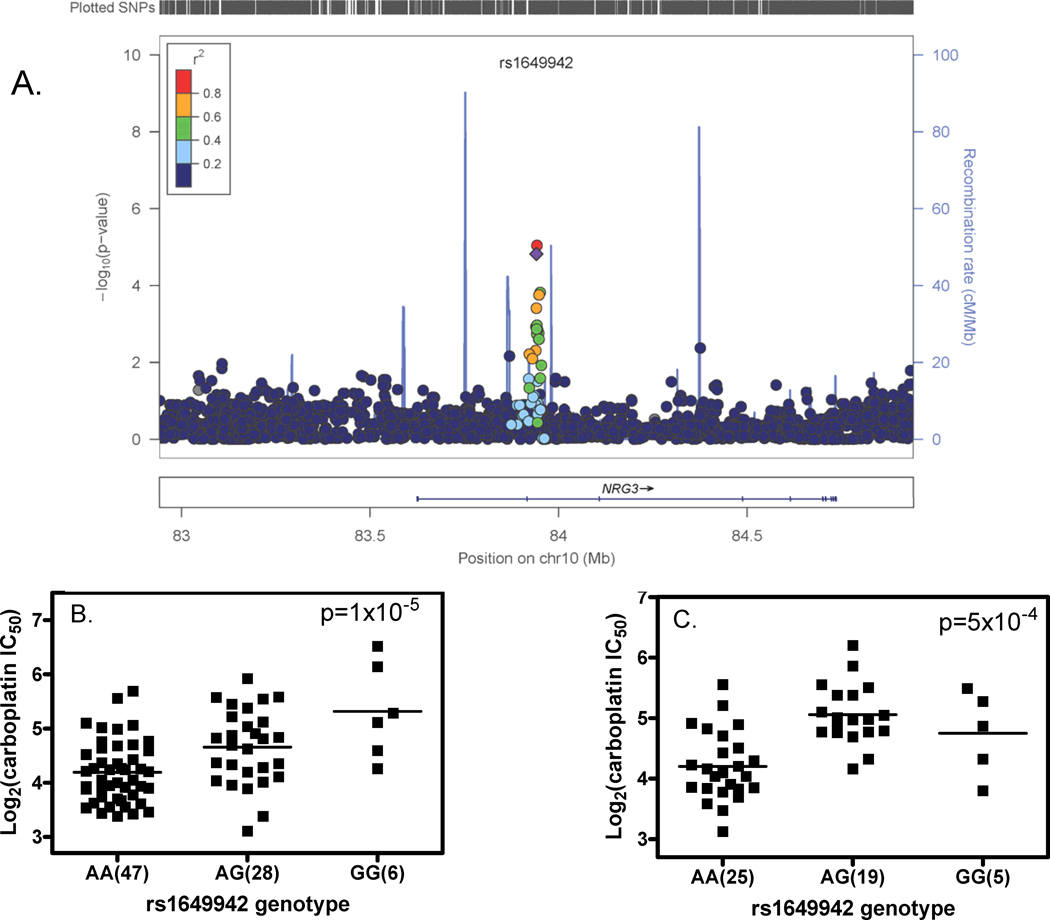
We successfully genotyped 17 of the 18 SNPs in the replication set. One SNP (rs1649942) was significantly associated with carboplatin sensitivity (Bonferroni adjusted p-value =8.5×10−3. Figure 1). The next SNP ordered by strength of association was rs12053210 (in complete LD with rs12614692) with unadjusted p-value = 1×10−2 and FDR = 9×10−2. This is not significant at the Bonferroni corrected 0.05 level and the effect in the replication set was opposite to that found in the discovery set (Supplementary Table 4).
Figure 1.
Relationships between SNP genotype and log2 transformed carboplatin IC50 in the rs1649942 region on chromosome 10. A) A local genomic region around SNP rs1649942, which is located in the intron 10 of neuregulin 3 gene (NRG3). Each dot represents a relationship between a SNP genotype and log2 transformed carboplatin IC50. The relationship is shown as –log10(p-value). Plots were made with LocusZoom (40) and the color of each dot represents the SNP’s linkage disequilibrium r2 in the CEU with the SNP of interest (rs1649942) labeled as a purple diamond; B) rs1649942 genotype and log2 transformed carboplatin IC50 association in the HapMap CEU discovery samples; C) rs1649942 genotype and log2 transformed carboplatin IC50 association in the replication samples. For figure 1B and 1C, each bar represents the mean log2IC50 value.
SNP rs1649942 was previously reported to be associated with cisplatin IC50 in HapMap CEU discovery samples (10) (Supplementary Fig2A) and was found to be associated with cisplatin IC50 in the replication samples (Supplementary Fig2B). This SNP is located within the intron of NRG3 (Figure 1A), has been previously shown to be a master regulator associated with the expression level of 39 genes at a p≤1×10−4 (23). In this study, using a more stringent cutoff (Bonferroni adjusted p-value<5×10−2), we found the expression levels of 18 genes associated with this SNP genotype. Of them, 10 are associated with carboplatin IC50 with the gene expression levels of 7 negatively correlated with carboplatin IC50 (ALDH2, CRIM1, KYNU, LOC100131869, OAS1, RAPGEF5, SLC2A5), suggesting the higher the gene expression the greater cellular sensitivity to carboplatin. The remaining 3 genes (BCR, PSTPIP2 and SHFM3P1), higher expression is associated with cellular resistance to carboplatin. In particular, we observed that the SNP was associated with the expression of ALDH2 and KYNU, at a stringent Bonferroni threshold. Furthermore, the SNP ranked in the top 2% and 5% respectively of all eQTLs for ALDH2 and KYNU respectively, at expression p-value < 10−4. Importantly, a genome-wide scan (using a publicly available genomic resource we created, http://www.scandb.org) revealed that no eQTL more predictive of expression for either gene showed an association with carboplatin IC50. In addition, rs1649942 has an integrated haplotype score (iHS) of 2.8, which indicates a shorter derived allele haplotype at the SNP locus, suggesting functional significance and recent positive selection in Caucasians.
Examination of genetic variants in ovarian cancer clinical samples
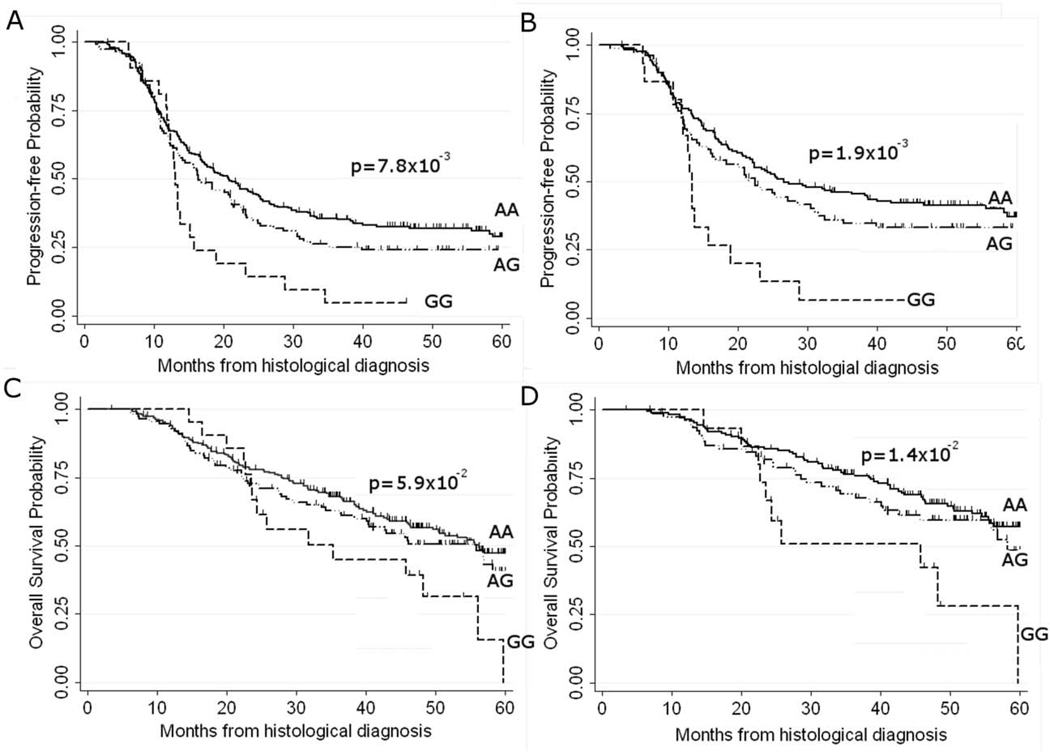
We genotyped rs1649942 in a two-phase validation analysis of 377 non-Hispanic white AOCS (phase 1) and 1,326 non-Hispanic white patients (phase 2) from six OCAC sites, TCGA and an additional 154 patients from the AOCS (see Supplementary Table 2). All genotype data conformed to Hardy Weinberg proportions (PHWE ≥ 0.1), and the minor allele frequency (MAF) for rs1649942 was 0.24 (site-specific range 0.19 to 0.27). In phase 1 analysis we observed a significant decrease in PFS associated with each additional copy of the minor allele of the rs1649942 SNP [adjusted HRper-allele=1.25 (95% CI:1.03–1.52), p=2.3×10−2]. This association with PFS was even more pronounced in a subset of women with optimally debulked tumours (residual disease ≤1cm) [adjusted HRper-allele=1.43 (95% CI:1.12–1.81), p=4×10−3]. Analysis of OS using right censoring at 5 years post-diagnosis to account for deaths unrelated to ovarian cancer also showed a significant association with this SNP in optimally debulked patients [adjusted HRper-allele=1.48 (95% CI:1.10–2.0), p=9×10−3] (Table 2 and Figure 2). In phase 2 validation, baseline median PFS was significantly different across sites (PLog-rank =7×10−4) ranging from 15–30 months (see Supplementary Figure 1), which we accounted for in summary estimates using the weighted meta-analysis approach. However, in the phase 2 analyses of an additional 1,326 non-Hispanic white patients from multiple sites, neither the site-specific estimates, nor the summary estimates from weighted meta-analysis, supported the associations observed between PFS or OS and the rs1649942 SNP in phase 1 analysis (see Supplementary Figure 3). In light of the null findings from phase 2 validation analysis, we re-analyzed phase 1 AOCS data using the same analytical methods as for phase 2, and observed a similarly significant association for PFS in all patients [adjusted HRper-allele=1.22 (95% CI: 0.98–1.52), p=7×10−2] and in patients with no residual disease [adjusted HRper-allele=1.81 (95% CI: 1.16–2.81), p=9×10−3], but no significant association for OS in either all patients or the subset with no residual disease (adj. Pper-allele >9×10−2) was observed. When we restricted phase 2 analysis to patients known to have had the standard doses of paclitaxel and carboplatin (n=776), we found no supporting evidence for the associations observed in phase 1. Likewise, analysis of all available patient data from the OCAC validation sites regardless of chemotherapy regimen (n=3,190) yielded no additional support for phase 1 associations.
Table 2.
Association between NRG3 rs1649942 genotypes and progression-free survival in phase 1 validation patients from the AOCS
| rs1649942 Genotype | Patients | Progression-free Survival | Overall Survival | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | aHR | 95% | CI | P | aHR | 95% | CI | P | |
| All Patients | ||||||||||
| AA | 242 | 64.19 | 1.00 | 1.00 | ||||||
| AG | 114 | 30.24 | 1.25 | (0.95 | – 1.63) | 1×10−1 | 1.13 | (0.81 | – 1.56) | 0.48 |
| GG | 21 | 5.57 | 1.57 | (0.98 | – 2.51) | 6×10−2 | 1.50 | (0.87 | – 2.58) | 0.14 |
| per-allele | 1.25 | (1.03 | – 1.52) | 2×10−2 | 1.18 | (0.94 | – 1.50) | 0.15 | ||
| Optimally Debulked Patients (residual disease≤1cm) | ||||||||||
| AA | 177 | 65.31 | 1.00 | 1.00 | ||||||
| AG | 79 | 29.15 | 1.37 | (0.98 | – 1.93) | 7×10−2 | 1.42 | (0.93 | – 2.17) | 1×10−1 |
| GG | 15 | 5.54 | 2.14 | (1.21 | – 3.78) | 9×10−3 | 2.32 | (1.17 | – 4.61) | 2×10−2 |
| per-allele | 1.43 | (1.12 | – 1.81) | 4×10−3 | 1.48 | (1.10 | – 2.00) | 9×10−3 | ||
| Sub-Optimally Debulked Patients (residual disease >1cm) | ||||||||||
| AA | 44 | 61.97 | 1.00 | 1.00 | ||||||
| AG | 23 | 32.39 | 0.96 | (0.55 | – 1.61) | 0.82 | 0.84 | (0.46 | – 1.56) | 0.59 |
| GG | 4 | 5.63 | 1.07 | (0.38 | – 3.05) | 0.89 | 0.89 | (0.27 | – 2.92) | 0.84 |
| per-allele | 0.99 | (0.65 | – 1.50) | 0.95 | 0.89 | (0.55 | – 1.43) | 0.64 | ||
Adjusted for FIGO stage and level of residual disease (nil, ≤1cm; >1cm & ≤2cm; >2cm)
Figure 2.
Kaplan-Meier plots for rs1649942 genotype effect on progression free survival (PFS) and overall survival (OS) probabilities in all phase 1 AOCS patients (n=377) and a subset of optimally debulked patients (n=271). Log rank p-values compare survival probabilities across genotypes, and tick marks indicate censoring events. A) rs1649942 genotype effect on PFS in all patients and B) optimally debulked patients; C) OS in all patients, and D) optimally debulked patients.
Relationship between rs1649942 and target gene expression
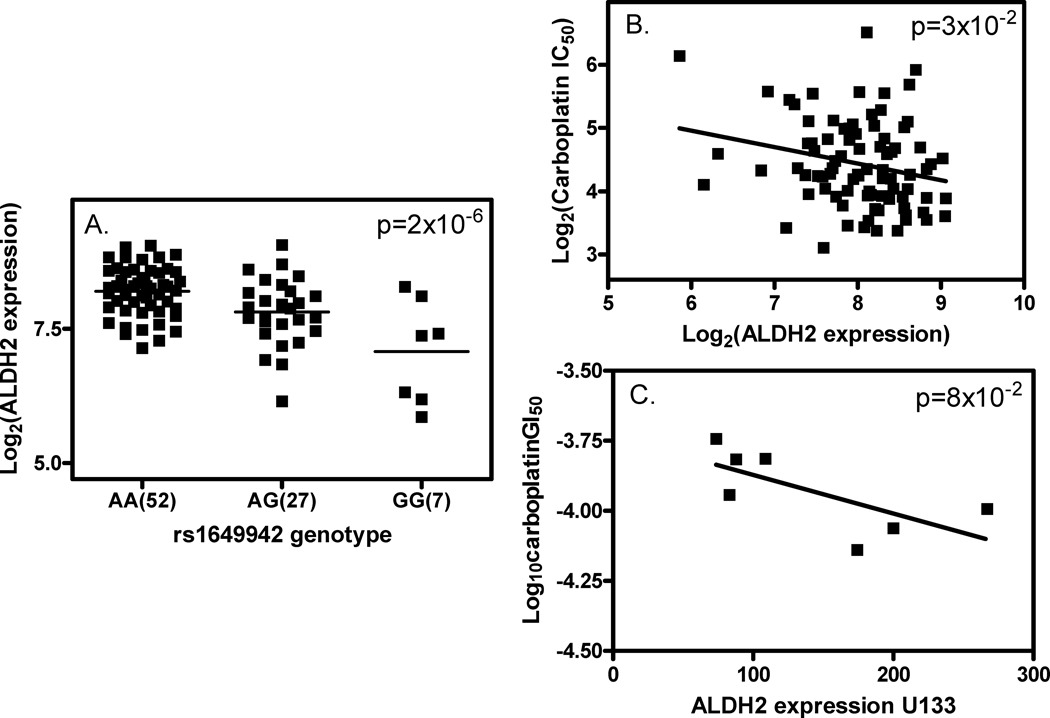
To evaluate the potential functional significance of this SNP, we examined the relationship between the SNP and its target gene expression in LCLs and NCI-60 cell lines. In the discovery set of LCLs, we found a significant association between the clinically validated SNP (rs1649942) and the baseline expression of 18 genes including ALDH2 (Fig3A, Supplementary Table 4) and KYNU. In addition, ten of the 18 gene expression traits are correlated with carboplatin IC50 (Supplementary Table 4). Figure 3B illustrates that increasing the expression of ALDH2, a target gene, confers greater carboplatin sensitivity in LCLs. Incidentally, ALDH2 expression also showed a borderline significant correlation with cisplatin sensitivity (IC50) in LCLs (p=8×10−2, Supplementary Figure 2C). In a set of 7 ovarian cancer cell lines as part of NCI-60 cancer cell line resource, using the GC180405 microarray_U133 array data, we found a borderline significant association between ALDH2 expression and log10GI50s of carboplatin (p=8×10−2, Figure 3C) and cisplatin (p=5×10−2, Supplementary Fig2D). This is in agreement with our LCL findings that higher gene expression confers greater platinum sensitivity. In addition, another gene (KYNU), whose expression is associated with rs1649942 genotype, was recently reported as one of the genes within an expression signature that predicted OS in ovarian cancer patients receiving platinum-based chemotherapy (24).
Figure 3.
Relationship between rs1649942, ALDH2 gene expression and carboplatin sensitivity. A) rs1649942 genotype and log2 transformed ALDH2 expression association in the HapMap CEU samples (n=86); B) correlation between log2 carboplatin IC50 and log2 transformed ALDH2 expression in the HapMap CEU samples; C) correlation between log10 transformed carboplatin GI50 and log2 transformed ALDH2 expression in 7 ovarian cancer cell lines as a subset of the NCI-60 cell panel using GC180405 microarray_U133 data.
Discussion
In this study, we used LCLs from the well-genotyped International HapMap collection and identified 18 unique SNPs that associate with carboplatin sensitivity from >2 million SNPs. One of these was replicated in a set of independent LCL samples (Bonferroni corrected p<5×10−2). The SNP of interest (rs1649942) shows a r2 of 0.20 or 0.23 with carboplatin IC50 in CEU discovery and validation samples, respectively, suggesting this SNP is explaining about 20% of the phenotypic variation. We found that this SNP is associated with PFS and OS in phase I analysis of 377 Australian ovarian cancer patients who received at least 4 cycles of carboplatin-based chemotherapy. However, in a larger, second phase of evaluation of patient samples, we did not replicate these findings. The potential mechanism of action this SNP in LCLs may be through its association with the expression of 18 target genes (e.g., ALDH2 and KYNU using a stringent Bonferroni cutoff). Ten of these target gene expression traits are also correlated with carboplatin sensitivity in LCLs.
There is a pressing need to identify germline variation that predicts response to standard therapy for advanced ovarian cancer (platinum plus taxane) since the 5-year survival rate is approximately 45%. In fact, ovarian cancer kills approximately 15,000 women in the United States every year, and more than 140,000 women worldwide (25). Thus, identifying those at risk for non-response to certain chemotherapy allows for the possibility of administering alternative chemotherapy and potentially improving treatment outcomes.
An alternative approach to “personalized medicine” is to identify a set of gene expression signatures instead of genetic variants. In fact, a 14-gene expression predictive model was developed to predict early relapse in women with advanced ovarian cancer and treated with platinum-taxol (26). However, evaluating gene expression in tumors of patients is cumbersome, variable and expensive. A candidate gene approach has also been attempted to identify genetic markers that predict ovarian cancer treatment outcomes but failed in producing unequivocal results (27). GWAS provides an unbiased approach to evaluate all genetic variation in the genome that may contribute to disease risks (28–30) and/or drug response (31). Therefore, we employed GWAS in a cell-based model to identify germline variants with clinical applicability. The in vitro model system could be applied to other toxic drugs that would be difficult, if not impossible, to study in non-diseased patients. GWAS identified SNPs in this study that would not have been likely “candidate SNPs” based on the drug’s pharmacokinetics, pharmacodynamics or mechanism of action.
In LCLs, the rs1649942 SNP is within the neuregulin 3 (NRG3) gene, which has been shown to activate the tyrosine phosphorylation of its cognate receptor, ERBB4, and is thought to influence neuroblast proliferation, migration and differentiation by signalling through ERBB4 (32, 33). SNPs within the NRG3 gene have been implicated in heart failure mortality (34); schizophrenia (35); and ADHD (36). However, the NRG3 gene itself was not well represented using the exon array. In efforts to interrogate the genomic region more closely, we used whole-genome sequence data from the 1000 Genomes project and identified 4 additional SNPs in moderate LD (r2 > 0.70) with our SNP in CEU; none showed more significant association with carboplatin IC50. Furthermore, we explored the possibility that the SNP distantly regulates other genes in the genome to achieve its effect. Indeed, rs1649942 genotype is strongly associated with more than 10 transcriptional expression traits, suggesting that it may be a genomic master regulator (23). A simple base pair change at this locus may produce a cascade of expression signal changes, resulting in phenotypic variation (in our case, patient survival post carboplatin treatment).
Interestingly, we found both the SNP (rs1649942) and one of its target genes (ALDH2, a mitochondrial isoform of aldehyde dehydrogenase) were also associated with sensitivity to cisplatin, another commonly used platinating agent, in our LCL model (10). A recent report showed the higher expression of ALDH1, a cytosolic isoform of the aldehyde dehydrogenases family, is associated with higher response to chemotherapy, longer disease-free survival and OS time in ovarian cancers (37). In agreement, we found that higher ALDH2 expression was significantly correlated to sensitivity to platinum-induced cytotoxicity in both LCLs and ovarian cancer cell lines. ALDH1 was not also identified in our model due to the lack of expression of this gene in HapMap CEU samples.
Despite its many advantages over other approaches, GWAS may suffer from a high rate of false discovery. Therefore, NCI and NHGRI have jointly published a set of guidelines suggested to be used in designing a replication study following GWAS (15). Our study sought to adhere closely to these guidelines and encompassed not only an independent set of in vitro replication samples, but in vivo clinical samples for validation as well. In our phase 1 in vivo study using 377 AOCS patients, the risk allele of rs1649942 was associated with a modest increased risk of disease progression and death following carboplatin-based chemotherapy, with an even greater genetic contribution for both PFS and OS among a subset of patients with optimally-debulked tumors. The reason for the greater effect in this subset is not entirely clear, but this result mirrors our previous observation that an association between PFS and the ABCB1 2677G > T/A SNP was only seen in women with minimal residual disease (18). Since clinical outcomes obtained from optimally-debulked patients may represent the ideal scenario in which to isolate effects due primarily to chemotherapy from the confounders associated with residual disease, the effect of rs1649942 in these particular patients is of interest but it should be noted that this result was based on small numbers of patients. There were no significant associations observed between rs1649942 genotype and factors related to prognosis in ovarian cancer, including patient age, stage, histology and residual disease, suggesting that the observed genetic effect on patient survival is likely to be related to its effect on chemotherapeutic response rather than to disease characteristics.
However, we did not replicate phase 1 findings in our phase 2 analysis, which differed to the phase 1 analyses in several ways. In phase 2 we categorized residual disease as ‘nil’ vs ‘any’ as opposed to ≤ or > 1cm so that we could include patients from sites which did not use this coding, and adjusted for grade and histology (in addition to stage which we used for the adjusted analyses in phase 1); we also included patients (n=550) presumed, rather than known, to have had standard doses of paclitaxel (175 or 135 mg/m2) and carboplatin (area under the curve, 5 or 6) in order to increase our power. However, when we re-analyzed the phase 1 data using the same analytical method as for phase 2, we obtained similar significant associations with rs1649942. When we restricted phase 2 analysis to patients with known doses as in phase 1 (n=776), we still found no association with this SNP. While phase 1 estimates were based on small numbers and may be false discovery, it is possible that failure to observe an association with the rs1649942 SNP in phase 2 analysis may reflect differences in clinical definitions across studies that cannot be adequately accounted for in the analysis, and low power to detect an association in the 776 patients whose treatment details were known. For example, the criteria used to define disease progression varied across studies and in some cohorts (TCGA) no consistent definition of progression was used. Time to progression was the clinical outcome measure used in this study, as the measurement of ‘response’ to primary chemotherapy in ovarian cancer is confounded by the fact that chemotherapy is combined with debulking surgery. A fall in CA125 cannot distinguish between the effects of chemotherapy and the effects of surgery, and imaging can only be used to assess response in patients with measurable disease remaining at the end of surgery (i.e., not in optimally debulked patients) (38). The clinical validation studies used self-reported ethnicity to determine the non-Hispanic whites. Since the cell-based finding on rs1649942 is specific to Caucasians, the use of self-reported ancestry is likely to include patients with differing ethnic backgrounds and potentially mask the Caucasian-specific association. This is particularly true in the second phase validation study, since patients were recruited from various sites across the world. Using ancestry informative markers to define ethnicity could potentially influence the final findings (39).
Interestingly, we have recently identified a suggestive association between this SNP and therapy-induced decreases in platelets in 60 head and neck cancer patients who underwent carboplatin-based induction therapy (11). Given this and the results of our in vitro experiments, we therefore cannot discount the possibility that this SNP may influence chemotherapy outcomes in ovarian cancer patients. Further analysis is warranted in larger, well-characterized clinical samples.
Given the obstacles to performing large, replicable pharmacogenetic studies aimed at discovering novel variants and the clinical confounders of such, the cell-based model we developed to identify genetic variants that may predict PFS and OS in ovarian cancer patients has important implications in the field of oncology. We acknowledge the limitations of using a cell-based model for pharmacogenomic discovery but the advantages compared to attempting to perform GWAS in a clinical trial are enormous, provided large cohorts of well characterized patients are available for validation. Cell-based models are much less expensive, many of the environmental confounders can be controlled and the effects of a single chemotherapeutic agent can be studied. Therefore, our cell-based approach provides a useful alternative tool aimed at identifying clinically relevant genotype-phenotype relationships through a genome-wide approach.
Statement of translational relevance.
One of the greatest challenges in anticancer agents pharmacogenomic markers discovery using a whole genome approach is identifying a relevant system. Although humans are the most relevant system for study, the whole genome approach requires large numbers of well-phenotyped patients treated with the same dosage regimen of a single chemotherapeutic drug. The studies are extremely expensive and require years to accrue for an adequately powered study. To this end, we have developed an in vitro model system that overcomes these challenges. Genome-wide germline genetic marker discovery and replication were conducted in these cell-based models and findings were validated in clinical settings. This study demonstrates the potential of this cell-based genome-wide approach to identify clinically important germline predictors of outcome following chemotherapy, but also the need for extensive validation in clinical samples.
Supplementary Material
Acknowledgments
We are grateful for excellent technical support provided by Steve Wisel in maintaining the cell lines. The full AOCS Study Group is listed on (http://www.aocstudy.org/). The results published here are in part based upon data generated by The Cancer Genome Atlas Pilot Project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA can be found at http://cancergenome.nih.gov/. We gratefully acknowledge the cooperation of all participating institutions and the contributions of the women who participated in this study.
Financial support: This Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org) study was supported by NIH/NIGMS grant U01GM61393 and data deposits are supported by U01GM61374 (http://pharmgkb.org/). This study was also supported by P50 CA125183 University of Chicago Breast Cancer SPORE grant (MED).
RSH received support from NIH/NIGMS grant K08GM089941, University of Chicago Cancer Center Support Grant (#P30 CA14599), and Breast Cancer SPORE Career Development Award.
YSF was one of the awardees of the Pritzker School of Medicine Experience in Research (PSOMER) program, supported by the National Heart Lung and Blood Institute (Grant NHLBI 2 T35 HL07764).
The Australian Ovarian Cancer Study was supported by the U.S. Army Medical Research and Materiel Command under DAMD17-01-1-0729, the National Health and Medical Research Council of Australia, Cancer Council Victoria, Cancer Council Queensland, Cancer Council New South Wales, Cancer Council South Australia, The Cancer Foundation of Western Australia and Cancer Council Tasmania. GCT is a Senior Principal Research Fellow of the NHMRC, and this work was supported by NHMRC funding. YL is funded by NHMRC grant 496675 and SM is supported by an NHMRC career development award.
The Mayo Clinic study is supported by R01 CA122443, P50 CA136393. SCOTROC biological studies were supported by Cancer Research UK C536/A6689.
Abbreviations
- LCLs
lymphoblastoid cell lines
- SNPs
single nucleotide polymorphisms
- GWAS
genome-wide association studies
- QTDT
quantitative transmission disequilibrium test
- LD
linkage disequilibrium
- AOCS
the Australian Ovarian Cancer Study
- OCAC
Ovarian Cancer Association Consortium
- TCGA
the Cancer Genome Atlas
- PFS
progression-free survival
- OS
overall survival
- FIGO
Fédération Internationale des Gynaecologistes et Obstetristes
- HR
hazard ratio
- MAF
minor allele frequency
- CEU
represents the Caucasian of Northern and Western European descents
- CEPH
represents Centre d’ Etude du Polymorphisme Humain sample collection
Footnotes
Conflict of interest: None
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra S. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 3.Shah N, Dizon D. New-generation platinum agents for solid tumors. Future Oncol. 2009;5:33–42. doi: 10.2217/14796694.5.1.33. [DOI] [PubMed] [Google Scholar]
- 4.Guppy A, Nathan P, Rustin G. Epithelial ovarian cancer: a review of current management. Clin Oncol (R Coll Radiol) 2005;17:399–411. doi: 10.1016/j.clon.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Welsh M, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, et al. Pharmacogenomic Discovery Using Cell-Based Models. Pharmacologic Reviews. 2009 doi: 10.1124/pr.109.001461. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. PNAS. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang RS, Duan S, Kistner EO, Bleibel WK, Delaney SM, Fackenthal DL, et al. Genetic Variants Contributing to Daunorubicin-Induced Cytotoxicity. Cancer Res. 2008;68:3161–3168. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang RS, Duan S, Kistner EO, Christine HM, Dolan ME. Genetic Variants Associated with Carboplatin-induced Cytotoxicity in Cell Lines derived from Africans. Molecular Cancer Therapeutics. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of Genetic Variants Contributing to Cisplatin-Induced Cytotoxicity using a Genome-wide Approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziliak D, O'Donnell PH, Im HK, Gamazon ER, Chen P, Delaney S, et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Translational Research. 2011;157:265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Molecular Cancer Therapeutics. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis G, Cardon L, Cookson W. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040072. e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NCI-NHGRI working group on replication in association studies. Replicating genotype-phenotype associations. Nature. 2007;447:655. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 16.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song H, Ramus SJ, Kjaer SK, DiCioccio RA, Chenevix-Trench G, Pearce CL, et al. Association between invasive ovarian cancer susceptibility and 11 best candidate SNPs from breast cancer genome-wide association study. Human Molecular Genetics. 2009;18:2297–2304. doi: 10.1093/hmg/ddp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnatty S, Beesley J, Paul J, Fereday S, Spurdle A, Webb P, et al. ABCB1 (MDR 1) polymorphisms and progression-free survival among women with ovarian cancer following paclitaxel/carboplatin chemotherapy. Clin Cancer Res. 2008;14:5594–5601. doi: 10.1158/1078-0432.CCR-08-0606. [DOI] [PubMed] [Google Scholar]
- 19.Vergote I, Rustin G, Eisenhauer E, Kristensen G, Pujade-Lauraine E, Parmar M, et al. Re: new guidelines to evaluate the response to treatment in solid tumors [ovarian cancer]. Gynecologic Cancer Intergroup. J Natl Cancer Inst. 2000;92:1534–1535. doi: 10.1093/jnci/92.18.1534. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Willer C, Ding J, Scheet P, Abecasis G. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd MR, Paull KD. Some practical consideration and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Des. 1995;34:91–109. [Google Scholar]
- 22.Weinstein JN. 'Omic' and hypothesis-driven research in the molecular pharmacology of cancer. Curr Opin Pharmacol. 2002;2:361–365. doi: 10.1016/s1471-4892(02)00185-6. [DOI] [PubMed] [Google Scholar]
- 23.Gamazon E, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic Drug Susceptibility Associated SNPs are Enriched in Expression Quantitative Trait Loci. Proc Natl Acad Sci USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crijns APG, Fehrmann RSN, Jong Sd, Gerbens F, Meersma GJ, Klip HG, et al. Survival-Related Profile, Pathways, and Transcription Factors in Ovarian Cancer. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000024. e1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown PO, Palmer C. The preclinical natural history of serous ovarian cancer: defining the target for early detection. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000114. e1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann L, Lu K, Linette G, Cliby W, Kalli K, Gershenson D, et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res. 2005;11:2149–2155. doi: 10.1158/1078-0432.CCR-04-1673. [DOI] [PubMed] [Google Scholar]
- 27.Marsh S, Paul J, King C, Gifford G, McLeod H, Brown R. Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: the Scottish Randomised Trial in Ovarian Cancer. J Clin Oncol. 2007;25:4528–4535. doi: 10.1200/JCO.2006.10.4752. [DOI] [PubMed] [Google Scholar]
- 28.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandhu M, Waterworth D, Debenham S, Wheeler E, Papadakis K, Zhao J, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000433. e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D, Sliwkowski M, Mark M, Frantz G, Akita R, Sun Y, et al. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Révillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat J. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann Oncol. 2008;19:73–80. doi: 10.1093/annonc/mdm431. [DOI] [PubMed] [Google Scholar]
- 34.Parsa A, Chang Y, Kelly R, Corretti M, Ryan K, Robinson S, et al. Hypertrophy-associated polymorphisms ascertained in a founder cohort applied to heart failure risk and mortality. Clin Transl Sci. 2011;4:17–23. doi: 10.1111/j.1752-8062.2010.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao W, Wang Y, Kleinman J, Lipska B, Hyde T, Weinberger D, et al. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci USA. 2010;107:15619–15624. doi: 10.1073/pnas.1005410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonuga-Barke E, Lasky-Su J, Neale B, Oades R, Chen W, Franke B, et al. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet. 2000;147B:1359–1368. doi: 10.1002/ajmg.b.30860. [DOI] [PubMed] [Google Scholar]
- 37.Chang B, Liu G, Xue F, Rosen D, Xiao L, Wang X, et al. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22:817–823. doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rustin G, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG) Int J Gynecol Cancer. 2011;21:419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 39.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, et al. Discerning the Ancestry of European Americans in Genetic Association Studies. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.0030236. e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruim R, Welch R, Sanna S, Teslovich T, Chines P, Gliedt T, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.