Abstract
Background
Heparanase degradation of heparan sulfate plays important roles in a number of pathological processes, including inflammation. In vitro experiments show that heparanase is capable of degrading heparin, a polysaccharide present in mast cells where it has a key role in promoting the storage of secretory granule compounds.
Objective
To investigate the functions of heparanase in mast cells.
Methods
Primarily cultured fetal skin-derived mast cells isolated from embryos and adult peritoneal mast cells were analyzed for storage and release of granule molecules in response to mast cell activation.
Results
Fetal skin-derived mast cells from heparanase-overexpressing mice contained substantially shorter heparin chains, and significantly less proteases than did the control cells. Conversely, fetal skin-derived mast cells lacking heparanase contained heparin of larger size and more proteases than did the control cells. Correspondingly, heparanase-overexpressing adult mast cells exhibited reduced release of heparin-bound proteases, a finding that could be attributed to spontaneous release of granular compounds. Heparanase was found to be upregulated in mast cells upon activation.
Conclusion
These findings reveal a novel function of heparanase in maintaining mast cell homeostasis through controlled degradation of heparin present in the mast cell secretory granules.
Keywords: Mast cell, heparin, heparanase, protease, allergy
Introduction
Mast cells (MCs) are found in most connective tissues and are characterized by their high content of secretory granules. In addition to the recognition as critical effector cells in allergic disorders and other IgE-dependent immune responses, recently, MCs have been strongly implicated in autoimmune disease. Activation of MCs can trigger degranulation, resulting in release of various preformed compounds that are stored in the granules, including histamine, cytokines, heparin and a range of MC-specific proteases 1, 2. Rodent MCs are classified as being of either mucosal (MMCs) or connective tissue type (CTMCs) MCs. MMCs are mainly located in mucosal surfaces, whereas CTMCs are present in, e.g., skin and the peritoneal cavity 3. These two MC subclasses have distinct secretory granule contents, in particular with regards to repertoire of MC proteases expressed and types of proteoglycans synthesized 4-7. For example, MMCs predominantly produce highly sulfated chondroitin sulfate 8, while CTMCs mainly produce heparin 3.
Heparin is a negatively charged glycosaminoglycan (GAG) composed of repeating disaccharide units of glucosamine and hexuronic acid (glucuronic or iduronic acid) that can be sulfated at various positions 9. The nascent heparin chain has an average molecular weight of 60-100 kDa 10. However, heparin recovered from various organs is often a mixture of Mr 5-30 kDa fragments, indicating in vivo degradation of macromolecular heparin. However, the mechanisms behind the in vivo processing of MC heparin, and the functional consequences of this processing, are not known. Although exogenously administered heparin has a wide clinical use as an anticoagulant, the fact that endogenous heparin is virtually non-detectable in blood argues against a major physiological role of heparin in the regulation of blood coagulation. Instead, previous studies have pointed to a key role for heparin in regulating MC granule composition, as shown by severely impaired protease and histamine storage in MCs with a defective heparin biosynthesis 11, 12.
Early studies revealed the presence of a heparin-cleaving endoglucuronidase in a murine mastocytoma 13. Later, an enzyme degrading heparan sulfate (HS), a polysaccharide structurally related to heparin, was identified in platelets, but it was assumed that the heparin- and HS-degrading activities detected in mastocytoma and platelets, respectively, were due to distinct enzymes 14. However, recent work indicates that mammalian cells express primarily one single heparanase, and it has been shown that this enzyme can cleave both HS and heparin in vitro 15-18. Heparanase specifically cleaves glycosidic bonds between glucuronic acid and glucosamine in heparin and HS, yielding fragments of variable size 18-20.
Overexpression of heparanase in mice results in extensive degradation of HS, while inactivation of the heparanase gene leads to accumulation of non-degraded HS, accompanied by diverse phenotypes that may be related to alterations in HS structure 21-25. However, the function of heparanase in MCs has not been reported previously. In this study, we investigated the effects of heparanase overexpression or absence on MCs. Our results point to a key role for heparanase in regulating the structure of MC heparin, indicating a modulatory effects of heparanase on MC functionality, as evidenced by modification of protease storage, granular morphology and ability to respond to MC-activating agents. Based on this study, heparanase emerges as a novel regulator of MC function.
Methods
Animals
Heparanase transgenic (hpa-tg) and knockout (hpa-KO) mice were generated as described 21, 24 and maintained in the animal facility at Biomedical Center, Uppsala, Sweden. The experiments were conducted in accordance with the local ethical regulations.
Preparation of FSMCs and isolation of peritoneal MCs
The FSMCs were prepared essentially as described 26. The cells were used when they reached >95% toluidine blue positivity. The peritoneal cells were collected from lavage of Hpa-tg and Ctr mice (12–18 weeks). Total cell number from each mouse was counted under phase contrast microscope. The MCs were isolated from the peritoneal lavage by FAC sorting after staining with FITC - conjugated anti-c-kit (CD117) antibody. The control samples stained with FITC-conjugated IgG. A minimum of 10,000 events (cells)/sample were counted.
Purification and analysis of heparin from FSMCs
Preparation of metabolically labeled heparin from FSMCs and structural analysis were performed as previously described 23, 27.
Characterization of MC
The MCs were characterized by staining with toluidine blue, May Grünwald/Giemsa, anti-c-kit, anti-heparanase and anti-heparin antibodies. The properties of the cells were examined by different microscopic techniques.
Analysis of protein and gene expression
Proteases and heparanase were determined by Western blot after separation on 12% SDS-PAGE. The results were analyzed by quantification of the band intensity with Image J. (NIH, USA). For gene expression, qPCR was applied using the primers shown in Table 1 (Online Repository). Samples were normalized using the mRNA level of GAPDH from each sample and data are reported as the ratio of treated cells/untreated cells or expression fold.
Table 1. Primers used in qPCR.
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| mHpse | TGTCCTGAACCTCCATAATGTC | TACGTATCCACTGGTTTCCTGA |
| mGAPDH | ACTCCACTCACGGCAAATTC | TCTCCATGGTGGTGAAGACA |
| mMCP-4 | GCCAAAGAGACTCCCTCTGTGATT | GCATCTCCGCGTCCATAAGATACA |
| mMCP-5 | TTGCCAGCCTGTGAGGAAA | TACAGACAGGCCAGATCGCAT |
| mMCP-6 | CATTGATAATGACGAGCCTCTCC | CATCTCCCGTGTAGAGGCCAG |
| MC-CPA | TGACAGGGAGAAGGTATTCCG | CCAAGGTTGACTGGATGGTCT |
Activation of MCs and analysis of degranulation
The MCs were activated by in vitro Ionomycin +PMA or anti-DNP-IgE + DNP-BSA stimulation, or by in vivo stimulation through intravenous injection of anti-DNP-IgE + DNP-BSA. β-hexosaminidase activity was determined as described 28. Substrate conversion was measured by absorbance at 405 nm (OD). Histamine was quantified using the Histamine Enzyme Immunoassay Kit (SPI Bio, France) according to the manufacturer's instruction. The values were normalized to the total MC number in each mouse.
The detailed description of Methods is presented in the Online Repository.
Data analysis and statistics
Statistical differences were determined by unpaired t-test.
Results
Heparanase is responsible for degradation of heparin in MCs
Fetal skin-derived mast cells (FSMCs) were established from E16.5 mouse embryos and were allowed to differentiate in vitro. Western blot analysis of cell lysates confirmed overexpression of heparanase in the heparanase transgenic (hpa-tg) FSMCs, and null expression of heparanase in hpa-KO FSMCs (see Fig. E1A and B in the Online Repository). Cytospin slides prepared from in vitro-matured cells were stained with toluidine blue at pH 2.5 and microscopic analysis showed a typical metachromatic staining of control FSMCs (Fig 1A). In comparison, hpa-tg FSMCs stained substantially weaker (Fig 1B), while hpa-KO FSMCs showed a more pronounced staining as compared with Ctr cells (Fig 1C). Toluidine blue is a cationic dye that binds strongly to negatively charged GAGs such as heparin. Hence, the altered toluidine blue staining in heparanase-overexpressing and heparanase-deficient FSMCs indicates a role for heparanase in regulating the properties of the sulfated GAG contents of FSMCs. In contrast to the dramatic change in toluidine blue staining, DAPI staining of nuclei did not reveal any alterations in hpa-KO or hpa-tg cells, as compared with Ctr FSMC (Fig. 1A-C, inserts).
Figure 1. Characterization of FSMCs.
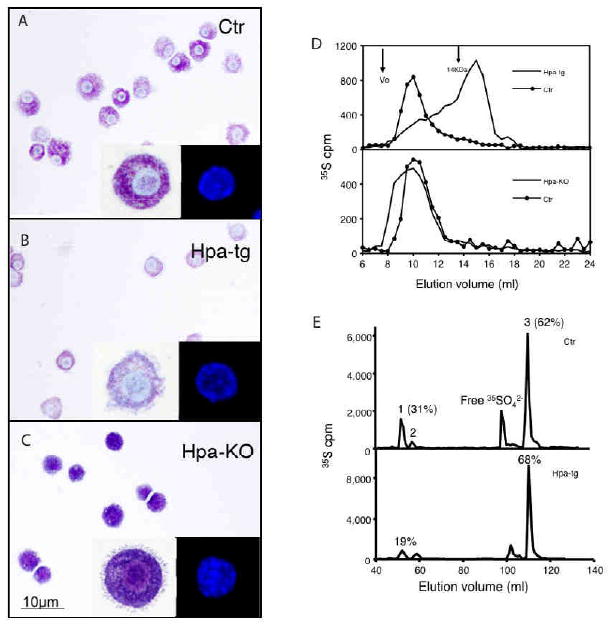
In vitro differentiated FSMCs from Ctr (A) hpa-tg (B), and hpa-KO (C) embryos were stained with toluidine blue. Magnification 40×. Inserts represent enlarged or DAPI-stained cells. (D) Gel chromatography analysis of metabolically 35S-labeled heparin from FSMCs. Upper panel: Ctr vs. hpa-tg; lower panel: Ctr vs. hpa-KO. (E) Analysis of disaccharides by anion-exchange HPLC. The numbered peaks represent: 1. -GlcA-GlcNS6S-; 2. -IdoA-GlcNS6S-; 3. -IdoA2S-GlcNS6S- sequences of the intact heparin chains. The percentages of the respective components in total recovered disaccharides are indicated for peaks 1 and 3. Upper panel: Ctr; lower panel: hpa-tg.
To further elucidate the effects of heparanase on GAGs, FSMCs were cultured in the presence of 35S-sulfate, followed by purification and characterization of metabolically labeled GAGs. Treatment of the GAGs with Chondroitinase ABC resulted in negligible degradation, indicating that FSMCs express low amounts of chondroitin sulfate. This is in accordance with our previous observations 27. In contrast, the purified 35S-labeled material was completely degraded by treatment with nitrous acid at pH 1.5, confirming that FSMCs predominantly produced heparin. Gel chromatography analysis revealed substantially shorter heparin chains derived from hpa-tg FSMCs than from Ctr cells, whereas hpa-KO FSMCs contained substantially longer heparin chains than material from Ctr counterparts (Fig 1D). Together, these findings demonstrate a role for heparanase in degrading and thereby controlling the size of heparin in MCs. The fine structure of the heparin was further assessed by analysis of its disaccharide composition. Deaminative cleavage (pH 1.5) of the 35S-labeled heparin resulted in complete degradation. The resultant disaccharides were collected and separated by anion-exchange HPLC (Partisil-10 SAX). The proportion of tri-sulfated disaccharide (peak 3), -IdoA2S-GlcNS6S-, did not show any appreciable difference when comparing hpa-tg and Ctr heparin (68% and 62%, respectively). However, the amount of disaccharide units (peak 2) containing the heparanase cleavage sequence, i.e. -GlcA-GlcNS6S, was significantly decreased in hpa-tg heparin, from 31% in Ctr to 19%, confirming substantial cleavage of heparin by heparanase (Fig 1E). There was no difference in disaccharide composition between the heparin samples from Ctr and hpa-KO FSMCs (data not shown).
Heparanase degradation of heparin impairs storage of proteases in FSMCs
Previous studies have shown that a major function of heparin is to promote the storage of MC-specific proteases within the MC secretory granules 11, 12. It was therefore of interest to assess whether the effect of heparanase on heparin molecular structure could lead to alterations in the storage of these proteases in MCs. Indeed, Western blot analysis revealed a substantial decrease in the amount of stored MC carboxypeptidase A (MC-CPA) as well as in the content of the tryptase mouse MC protease 6 (mMCP-6), and an almost complete loss of the chymase mMCP-5 in hpa-tg FSMCs in comparison with the Ctr counterparts (Fig 2A-C; left panels). Conversely, elevated storage of these proteases was observed in hpa-KO FSMCs (Fig 2A-C; right panels).
Figure 2.
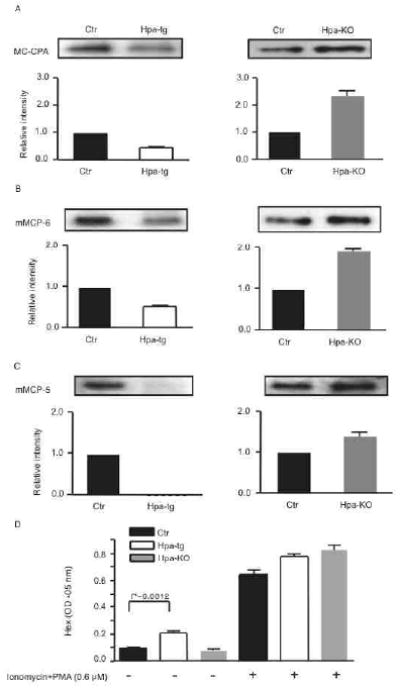
Protease storage and ß-hexosaminidase release in FSMCs.
Western blot of MC-CPA (A), mMCP-6 (B), or mMCP-5 (C) in cell lysates. Data show the ratio of band intensity/total protein amount. The respective levels in Ctr cells are defined as 1. Total protein loaded for detection of MC-CPA: 0.1 μg; mMCP-5: 0.4 μg and mMCP-6: 0.2 μg. (D) ß-hexosaminidase (Hex) activity in the conditioned media of FSMCs cultured with or without Ionomycin+PMA stimulation. The data represent three independent experiments (mean±SD).
To investigate whether heparanase has a role in regulating FSMC degranulation events, the release of ß-hexosaminidase in response to Ionomycin + phorbol myristate acetate (PMA) activation was assessed. Interestingly, a significantly higher basal release of ß-hexosaminidase was observed in cultures of hpa-tg cells as compared to Ctr counterparts (Fig 2D), suggesting that overexpression of heparanase impaired the integrity of secretory granules, leading to leakage of granule mediators into the extracellular space. Ionomycin + PMA stimulation resulted in substantial release of ß-hexosaminidase from Ctr cells (7-fold increase over non-stimulated cells), and the hpa-KO cells responded even more vividly (9.4-fold increase). In contrast, hpa-tg FSMCs responded poorly to Ionomycin + PMA stimulation, with only a 2.7-fold increase of ß-hexosaminidase released into the cell culture medium upon stimulation.
Overexpression of heparanase perturbs the morphology of peritoneal MCs
To verify that the findings in FSMCs were not an embryonic development-associated phenomenon, we extended the investigation to adult mice. Because peritoneal MCs are characterized as CTMCs and are known to produce heparin, we analyzed MCs from peritoneal lavage of the transgenic mice along with controls. May Grünwald/Giemsa staining revealed the expected presence of strongly stained MCs in lavage from Ctr animals (indicated by arrows in Fig 3A). In comparison, MCs present in peritoneal lavage from hpa-tg mice stained substantially weaker (Fig 3B, arrows), i.e. resembling the difference observed when comparing hpa-tg and Ctr FSMCs (see Fig 1A, B). Moreover, hpa-tg MCs appeared smaller than Ctr cells, as shown both by May-Grünwald/Giemsa staining (Fig 3A, B) and by staining for the MC cell surface marker c-kit (Fig 3C, D). As shown by Western blot analysis, the total level of c-kit was not affected by the overexpression of heparanase (Fig. E1D in the Online Repository). Further examination by transmission electron microscopy revealed striking morphological effects of heparanase overexpression on granule ultrastructure, with MCs from hpa-tg mice containing smaller granules than did corresponding Ctr cells. Moreover, whereas the granules in Ctr cells were rounded, the granules in hpa-tg MCs were irregularly shaped (Fig 3E, F). In contrast, the morphologies of Ctr and hpa-KO peritoneal MCs was indistinguishable (data not shown).
Figure 3.
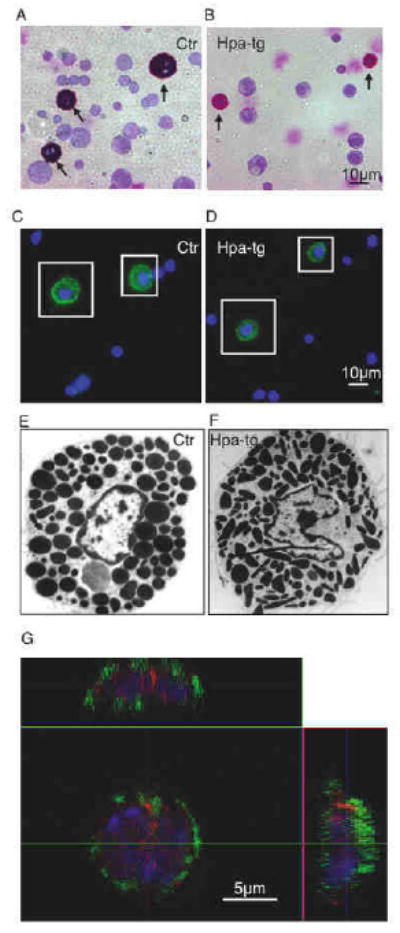
Morphological examination of peritoneal MCs.
Peritoneal cells were stained with May Grünwald/Giemsa: (A), Ctr; (B), hpa-tg (arrows point to MCs); or with FITC-conjugated anti-c-kit antibody: (C), Ctr; (D), hpa-tg; MCs are illustrated by squares. Original magnification 40×. (E, F) TEM analysis of Ctr (E) and hpa-tg (F) peritoneal MCs. Original magnification 3000×. (G) Hpa-tg MCs double immunostained for c-kit (green) and heparanase (red) examined by confocal microscopy (z-scan). The nuclei were stained with DAPI (blue).
Overexpression of heparanase in hpa-tg MC was confirmed by Western blot analysis of the c-kit positive cells isolated by FACS (Fig. E1C in the Online Repository). Note that heparanase was below the level of detection in Ctr peritoneal MCs by the method applied, in agreement with our early notion that the enzyme is expressed at a low level in normal tissues 23. Immunocytostaining with anti-heparanase antibodies gave, in contrast to the surface staining of c-kit, strong granule-shaped intracellular signals in hpa-tg MCs (Fig. 3G); in comparison, Ctr MCs had substantially weaker signals for heparanase staining, confirming the low level of the enzyme in normal MC cells (Fig. E2A in the Online Repository).
Overexpression of heparanase results in distressed MC degranulation
To determine whether the morphological disturbances of the granules in hpa-tg MCs have functional implications, we examined the degranulation properties of peritoneal lavage cells upon activation. Analysis of ß-hexosaminidase release revealed, similar to the findings in FSMCs, a higher baseline release of ß-hexosaminidase in hpa-tg than in Ctrl cells (Fig 4A, B). Moreover, in response to Ionomycin +PMA, but not in response to IgE-mediated activation, hpa-tg cells released more ß-hexosaminidase than did Ctr cells.
Figure 4.
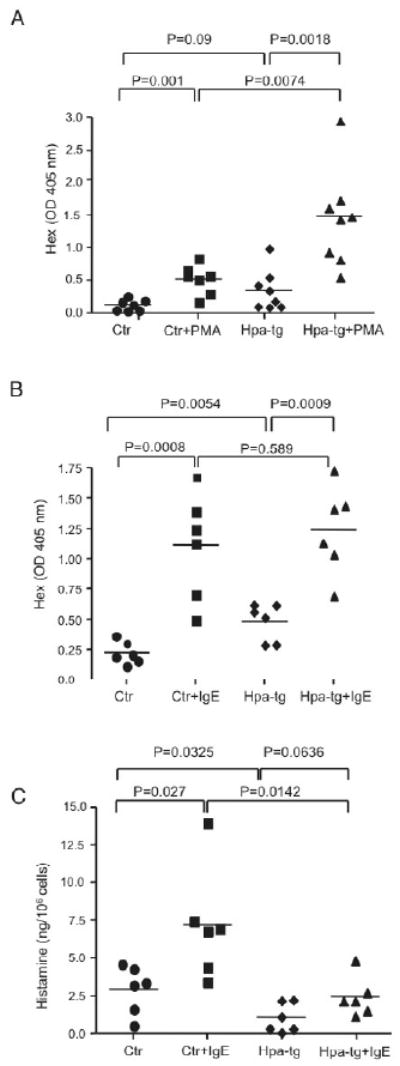
Effect of heparanase overexpression on MC degranulation.
Release of ß-hexosaminidase by peritoneal cells upon (A) Ionomycin + PMA or (B) anti-DNP-IgE + DNP-BSA treatment. The data are expressed as OD405nm values (ß-hexosaminidase activity) in the medium before and after activation. (C) Release of histamine by peritoneal cells upon anti-DNP-IgE + DNP-BSA treatment. Each symbol represents data from an individual animal.
To further examine the effect of heparanase overexpression on MC degranulation events, we measured the histamine release in response to IgE-mediated peritoneal cell activation. As shown in Fig 4C, the baseline release of histamine was lower in hpa-tg as compared to Ctr cells. Moreover, the release of histamine in response to IgE-mediated activation was profoundly lower in hpa-tg as compared with Ctr cells. A likely explanation for the differential effects of heparanase overexpression on ß-hexosaminidase vs. histamine release may lie within their differential dependence on heparin for storage; whereas ß-hexosaminidase is reported to be heparin-independent 29, histamine is known to be strongly dependent on heparin 30.
Next, the effect of heparanase overexpression on MC protease storage and release was assessed by Western blot analysis. Quantification of the band intensities normalized against number of MCs showed a significantly reduced storage of MC-CPA in hpa-tg peritoneal cells as compared with Ctr; in comparison, the reduction of mMCP-5 was modest (Fig 5A, B). Upon IgE-mediated activation, in average about 20% of the total MC-CPA and total mMCP-5 were lost (released) from Ctr cells. In contrast, IgE-activated hpa-tg cells released averagely about 3% of the total MC-CPA and essentially no release of mMCP-5. The reduced storage and release of proteases in hpa-tg cells were not due to a reduced expression of the proteases; on the contrary, expression of these proteases at the mRNA level was upregulated in hpa-tg MCs (Fig. E4 in the Online Repository).
Figure 5.
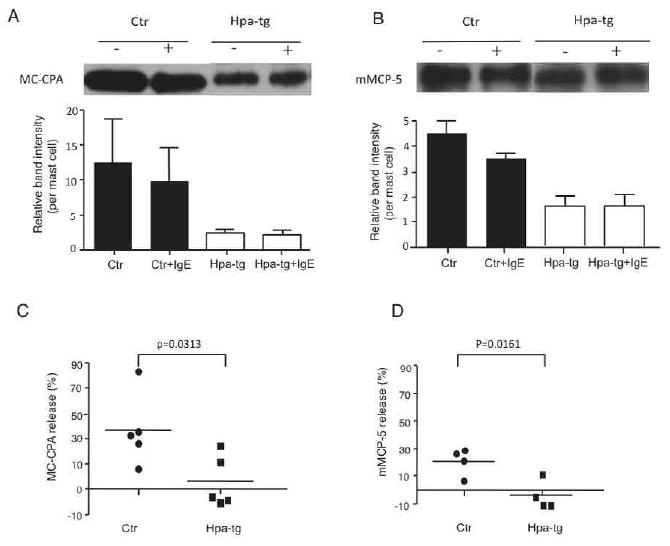
Detection of proteases in the peritoneal MCs.
Representative Western blot for MC-CPA (A) and mMCP-5 (B) in non-treated (-) or IgE-activated (+) peritoneal MCs. The bands were quantified by Image J and normalized to the MC number in each mouse (n=4-5); the average relative band intensity is shown. Release of MC-CPA (C) and (D) mMCP-5 in response to IgE cross-linking is calculated as: (non-activated- activated)/non-activated ×100%.
MC activation induces heparanase expression
The finding that heparanase modulates MC function prompted us to examine whether heparanase expression is altered in MCs in response to activation. First, FSMCs isolated from Ctr embryos were stimulated with Ionomycin + PMA, followed by total RNA extraction and analysis of heparanase expression using quantitative real time RT-PCR. Indeed, MC activation caused a robust upregulation of heparanase expression, in particular at early time points after MC activation (Fig 6A). Next, we examined the heparanase expression in MCs following IgE-mediated activation in vivo. Adult mice sensitized with anti-dinitrophenyl (DNP) IgE were challenged by intravenous injection of DNP-BSA conjugate. Total peritoneal cells were collected from treated and non-treated mice (n=7). The c-kit+ cells (MCs) were collected by FAC sorting for preparation of total RNA. Quantitative real time RT-PCR revealed that in vivo IgE-mediated activation caused a minor upregulation of heparanase expression in peritoneal MCs (Fig 6B).
Figure 6.
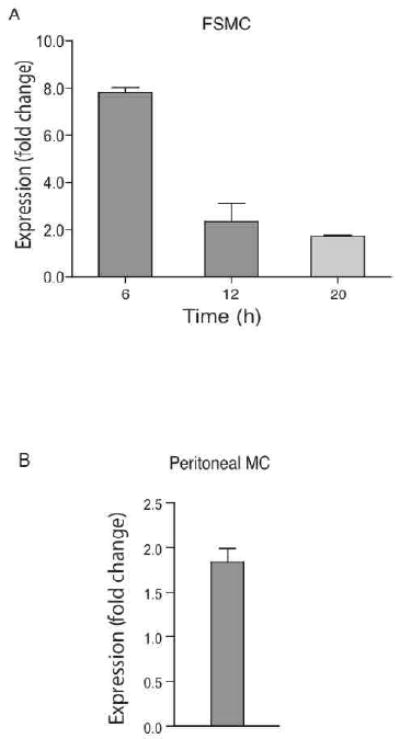
Effect of MC activation on heparanase expression. (A) Quantitative RT-PCR analysis of heparanase expression in FSMCs (from 6 embryos) at the time points indicated after activation with Ionomycin+PMA. (B) MCs were activated in vivo (7 mice/group) by anti-DNP-IgE +DNP-BSA treatment. The Q-PCR signals of heparanase were standardized against that of GAPDH (defined as 1). The error bars in (A) represent the average of duplicate samples and in (B) indicates standard deviation from 3 independent experiments performed in triplicates.
Discussion
Heparanase has been shown to specifically cleave the glycosidic bond between glucuronic acid and glucosamine residues in heparan sulfate (HS). Recognized for its upregulated expression in some pathological conditions, e.g. inflammatory reactions and metastatic tumors, heparanase has recently attracted much attention in the context of modulation of HS structure and function 23, 25, 31-33. However, although recombinant heparanase has been shown to cleave both HS and heparin in vitro 19, and is able to degrade macromolecular heparin isolated from rat skin 18, direct evidence showing that heparanase can process MC heparin in vivo has not been reported. Moreover, the potential functional implications of MC heparin processing by heparanase have not been addressed previously.
The generation of transgenic mice overexpressing or lacking heparanase 21, 24 has provided excellent models to examine heparanase function in vivo. As FSMCs have been shown to resemble CTMCs and produce heparin 27, we used FSMCs to study the impact of heparanase on heparin processing and MC function. As expected, in vitro-matured Ctr and hpa-KO embryonic FSMCs stained positively with toluidine blue; in comparison, hpa-tg FSMCs stained considerably weaker (Fig 1A-C). As toluidine blue stains negatively charged GAGs, the weaker staining of hpa-tg FSMCs reflects an alteration of their GAG content. Indeed, analysis of metabolically 35S-labeled heparin showed an accumulation of dramatically shortened chains in hpa-tg cells (Fig 1D), indicating an extensive fragmentation of the polysaccharide by overexpressed heparanase. Conversely, the heparin chains in hpa-KO FSMCs were non-degraded. Moreover, co-staining of heparanase with heparin in connective tissue (intestine) MC of hpa-tg mouse (Fig. E2B provides additional evidence for the intracellular degradation of heparin by heparanase. Together, these findings clearly demonstrate that heparanase is responsible for processing of heparin within MC. Moreover, the decreased proportion of -GlcA-GlcNS6S-disaccharide units in heparin isolated from hpa-tg FSMCs provided additional molecular evidence for heparanase-catalyzed heparin cleavage (Fig 1E). It should be noted that a reduction of this disaccharide unit was also observed in HS isolated from tissues overexpressing heparanase 23, suggesting that heparanase has a dual role in regulating the size of both heparin and HS in vivo. This notion is also in agreement with our earlier observations that heparanase can degrade heparin in vitro 18.
Various proteases and other types of preformed granule compounds may account for up to 50% of the total weight of a mature MC 2. Murine MCs express the exopeptidase MC-CPA and several MC-specific serine proteases, and large amounts of these proteases can be released when MC degranulation is induced 2, 7, 34. Recent investigations, involving the assessment of mouse stains lacking individual MC proteases in different animal models of disease, have shown that these enzymes may account for many of the biological functions attributed to MCs 2. Hence, mechanisms that are involved in the regulation of MC protease storage are likely to have profound impacts on MC function in vivo.
To examine the impact of heparanase on storage and release of MC proteases, we determined the cellular contents of selected proteases before and after MC activation. Interestingly, the storage of three main CTMC proteases, i.e. MC-CPA, mMCP-5 and mMCP-6, was reduced in hpa-tg FSMCs, indicating that optimal protease storage cannot occur if the granule heparin is subject to substantial degradation by heparanase. This notion was further substantiated by the finding that heparanase-deficient FSMCs contained higher amounts of these proteases than did Ctr cells, suggesting that MCs containing “oversized” heparin chains can accommodate super-optimal levels of MC proteases (Fig 2A-C). Thus, the storage of MC proteases may be regulated at the level of heparanase expression/activity, with induction of heparanase leading to reduced protease storage, in turn resulting in attenuated MC responses. Conversely, reduced heparanase expression may lead to super-optimal protease storage, possibly leading to exaggerated MC responses. An effect of heparanase overexpression was also seen in adult peritoneal MCs, which showed striking morphological alterations (Fig 3) accompanied by reduced ability to store MC proteases (Fig 5) and to release histamine upon activation (Fig 4C). Interestingly, elimination of heparanase appeared to have minimal effects on adult peritoneal MCs (data not shown). A possible explanation for this finding may be that heparanase has profound effects on MCs during their developmental stages, but less effects on normal mature cells. In line with this notion, heparanase protein is detectable in FSMCs, but is undetectable in peritoneal MCs from adult mice (Fig. E1A-C in the Online Repository).
Intriguingly, whereas the storage of MC-CPA and mMCP-6 was affected to a relatively modest extent by the absence or overexpression of heparanase in FSMCs, mMCP-5 storage was dramatically reduced in hpa-tg cells. Although we cannot with certainty explain the mechanism behind this finding, it is notable that mMCP-5, but not MC-CPA or mMCP-6, contains two distinct patches of positively charged regions exposed on the protein surface 35. Possibly, optimal storage of mMCP-5 requires that both of these regions are bound to the same heparin chain, and thus that optimal mMCP-5 storage requires long-chain heparin, whereas the heparin chain length is less critical in promotion of MC-CPA and mMCP-6 storage. Indeed, previous studies have shown that optimal interaction of MC chymase with heparin requires a high heparin chain length 36.
Together, the findings presented here suggest that heparanase, by regulating the size of MC heparin can have an important indirect role in the regulation of MC protease storage and in controlling MC degranulation processes. Considering that MC proteases have important functions in association with a number of pathological settings, including allergic reactions 2, 37-39, heparanase-mediated regulation of MC protease storage/release may thus have profound consequences in various types of disease. Conceivably, induction or repression of heparanase expression in the context of a given pathological stimulation could lead to effects on the storage and release of the MC secretory granule mediators. Clearly, this modulation leads to altered MC-driven responses.
Supplementary Material
Acknowledgments
We are grateful to Prof. Ulf Lindahl (Uppsala University) for the valuable discussions in the process of this study.
Findings: The Swedish Medical Research Council (K2009-67X-21128-01-3), Swedish Cancer Research Foundation (09 0717), Polysackaridforskning Foundation (Uppsala), Sweden and National Institutes of Health (NIH) grant CA106456 (I.V.).
Abbreviations used
- MC
mast cell
- IgE
immunoglobulin E
- FCMC
Fetal skin-derived mast cell
- mMCP
mouse mast cell protease
- mCPA
mouse caboxylpeptidase
- HS
heparan sulfate
- Ctr
control
- Hpa-tg
transgenic mice overexpressing heparanase
- Hpa-KO
heparanase knockout mice
- PMA
phorbol myristate acetate
- DNP
2,4-dinitrophenol
Footnotes
Disclosure of potential conflict of interest: The authors declared there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lundequist A, Pejler G. Biological implications of preformed mast cell mediators. Cell Mol Life Sci. 2010 Nov 11; doi: 10.1007/s00018-010-0587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pejler G, Ronnberg E, Waern I, Wernersson S. Mast cell proteases: multifaceted regulators of inflammatory disease. Blood. 2010 Jun 17;115(24):4981–90. doi: 10.1182/blood-2010-01-257287. [DOI] [PubMed] [Google Scholar]
- 3.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997 Oct;77(4):1033–79. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 4.Stevens RL. Mast cell proteoglycans. Prog Clin Biol Res. 1989;297:131–43. discussion 43-4. [PubMed] [Google Scholar]
- 5.Welle M. Development, significance, and heterogeneity of mast cells with particular regard to the mast cell-specific proteases chymase and tryptase. J Leukoc Biol. 1997 Mar;61(3):233–45. doi: 10.1002/jlb.61.3.233. [DOI] [PubMed] [Google Scholar]
- 6.Miller HR, Pemberton AD. Tissue-specific expression of mast cell granule serine proteinases and their role in inflammation in the lung and gut. Immunology. 2002 Apr;105(4):375–90. doi: 10.1046/j.1365-2567.2002.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pejler G, Abrink M, Ringvall M, Wernersson S. Mast cell proteases. Adv Immunol. 2007;95:167–255. doi: 10.1016/S0065-2776(07)95006-3. [DOI] [PubMed] [Google Scholar]
- 8.Enerbäck L, Kolset SO, Kusche M, Hjerpe A, Lindahl U. Glycosaminoglycans in rat mucosal mast cells. Biochem J. 1985;227:661–8. doi: 10.1042/bj2270661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl U. Biosynthesis of Heparin and related polysaccharides. In: Lane DA, Lindahl U, editors. HEPARIN Chemical and Biological Properties, Clinical Applications. First. London: Edward Arnold; 1989. pp. 159–89. [Google Scholar]
- 10.Horner AA. Macromolecular heparin from rat skin. Isolation, characterization, and depolymerization with ascorbate. J Biol Chem. 1971;246(1):231–9. [PubMed] [Google Scholar]
- 11.Humphries DE, Wong GW, Friend DS, Gurish MF, Qiu WT, Huang C, et al. Heparin is essential for the storage of specific granule proteases in mast cells. Nature. 1999;400:769–72. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]
- 12.Forsberg E, Pejler G, Ringvall M, Lunderius C, Tomasini-Johansson B, Kusche-Gullberg M, et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400(6746):773–6. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 13.Ogren S, Lindahl U. Cleavage of macromolecular heparin by an enzyme from mouse mastocytoma. J Biol Chem. 1975;250(7):2690–7. [PubMed] [Google Scholar]
- 14.Thunberg L, Backstrom G, Wasteson A, Robinson HC, Ogren S, Lindahl U. Enzymatic depolymerization of heparin-related polysaccharides Substrate specificities of mouse mastocytoma and human platelet endo- beta-D-glucuronidases. J Biol Chem. 1982;257(17):10278–82. [PubMed] [Google Scholar]
- 15.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5(7):803–9. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274(34):24153–60. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- 17.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5(7):793–802. doi: 10.1038/10518. see comments. [DOI] [PubMed] [Google Scholar]
- 18.Gong F, Jemth P, Escobar Galvis ML, Vlodavsky I, Horner A, Lindahl U, et al. Processing of macromolecular heparin by heparanase. J Biol Chem. 2003 Sep 12;278(37):35152–8. doi: 10.1074/jbc.M300925200. [DOI] [PubMed] [Google Scholar]
- 19.Pikas DS, Li JP, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 1998;273(30):18770–7. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- 20.Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010 May 7;285(19):14504–13. doi: 10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zcharia E, Metzger S, Chajek-Shaul T, Aingorn H, Elkin M, Friedmann Y, et al. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. Faseb J. 2004 Feb;18(2):252–63. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- 22.Zcharia E, Zilka R, Yaar A, Yacoby-Zeevi O, Zetser A, Metzger S, et al. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. FASEB J. 2005 Feb;19(2):211–21. doi: 10.1096/fj.04-1970com. [DOI] [PubMed] [Google Scholar]
- 23.Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, et al. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 2007 Dec;3(12):773–8. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 24.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, et al. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS ONE. 2009;4(4):e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waern I, Jia J, Pejler G, Zcharia E, Vlodavsky I, Li JP, et al. Accumulation of Ym1 and formation of intracellular crystalline bodies in alveolar macrophages lacking heparanase. Mol Immunol. 2010 Apr;47(7-8):1467–75. doi: 10.1016/j.molimm.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Feyerabend TB, Hausser H, Tietz A, Blum C, Hellman L, Straus AH, et al. Loss of histochemical identity in mast cells lacking carboxypeptidase A. Mol Cell Biol. 2005 Jul;25(14):6199–210. doi: 10.1128/MCB.25.14.6199-6210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feyerabend TB, Li JP, Lindahl U, Rodewald HR. Heparan sulfate C5-epimerase is essential for heparin biosynthesis in mast cells. Nat Chem Biol. 2006 Apr;2(4):195–6. doi: 10.1038/nchembio777. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz LB, Austen KF, Wasserman SI. Immunologic release of beta-hexosaminidase and beta-glucuronidase from purified rat serosal mast cells. J Immunol. 1979 Oct;123(4):1445–50. [PubMed] [Google Scholar]
- 29.Henningsson F, Hergeth S, Cortelius R, Abrink M, Pejler G. A role for serglycin proteoglycan in granular retention and processing of mast cell secretory granule components. FEBS J. 2006 Nov;273(21):4901–12. doi: 10.1111/j.1742-4658.2006.05489.x. [DOI] [PubMed] [Google Scholar]
- 30.Ringvall M, Ronnberg E, Wernersson S, Duelli A, Henningsson F, Abrink M, et al. Serotonin and histamine storage in mast cell secretory granules is dependent on serglycin proteoglycan. J Allergy Clin Immunol. 2008 Apr;121(4):1020–6. doi: 10.1016/j.jaci.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 31.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007 May;151(1):1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiegel A, Zcharia E, Vagima Y, Itkin T, Kalinkovich A, Dar A, et al. Heparanase regulates retention and proliferation of primitive Sca-1+/c-Kit+/Lin- cells via modulation of the bone marrow microenvironment. Blood. 2008 May 15;111(10):4934–43. doi: 10.1182/blood-2007-10-116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massena S, Christoffersson G, Hjertstrom E, Zcharia E, Vlodavsky I, Ausmees N, et al. A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood. 2010 Jun 8;116(11):1924–31. doi: 10.1182/blood-2010-01-266072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens RL, Adachi R. Protease-proteoglycan complexes of mouse and human mast cells and importance of their beta-tryptase-heparin complexes in inflammation and innate immunity. Immunol Rev. 2007 Jun;217:155–67. doi: 10.1111/j.1600-065X.2007.00525.x. [DOI] [PubMed] [Google Scholar]
- 35.Sali A, Matsumoto R, McNeil HP, Karplus M, Stevens RL. Three-dimensional models of four mouse mast cell chymases. Identification of proteoglycan binding regions and protease-specific antigenic epitopes. J Biol Chem. 1993 Apr 25;268(12):9023–34. [PubMed] [Google Scholar]
- 36.Pejler G, Maccarana M. Interaction of heparin with rat mast cell protease 1. J Biol Chem. 1994 May 20;269(20):14451–6. [PubMed] [Google Scholar]
- 37.Marshall JS. Mast-cell responses to pathogens. Nat Rev Immunol. 2004 Oct;4(10):787–99. doi: 10.1038/nri1460. [DOI] [PubMed] [Google Scholar]
- 38.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008 Jul 24;454(7203):445–54. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010 Jun;10(6):440–52. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


