Abstract
RNA-seq technologies are now replacing microarrays for profiling gene expression. Here we describe a robust RNA-seq strategy for multiplex analysis of RNA samples based on deep sequencing. First, an oligo-dT linked to an adaptor sequence is used to prime cDNA synthesis. Upon solid phase selection, second strand synthesis is initiated using a random primer linked to another adaptor sequence. Finally, the library is released from the beads and amplified using a bar-coded primer together with a common primer. This method, referred to as Multiplex Analysis of PolyA-linked Sequences (MAPS), preserves strand information, permits rapid identification of potentially new polyadenylation sites, and profiles gene expression in a highly cost effective manner. We have applied this technology to determine the transcriptome response to knockdown of the RNA binding protein TLS, and compared the result to current microarray technology, demonstrating the ability of MAPS to robustly detect regulated gene expression.
Keywords: RNA-seq, multiplexing strategy, gene expression profiling, translocation-in-liposarcoma
Introduction
RNA-seq has the obvious advantage over microarray in profiling gene expression, in that it generates digital information of individual annotated genes with literally unlimited dynamic range. In addition, RNA-seq has the ability to comprehensively detect novel transcripts and mRNA variants resulting from alternative promoter usages, splice sites, and polyadenylation.
Many RNA-seq protocols have been developed in recent years (for review, see [1, 2]). Standard RNA-seq procedure uses fragmented RNA to prepare dsDNA followed by adaptor ligation [3]. This method minimizes a bias towards the 3′ end of genes by converting poly(A+) RNA to cDNA. However, this approach provides no strand information, which is crucial for detection of anti-sense transcripts from genes within genes or transcription within overlapping genic regions in opposite directions. To overcome these shortcomings, various strategies have been developed to preserve the strand information, including (1) the use of different adaptors at the 5′ and 3′ ends [4, 5], (2) 3′ end polyA tailing [6], (3) double-random priming with distinct adaptors [7, 8], and (4) dUTP marking of the 2nd strand followed by selective degradation of the strand after linker ligation in order to sequence only the 1st strand [9].
While these methods have the ability to detect structural variations in mRNA, the counts generated depend on both abundance and length of individual transcripts. As a result, rare but long transcripts are more easily quantified than rare but short transcripts; the latter would require a much higher overall tag density to detect. For the purpose of gene expression profiling, the alternative approach focuses on the 3′ end of each gene which, like the original SAGE technology, is referred to 3′-tag digital gene expression [10, 11]. Key steps of the method include cleavage of dsDNA with a frequent restriction enzyme, adaptor ligation, and affinity purification of biotinylated oligo-dT initially used to prime cDNA synthesis. It has been demonstrated that this approach is more robust in detecting low abundant mRNAs [10]. Another obvious advantage of this method is its ability to systematically identify polyadenylation sites, which has been applied to C. Elegans [11].
Here we describe a much simpler version of the 3′-tag digital gene expression approach, which we refer to as Multiplex Analysis of PolyA-linked Sequences (MAPS). This method, which is modified from our original double-random priming strategy [7], uses biotinylated oligo-dT directly linked to a specific sequencing adaptor to prime cDNA synthesis. This is followed by second strand synthesis using a random primer attached to a second adaptor. Using this technology, we detected >10,000 previously unannotated polyadenylation sites in HeLa cells and characterized the transcriptional response to knockdown of the Pol II-associated RNA binding protein, TLS in comparison with microarray. Our analysis has demonstrated the robustness of MAPS in studying regulated gene expression.
Results and Discussion
Experimental design for Multiplex Analysis of Poly(A)-linked Sequences (MAPS)
Our strategy, depicted in Fig. 1, employs an oligo-dT linked to a specific sequencing primer (B) corresponding to the P7 sequence anchored on the surface of Illumina flowcells to prime cDNA synthesis. The 3′ end of the primer contains two random nucleotides in order to ensure cDNA synthesis from the beginning of the poly(A) tail. The primer also carries a biotin moiety for solid phase selection. Excess oligo-dT is removed after cDNA synthesis and the 3′ end of cDNA is blocked by using terminal transferase in the presence of ddNTP. Following biotin selection, second strand synthesis is performed directly on magnetic beads using a random primer (8X) linked to a specific sequencing adaptor. After washing, the second strand is released from the beads by heat and eluted products are amplified using Primer B in combination with a bar-coded primer. The bar-coded primer consists of the Illumina sequencing primer (A), which enables the amplified sequence to anneal to the P5 primer on the Illumina flowcell, followed by a 4-nucleotide coded region (address), and the sequencing adaptor. The PCR products, while approximately within the same size region (see below), are size selected from acrylamide gel to minimize short fragments, quantified, and loaded onto an Illumina flowcell for deep sequencing.
Figure 1.
Multiplex Analysis of PolyA-linked Sequences (MAPS). Total RNA is reverse transcribed using a primer containing oligo-dTNN linked to Primer B sequencing primer with a biotin in the 5′ end. The cDNA is captured and extended using a random primer attached to a sequencing primer. The cDNA sequence is PCR amplified using a primer containing sequencing primer A, a specific address (bar-code), and the sequencing adaptor in combination with Primer B. The PCR product is gel purified and sequenced first with the sequencing adaptor to determine the cDNA sequence, and second with Primer A to determine the unique bar-code.
Two rounds of sequencing are performed, starting with the insert, using the sequencing adaptor for 32 to 34 cycles of sequencing. The products are next stripped off the surface of the flowcell and a second round of sequencing is performed using primer A to identify the bar-coded region. In our hands, both sequencing reactions can be accomplished with the standard 36 cycle kit (because of extra sequencing reagents included in the kit). The sequences of bar-coded regions allow the assignment of individual samples in a multiplex run. We have successfully decoded >99% of the sequences using this method.
We build in two key features in the MAPS strategy. First, second strand synthesis is initiated by simple random priming. This method is much simpler than dsDNA synthesis followed by restriction digestion and linker ligation, which is currently used in the 3′-tag digital gene expression protocol. Although in theory random priming can take place anywhere on the first strand cDNA, the following PCR reaction ensures biased amplification of short fragments from the 3′ end of expressed genes.
The second feature of the MAPS strategy is obviously the ease of multiplexing samples using individual bar-coded primers. Our strategy is distinct from standard protocols using bar-coded adaptors during library construction because it ensures all library construction is performed under uniform conditions without the concern of differences in quality among different bar-coded adaptors. In our applications, we routinely multiplex 6 reactions, typically 3 biological repeats of control and 3 treated samples in one lane of the Illumina GAII Analyzer. Multiplexing 6 reactions in a single lane permits the use of simple statistical analysis between differentially expressed genes. This level of multiplexing also achieves the economy of genome-wide analysis, as the cost for transcriptome analysis is ~$100 per sample, which is comparable to current microarray platforms. In future applications using HiSeq2000, it should be possible to multiplex 18 or more samples per lane, thus dramatically reducing the cost of gene expression profiling to a mere $35 per sample.
Detection of known and potential new polyadenylation sites in the human genome
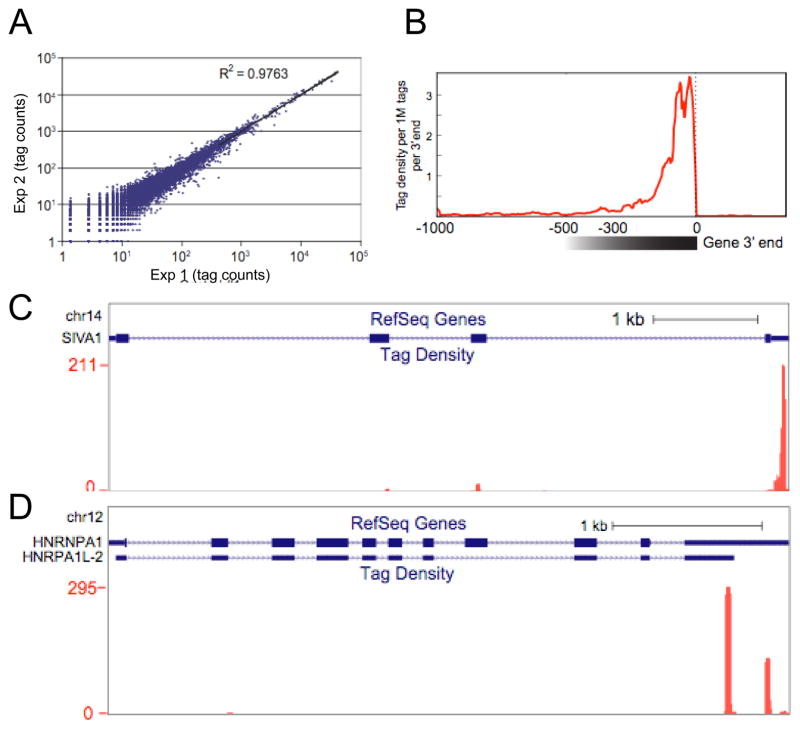
We first applied the MAPS strategy to total RNA isolated from HeLa cells, demonstrating the high reproducibility of this approach (Fig. 2A). From three biological repeats, we obtained ~15M tags, among which ~10M were uniquely mapped to the human genome (the control raw in Table 1). We removed potential internal priming events [12], resulting in ~8.3M tags, which were used for further analysis. As expected, the tags were clustered at the 3′ end of genes, ~300nt upstream of the polyA site (Fig. 2B). The two gene examples provided clearly indicate that MAPS is able to detect both unique and alternative polyadenylation sites (Fig. 2C and 2D).
Figure 2.
Identification of the 3′ end-linked sequence tags by MAPS. (A) Reproducibility of MAPS. (B) Mapping tags to the distal polyA site of annotated genes. The data also reveal the location of tags within a 300nt window upstream of the polyA site in most cases. (C and D) Examples of tags at a single (C) and double (D) 3′ ends.
Table 1.
Statistics of sequencing data
| Sample | Total Decoded | After Remove Adaptor & PolyA, Length>=16 | Mappable | Unique Mapped | No Internal Priming |
|---|---|---|---|---|---|
| TLS KD | 13537523 | 12892715 | 11984201 | 9323591 | 7939799 |
| Control | 16394011 | 14936999 | 13366509 | 10057514 | 8322360 |
We next clustered the tags based on annotated polyA sites, resulting in 8,859 clusters (Table 2, control). By scanning against 13 polyA signal variant motifs [13], we found that 7547 (85.2%) clusters contain a known polyA signal within the cluster, with remaining 1312 (14.7%) clusters lacking a canonical polyA signal. This rate is similar to that found in C. elegans [11]. Strikingly, we also detected 12,532 potential new polyA sites in the human genome, ~80% of which are also associated with a polyA signal (Table 2). This observation suggests that there are a huge number of polyA sites, either within known genes or associated with new transcripts, that remain to be annotated in the human genome. Therefore, our method can be used not only to detect those signals, but also analyze differential polyadenylation in future studies.
Table 2.
Association of tag clusters with known and potential new polyA signals
| Control: | ||
| Clusters | With polyA signal | Without polyA signal |
| With knownGene 3′end | 7547 (85.2%) | 1312 (14.7%) |
| Other | 9966 (79.5%) | 2566 (20.5%) |
| TLS KD: | ||
| Clusters | With polyA signal | Without polyA signal |
| With knownGene 3′end | 5772 (85.8%) | 952 (14.2%) |
| Other | 6002 (77.6%) | 1728 (22.4%) |
Profiling FUS/TLS-regulated gene expression
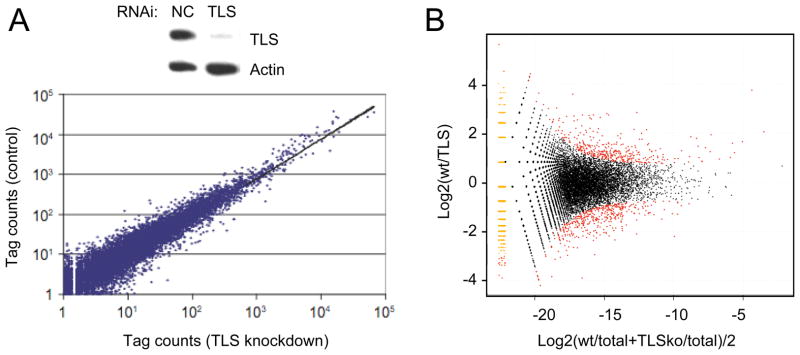
We next applied the MAPS strategy to profile gene expression in response to knockdown of FUS/TLS in HeLa cells. FUS/TLS is an RNA binding protein originally identified as a frequent chromosome translocation partner in liposarcomas [14–19]. Most recently, FUS/TLS was linked to Amyotrophic Lateral Sclerosis (ALS) [27, 28]. We used RNAi to achieve efficient knockdown of FUS/TLS in HeLa cells (Fig. 3A). Both control and specific RNAi-treated samples were subjected to library preparation according to MAPS followed by deep sequencing in a single lane on an Illumina flowcell. After filtering tags likely resulting from internal priming, we obtained ~8M uniquely mapped tags under FUS/TLS knockdown conditions. This result is comparable to those from control siRNA-treated cells (Table 1). Similar to control cells, FUS/TLS knockdown samples were highly reproducible when comparing between biological repeats (data not shown), and again, more than half of tag clusters were mapped outside known polyA sites (Table 2). By comparing between control and specific RNAi-treated cells, we identified several hundred genes that were either up- or down-regulated in response to in vivo depletion of FUS/TLS (Fig. 3B).
Figure 3.
Differential gene expression in response to FUS/TLS knockdown. (A) Top panel: Western gel comparing FUS/TLS and Actin protein levels after treatment with control and FUS/TLS siRNA; lower panel: Scatter plot compares control and FUS/TLS siRNA-treated samples. (B) Detection of FUS/TLS regulated genes on the M/A plot. Differentially regulated genes (FDR<0.05) are labeled red.
Comparison with microarrays
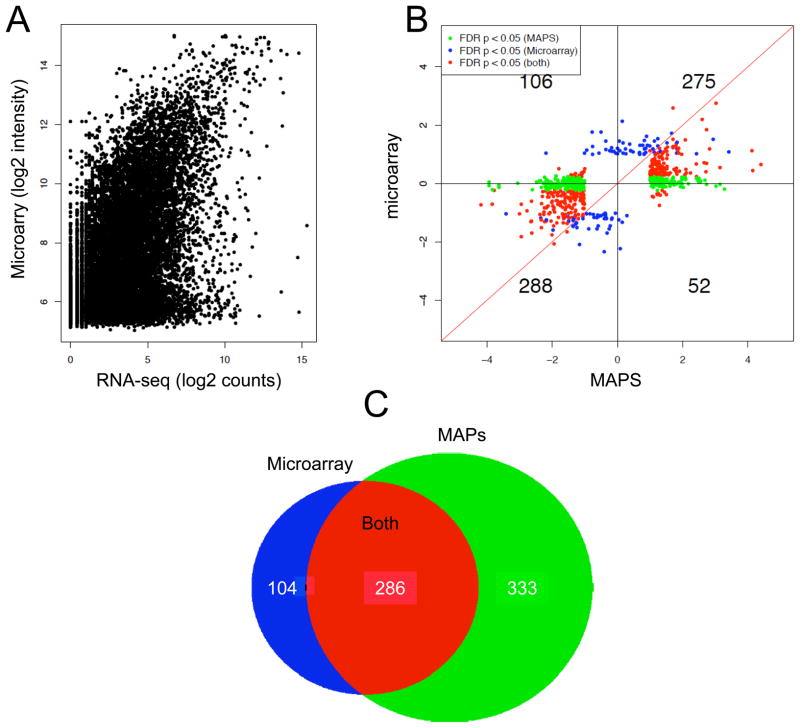
In order to validate the data generated by MAPS on the global scale, we performed microarray analysis on the same sets of samples from control and FUS/TLS RNAi-treated cells (Fig. 4A). We observed a similar pattern to previously documented comparisons of RNA-seq and microarray platforms [29]. As noted before, microarray intensity appears larger than RNA-seq counts, likely due to elevated background on the microarray. We next compared differentially regulated genes, demanding FDR=<0.05 and fold change (FC)=>2 (Fig. 4B). While the two platforms generally agree with one another in terms of detecting similar trend changes for both up- and down-regulated genes, it is clear that a large number of genes that scored quantitative differences failed to show differences on the microarray platform (green dots). As a result, the RNA-seq data revealed more quantitative differences than the microarray (Fig. 4C). We also noted that, although there were a number of genes (104) that were only identified as differentially expressed according to the microarray (blue dots), those genes tended to show differences slightly above the fold cut-off (Fig. 4B). Previous analyses show similar patterns of agreement and discrepancy between microarray and RNA-seq platforms [29]. We have also tested the reproducibility and multiplex capacity of MAPS technology. To do this, we compared the same samples and multiplexed three or six samples at once in a lane (Supplementary Fig. 1). The number of reads per gene is slightly reduced when multiplexing at a higher level, but importantly, the relative amount of gene is virtually identical. The ability of MAPS to detect more differentially expressed genes than microarray suggests that the digital method is more suitable for quantitative analysis of regulated gene expression. Further, the ability of MAPS to effectively multiplex with six samples in a highly reproducible manner illustrates the power and monetary benefit of this technology.
Figure 4.
Comparison between MAPS and microarray. (A) Comparison between signal intensity detected by micorarray and count by MAPS on individual annotated genes. (B) Comparison between differentially regulated genes by FUS/TLS identified by micorarray and MAPS based on FDR=<0.05 and fold change (FC) =>2. (C) Venn diagram of differentially regulated genes uniquely or commonly detected by the two methods.
Conclusions
We describe a multiplex strategy for profiling gene expression. The procedure is much simpler than existing 3′-tag digital gene expression analyses and is highly cost effective compared to microarray-based approaches. The MAPS strategy provides strand information, thus permitting accurate assignment of sequence tags from the transcription of both sense and antisense strands within many gene loci. In addition to quantitative analysis of gene expression, our method can also be applied to detect new polyadenylation sites. We suggest a large of number of these sites identified have yet to be annotated. Although, some of these new polyA-containing transcripts correspond to non-coding RNAs expressed at low levels, their biological importance is appreciated. We have applied this technology to study regulated gene expression by FUS/TLS, a RNA binding protein implicated in diverse biological processes, illustrating that this method can be applied to study a multitude of different specific conditions. MAPS is a simple technique that enables high quality, accurate and digital gene expression data at an economical cost.
Materials and Methods
Cell culture and RNA isolation
HeLa cells cultured in DMEM plus 10% FBS were transfected using Lipofectamine 2000 (Invitrogen) with 50pmol/ml of either TLS/FUS siRNA 5′-ACAGCCCATGATTAATTTGTA-3′ or control siRNA for 64 hours. The cells were subsequently harvested using Trizol followed by an Ambion RNA easy clean up. The resulting RNA was split into two samples, one for microarray analysis and the other for RNA-seq.
Multiplex RNA-seq
Total RNA (1–3ug) was added to 1ul of 50uM Bio-B-TNN (Biotin-5′-CAAGCAGAAGACGGCATACGAGTTTTTTTTTTTTTTTTTTTTNV-3′, note that the sequence at the 5′ portion corresponds to Primer B or P7 on the Illumina flowcell and that V in the last position represents A, G, or C, but not T), 1ul of 10mM dNTPs and distilled water to a total of 13ul. The mixture was heated for 5 minutes at 65°C, and subsequently incubated on ice for at least 1 minute. After the samples were cooled, 4ul of 5X First-strand buffer (Invitrogen), 1ul of 0.1M DTT, 1ul RNase Inhibitor (Invitrogen), and 1ul Superscript III RT (Invitrogen) were added to the reaction. The reaction was incubated at 50°C for 60 minutes. To inactivate the reaction, 80ul of water was added and the sample was heated at 70°C for 15minutes. To concentrate and remove free biotinylated oligos, the reaction along with 500ul of Qiagen PCR purification buffer (Qiagen PCR Purification Kit Cat # 28104) was added to the column. After washing the Qiagen column once with the binding buffer and twice with wash buffer, the cDNA was then eluted with 50ul of Qiagen elution buffer (Tris-HCl pH 8.5).
The eluted 50ul sample was added to a mixture of 15ul of 10X terminal transferase buffer (NEB cat #M0315S), 3ul of 130uM ddNTP mix, 80 ul of distilled water, and 2ul of terminal transferase (NEB cat #M0315S) to block the first strand. The reaction was incubated at 37°C for 1hr. EDTA was added to the final concentration of 20mM to stop the reaction. To the resulting primer extension product, 5ul Streptavidin-coated magnetic beads was added (Invitrogen (Dynabeads My One Streptavidin C1, Cat #65001), and the sample incubated at room temperature for 20 minutes. The collected beads were washed first with 100ul NaOH (0.1M) by pipetting up and down, and incubated for 5 minutes at room temperature. The beads were washed with water twice, then 1ul of sequencing Primer A-Random (100uM) (5′-GCTGATGCTACGACCACAGGNNNNNNN-3′, note that the specific sequence at the 5′ portion corresponds to the primer for sequencing on the Illumina flowcell) was added to the beads along with 5ul of 10X standard Taq DNA polymerase buffer (NEB), 1ul 10mM dNTPs, and water up to 49ul. The reaction was initiated by adding 1ul of Taq DNA polymerase (5ul, NEB) and incubated at 25°C for 1hr, 72°C for 30 seconds to extend, and then 75°C for 5 minutes. EDTA was added up to a final concentration of 10mM to stop the reaction. The beads were washed twice with 150ul of wash buffer (10mM Tris, pH 7.5, 1mM EDTA, 0.1% Tween-80 or Triton-X 100) and re-suspended in 20ul water and heated for 5 minutes at 95°C to release the products extended from the random primer.
The released DNA was amplified for 18 cycles using a common primer (Primer B: 5′-CAAGCAGAAGACGGCATACGAG-3′) in combination with a bar-coded PCR primer (5′-AATGATACGGCGACCACCGAGA-TXXXX-GCTGATGCTACGACCACAGG-3′, note that the specific sequence at the 5′ portion corresponds to Primer A or P5 on the Illlumon flowcell and that the specific sequence after the bar-code region corresponds to the sequencing primer).
The PCR products were precipitated with EtOH in the presence of glycogen (1:100), and resuspended 10ul water. The PCR products were fractionated on a 10% acrylamide gel. DNA fragments in the 150–350nt range were cut out, eluted with the elution buffer (10mM Tris-pH7.5, 1mM EDTA, 0.1% Triton X-100), and subjected to sequencing on the Illumina GAII Gene Analyzer.
Analysis of RNA-seq data
The analysis was done by using in-house made scripts, which used bx-python (http://bx-python.trac.bx.psu.edu), kent source [30], and BEDTools [31]. Fastq files were aligned with Bowtie [32] to merged transcript sequences from the UCSC Known Gene Database [33, 34]. The parameter for the mapping program Bowtie was “--solexa1.3-quals -l25 -n2 -e 200 -m1 --best --strata --trim5 4” [32]. The mapping started at the 5th position of each tag to increase higher rate and up to 2 mismatches were allowed. The unmapped tags in the first round were then mapped to the genome (hg18). The internal priming was checked for each tag in its 300nt downstream sequence to see whether there was at least one polyA stretch (consecutive 8 As or 9 As in a 10nt window) [12]. Differential expression between TLS knock-down and control was assessed using the edgeR package in R [35].
Microarray analysis and comparison with MAPS
Total RNA was isolated from control and TLS knockdown samples in triplicate and applied to Illumina BeadChip (HumanHT-12 v4) containing ~48K probes. Array hybridization was performed at the UCSD Core facility according to manufacturer’s protocol. Quantile normalization was performed in R using the lumi package [36]. Differential expression was measured in R using the limma package [37]. A False Discovery Rate of 0.05 was applied to the resulting p-values to correct for multiple-hypothesis testing.
Data from RNA-seq and microarrays were merged on the Gene Symbol. For comparison of control gene expression, 1 was added to the RNA-seq counts before taking the log in order to avoid log of 0. For comparison of differentially expressed genes, we limited the analysis to genes that had at least one tag in one replicate under each condition.
Supplementary Material
Acknowledgments
We thank members of the Fu lab for technical help and stimulating discussion during the course of this study. We acknowledge early contribution of Ravi Dinakar to this project. This work was supported by a training grant (PI: Dan Donoghue) from NCI to KFW (T32 CA009523) and NIH grants (GM049369 and HG004659) to XDF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Nat Methods. 2010;7:709–15. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 4.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamanova L, Andrews RM, James KD, Sheridan EM, Ellis PD, Langford CF, Ost TW, Collins JE, Turner DJ. Nat Methods. 2010;7:130–2. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. Nat Protoc. 2010;5:516–35. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Lovci MT, Kwon YS, Rosenfeld MG, Fu XD, Yeo GW. Proc Natl Acad Sci U S A. 2008;105:20179–84. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, Raymond CK. Nat Methods. 2009;6:647–9. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 9.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, Kocher JP. BMC Genomics. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, Attie O, Chen K, Salehi-Ashtiani K, Vidal M, Harkins TT, Bouffard P, Suzuki Y, Sugano S, Kohara Y, Rajewsky N, Piano F, Gunsalus KC, Kim JK. Science. 2010;329:432–5. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaudoing E, Gautheret D. Genome Res. 2001;11:1520–6. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez F, Granjeaud S, Ara T, Ghattas B, Gautheret D. Rna. 2006;12:1794–801. doi: 10.1261/rna.136206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletchers CD, Aurias A, Thomas G. Nat Genet. 1993;4:341–5. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 15.Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, et al. Embo J. 1993;12:4481–7. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters BL, Panagopoulos I, Allen EF. Cancer Genet Cytogenet. 2000;121:109–16. doi: 10.1016/s0165-4608(00)00237-5. [DOI] [PubMed] [Google Scholar]
- 17.Panagopoulos I, Hoglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Oncogene. 1996;12:489–94. [PubMed] [Google Scholar]
- 18.Panagopoulos I, Mandahl N, Ron D, Hoglund M, Nilbert M, Mertens F, Mitelman F, Aman P. Cancer Res. 1994;54:6500–3. [PubMed] [Google Scholar]
- 19.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. Cancer Res. 1994;54:2865–8. [PubMed] [Google Scholar]
- 20.Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, de Rooij DG, Akhmedov A, Ashley T, Ron D. Embo J. 2000;19:453–62. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. Embo J. 1996;15:5022–31. [PMC free article] [PubMed] [Google Scholar]
- 23.Chansky HA, Hu M, Hickstein DD, Yang L. Cancer Res. 2001;61:3586–90. [PubMed] [Google Scholar]
- 24.Yang L, Embree LJ, Hickstein DD. Mol Cell Biol. 2000;20:3345–54. doi: 10.1128/mcb.20.10.3345-3354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Embree LJ, Tsai S, Hickstein DD. J Biol Chem. 1998;273:27761–4. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 26.Meissner M, Lopato S, Gotzmann J, Sauermann G, Barta A. Exp Cell Res. 2003;283:184–95. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 27.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 28.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. Genome Res. 2008;18:1509–17. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. Nucleic Acids Res. 2004;32:D493–6. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. Bioinformatics. 2006;22:1036–46. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du P, Kibbe WA, Lin SM. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 37.Smyth GK. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.