Abstract
Enzymatic degradation of plant polysaccharides has many industrial applications, such as within the paper, food, and feed industry and for sustainable production of fuels and chemicals. Cellulose, hemicelluloses, and pectins are the main components of plant cell wall polysaccharides. These polysaccharides are often tightly packed, contain many different sugar residues, and are branched with a diversity of structures. To enable efficient degradation of these polysaccharides, fungi produce an extensive set of carbohydrate-active enzymes. The variety of the enzyme set differs between fungi and often corresponds to the requirements of its habitat. Carbohydrate-active enzymes can be organized in different families based on the amino acid sequence of the structurally related catalytic modules. Fungal enzymes involved in plant polysaccharide degradation are assigned to at least 35 glycoside hydrolase families, three carbohydrate esterase families and six polysaccharide lyase families. This mini-review will discuss the enzymes needed for complete degradation of plant polysaccharides and will give an overview of the latest developments concerning fungal carbohydrate-active enzymes and their corresponding families.
Keywords: Polysaccharides, Plant biomass, Fungal enzymes, Aspergillus, Trichoderma
Introduction
Plant polysaccharides have applications in many industrial sectors, such as biofuel, pulp and paper, and food and feed. Cellulose, hemicelluloses, and pectin are the main components of plant cell walls representing up to 70% of the biomass (Jorgensen et al. 2007). Of the three, cellulose is the least complex polymer with a linear structure of β-1,4-linked d-glucose residues. The long glucose chains are tightly bundled together in microfibrils and are non-covalently linked together by hemicelluloses (Kolpak and Blackwell 1976; Carpita and Gibeaut 1993).
Hemicelluloses are classified according to the main sugar in the backbone of the polymer, i.e., xylan (β-1,4-linked d-xylose), mannan (β-1,4-linked d-mannose), or xyloglucan (β-1,4-linked d-glucose). The backbone of hemicelluloses has many branches composed of monomers such as d-galactose, d-xylose, l-arabinose, and d-glucuronic acid. The precise composition of hemicellulose is strongly dependent on the plant species and tissue (Scheller and Ulvskov 2010). For instance, hard wood xylans often have d-glucuronic acid attached to their backbone, whereas l-arabinose is the most common branching residue in cereal xylans (de Vries and Visser 2001). Moreover, hemicelluloses are often acetylated and to a lesser extent ester-linked with feruloyl or p-coumaroyl residues (Ebringerova et al. 1990; Xu et al. 2010).
Pectin is less prominently present in most plant biomass compared to cellulose and hemicellulose. However, some plant biomass types (e.g., citrus peels) are very rich in pectin (Angel Siles Lopez et al. 2010; Ridley et al. 2001; Grohmann and Bothast 1994). The backbone of pectin consists mainly of alpha-1,4-linked d-galacturonic acid residues that can be methyl-esterified or substituted with acetyl groups. Pectins are classified in three general groups, homogalacturonan (linear polymer), xylogalacturonan (branched by β-1,3-linked d-xylose), and rhamnogalacturonan (Ridley et al. 2001; Wong 2008; Caffall and Mohnen 2009). The latter polysaccharide is the most complex pectin structure. Its backbone consists of alternating l-rhamnose and d-galacturonic acid residues, while branches with β-1,4-linked d-galactose and different α-linked l-arabinose residues are connected to the l-rhamnose residues (Ridley et al. 2001; Wong 2008).
In nature, fungi play a central role in the degradation of plant biomass. Plant-biomass-degrading fungi produce an extensive set of carbohydrate-active enzymes specifically dedicated to degrade plant polysaccharides. However, these sets differ between fungal species. For instance, Trichoderma reesei has a highly efficient set of enzymes involved in cellulose degradation (Martinez et al. 2008; Kubicek et al. 2011), while Aspergillus species produce many enzymes to degrade pectin (Martens-Uzunova and Schaap 2009). The industrial importance of polysaccharide-degrading enzymes and the availability of many fungal genomes have strongly deepened our understanding of fungal biodiversity with respect to plant cell wall degradation. This mini-review will give an overview of the latest developments and insights into fungal enzymes involved in plant polysaccharide degradation.
Dedicated fungal toolboxes for the degradation of specific plant polysaccharides
Carbohydrate-active enzymes can be organized in different families based on amino acid sequence of the structurally related catalytic modules (www.cazy.org) (Cantarel et al. 2009; Henrissat 1991). Fungal enzymes involved in plant polysaccharide degradation are assigned to at least 35 glycoside hydrolase (GH) families, three carbohydrate esterase (CE) families, and six polysaccharide lyase (PL) families (Battaglia et al. 2011; Coutinho et al. 2009). Even though enzymes within the same family share sequence similarity, some families can contain multiple activities. For example, GH5 contains many catalytic activities, including exoglucanases, endoglucanases, and endomannanases (Dias et al. 2004). In addition, a specific enzyme activity can be present in several CAZy families. This is important for efficient degradation of plant polysaccharides as enzymes of each family have often complementary substrate specificity. For instance, endoxylanases in GH10 have lower substrate specificity and can degrade xylan backbones with many substitutions, while GH11 endoxylanases have higher substrate specificity with preference for unsubstituted xylan chains (Pollet et al. 2010; Biely et al. 1997).
Annotation of carbohydrate-active enzymes has been done for many fungal genomes (Pel et al. 2007; Espagne et al. 2008; Battaglia et al. 2011; Ohm et al. 2010; Martinez et al. 2004; Martinez et al. 2008). As an illustration, Table 1 shows a comparison of carbohydrate-active enzymes involved in plant polysaccharide degradation of 13 fungal genomes, including industrial fungi such as Aspergillus oryzae, Aspergillus niger, Penicillium chrysogenum, T. reesei, and Saccharomyces cerevisiae. Most apparent from this table is the correlation between habitat and the amount of carbohydrate-active enzymes. For example, the Saccharomycete S. cerevisiae does not require extracellular enzymes for polysaccharide degradation to survive in its natural niches like surfaces of rotting fruits (Liti et al. 2009; Cherry et al. 1997). This fungus has therefore hardly any carbohydrate-active enzymes. Another Saccharomycete Pichia stipitis can be found, among other places, in the guts of termites that inhabit and degrade white-rotted hardwood (Jeffries et al. 2007). The genome of this fungus contains only a few β-glucosidases and β-mannosidases to degrade glucan and mannan oligosaccharides, which are present in the termite guts (Jeffries et al. 2007). In contrast to both Saccharomycetes, the filamentous fungi in Table 1 have a lifestyle involving degradation of plant biomass and feeding from plant polysaccharides. The genomes of these fungi therefore contain many more genes encoding carbohydrate-active enzymes. For example, the saprobe A. niger has 178 putative GH enzymes, nine putative CE enzymes, and 13 putative PL enzymes involved in plant biomass degradation (Coutinho et al. 2009). However, there is also variation within this group of fungi. For instance, Aspergilli have a large number of enzymes involved in pectin degradation, in contrast to the cellulose-degrading specialist T. reesei and the lignin-degrader Phanerochaete chrysosporium. The Zygomycete Rhizopus oryzae has a different set of carbohydrate-active enzymes compared to the other filamentous fungi (Battaglia et al. 2011). For instance, the difficulty of this fungus to grow on xylan substrates is reflected in its absence of genes required for xylan degradation. This fungus has therefore also been described as a fast grower on easily accessible and digestible substrates (Richardson 2009).
Table 1.
Number of putativegenes involved plant polysaccharide degradation of 13 fungal genomes based on the CAZy database (http://www.cazy.org)
| Total putative genes involved in plant polysaccharide degradation | AN | An | Ao | PC | Fg | Nc | Pa | Tr | Ps | SC | Pc | Sc | Ro | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glycoside hydrolases (GHs) | 194 | 178 | 207 | 158 | 190 | 119 | 161 | 113 | 33 | 29 | 140 | 183 | 70 | |||
| Carbohydrate esterases (CEs) | 10 | 9 | 15 | 7 | 17 | 10 | 17 | 6 | 2 | 1 | 11 | 14 | 9 | |||
| Polysaccharide lyases (PLs) | 20 | 13 | 20 | 9 | 20 | 3 | 7 | 0 | 0 | 0 | 0 | 13 | 0 | |||
| Substrate | Enzyme activity | Code | CAZy fam. | AN | An | Ao | PC | Fg | Nc | Pa | Tr | Ps | SC | Pc | Sc | Ro |
| Cellulose | β-1,4-endoglucanase | EGL | GH5,7,12,45 | 20 (7) | 15 (6) | 20 (5) | 18 | 20 | 14 | 22 (10) | 13 | 4 (0) | 5 (0) | 31 (5) | 22 (4) | 12 (5) |
| Cellobiohydrolase | CBH | GH6,7 | 5 (4) | 4 (4) | 4 (3) | 3 | 3 | 8 | 10 (6) | 3 | 0 (0) | 0 (0) | 10 (7) | 3 (3) | 0 (0) | |
| β-1,4-glucosidase | BGL | GH1,3 | 22 (19) | 20 (15) | 21 (18) | 20 | 24 | 10 | 12 (9) | 15 | 5 (4) | 0 (0) | 13 (9) | 15 (12) | 6 (5) | |
| Xyloglucan | Xyloglucan β-1,4-endoglucanase | XEG | GH12,74 | 3 | 4 | 4 | 3 | 5 | 2 | 3 | 3 | 0 | 0 | 6 | 2 | 0 |
| α-arabinofuranosidase | ABF | GH51,54 | 4 | 5 | 4 | 4 | 3 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 0 | |
| α-xylosidase | AXL | GH31 | 10 | 7 | 10 | 11 | 8 | 5 | 5 | 4 | 3 | 1 | 6 | 4 | 3 | |
| α-fucosidase | AFC | GH29,95 | 3 | 3 | 3 | 1 | 3 | 0 | 0 | 4 | 0 | 0 | 1 | 3 | 0 | |
| α-1,4-galactosidase | AGL | GH27,36 | 7 | 7 | 6 | 3 | 5 | 1 | 3 | 10 | 0 | 0 | 3 | 1 | 6 | |
| β-1,4-galactosidase | LAC | GH2,35 | 14 (7) | 11 (5) | 14 (8) | 8 | 14 | 7 | 8 (4) | 8 | 2 (1) | 0 (0) | 5 (2) | 8 (4) | 1 (1) | |
| Xylan | β-1,4-endoxylanase | XLN | GH10,11 | 5 | 5 | 8 | 4 | 8 | 6 | 14 | 5 | 1 | 0 | 7 | 6 | 0 |
| β-1,4-xylosidase | BXL | GH3,43 | 37 (11) | 28 (6) | 38 (11) | 31 | 38 | 16 | 21 (5) | 15 | 5 (1) | 0 (0) | 15 (2) | 31 (2) | 8 (0) | |
| α-arabinofuranosidase | ABF | GH51,54 | 4 | 5 | 4 | 4 | 3 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 0 | |
| Arabinoxylan arabinofuranohydrolase | AXH | GH62 | 2 | 1 | 2 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | |
| α-glucuronidase | AGU | GH67,115 | 2 | 1 | 5 | 1 | 3 | 2 | 4 | 2 | 1 | 0 | 1 | 2 | 0 | |
| α-1,4-galactosidase | AGL | GH27,36 | 7 | 7 | 6 | 3 | 5 | 1 | 3 | 10 | 0 | 0 | 3 | 1 | 6 | |
| β-1,4-galactosidase | LAC | GH2,35 | 14 (7) | 11 (5) | 14 (8) | 8 | 14 | 7 | 8 (4) | 8 | 2 (1) | 0 (0) | 5 (2) | 8 (4) | 1 (1) | |
| Acetyl xylan/feruloyl esterase | AXE/FAE | CE1 | 4 | 5 | 5 | 2 | 5 | 7 | 14 | 3 | 1 | 1 | 5 | 9 | 0 | |
| Galactomannan | β-1,4-endomannanase | MAN | GH5,26 | 18 (8) | 11 (2) | 14 (3) | 14 | 13 | 8 | 13 (2) | 8 | 4 (0) | 5 (0) | 20 (4) | 19 (2) | 7 (1) |
| β-1,4-mannosidase | MND | GH2 | 10 (3) | 6 (3) | 7 (3) | 6 | 11 | 5 | 7 (1) | 7 | 2 (1) | 0 (0) | 2 (2) | 4 (3) | 0 (0) | |
| β-1,4-galactosidase | LAC | GH2,35 | 14 (7) | 11 (5) | 14 (8) | 8 | 14 | 7 | 8 (4) | 10 | 2 (1) | 0 (0) | 5 (2) | 8 (4) | 1 (1) | |
| α-1,4-galactosidase | AGL | GH27,36 | 7 | 7 | 6 | 3 | 5 | 1 | 3 | 8 | 0 | 0 | 3 | 1 | 6 | |
| Pectin | Endo-/exo-(rhamno)galacturonases | GH28 | 10 | 22 | 20 | 5 | 6 | 2 | 0 | 4 | 0 | 1 | 4 | 3 | 18 | |
| α-rhamnosidase | RHA | GH78 | 9 | 8 | 8 | 5 | 7 | 0 | 1 | 1 | 0 | 0 | 1 | 3 | 0 | |
| α-arabinofuranosidase | ABF | GH51,54 | 4 | 5 | 4 | 4 | 3 | 2 | 1 | 2 | 0 | 0 | 2 | 2 | 0 | |
| Endoarabinanase | ABN | GH43 | 18 (4) | 11 (5) | 20 (5) | 14 | 17 | 7 | 10 (5) | 2 | 0 (0) | 0 (0) | 4 (2) | 19 (12) | 2 (2) | |
| Exoarabinanase | ABX | GH93 | 2 | 1 | 3 | 2 | 2 | 2 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | |
| β-1,4-endogalactanase | GAL | GH53 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | |
| Unsaturated glucuronyl hydrolase | UGH | GH88 | 3 | 1 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |
| Unsat.-rhamnogalacturonan hydrolase | URH | GH105 | 4 | 2 | 2 | 2 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | |
| β-1,4-xylosidase | BXL | GH3,43 | 37 (11) | 28 (6) | 38 (11) | 31 | 38 | 16 | 21 (5) | 14 | 5 (1) | 0 (0) | 15 (2) | 31 (2) | 8 (0) | |
| β-1,4-galactosidase | LAC | GH2,35 | 14 (7) | 11 (5) | 14 (8) | 8 | 14 | 7 | 8 (4) | 8 | 2 (1) | 0 (0) | 5 (2) | 8 (4) | 1 (1) | |
| Pectin lyase | PEL | PL1 | 9 | 6 | 12 | 5 | 9 | 1 | 4 | 0 | 0 | 0 | 0 | 5 | 0 | |
| Pectate lyase | PLY | PL1,3,9 | 15 | 10 | 16 | 6 | 17 | 2 | 6 | 0 | 0 | 0 | 0 | 10 | 0 | |
| Rhamnogalacturonan lyase | RGL | PL4,11 | 5 | 3 | 4 | 3 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | |
| Pectin methyl esterase | PME | CE8 | 3 | 1 | 5 | 2 | 6 | 1 | 1 | 0 | 0 | 0 | 2 | 2 | 6 | |
| Rhamnogalacturonan acetyl esterase | RGAE | CE12 | 2 | 2 | 4 | 2 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | |
| Feruloyl esterase | FAE | CE1 | 4 | 5 | 5 | 2 | 5 | 7 | 14 | 3 | 1 | 1 | 5 | 9 | 0 | |
| Starch | α-amylase | AMY | GH13 | 13 | 16 | 17 | 16 | 8 | 10 | 9 | 5 | 5 | 9 | 9 | 13 | 4 |
| glucoamylase | GLA | GH15 | 2 | 2 | 3 | 3 | 3 | 2 | 3 | 2 | 1 | 1 | 2 | 3 | 6 | |
| α-1,4-glucosidase | AGD | GH31 | 10 | 7 | 10 | 11 | 8 | 5 | 5 | 4 | 3 | 1 | 6 | 4 | 3 | |
| Inulin | Inulinase | INU | GH32 | 2 | 4 | 4 | 7 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
Total values obtained per enzyme activity correspond to the theoretical maximum number of active enzymes that could be obtained by adding up all the members of the concerned families. The values between brackets indicate a more precise estimate as taking into consideration the functional assignments based on sequence similarities to biochemically characterized enzymes. These values are to be taken as a maximum rather than as absolute values. AN = Aspergillus nidulans A4 (Coutinho et al. 2009), An = Aspergillus niger CBS513.88 (Coutinho et al. 2009), Ao = Aspergillus oryzae RIB4 (Coutinho et al. 2009), PC = Penicillium chrysogenum Wisconsin 54–1255 (van den Berg et al. 2008), Fg = Fusarium graminearum/Gibberella zeae PH-1 (Cuomo et al. 2007), Nc = Neurospora crassa OR74A (Galagan et al. 2003), Pa = Podospora anserina S mat + (Espagne et al. 2008), Tr = Trichoderma reesei/Hypocrea jecorina QM6A (Martinez et al. 2008), Ps = Pichia stipitis CBS6054 (Jeffries et al. 2007), SC = Saccharomyces cerevisiae S288C (Cherry et al. 1997), Pc = Phanerochaete chrysosporium RP78 (Martinez et al. 2004), Sc = Schizophyllum commune H4-8 (Ohm et al. 2010), Ro = Rhizopus oryzae 99–880 (Battaglia et al. 2011)
To have a better impression of the latest developments regarding fungal carbohydrate-active enzymes, the following sections will discuss the enzymes needed for each of the main polysaccharides: cellulose, hemicellulose, and pectin. Furthermore, as an illustration of a fungal enzyme set, each section will show the genes encoding characterized and putative polysaccharide-degrading enzymes of A. niger.
Cellulose degradation
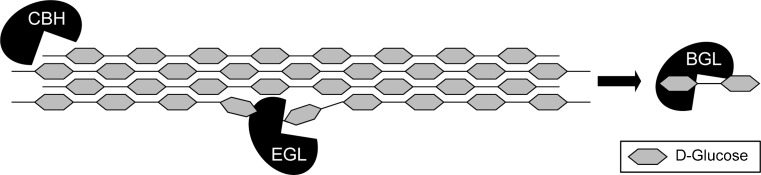
Cellulose degradation requires three classes of enzymes, β-1,4-endoglucanases (EGL), exoglucanases/cellobiohydrolases (CBH), and β-glucosidase (BGL), which are divided over eight GH families (Fig. 1; Vlasenko et al. 2010; de Vries et al. 2011). For example, A. niger has five EGLs within GH families 5 and 12, four CBHs in families 6 and 7, and 13 BGLs in families 1 and 3 (Table 2). In comparison, one of the most efficient cellulose-degrading fungi T. reesei has five characterized EGLs within GH families 5, 7, 12, and 45, two highly expressed CBHs in families 6 and 7, and two characterized BGLs in families 1 and 3 (Martinez et al. 2008; Kubicek et al. 2011). Although T. reesei does not have the biggest number of cellulases, its set of enzymes is very efficient in breaking down cellulose by acting synergistically (Ward et al. 1993). The family numbers are also used in the nomenclature of hydrolytic enzymes (Henrissat et al. 1998). For example, the first three letters of endoglucanase Cel5A of T. reesei refer to its substrate cellulose, the number to glycoside hydrolase family 5 and the last capital letter indicates that this was the first enzyme reported of this family with this activity. This review will mainly refer to the original functionally based nomenclature. We will therefore use for instance EGII instead of Cel5A, based on the consideration that these names were mainly used in database depositions and give a clearer indication of function.
Fig. 1.
Schematic structure of cellulose with cellulolytic enzymes. BGL β-glucosidase, CBH cellobiohydrolase, EGL β-1,4-endoglucanase
Table 2.
Genes encoding characterized and putative enzymes of Aspergillus niger CBS513.88 involved in cellulose degradation (Coutinho et al. 2009)
| Enzyme class | Code | CAZy families | Genes of characterized and putative enzymes | Reference |
|---|---|---|---|---|
| β-1,4-endoglucanase | EGL | GH5 | An07g08950 (eglB), An01g11670, An16g06800 | (van Peij et al. 1998) |
| GH7 & GH45 | – | |||
| GH12 | An14g02670 (eglA), An01g03340, An03g05380 | (van Peij et al. 1998) | ||
| Cellobiohydrolase | CBH | GH6 | An08g01760, An12g02220 | |
| GH7 | An07g09330 (cbhA), An01g11660 (cbhB) | (Gielkens et al. 1999) | ||
| β-1,4-glucosidase | BGL | GH1 | An11g02100, An04g03170, An03g03740, An02g08600 | |
| GH3 | An18g03570 (bgl1), An07g09760, An08g08240, An11g00200, An14g01770, An17g00520, An03g05330, An06g02040, An15g01890, An11g06090, An15g04800 | (Dan et al. 2000; Pel et al. 2007) |
The genes with names between brackets are biochemically characterized and their references are given in the last column
EGI (GH7) and EGII (GH5) are the most abundantly produced of the five EGLs of T. reesei (Foreman et al. 2003; Vlasenko et al. 2010). Both enzymes have a carbohydrate-binding module which greatly enhances the efficiency in degradation of cellulose microfibrils by binding to cellulose (Beckham et al. 2010; Guillen et al. 2010). EGIII, from GH12, is expressed at a lower level than EGI and EGII, but has a broad activity against cellulose, β-1,3-1,4-glucan, xyloglucan, and xylan (Sprey and Bochem 1993; Eriksson et al. 2002). Two proteins from GH61 were expressed in the presence of cellulose and activity has been measured against β-glucan (Saloheimo et al. 1997; Foreman et al. 2003). However, the enzymes within GH61 have recently shown not to be glycoside hydrolases (Harris et al. 2010). Although they are involved in improving the efficiency of degrading tightly packed cellulose microfibrils, the precise function of GH61 enzymes is still unknown (Harris et al. 2010; Vaaje-Kolstad et al. 2010).
The two highly expressed CBHs of T. reesei, CBHI and CBHII, belong to families 7 and 6, respectively. These are considered to work synergistically and have preference for the reducing or non-reducing end, respectively, of the cellulose chains (Nutt et al. 1998). CBHs are also considered to be important for the hydrolysis of the crystalline parts of cellulose (Liu et al. 2011). These CBHs of T. reesei are highly sensitive to product inhibition, in particular by cellobiose, which might explain the need for a high amount of CBHs in an effective fungal cellulase enzyme mix (Bezerra and Dias 2005; Holtzapple et al. 1990; Kristensen et al. 2009).
After endo- and exo-cleaving of cellulose, BGLs belonging to GH families 1 and 3 degrade the remaining oligosaccharides to monomeric glucose. BGLs are a group with very diverse properties and cellular location, although most BGLs belong to GH3 and have a similar retaining mechanism (Decker et al. 2001; Saloheimo et al. 2002). The two known BGLs of T. reesei, BGI and BGII, are produced at low levels (Reczey et al. 1998) and subject to strong product inhibition (Chen et al. 1992). These characteristics hinder the function of T. reesei for extensive in vitro saccharification of cellulose. Therefore, in industrial applications, T. reesei cellulase mixtures are often supplemented with BGLs from Aspergilli, which are highly expressed and more glucose tolerant (Decker et al. 2000; Reczey et al. 1998; Riou et al. 1998). Another strategy to convert cellulose into a sustainable product is to express BGLs in the fermentation host like S. cerevisiae (Li et al. 2010; Ha et al. 2011). This way, oligosaccharides like cellobiose are directly fermented into fuels or chemicals.
Hemicellulose degradation
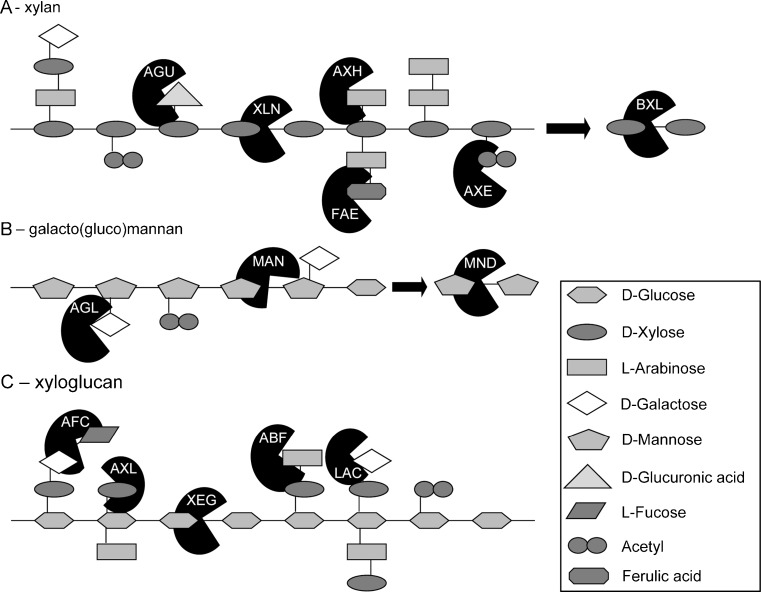
Hemicellulose, the second most abundant plant polysaccharide, is a group of complex structures composed of different residues with three kinds of backbones and many different branches (Fig. 2). The three backbones of the corresponding group of hemicelluloses are hydrolysed by a specific set of dedicated carbohydrate-active enzymes: β-1,4-endoxylanase and β-1,4-xylosidase for xylan, xyloglucan-active β-1,4-endoglucanase and β-1,4-glucosidase for xyloglucan, and β-1,4-endomannanase and β-1,4-mannosidase for (galacto-) mannan (Table 3) (de Vries and Visser 2001).
Fig. 2.
a–c Schematic structure of three hemicelluloses, xylan, galacto(gluco)mannan, and xyloglucan, with hemicellulolytic enzymes. ABF α-arabinofuranosidase, AFC α-fucosidase, AGL α-1,4-galactosidase, AGU α-glucuronidase, AXE acetyl (xylan) esterase, AXH arabinoxylan α-arabinofuranohydrolase, AXL α-xylosidase, BXL β-1,4-xylosidase, FAE feruloyl esterase, LAC β-1,4-galactosidase, MAN β-1,4-endomannanase, MND β-1,4-mannosidase, XEG xyloglucan-active β-1,4-endoglucanase, XLN β-1,4-endoxylanase
Table 3.
Genes encoding characterized and putative enzymes of Aspergillus niger CBS513.88 involved in the degradation of the three hemicellulose backbones (Coutinho et al. 2009)
| Enzyme class | Code | CAZy families | Genes of characterized and putative enzymes | Reference |
|---|---|---|---|---|
| Xyloglucan-active | XEG | GH12 | An14g02670 (eglA), An01g03340, An03g05380 | (van Peij et al. 1998) |
| β-1,4-endoglucanase | GH74 | An01g01870 (eglC) | (Hasper et al. 2002) | |
| β-1,4-endoxylanase | XLN | GH10 | An03g00940 (xynA) | (Krengel and Dijkstra 1996) |
| GH11 | An01g00780 (xynB), An01g14600, An14g07390, An15g04550 | (Levasseur et al. 2005) | ||
| β-1,4-xylosidase | BXL | GH3 | An01g09960 (xynD), An17g00300 | (van Peij et al. 1997) |
| GH43 | An11g03120, An02g00140, An08g10780, An08g01900 | |||
| β-1,4-endomannanase | MAN | GH5 | An05g01320 (manA) | (Ademark et al. 1998) |
| GH26 | An15g07760 | |||
| β-1,4-mannosidase | MND | GH2 | An11g06540 (mndA), An12g01850, An01g06630 | (Ademark et al. 2001) |
The genes with names between brackets are biochemically characterized and their references are given in the last column
The fungal β-1,4-endoxylanases belong to GH families 10 and 11 (Polizeli et al. 2005). Similar to enzymes within families GH1, GH2, and GH5, endoxylanases of GH10 have a TIM-barrel fold at their catalytic domain in contrast to GH11 which has a β-jelly roll structure at its catalytic domain (Henrissat and Bairoch 1993; Pollet et al. 2010). As a consequence, the two groups of endoxylanases differ from each other in substrate specificity (Biely et al. 1997). GH10 endoxylanases have in general a broader substrate specificity then endoxylanases of family GH11. Specifically, GH10 enzymes not only degrade linear chains of 1,4-linked d-xylose residues, but also xylan backbones with a high degree of substitutions and smaller xylo-oligosaccharides (Biely et al. 1997; Vardakou et al. 2003; Pollet et al. 2010). GH10 endoxylanases are therefore important for the complete degradation of substituted xylans. Although, in principal, less accessory enzymes are required with a higher amount of GH10 endoxylanases, fungal genomes have no clear correlation between the amount of GH10 endoxylanases and number of accessory enzymes.
The released xylo-oligosaccharides are degraded by β-xylosidases. Most fungal β-xylosidases belong to GH3 (Mozolowski and Connerton 2009), but several putative β-xylosidases are assigned to GH43 (e.g., in Penicillium herquei and A. oryzae (Ito et al. 2003; Machida et al. 2005)). GH3 is a conserved family containing mainly BGLs. As shown in T. reesei and bacteria, β-xylosidases of GH3 contain the conserved Asp-311 residue but miss the other active-site Asp-120 residue of β-glucosidases, which might explain the discrimination between β-xylosidases or β-glucosidases within this family (Margolles-Clark et al. 1996; Dodd et al. 2010).
Xyloglucan-active endoglucanases, also referred to as xyloglucanases, belong to GH families 12 and 74 (Grishutin et al. 2004). The main difference between enzymes of GH12 and GH74 is their retaining and inverting mechanism, respectively (Gilbert et al. 2008). The specific modes of action of the different xyloglucanases were recently elucidated (Desmet et al. 2007; Powlowski et al. 2009; Yaoi et al. 2009; Master et al. 2008). For instance, in contrary to the GH74 xyloglucanase from T. reesei, the GH12 enzyme from A. niger does not cleave at branched glucose residues and prefers xylogluco-oligosaccharides containing more than six glucose residues with, at least, one non-branched glucose residue (Master et al. 2008; Desmet et al. 2007).
Endomannanases, involved in the degradation of mannan polysaccharides, belong to GH families 5 and 26. However, fungal endomannanases are predominately present in GH5. The GH5 endomannanases from A. niger and T. reesei both show substrate specificity towards manno-oligosaccharides with more than three d-mannose residues (Tenkanen et al. 1997; Do et al. 2009). Like many other fungal carbohydrate-active enzymes, some endomannanases have a carbohydrate-binding module (mainly CBM1) which promotes the association of the enzyme with the substrate (Herve et al. 2010; Pham et al. 2010; Boraston et al. 2004). The released mannobiose and mannotriose are further degraded by β-1,4-mannosidases belonging to GH family 2 (Ademark et al. 2001).
To completely degrade hemicellulose, all substitutions on the hemicellulose backbones have to be released. This requires at least nine different enzyme activities divided over at least 12 GH families and four CE families (Table 4).
Table 4.
Genes encoding characterized and putative enzymes of Aspergillus niger CBS513.88 involved in the degradation of the substitutions on the hemicellulose backbones (Coutinho et al. 2009)
| Enzyme class | Code | CAZy families | Genes of characterized and putative enzymes | Reference |
|---|---|---|---|---|
| α-arabinofuranosidase | ABF | GH51 | An01g00330 (abfA), An08g01710, An09g00880 | (Flipphi et al. 1993c) |
| GH54 | An15g02300 (abfB) | (Flipphi et al. 1993b) | ||
| α-xylosidase | AXL | GH31 | – | |
| α-fucosidase | AFC | GH29 | An13g02110 | |
| GH95 | An16g02760, An16g00540 | |||
| α-1,4-galactosidase | AGL | GH27 | An06g00170 (aglA), An02g11150 (aglB), An01g01320, An14g01800, An11g06330 | (den Herder et al. 1992; de Vries et al. 1999) |
| GH36 | An09g00260 (aglC), An04g02700, An09g00270 | (Ademark et al. 2001) | ||
| β-1,4-galactosidase | LAC | GH2 | – | |
| GH35 | An01g12150 (lacA), An01g10350, An07g04420, An06g00290, An14g05820 | (Kumar et al. 1992) | ||
| Arabinoxylan α-arabinofuranohydrolase | AXH | GH62 | An03g00960 (axhA) | (Gielkens et al. 1997) |
| α-glucuronidase | AGU | GH67 | An14g05800 (aguA) | (de Vries et al. 2002c) |
| GH115 | – | |||
| Acetyl (xylan) esterase | AXE | CE1 | An12g05010 (axeA) | (van Peij et al. 1998) |
| Feruloyl/p-coumaroyl esterase | FAE | CE1 | An09g00120 (faeA), An12g10390 (faeB), An12g02550 | (de Vries et al. 1997; de Vries et al. 2002d) |
The genes with names between brackets are biochemically characterized and their references are given in the last column
l-Arabinose, a common residue in hemicellulose, is cleaved from arabinose-substituted xyloglucan and (arabino-)xylan by α-arabinofuranosidases and arabinoxylan arabinofuranohydrolases. Fungal α-arabinofuranosidases are mainly found in GH families 51 and 54, although some bifunctional enzymes from GH3 and GH43 were described to have α-arabinofuranosidase activity (Saha 2000; Ravanal et al. 2010). The difference in substrate specificity between GH51 and GH54 enzymes is illustrated by the two main arabinofuranosidases of A. niger. AbfA (GH51) releases l-arabinose from arabinan and sugar beet pulp, while AbfB (GH54) also releases l-arabinose from xylan (de Vries and Visser 2001). Arabinofuranosidases within GH54 are also described to have a carbohydrate-binding module (CBM 42) with specific binding to arabinofuranose side chains of hemicellulose (Miyanaga et al. 2004; Miyanaga et al. 2006). The arabinoxylan arabinofuranohydrolases from GH62 act specifically against the α-1,2- or α-1,3-linkage of the l-arabinose residues of arabinoxylan (Verbruggen et al. 1998), but are also sensitive to the substitutions of adjacent d-xylose residues. For instance, AxhA from A. niger is not able to release arabinobiose from xylan or substituted l-arabinose from d-xylose residues adjacent to d-glucuronic acid residues (Verbruggen et al. 1998; Sakamoto et al. 2011).
d-Xylose residues with α-linkages are released from the xyloglucan backbone by α-xylosidases. Of two α-xylosidases in Aspergillus flavus, AxlI hydrolyzes xyloglucan oligosaccharides and AxlII is most active on p-nitrophenyl α-l-xylose residues and does not hydrolyze xyloglucan (Yoshikawa et al. 1993, 1994). α-Xylosidases were suggested to belong to family GH31 based on genome analysis within Aspergilli which was proven by overexpression in Pichia pastoris (Bauer et al. 2006; de Vries et al. 2005). This GH31 family contains mainly enzymes with α-glucosidase activity and has only a limited number of characterized α-xylosidases (de Vries et al. 2005).
l-Fucose residues in xyloglucan branches are released by α-fucosidases belonging to GH29 and GH95. Several α-fucosidases of plants have been identified to degrade xyloglucan (Leonard et al. 2008; Ishimizu et al. 2007; Minic and Jouanin 2006). Nevertheless, only A. niger, Aspergillus nidulans, and Penicillium multicolor have been reported to produce an α-fucosidase which release l-fucose residues similar to the fucose-linkages in xyloglucan (Ajisaka et al. 1998; Ajisaka and Shirakabe 1992; Bauer et al. 2006).
Alpha-linked d-galactose residues are released from hemicellulose, e.g., xylan and galactomannans, by α-galactosidases belonging to GH27 and GH36. The α-galactosidases of GH27 and GH36 both act via a double-displacement mechanism and are considered to have a common evolutionary origin (Rigden 2002). Some enzymes of GH27 also showed α-N-acetylgalactosaminidase activity and it is therefore argued that some GH27 α-galactosidases are not involved in hemicellulose degradation (Kulik et al. 2010). The GH36 α-galactosidases are often larger in size and are active against mono- and oligosaccharides, such as melibiose and raffinose (Ademark et al. 2001). The presence of terminal β-linked d-galactose residues in some hemicelluloses, e.g., xylan, xyloglucan, and galactoglucomannans, suggested that β-galactosidases (GH2 and GH35) also play a role in degradation of hemicelluloses (Sims et al. 1997). This was proven by the increased release of d-galactose residues from wheat flour after supplementing LacA of A. niger to an enzyme cocktail (de Vries et al. 2000). Bga1, a β-galactosidase of T. reesei, showed broad substrate specificity with also activity against polymeric galactans (Gamauf et al. 2007). Nevertheless, the fast majority of studies concerning β-galactosidases are focused on their activity against lactose.
d-Glucuronic acid residues from polymeric xylan are released by α-glucuronidases belonging to GH67 and the newly identified family GH115 (Ryabova et al. 2009; Chong et al. 2011). Like many other carbohydrate-active enzyme families, the difference between α-glucuronidases from both families lays in their substrate specificity. GH67 α-glucuronidases are active on short oligosaccharides, while some of the GH115 α-glucuronidases are active on polymeric xylan (Chong et al. 2011; Tenkanen and Siika-aho 2000). The GH115 α-glucuronidase from Pichia stipitis showed however higher activity against short oligosaccharides, which corresponds with its ability to degrade oligosaccharides within their environment (Kolenova et al. 2010). Remarkably, GH67 α-glucuronidases are found exclusively within Ascomycetes, while GH115 α-glucuronidases are present in Ascomycetes and Basidiomycetes (Chong et al. 2011).
Acetyl residues from xylan chains are released by acetylxylan esterases belonging to CE families 1, 4, 5, and 16 (Biely et al. 2011). The presence of acetylxylan esterases is essential for efficient degradation of the xylan backbone by endoxylanases. For instance, only in the presence of A. niger acetylxylan esterase, birchwood xylan can be degraded by the three endoxylanases and the β-xylosidase of A. niger (Kormelink et al. 1993). The main difference between the CE families is their preference for hydrolyzing the different O-linked acetyl groups. CE families 1, 4, and 5 have a strong preference for 2-O-linked residues, the most common linkage in hemicellulose, while CE16 prefers 3-O- and 4-O-linked residues (Li et al. 2008; Biely et al. 2011). There has also been a description of acetyl esterase of A. oryzae active against acetyl residues attached to galactomannan chains (Tenkanen et al. 1995). However, the gene encoding this specific enzyme has not yet been identified nor characterized in other fungi.
p-Coumaric acid and ferulic acid, the two cinnamic acids present in xylan, are removed by feruloyl/p-coumaroyl esterases. Most of these esterases have not been assigned to CE families. However, several classifications have been reported for these enzymes based on sequence similarity and substrate specificity (Crepin et al. 2004; Olivares-Hernandez et al. 2010; Benoit et al. 2008). One particular group of esterases, belonging to Aspergillus and Penicillium sp., has preference for substrates with methoxy substituents (Koseki et al. 2009; Kroon et al. 1997). For example, FaeA of A. niger prefers substrates with a methoxy group at position three, such as ferulic acid (Benoit et al. 2007; Kroon et al. 1997). FaeB of A. niger belongs to another group of esterases with preference for substrates containing one or two hydroxyl substitutions, such as p-coumaric acid (de Vries et al. 1997; Koseki et al. 2009; Kroon et al. 1997).
Pectin degradation
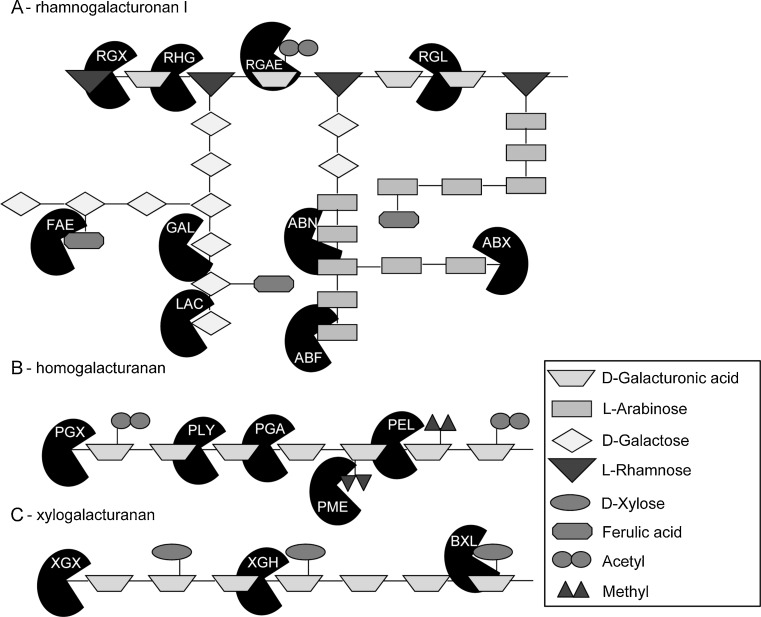
The degradation of pectin backbones (Fig. 3) requires two classes of enzymes: glycoside hydrolases and polysaccharide lyases (PL; Table 5). A large part of the fungal glycoside hydrolases involved in the degradation of the pectin backbone belongs to GH family 28 (Martens-Uzunova and Schaap 2009). These GH28 enzymes can be divided in groups according to the specific pectin region they attack: endo- and exo-polygalacturonase (GH28) cleave the backbone of the smooth regions, while the more intricate, hairy regions are further attacked by endo- and exo-rhamnogalacturonase (GH28), xylogalacturonase (GH28), α-rhamnosidases (GH78), unsaturated glucuronyl hydrolases (GH88), and unsaturated rhamnogalacturonan hydrolases (GH105).
Fig. 3.
a–c Schematic structures of three pectins, rhamnogalacturonan I, homogalacturonan, xylogalacturonan, with pectinolytic enzymes. ABF α-arabinofuranosidase, ABN endoarabinanase, ABX exoarabinanase, BXL β-1,4-xylosidase, FAE feruloyl esterase, GAL β-1,4-endogalactanase, LAC β-galactosidase, PEL pectin lyase, PLY pectate lyase, PGA endopoly-galacturonase, PGX exo-polygalacturonase, PME pectin methyl esterase, RGAE rhamnogalacturonan acetyl esterase, RGL rhamnogalacturonan lyase, RHG endorham-nogalacturonase, RGX exorham-nogalacturonase, XGH endoxylo-galacturonase, XGX exoxylogalacturonase. α-Rhamnosidase (RHA), unsaturated rhamnogalacturonase (URH), and unsaturated glucuronyl hydrolase (UGH) are not depicted in this figure
Table 5.
Genes encoding characterized and putative enzymes of Aspergillus niger CBS513.88 involved in the degradation of pectin (Coutinho et al. 2009; Martens-Uzunova and Schaap 2009)
| Enzyme class | Code | CAZy families | Genes of characterized and putative enzymes | Reference |
|---|---|---|---|---|
| Endopoly-galacturonase | PGA | GH28 | An16g06990 (pgaA), An02g04900 (pgaB), An05g02440 (pgaC), An09g03260 (pgaD), An01g14670 (pgaE), An01g11520 (pgaI), An15g05370 (pgaII) | (Parenicova et al. 2000b; Parenicova et al. 2000a; Bussink et al. 1992; Parenicova et al. 1998; Kester and Visser 1990) |
| Exopoly-galacturonase | PGX | GH28 | An11g04040 (pgxA), An03g06740 (pgxB), An02g12450 (pgxC), An12g07500 (pgaX) | (Martens-Uzunova et al. 2006) |
| Endorhamno-galacturonase | RHG | GH28 | An12g00950 (rhgA), An14g04200 (rhgB), An06g02070, An11g06320, An11g08700, An07g01000 | (Suykerbuyk et al. 1997) |
| Exorhamno-galacturonase | RGX | GH28 | An01g14650 (rgxA), An03g02080 (rgxB), An18g04810 (rgxC) | (Martens-Uzunova et al. 2006) |
| Endoxylo-galacturonase | XGH | GH28 | An04g09700 (xghA) | (van der Vlugt-Bergmans et al. 2000) |
| α-rhamnosidase | RHA | GH78 | An15g04530, An01g06620, An10g00290, An08g09140, An12g05700, An07g00240, An04g09070, An18g04800 | |
| Endoarabinanase | ABN | GH43 | An09g01190 (abnA), An16g02730, An02g01400, An07g04930, An02g10550 | (Flipphi et al. 1993a) |
| Exoarabinanase | ABX | GH93 | 49311a | |
| β-1,4-endogalactanase | GAL | GH53 | An18g05940 (galA), An16g06590 | (de Vries et al. 2002b) |
| Unsaturated glucuronyl hydrolase | UGH | GH88 | An01g01340 | |
| Unsaturated rhamnogalacturonase | URH | GH105 | An14g05340, An14g02920 | |
| Pectin methyl esterase | PME | CE8 | An03g06310 (pmeA),An04g09690, An02g12505 | (Khanh et al. 1991) |
| Rhamnogalacturonanacetyl esterase | RGAE | CE12 | An09g02160 (rgaeA), An04g09360 | (de Vries et al. 2000) |
| Pectin lyase | PEL | PL1 | An14g04370 (pelA), An03g00190 (pelB), An11g04030 (pelC), An19g00270 (pelD), An15g07160 (pelF) | (Harmsen et al. 1990; Kusters-van Someren et al. 1992; Gysler et al. 1990; de Vries et al. 2002a) |
| Pectate lyase | PLY | PL1 | An10g00870 (plyA) | (Benen et al. 2000) |
| PL3 and PL9 | – | |||
| Rhamnogalacturonan | RGL | PL4 | An14g01130 (rglA), An11g00390 | (de Vries et al. 2002a) |
| Lyase | PL11 | – |
The genes with names between brackets are biochemically characterized and their references are given in the last column
aExoarabinanase has only been identified in A. niger isolate ATCC1015
Endo- and exo-polygalacturonases of GH28 in general cleave the α-1,4-glycosidic bonds between the α-galacturonic acids. The genome of A. niger contains seven endopolygalacturonases, each of them exhibiting distinct kinetic properties, substrate methylation sensitivity and mode of action (Martens-Uzunova and Schaap 2009; Benen et al. 2000; Bussink et al. 1991; Parenicova et al. 1998, 2000a). For instance, although the structures of PgaI and PgaII are highly similar, only PgaI has enzyme processivity due to a narrower substrate binding cleft and the presence of an arginine at position 96 (van Santen et al. 1999; van Pouderoyen et al. 2003). The genome of A. niger contains four potential exopolygalacturonases: PgaX, PgxA, PgxB, and PgxC (Martens-Uzunova et al. 2006). Of these, PgxB prefers homogalacturonan as a substrate, while PgxC has high activity against homogalacturonan and xylogalacturonan (Martens-Uzunova et al. 2006). PgxA has a low activity against homogalacturonan and is more active against xylogalacturonan. Therefore, PgxA very likely represents a new kind of exoxylogalacturanase (Martens-Uzunova et al. 2006). An endo-acting xylogalacturanase, XghA, was already described to specifically degrade the xylogalacturonan present in the hairy regions (van der Vlugt-Bergmans et al. 2000). Also within R. oryzae, 15 of the 18 putative GH28 polygalacturonases showed different substrate specificity for polygalacturonan, trigalacturonan, and digalacturonan, and their activity against polygalacturonan ranged from less than 1 to 200 μmol/min/mg protein (Mertens and Bowman 2011). These different characteristics between polygalacturonases of A. niger and R. oryzae shows the requirement for a large number of isoenzymes to concertedly act on complex plant polysaccharides.
Another important group of GH28 are the rhamnogalacturonan hydrolases, which employ either an endo- or exolytic mechanism to cleave the α-1,2-glycosidic bonds formed between d-galacturonic acid and l-rhamnose residues in the hairy regions (Kofod et al. 1994; Suykerbuyk et al. 1995). A. niger has two characterized (RhgA and RhgB) and four putative endorhamnogalacturonanases (Martens-Uzunova and Schaap 2009; Suykerbuyk et al. 1997). The pattern of reaction products produced after degradation of modified hairy regions with either RhgA or RhgB is quite different, suggesting that each enzyme acts on a structurally different region of the substrate (Suykerbuyk et al. 1997). Exorhamnogalacturonases release d-galacturonic acid residues from the non-reducing end of rhamnogalacturonan chains but not from homogalacturonans (Mutter et al. 1998). Sequence analysis of the A. niger genome indicates the presence of three genes, RgxA, RgxB, and RgxC, encoding putative exorhamnogalacturonases (Martens-Uzunova et al. 2006).
Hydrolysis of the pectin backbone also requires enzymes from other GH families: α-rhamnosidases (GH78), unsaturated glucuronyl hydrolases (GH88), and unsaturated rhamnogalacturonan hydrolases (GH105). As several of these enzymes and families have only recently been described, little biochemical characterization has been performed on them (see Table 5 for the putative genes of A. niger). For example, characterization of unsaturated glucuronyl hydrolases and unsaturated rhamnogalacturonan hydrolases is lacking in fungi, although several putative enzymes have been identified.
Pectin and pectate lyases both cleave, via a β-elimination mechanism, the α-1,4-linked d-galacturonic acid residues within the smooth regions of pectin (Lombard et al. 2010). A comparison between the structures of pectin and pectate lyases has indicated that both lyases most likely descended from a common ancestor enzyme (Mayans et al. 1997; Vitali et al. 1998). However, both types of lyases have important differences in their active site. As a consequence, pectin lyases attack preferentially heavily methyl-esterified substrates and have their optimum pH around 5.5 (Mayans et al. 1997). In contrast, pectate lyases favor lower degrees of esterification, have their optimum pH around 8.5, and require Ca2+ for their activity (Mayans et al. 1997). Currently, all characterized pectin lyases belong to the PL1 family, while the fungal pectate lyases belong to PL1, PL3, and PL9. As an example, six pectin lyases and only one pectate lyase have been identified and partially characterized in A. niger (Benen et al. 2000; Harmsen et al. 1990; Gysler et al. 1990). In contrast, A. nidulans has only two pectin lyases, five identified pectate lyases, and another six putative pectate lyases (Galagan et al. 2005; Bauer et al. 2006), suggesting significant differences between these fungi (Coutinho et al. 2009).
Rhamnogalacturonan lyases differ substantially in their structure from pectin and pectate lyases, and cleave within the hairy regions of pectin. This group of lyases belongs to two families, PL4 and PL11, where PL4 lyases have a much lower optimum pH then PL11 lyases (Jensen et al. 2010). The PL4 rhamnogalacturonan lyase from Aspergillus aculeatus showed that cleavage preferably occurs four residues from l-rhamnose at the reducing end and is severely affected by the presence of acetyl groups in the backbone of rhamnogalacturonan (Mutter et al. 1998; de Vries et al. 2000). Therefore, the cooperative action of rhamnogalacturonan acetyl-esterases is required for efficient degradation (de Vries et al. 2000).
The pectin structures xylogalacturonan and rhamnogalacturonan also require accessory enzymes to remove the side chains and provide access for the main-chain hydrolysing pectinolytic enzymes. Of these, α-arabinofuranosidases (GH51 and GH54), β-galactosidases (GH2 and GH35), and β-xylosidases (GH3 and GH43) are also needed for hemicellulose degradation, while endoarabinanases (GH43), exoarabinanases (GH93), β-endogalactanases (GH53), and several esterases (CE8, CE12, and CE13) are specific for pectin degradation (Martens-Uzunova and Schaap 2009).
Future prospects
Many industrial processes make use of relative easy accessible sugar residues within cellulose or other plant polysaccharides. In industrial processes, the monomeric sugars within these structures are released from the plant material by a limited set of enzymes, utilizing only a small fraction of the available biodiversity of fungal enzymes. For instance, a minimal mixture of cellobiohydrolases (CBHI and CBHII) and endoglucanase (EGI) from T. reesei and β-glucosidase from A. niger is sufficient to efficiently degrade pure cellulose (Sternberg et al. 1977; Rosgaard et al. 2007a). Exploitation of the available sugars within the intricate fractions of plant biomass involves a more elaborate process. For example, plant polysaccharides have to be separated from the present lignin compounds by chemical, physical, or biological pretreatment (Hendriks and Zeeman 2009; Martinez et al. 2009). And, as described in the previous sections, releasing the sugars from the different plant polysaccharides requires a more complex set of carbohydrate-active enzymes. In addition to a number of cellulases, efficient enzymatic hydrolysis of pretreated plant biomass also needs synergistic activity of, at least, several endoxylanases, β-xylosidase, α-arabinofuranosidase, and acetyl esterase (Kormelink et al. 1993; Alvira et al. 2011; Gao et al. 2010). The precise mixture of hydrolytic enzymes also depends on the method of pretreatment and type of plant substrate (Rosgaard et al. 2007b; Saha and Cotta 2010). Nearly complete hydrolysis of specific xylan and pectin fractions can still be achieved with a relative small sets of enzymes (de Vries et al. 2000), but this is not the case for more crude biomass fractions. For this reason, a better understanding of plant polysaccharide degradation will help to design an enzyme mixture which can efficiently degrade a wide range of substrates.
The division of carbohydrate-active enzymes into different families based on modules of amino acid conservation (www.cazy.org) can help with a better understanding of the enzymatic repertoire of different fungi. In recent years, new families have been described and new activities have been assigned to known families (Martens-Uzunova et al. 2006; Cantarel et al. 2009; Li et al. 2008; Chong et al. 2011; Harris et al. 2010). Nevertheless, more knowledge is required to fully profit from this large database of information. The biggest limitation is the low coverage of biochemical information on specific enzyme classes. This is illustrated by the small amount of enzymatically characterized proteins that has been added to the CAZy database in the last decade in contrast to the large quantity of putative carbohydrate-active enzymes of sequenced fungal genomes (Cantarel et al. 2009). As a consequence, some enzyme activities are putatively assigned to families, but do not have biochemical support. For example, none of the many putative fungal unsaturated glucuronyl hydrolases (GH88) and unsaturated rhamnogalacturonan hydrolases (GH105) have been biochemically characterized. Another challenge is the large carbohydrate-active enzyme families containing enzymes with different enzyme classes. With increasing biochemical information, some families like GH2, GH3, GH5, GH28, or GH43 could be split up in smaller and better defined sub-families according to their hydrolytic function. This will allow a better predictive value of future fungal genome annotations, and thereby will lead to an even larger toolbox to access for industrial purposes.
Acknowledgments
JvdB was supported by a grant of the The Netherlands Organisation for Scientific Research (NWO) of the China–Netherlands Joint Scientific Thematic Research Programme (jstp.10.005) to RPdV.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ademark P, Varga A, Medve J, Harjunpaa V, Drakenberg T, Tjerneld F, Stalbrand H. Softwood hemicellulose-degrading enzymes from Aspergillus niger: purification and properties of a beta-mannanase. J Biotechnol. 1998;63(3):199–210. doi: 10.1016/S0168-1656(98)00086-8. [DOI] [PubMed] [Google Scholar]
- Ademark P, de Vries RP, Hagglund P, Stalbrand H, Visser J. Cloning and characterization of Aspergillus niger genes encoding an alpha-galactosidase and a beta-mannosidase involved in galactomannan degradation. Eur J Biochem. 2001;268(10):2982–2990. doi: 10.1046/j.1432-1327.2001.02188.x. [DOI] [PubMed] [Google Scholar]
- Ajisaka K, Shirakabe M. Regioselective synthesis of alpha-l-fucosyl-containing disaccharides by use of alpha-l-fucosidases of various origins. Carbohydr Res. 1992;224:291–299. doi: 10.1016/0008-6215(92)84115-9. [DOI] [PubMed] [Google Scholar]
- Ajisaka K, Fujimoto H, Miyasato M. An alpha-l-fucosidase from Penicillium multicolor as a candidate enzyme for the synthesis of alpha (1–>3)-linked fucosyl oligosaccharides by transglycosylation. Carbohydr Res. 1998;309(1):125–129. doi: 10.1016/S0008-6215(98)00112-8. [DOI] [PubMed] [Google Scholar]
- Alvira P, Negro MJ, Ballesteros M. Effect of endoxylanase and alpha-l-arabinofuranosidase supplementation on the enzymatic hydrolysis of steam exploded wheat straw. Bioresour Technol. 2011;102(6):4552–4558. doi: 10.1016/j.biortech.2010.12.112. [DOI] [PubMed] [Google Scholar]
- Angel Siles Lopez J, Li Q, Thompson IP. Biorefinery of waste orange peel. Crit Rev Biotechnol. 2010;30(1):63–69. doi: 10.3109/07388550903425201. [DOI] [PubMed] [Google Scholar]
- Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genomics. 2011;12:38. doi: 10.1186/1471-2164-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci U S A. 2006;103(30):11417–11422. doi: 10.1073/pnas.0604632103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham GT, Matthews JF, Bomble YJ, Bu L, Adney WS, Himmel ME, Nimlos MR, Crowley MF. Identification of amino acids responsible for processivity in a Family 1 carbohydrate-binding module from a fungal cellulase. J Phys Chem B. 2010;114(3):1447–1453. doi: 10.1021/jp908810a. [DOI] [PubMed] [Google Scholar]
- Benen JA, Kester HC, Parenicova L, Visser J. Characterization of Aspergillus niger pectate lyase A. Biochemistry. 2000;39(50):15563–15569. doi: 10.1021/bi000693w. [DOI] [PubMed] [Google Scholar]
- Benoit I, Coutard B, Oubelaid R, Asther M, Bignon C. Expression in Escherichia coli, refolding and crystallization of Aspergillus niger feruloyl esterase A using a serial factorial approach. Protein Expr Purif. 2007;55(1):166–174. doi: 10.1016/j.pep.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Benoit I, Danchin EG, Bleichrodt RJ, de Vries RP. Biotechnological applications and potential of fungal feruloyl esterases based on prevalence, classification and biochemical diversity. Biotechnol Lett. 2008;30(3):387–396. doi: 10.1007/s10529-007-9564-6. [DOI] [PubMed] [Google Scholar]
- Bezerra RM, Dias AA. Enzymatic kinetic of cellulose hydrolysis: inhibition by ethanol and cellobiose. Appl Biochem Biotechnol. 2005;126(1):49–59. doi: 10.1007/s12010-005-0005-5. [DOI] [PubMed] [Google Scholar]
- Biely P, Vrsanska M, Tenkanen M, Kluepfel D. Endo-beta-1,4-xylanase families: differences in catalytic properties. J Biotechnol. 1997;57(1–3):151–166. doi: 10.1016/S0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- Biely P, Mastihubova M, Tenkanen M, Eyzaguirre J, Li XL, Vrsanska M. Action of xylan deacetylating enzymes on monoacetyl derivatives of 4-nitrophenyl glycosides of beta-d-xylopyranose and alpha-l-arabinofuranose. J Biotechnol. 2011;151(1):137–142. doi: 10.1016/j.jbiotec.2010.10.074. [DOI] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382(Pt 3):769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink HJ, Brouwer KB, de Graaff LH, Kester HC, Visser J. Identification and characterization of a second polygalacturonase gene of Aspergillus niger. Curr Genet. 1991;20(4):301–307. doi: 10.1007/BF00318519. [DOI] [PubMed] [Google Scholar]
- Bussink HJ, Buxton FP, Fraaye BA, de Graaff LH, Visser J. The polygalacturonases of Aspergillus niger are encoded by a family of diverged genes. Eur J Biochem. 1992;208(1):83–90. doi: 10.1111/j.1432-1033.1992.tb17161.x. [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344(14):1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3(1):1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Hayn M, Esterbauer H. Purification and characterization of two extracellular beta-glucosidases from Trichoderma reesei. Biochim Biophys Acta. 1992;1121(1–2):54–60. doi: 10.1016/0167-4838(92)90336-C. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(6632 Suppl):67–73. doi: 10.1038/387s067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SL, Battaglia E, Coutinho PM, Henrissat B, Tenkanen M, de Vries RP. The alpha-glucuronidase Agu1 from Schizophyllum commune is a member of a novel glycoside hydrolase family (GH115) Appl Microbiol Biotechnol. 2011;90(4):1323–1332. doi: 10.1007/s00253-011-3157-y. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Andersen MR, Kolenova K, vanKuyk PA, Benoit I, Gruben BS, Trejo-Aguilar B, Visser H, van Solingen P, Pakula T, Seiboth B, Battaglia E, Aguilar-Osorio G, de Jong JF, Ohm RA, Aguilar M, Henrissat B, Nielsen J, Stalbrand H, de Vries RP. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(Suppl 1):S161–S169. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Crepin VF, Faulds CB, Connerton IF. Functional classification of the microbial feruloyl esterases. Appl Microbiol Biotechnol. 2004;63(6):647–652. doi: 10.1007/s00253-003-1476-3. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, Baker SE, Rep M, Adam G, Antoniw J, Baldwin T, Calvo S, Chang YL, Decaprio D, Gale LR, Gnerre S, Goswami RS, Hammond-Kosack K, Harris LJ, Hilburn K, Kennell JC, Kroken S, Magnuson JK, Mannhaupt G, Mauceli E, Mewes HW, Mitterbauer R, Muehlbauer G, Munsterkotter M, Nelson D, O’Donnell K, Ouellet T, Qi W, Quesneville H, Roncero MI, Seong KY, Tetko IV, Urban M, Waalwijk C, Ward TJ, Yao J, Birren BW, Kistler HC. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317(5843):1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- Dan S, Marton I, Dekel M, Bravdo BA, He S, Withers SG, Shoseyov O. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger beta-glucosidase. J Biol Chem. 2000;275(7):4973–4980. doi: 10.1074/jbc.275.7.4973. [DOI] [PubMed] [Google Scholar]
- de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001;65(4):497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Michelsen B, Poulsen CH, Kroon PA, van den Heuvel RH, Faulds CB, Williamson G, van den Hombergh JP, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63(12):4638–4644. doi: 10.1128/AEM.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, van den Broeck HC, Dekkers E, Manzanares P, de Graaff LH, Visser J. Differential expression of three alpha-galactosidase genes and a single beta-galactosidase gene from Aspergillus niger. Appl Environ Microbiol. 1999;65(6):2453–2460. doi: 10.1128/AEM.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Kester HC, Poulsen CH, Benen JA, Visser J. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr Res. 2000;327(4):401–410. doi: 10.1016/S0008-6215(00)00066-5. [DOI] [PubMed] [Google Scholar]
- de Vries RP, Jansen J, Aguilar G, Parenicova L, Joosten V, Wulfert F, Benen JA, Visser J. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 2002;530(1–3):41–47. doi: 10.1016/S0014-5793(02)03391-4. [DOI] [PubMed] [Google Scholar]
- de Vries RP, Parenicova L, Hinz SW, Kester HC, Beldman G, Benen JA, Visser J. The beta-1,4-endogalactanase A gene from Aspergillus niger is specifically induced on arabinose and galacturonic acid and plays an important role in the degradation of pectic hairy regions. Eur J Biochem. 2002;269(20):4985–4993. doi: 10.1046/j.1432-1033.2002.03199.x. [DOI] [PubMed] [Google Scholar]
- de Vries RP, van de Vondervoort PJ, Hendriks L, van de Belt M, Visser J. Regulation of the alpha-glucuronidase-encoding gene (aguA) from Aspergillus niger. Mol Genet Genomics. 2002;268(1):96–102. doi: 10.1007/s00438-002-0729-7. [DOI] [PubMed] [Google Scholar]
- de Vries RP, vanKuyk PA, Kester HC, Visser J. The Aspergillus niger faeB gene encodes a second feruloyl esterase involved in pectin and xylan degradation and is specifically induced in the presence of aromatic compounds. Biochem J. 2002;363(Pt 2):377–386. doi: 10.1042/bj3630377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, van Grieken C, VanKuyk PA, Wosten HA. The value of genome sequences in the rapid identification of novel genes encoding specific plant cell wall degrading enzymes. Curr Genomics. 2005;6:157–187. doi: 10.2174/1389202053971974. [DOI] [Google Scholar]
- de Vries RP, Nadal M, van den Brink J, Vivas-Duarte DA, Stålbrand H. Fungal degradation of plant oligo- and polysaccharides. In: Grunwald P, editor. Carbohydrate modifying enzymes and microorganisms. Singapore: Pan Stanford publishing Pte. Ltd.; 2011. [Google Scholar]
- Decker CH, Visser J, Schreier P. Beta-glucosidases from five black Aspergillus species: study of their physico-chemical and biocatalytic properties. J Agric Food Chem. 2000;48(10):4929–4936. doi: 10.1021/jf000434d. [DOI] [PubMed] [Google Scholar]
- Decker CH, Visser J, Schreier P. Beta-glucosidase multiplicity from Aspergillus tubingensis CBS 643.92: purification and characterization of four beta-glucosidases and their differentiation with respect to substrate specificity, glucose inhibition and acid tolerance. Appl Microbiol Biotechnol. 2001;55(2):157–163. doi: 10.1007/s002530000462. [DOI] [PubMed] [Google Scholar]
- den Herder IF, Rosell AM, van Zuilen CM, Punt PJ, van den Hondel CA. Cloning and expression of a member of the Aspergillus niger gene family encoding alpha-galactosidase. Mol Gen Genet. 1992;233(3):404–410. doi: 10.1007/BF00265437. [DOI] [PubMed] [Google Scholar]
- Desmet T, Cantaert T, Gualfetti P, Nerinckx W, Gross L, Mitchinson C, Piens K. An investigation of the substrate specificity of the xyloglucanase Cel74A from Hypocrea jecorina. FEBS J. 2007;274(2):356–363. doi: 10.1111/j.1742-4658.2006.05582.x. [DOI] [PubMed] [Google Scholar]
- Dias FM, Vincent F, Pell G, Prates JA, Centeno MS, Tailford LE, Ferreira LM, Fontes CM, Davies GJ, Gilbert HJ. Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J Biol Chem. 2004;279(24):25517–25526. doi: 10.1074/jbc.M401647200. [DOI] [PubMed] [Google Scholar]
- Do BC, Dang TT, Berrin JG, Haltrich D, To KA, Sigoillot JC, Yamabhai M. Cloning, expression in Pichia pastoris, and characterization of a thermostable GH5 mannan endo-1,4-beta-mannosidase from Aspergillus niger BK01. Microb Cell Fact. 2009;8:59. doi: 10.1186/1475-2859-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Kiyonari S, Mackie RI, Cann IK. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J Bacteriol. 2010;192(9):2335–2345. doi: 10.1128/JB.01654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebringerova A, Hromadkova Z, Petrakova E, Hricovini M. Structural features of a water-soluble l-arabino-d-xylan from rye bran. Carbohydr Res. 1990;198(1):57–66. doi: 10.1016/0008-6215(90)84276-Z. [DOI] [PubMed] [Google Scholar]
- Eriksson T, Karlsson J, Tjerneld F. A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (cel7A) and endoglucanase I (cel7B) of Trichoderma reesei. Appl Biochem Biotechnol. 2002;101(1):41–60. doi: 10.1385/ABAB:101:1:41. [DOI] [PubMed] [Google Scholar]
- Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury JM, Segurens B, Poulain J, Anthouard V, Grossetete S, Khalili H, Coppin E, Dequard-Chablat M, Picard M, Contamine V, Arnaise S, Bourdais A, Berteaux-Lecellier V, Gautheret D, de Vries RP, Battaglia E, Coutinho PM, Danchin EG, Henrissat B, Khoury RE, Sainsard-Chanet A, Boivin A, Pinan-Lucarre B, Sellem CH, Debuchy R, Wincker P, Weissenbach J, Silar P. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9(5):R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flipphi MJ, Panneman H, van der Veen P, Visser J, de Graaff LH. Molecular cloning, expression and structure of the endo-1,5-alpha-l-arabinase gene of Aspergillus niger. Appl Microbiol Biotechnol. 1993;40(2–3):318–326. doi: 10.1007/BF00170387. [DOI] [PubMed] [Google Scholar]
- Flipphi MJ, van Heuvel M, van der Veen P, Visser J, de Graaff LH. Cloning and characterization of the abfB gene coding for the major alpha-l-arabinofuranosidase (AbfB) of Aspergillus niger. Curr Genet. 1993;24(6):525–532. doi: 10.1007/BF00351717. [DOI] [PubMed] [Google Scholar]
- Flipphi MJ, Visser J, van der Veen P, de Graaff LH. Cloning of the Aspergillus niger gene encoding alpha-l-arabinofuranosidase A. Appl Microbiol Biotechnol. 1993;39(3):335–340. doi: 10.1007/BF00192088. [DOI] [PubMed] [Google Scholar]
- Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, Meerman HJ, Mitchell T, Mitchinson C, Olivares HA, Teunissen PJ, Yao J, Ward M. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278(34):31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422(6934):859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438(7071):1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gamauf C, Marchetti M, Kallio J, Puranen T, Vehmaanperä J, Allmaier G, Kubicek CP, Seiboth B. Characterization of the bga1-encoded glycoside hydrolase family 35 beta-galactosidase of Hypocrea jecorina with galacto-beta-d-galactanase activity. FEBS J. 2007;274(7):1691–1700. doi: 10.1111/j.1742-4658.2007.05714.x. [DOI] [PubMed] [Google Scholar]
- Gao D, Uppugundla N, Chundawat SP, Yu X, Hermanson S, Gowda K, Brumm P, Mead D, Balan V, Dale BE. Hemicellulases and auxiliary enzymes for improved conversion of lignocellulosic biomass to monosaccharides. Biotechnol Biofuels. 2010;4:5. doi: 10.1186/1754-6834-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielkens MM, Visser J, de Graaff LH. Arabinoxylan degradation by fungi: characterization of the arabinoxylan-arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr Genet. 1997;31(1):22–29. doi: 10.1007/s002940050172. [DOI] [PubMed] [Google Scholar]
- Gielkens MM, Dekkers E, Visser J, de Graaff LH. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1999;65(10):4340–4345. doi: 10.1128/AEM.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HJ, Stalbrand H, Brumer H. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11(3):338–348. doi: 10.1016/j.pbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Grishutin SG, Gusakov AV, Markov AV, Ustinov BB, Semenova MV, Sinitsyn AP. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim Biophys Acta. 2004;1674(3):268–281. doi: 10.1016/j.bbagen.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Bothast RJ (1994) Pectin rich residues generated by processing of citrus fruits, apples, and sugar beets. Enzymatic hydrolysis and biological conversion to value-added products. In: Enzymatic Conversion of Biomass for Fuels Production. Oxford University Press, Oxford, pp 372–390
- Guillen D, Sanchez S, Rodriguez-Sanoja R. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol. 2010;85(5):1241–1249. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- Gysler C, Harmsen JA, Kester HC, Visser J, Heim J. Isolation and structure of the pectin lyase D-encoding gene from Aspergillus niger. Gene. 1990;89(1):101–108. doi: 10.1016/0378-1119(90)90211-9. [DOI] [PubMed] [Google Scholar]
- Ha SJ, Galazka JM, Kim SR, Choi JH, Yang X, Seo JH, Glass NL, Cate JH, Jin YS. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci U S A. 2011;108(2):504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen JA, Kusters-van Someren MA, Visser J. Cloning and expression of a second Aspergillus niger pectin lyase gene (pelA): indications of a pectin lyase gene family in A. niger. Curr Genet. 1990;18(2):161–166. doi: 10.1007/BF00312604. [DOI] [PubMed] [Google Scholar]
- Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49(15):3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJ, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68(4):1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks AT, Zeeman G. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol. 2009;100(1):10–18. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293(3):781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Teeri TT, Warren RA. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425(2):352–354. doi: 10.1016/S0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- Herve C, Rogowski A, Blake AW, Marcus SE, Gilbert HJ, Knox JP. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A. 2010;107(34):15293–15298. doi: 10.1073/pnas.1005732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzapple M, Cognata M, Shu Y, Hendrickson C. Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol Bioeng. 1990;36(3):275–287. doi: 10.1002/bit.260360310. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Hashimoto C, Takeda R, Fujii K, Hase S. A novel alpha1,2-l-fucosidase acting on xyloglucan oligosaccharides is associated with endo-beta-mannosidase. J Biochem. 2007;142(6):721–729. doi: 10.1093/jb/mvm186. [DOI] [PubMed] [Google Scholar]
- Ito T, Yokoyama E, Sato H, Ujita M, Funaguma T, Furukawa K, Hara A. Xylosidases associated with the cell surface of Penicillium herquei IFO 4674. J Biosci Bioeng. 2003;96(4):354–359. doi: 10.1016/S1389-1723(03)90136-8. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H, Jin YS, Passoth V, Richardson PM. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25(3):319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- Jensen MH, Otten H, Christensen U, Borchert TV, Christensen LL, Larsen S, Leggio LL. Structural and biochemical studies elucidate the mechanism of rhamnogalacturonan lyase from Aspergillus aculeatus. J Mol Biol. 2010;404(1):100–111. doi: 10.1016/j.jmb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Jorgensen H, Vibe-Pedersen J, Larsen J, Felby C. Liquefaction of lignocellulose at high-solids concentrations. Biotechnol Bioeng. 2007;96(5):862–870. doi: 10.1002/bit.21115. [DOI] [PubMed] [Google Scholar]
- Kester HC, Visser J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem. 1990;12(2):150–160. [PubMed] [Google Scholar]
- Khanh NQ, Ruttkowski E, Leidinger K, Albrecht H, Gottschalk M. Characterization and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene. 1991;106(1):71–77. doi: 10.1016/0378-1119(91)90567-U. [DOI] [PubMed] [Google Scholar]
- Kofod LV, Kauppinen S, Christgau S, Andersen LN, Heldt-Hansen HP, Dorreich K, Dalboge H. Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J Biol Chem. 1994;269(46):29182–29189. [PubMed] [Google Scholar]
- Kolenova K, Ryabova O, Vrsanska M, Biely P. Inverting character of family GH115 alpha-glucuronidases. FEBS Lett. 2010;584(18):4063–4068. doi: 10.1016/j.febslet.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Kolpak FJ, Blackwell J. Determination of the structure of cellulose II. Macromolecules. 1976;9(2):273–278. doi: 10.1021/ma60050a019. [DOI] [PubMed] [Google Scholar]
- Kormelink FJ, Gruppen H, Vietor RJ, Voragen AG. Mode of action of the xylan-degrading enzymes from Aspergillus awamori on alkali-extractable cereal arabinoxylans. Carbohydr Res. 1993;249(2):355–367. doi: 10.1016/0008-6215(93)84100-K. [DOI] [PubMed] [Google Scholar]
- Koseki T, Fushinobu S, Ardiansyah SH, Komai M. Occurrence, properties, and applications of feruloyl esterases. Appl Microbiol Biotechnol. 2009;84(5):803–810. doi: 10.1007/s00253-009-2148-8. [DOI] [PubMed] [Google Scholar]
- Krengel U, Dijkstra BW. Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. J Mol Biol. 1996;263(1):70–78. doi: 10.1006/jmbi.1996.0556. [DOI] [PubMed] [Google Scholar]
- Kristensen JB, Felby C, Jorgensen H. Yield-determining factors in high-solids enzymatic hydrolysis of lignocellulose. Biotechnol Biofuels. 2009;2(1):11. doi: 10.1186/1754-6834-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon PA, Faulds CB, Brezillon C, Williamson G. Methyl phenylalkanoates as substrates to probe the active sites of esterases. Eur J Biochem. 1997;248(1):245–251. doi: 10.1111/j.1432-1033.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Doehren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gomez-Rodriguez EY, Gruber S, Han C, Henrissat B, Hermosa R, Hernandez-Onate M, Karaffa L, Kosti I, Le Crom S, Lindquist E, Lucas S, Lubeck M, Lubeck PS, Margeot A, Metz B, Misra M, Nevalainen H, Omann M, Packer N, Perrone G, Uresti-Rivera EE, Salamov A, Schmoll M, Seiboth B, Shapiro H, Sukno S, Tamayo-Ramos JA, Tisch D, Wiest A, Wilkinson HH, Zhang M, Coutinho PM, Kenerley CM, Monte E, Baker SE, Grigoriev IV. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12(4):R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik N, Weignerova L, Filipi T, Pompach P, Novak P, Mrazek H, Slamova K, Bezouska K, Kren V, Ettrich R. The alpha-galactosidase type A gene aglA from Aspergillus niger encodes a fully functional alpha-N-acetylgalactosaminidase. Glycobiology. 2010;20(11):1410–1419. doi: 10.1093/glycob/cwq105. [DOI] [PubMed] [Google Scholar]
- Kumar V, Ramakrishnan S, Teeri TT, Knowles JK, Hartley BS. Saccharomyces cerevisiae cells secreting an Aspergillus niger beta-galactosidase grow on whey permeate. Biotechnology (N Y) 1992;10(1):82–85. doi: 10.1038/nbt0192-82. [DOI] [PubMed] [Google Scholar]
- Kusters-van Someren M, Flipphi M, de Graaff L, van den Broeck H, Kester H, Hinnen A, Visser J. Characterization of the Aspergillus niger pelB gene: structure and regulation of expression. Mol Gen Genet. 1992;234(1):113–120. doi: 10.1007/BF00272352. [DOI] [PubMed] [Google Scholar]
- Leonard R, Pabst M, Bondili JS, Chambat G, Veit C, Strasser R, Altmann F. Identification of an Arabidopsis gene encoding a GH95 alpha1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry. 2008;69(10):1983–1988. doi: 10.1016/j.phytochem.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Asther M, Record E. Overproduction and characterization of xylanase B from Aspergillus niger. Can J Microbiol. 2005;51(2):177–183. doi: 10.1139/w04-116. [DOI] [PubMed] [Google Scholar]
- Li XL, Skory CD, Cotta MA, Puchart V, Biely P. Novel family of carbohydrate esterases, based on identification of the Hypocrea jecorina acetyl esterase gene. Appl Environ Microbiol. 2008;74(24):7482–7489. doi: 10.1128/AEM.00807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Du J, Sun J, Galazka JM, Glass NL, Cate JH, Yang X, Zhao H. Overcoming glucose repression in mixed sugar fermentation by co-expressing a cellobiose transporter and a beta-glucosidase in Saccharomyces cerevisiae. Mol Biosyst. 2010;6(11):2129–2132. doi: 10.1039/c0mb00063a. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Baker JO, Zeng Y, Himmel ME, Haas T, Ding SY. Cellobiohydrolase hydrolyzes crystalline cellulose on hydrophobic faces. J Biol Chem. 2011;286(13):11195–11201. doi: 10.1074/jbc.M110.216556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438(7071):1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Margolles-Clark E, Saloheimo M, Siika-aho M, Penttila M. The alpha-glucuronidase-encoding gene of Trichoderma reesei. Gene. 1996;172(1):171–172. doi: 10.1016/0378-1119(96)00167-9. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Schaap PJ. Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet Biol. 2009;46(Suppl 1):S170–S179. doi: 10.1016/j.fgb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Zandleven JS, Benen JA, Awad H, Kools HJ, Beldman G, Voragen AG, Van den Berg JA, Schaap PJ. A new group of exo-acting family 28 glycoside hydrolases of Aspergillus niger that are involved in pectin degradation. Biochem J. 2006;400(1):43–52. doi: 10.1042/BJ20060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22(6):695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26(5):553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Martinez AT, Ruiz-Duenas FJ, Martinez MJ, Del Rio JC, Gutierrez A. Enzymatic delignification of plant cell wall: from nature to mill. Curr Opin Biotechnol. 2009;20(3):348–357. doi: 10.1016/j.copbio.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Master ER, Zheng Y, Storms R, Tsang A, Powlowski J. A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem J. 2008;411(1):161–170. doi: 10.1042/BJ20070819. [DOI] [PubMed] [Google Scholar]
- Mayans O, Scott M, Connerton I, Gravesen T, Benen J, Visser J, Pickersgill R, Jenkins J. Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure. 1997;5(5):677–689. doi: 10.1016/S0969-2126(97)00222-0. [DOI] [PubMed] [Google Scholar]
- Mertens JA, Bowman MJ. Expression and characterization of fifteen Rhizopus oryzae 99–880 polygalacturonase enzymes in Pichia pastoris. Curr Microbiol. 2011;62(4):1173–1178. doi: 10.1007/s00284-010-9842-8. [DOI] [PubMed] [Google Scholar]
- Minic Z, Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem. 2006;44(7–9):435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. Crystal structure of a family 54 alpha-l-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J Biol Chem. 2004;279(43):44907–44914. doi: 10.1074/jbc.M405390200. [DOI] [PubMed] [Google Scholar]
- Miyanaga A, Koseki T, Miwa Y, Mese Y, Nakamura S, Kuno A, Hirabayashi J, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. The family 42 carbohydrate-binding module of family 54 alpha-l-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose. Biochem J. 2006;399(3):503–511. doi: 10.1042/BJ20060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozolowski GA, Connerton IF. Characterization of a highly efficient heterodimeric xylosidase from Humicola insolens. Enzyme Microb Technol. 2009;45(6–7):436–442. doi: 10.1016/j.enzmictec.2009.08.011. [DOI] [Google Scholar]
- Mutter M, Beldman G, Pitson SM, Schols HA, Voragen AG. Rhamnogalacturonan alpha-d-galactopyranosyluronohydrolase. An enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions of pectin. Plant Physiol. 1998;117(1):153–163. doi: 10.1104/pp.117.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt A, Sild V, Pettersson G, Johansson G. Progress curves—a mean for functional classification of cellulases. Eur J Biochem. 1998;258(1):200–206. doi: 10.1046/j.1432-1327.1998.2580200.x. [DOI] [PubMed] [Google Scholar]
- Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kues U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FW, vanKuyk PA, Horton JS, Grigoriev IV, Wosten HA. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28(9):957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- Olivares-Hernandez R, Sunner H, Frisvad JC, Olsson L, Nielsen J, Panagiotou G. Combining substrate specificity analysis with support vector classifiers reveals feruloyl esterase as a phylogenetically informative protein group. PLoS One. 2010;5(9):e12781. doi: 10.1371/journal.pone.0012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, Benen JA, Kester HC, Visser J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur J Biochem. 1998;251(1–2):72–80. doi: 10.1046/j.1432-1327.1998.2510072.x. [DOI] [PubMed] [Google Scholar]
- Parenicova L, Benen JA, Kester HC, Visser J. pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem J. 2000;345(Pt 3):637–644. doi: 10.1042/bj3450637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, Kester HC, Benen JA, Visser J. Characterization of a novel endopolygalacturonase from Aspergillus niger with unique kinetic properties. FEBS Lett. 2000;467(2–3):333–336. doi: 10.1016/S0014-5793(00)01173-X. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25(2):221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Pham TA, Berrin JG, Record E, To KA, Sigoillot JC. Hydrolysis of softwood by Aspergillus mannanase: role of a carbohydrate-binding module. J Biotechnol. 2010;148(4):163–170. doi: 10.1016/j.jbiotec.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Polizeli ML, Rizzatti AC, Monti R, Terenzi HF, Jorge JA, Amorim DS. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;67(5):577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- Pollet A, Delcour JA, Courtin CM. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit Rev Biotechnol. 2010;30(3):176–191. doi: 10.3109/07388551003645599. [DOI] [PubMed] [Google Scholar]
- Powlowski J, Mahajan S, Schapira M, Master ER. Substrate recognition and hydrolysis by a fungal xyloglucan-specific family 12 hydrolase. Carbohydr Res. 2009;344(10):1175–1179. doi: 10.1016/j.carres.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Ravanal MC, Callegari E, Eyzaguirre J. Novel bifunctional alpha-l-arabinofuranosidase/xylobiohydrolase (ABF3) from Penicillium purpurogenum. Appl Environ Microbiol. 2010;76(15):5247–5253. doi: 10.1128/AEM.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczey K, Brumbauer A, Bollok M, Szengyel Z, Zacchi G. Use of hemicellulose hydrolysate for beta-glucosidase fermentation. Appl Biochem Biotechnol. 1998;70–72:225–235. doi: 10.1007/BF02920139. [DOI] [PubMed] [Google Scholar]
- Richardson M. The ecology of the Zygomycetes and its impact on environmental exposure. Clin Microbiol Infect. 2009;15(Suppl 5):2–9. doi: 10.1111/j.1469-0691.2009.02972.x. [DOI] [PubMed] [Google Scholar]
- Ridley BL, O’Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57(6):929–967. doi: 10.1016/S0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- Rigden DJ. Iterative database searches demonstrate that glycoside hydrolase families 27, 31, 36 and 66 share a common evolutionary origin with family 13. FEBS Lett. 2002;523(1–3):17–22. doi: 10.1016/S0014-5793(02)02879-X. [DOI] [PubMed] [Google Scholar]
- Riou C, Salmon JM, Vallier MJ, Gunata Z, Barre P. Purification, characterization, and substrate specificity of a novel highly glucose-tolerant beta-glucosidase from Aspergillus oryzae. Appl Environ Microbiol. 1998;64(10):3607–3614. doi: 10.1128/AEM.64.10.3607-3614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosgaard L, Pedersen S, Langston J, Akerhielm D, Cherry JR, Meyer AS. Evaluation of minimal Trichoderma reesei cellulase mixtures on differently pretreated Barley straw substrates. Biotechnol Prog. 2007;23(6):1270–1276. doi: 10.1021/bp070329p. [DOI] [PubMed] [Google Scholar]
- Rosgaard L, Pedersen S, Meyer AS. Comparison of different pretreatment strategies for enzymatic hydrolysis of wheat and barley straw. Appl Biochem Biotechnol. 2007;143(3):284–296. doi: 10.1007/s12010-007-8001-6. [DOI] [PubMed] [Google Scholar]
- Ryabova O, Vrsanska M, Kaneko S, van Zyl WH, Biely P. A novel family of hemicellulolytic alpha-glucuronidase. FEBS Lett. 2009;583(9):1457–1462. doi: 10.1016/j.febslet.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Saha BC. Alpha-l-arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv. 2000;18(5):403–423. doi: 10.1016/S0734-9750(00)00044-6. [DOI] [PubMed] [Google Scholar]
- Saha BC, Cotta MA. Comparison of pretreatment strategies for enzymatic saccharification and fermentation of barley straw to ethanol. N Biotechnol. 2010;27(1):10–16. doi: 10.1016/j.nbt.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Ogura A, Inui M, Tokuda S, Hosokawa S, Ihara H, Kasai N. Identification of a GH62 alpha-l-arabinofuranosidase specific for arabinoxylan produced by Penicillium chrysogenum. Appl Microbiol Biotechnol. 2011;90(1):137–146. doi: 10.1007/s00253-010-2988-2. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Nakari-Setala T, Tenkanen M, Penttila M. cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur J Biochem. 1997;249(2):584–591. doi: 10.1111/j.1432-1033.1997.00584.x. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Kuja-Panula J, Ylosmaki E, Ward M, Penttila M. Enzymatic properties and intracellular localization of the novel Trichoderma reesei beta-glucosidase BGLII (cel1A) Appl Environ Microbiol. 2002;68(9):4546–4553. doi: 10.1128/AEM.68.9.4546-4553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Sims IM, Craik DJ, Bacic A. Structural characterisation of galactoglucomannan secreted by suspension-cultured cells of Nicotiana plumbaginifolia. Carbohydr Res. 1997;303(1):79–92. doi: 10.1016/S0008-6215(97)00144-4. [DOI] [PubMed] [Google Scholar]
- Sprey B, Bochem HP. Formation of cross-fractures in cellulose microfibril structure by an endoglucanase-cellobiohydrolase complex from Trichoderma reesei. FEMS Microbiol Lett. 1993;106(3):239–243. doi: 10.1111/j.1574-6968.1993.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Sternberg D, Vijayakumar P, Reese ET. beta-Glucosidase: microbial production and effect on enzymatic hydrolysis of cellulose. Can J Microbiol. 1977;23(2):139–147. doi: 10.1139/m77-020. [DOI] [PubMed] [Google Scholar]
- Suykerbuyk ME, Schaap PJ, Stam H, Musters W, Visser J. Cloning, sequence and expression of the gene coding for rhamnogalacturonase of Aspergillus aculeatus; a novel pectinolytic enzyme. Appl Microbiol Biotechnol. 1995;43(5):861–870. doi: 10.1007/BF02431920. [DOI] [PubMed] [Google Scholar]
- Suykerbuyk ME, Kester HC, Schaap PJ, Stam H, Musters W, Visser J. Cloning and characterization of two rhamnogalacturonan hydrolase genes from Aspergillus niger. Appl Environ Microbiol. 1997;63(7):2507–2515. doi: 10.1128/AEM.63.7.2507-2515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenkanen M, Siika-aho M. An alpha-glucuronidase of Schizophyllum commune acting on polymeric xylan. J Biotechnol. 2000;78(2):149–161. doi: 10.1016/S0168-1656(99)00240-0. [DOI] [PubMed] [Google Scholar]
- Tenkanen M, Thornton J, Viikari L. An acetylglucomannan esterase of Aspergillus oryzae; purification, characterization and role in the hydrolysis of O-acetyl-galactoglucomannan. J Biotechnol. 1995;42(3):197–206. doi: 10.1016/0168-1656(95)00080-A. [DOI] [PubMed] [Google Scholar]
- Tenkanen M, Makkonen M, Perttula M, Viikari L, Teleman A. Action of Trichoderma reesei mannanase on galactoglucomannan in pine kraft pulp. J Biotechnol. 1997;57(1–3):191–204. doi: 10.1016/S0168-1656(97)00099-0. [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VG. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martin JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, van der Klei IJ, van Peij NN, Veenhuis M, von Dohren H, Wagner C, Wortman J, Bovenberg RA. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26(10):1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- van der Vlugt-Bergmans CJ, Meeuwsen PJ, Voragen AG, van Ooyen AJ. Endo-xylogalacturonan hydrolase, a novel pectinolytic enzyme. Appl Environ Microbiol. 2000;66(1):36–41. doi: 10.1128/AEM.66.1.36-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peij NN, Brinkmann J, Vrsanska M, Visser J, de Graaff LH. beta-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245(1):164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64(10):3615–3619. doi: 10.1128/AEM.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pouderoyen G, Snijder HJ, Benen JA, Dijkstra BW. Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger. FEBS Lett. 2003;554(3):462–466. doi: 10.1016/S0014-5793(03)01221-3. [DOI] [PubMed] [Google Scholar]
- van Santen Y, Benen JA, Schroter KH, Kalk KH, Armand S, Visser J, Dijkstra BW. 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J Biol Chem. 1999;274(43):30474–30480. doi: 10.1074/jbc.274.43.30474. [DOI] [PubMed] [Google Scholar]
- Vardakou M, Katapodis P, Samiotaki M, Kekos D, Panayotou G, Christakopoulos P. Mode of action of family 10 and 11 endoxylanases on water-unextractable arabinoxylan. Int J Biol Macromol. 2003;33(1–3):129–134. doi: 10.1016/S0141-8130(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Verbruggen MA, Beldman G, Voragen AG. Enzymic degradation of sorghum glucuronoarabinoxylans leading to tentative structures. Carbohydr Res. 1998;306(1–2):275–282. doi: 10.1016/S0008-6215(97)10065-9. [DOI] [PubMed] [Google Scholar]
- Vitali J, Schick B, Kester HC, Visser J, Jurnak F. The tree-dimensional structure of Aspergillus niger pectin lyase B at 1.7-A resolution. Plant Physiol. 1998;116(1):69–80. doi: 10.1104/pp.116.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasenko E, Schulein M, Cherry J, Xu F. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour Technol. 2010;101(7):2405–2411. doi: 10.1016/j.biortech.2009.11.057. [DOI] [PubMed] [Google Scholar]
- Ward M, Wu S, Dauberman J, Weiss G, Larenas E, Bower B, Rey M, Clarkson K, Bott R. Biochemistry and genetics of cellulose degradation. New York: Academic Press; 1993. [Google Scholar]
- Wong D. Enzymatic deconstruction of backbone structures of the ramified regions in pectins. Protein J. 2008;27(1):30–42. doi: 10.1007/s10930-007-9105-0. [DOI] [PubMed] [Google Scholar]
- Xu C, Leppanen AS, Eklund P, Holmlund P, Sjoholm R, Sundberg K, Willfor S. Acetylation and characterization of spruce (Picea abies) galactoglucomannans. Carbohydr Res. 2010;345(6):810–816. doi: 10.1016/j.carres.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Yaoi K, Kondo H, Hiyoshi A, Noro N, Sugimoto H, Tsuda S, Miyazaki K. The crystal structure of a xyloglucan-specific endo-beta-1,4-glucanase from Geotrichum sp. M128 xyloglucanase reveals a key amino acid residue for substrate specificity. FEBS J. 2009;276(18):5094–5100. doi: 10.1111/j.1742-4658.2009.07205.x. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Yamamoto K, Okada S. Isolation of Aspergillus flavus MO-5 producing two types of intracellular alpha-d-xylosidases: purification and characterization of alpha-d-xylosidase I. Biosci Biotechnol Biochem. 1993;57(8):1275–1280. doi: 10.1271/bbb.57.1275. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Yamamoto K, Okada S. Classification of some alpha-glucosidases and alpha-xylosidases on the basis of substrate specificity. Biosci Biotechnol Biochem. 1994;58(8):1392–1398. doi: 10.1271/bbb.58.1392. [DOI] [PubMed] [Google Scholar]