Desmoplakin recruits the centrosomal protein Lis1 to the epidermal cell cortex, where it regulates cortical microtubule organization and desmosome stability.
Abstract
Desmosomes are cell–cell adhesion structures that integrate cytoskeletal networks. In addition to binding intermediate filaments, the desmosomal protein desmoplakin (DP) regulates microtubule reorganization in the epidermis. In this paper, we identify a specific subset of centrosomal proteins that are recruited to the cell cortex by DP upon epidermal differentiation. These include Lis1 and Ndel1, which are centrosomal proteins that regulate microtubule organization and anchoring in other cell types. This recruitment was mediated by a region of DP specific to a single isoform, DPI. Furthermore, we demonstrate that the epidermal-specific loss of Lis1 results in dramatic defects in microtubule reorganization. Lis1 ablation also causes desmosomal defects, characterized by decreased levels of desmosomal components, decreased attachment of keratin filaments, and increased turnover of desmosomal proteins at the cell cortex. This contributes to loss of epidermal barrier activity, resulting in completely penetrant perinatal lethality. This work reveals essential desmosome-associated components that control cortical microtubule organization and unexpected roles for centrosomal proteins in epidermal function.
Introduction
Desmosomes are essential cell–cell adhesion structures that provide strength to tissues that experience mechanical stress. Mutations in genes encoding desmosomal components lead to a range of disorders in the skin and heart. In the epidermis, these range from relatively mild skin thickening to severe blistering, resulting in lethality (Vasioukhin et al., 2001b; Jonkman et al., 2005; Thomason et al., 2010). These defects are caused by the loss of intermediate filament attachment to the desmosome, which is largely provided by the desmosomal linker protein desmoplakin (DP; Bornslaeger et al., 1996; Gallicano et al., 1998; Vasioukhin et al., 2001b).
DP is also necessary for microtubule organization in the epidermis (Lechler and Fuchs, 2007). Proliferative cells of the basal epidermis have an apical centrosome that acts as a strong microtubule-organizing center. When these cells differentiate, the centrosome loses its microtubule-organizing center activity, and the microtubules relocalize to the cell cortex (Lechler and Fuchs, 2007). Although the reorganization of microtubules into noncentrosomal arrays occurs in many differentiated cells, the mechanisms underlying this remain poorly understood (Bartolini and Gundersen, 2006). Functionally, DP recruits the centrosomal protein ninein to the desmosome (Lechler and Fuchs, 2007). Ninein is required for microtubule minus end anchoring at the centrosome (Dammermann and Merdes, 2002), leading to the hypothesis that ninein functions downstream of desmosomes to reorganize microtubules to the cell cortex. This process may be conserved in other differentiating cell types, as ninein is known to relocalize from the centrosome to sites of noncentrosomal microtubule arrays in both neurons and pillar cells of the inner ear (Mogensen et al., 2000; Baird et al., 2004). However, neither the functional role of ninein nor the localization of other centrosomal proteins to the desmosome has been reported. In addition to ninein, the microtubule plus tip–binding protein CLIP170 also localizes to desmosomes. Immunoelectron microscopic analysis places CLIP170 on the cytoplasmic side of the desmosomal plaque (Wacker et al., 1992). Neither of these proteins requires microtubules for localization to the desmosome, suggesting that they function downstream of the desmosome to control microtubule organization.
Although microtubule anchoring at the centrosome is poorly understood, proteins other than ninein have been implicated in this function. These include γ-tubulin ring complex components, PCM-1, centrin, dynein/dynactin, TACC3/maskin, and Nde1/Ndel1 (Quintyne et al., 1999; Dammermann and Merdes, 2002; Guo et al., 2006; Albee and Wiese, 2008). How these proteins affect centrosomal microtubule anchoring and whether their effects are secondary to nucleation and/or centrosome structure defects are not completely understood. In contrast to centrosomal arrays, noncentrosomal microtubule arrays are even less well characterized in animal cells. Plant cells have robust cortical microtubules, which are nucleated and organized by γ-tubulin–containing complexes (Murata et al., 2005). This is not the case in several mammalian cell types, such as differentiated epidermis and pillar cells of the inner ear, where γ-tubulin localization does not correlate with noncentrosomal arrays of microtubules (Mogensen et al., 2000; Lechler and Fuchs, 2007). In these cell types, only ninein localization has been closely correlated with microtubule arrays. Meanwhile, in simple epithelial cells, the noncentrosomal protein Nezha plays a similar role by linking microtubule minus ends to adherens junctions (Meng et al., 2008). Together, these findings suggest a variety of ways in which noncentrosomal microtubule arrays may be generated. As different cell types adopt different noncentrosomal microtubule arrangements, different molecular players may be involved in each case.
Ndel1 and Lis1 are excellent candidates for molecular regulators of noncentrosomal microtubule arrays. Both of these proteins localize to centrosomes in some cell types/conditions, and Ndel1 plays a role in microtubule anchoring at that site (Sasaki et al., 2000; Guo et al., 2006). Lis1 is best known for its role in cortical migration and development in the brain (Wynshaw-Boris, 2007). Functionally, Lis1 can directly interact with and stabilize microtubules as well as regulate dynein motor activity (Sapir et al., 1997; Faulkner et al., 2000; Coquelle et al., 2002). Lis1 is widely expressed in cells other than neurons, though its function has not been characterized in the epidermis.
Here, we demonstrate that Ndel1 and Lis1 are centrosomal proteins in proliferative epidermal cells that relocalize to desmosomes upon differentiation. Genetic loss of Lis1 in the epidermis leads not only to microtubule organization defects but also to severe loss of desmosome stability and epidermal barrier activity, resulting in lethality shortly after birth.
Results
We previously reported that DP recruits the centrosomal protein ninein to desmosomes in differentiated epidermis, suggesting a functional link between desmosomes and microtubule organization. To determine whether DP can recruit other centrosomal proteins in addition to ninein, we examined the localization pattern of several centrosomal proteins in wild-type (WT) keratinocytes. Most centrosomal proteins that we examined (γ-tubulin, pericentrin, centrin, and ODF2) did not relocalize to the desmosome, suggesting specificity of recruitment (unpublished data). However, we were particularly interested in proteins like ninein that are associated with centrosome maturation and microtubule anchoring. Nde1/Ndel1 are homologous proteins that have been shown to specifically localize to the mother centriole and contribute to microtubule-anchoring activity (Guo et al., 2006). Both proteins are expressed in keratinocytes (unpublished data). Using both laboratory-generated and commercially available antibodies, we assayed the localization of these proteins (Figs. 1 [A–C and G] and S1). The two antibodies we generated recognized a predominant band at ∼39 kD, consistent with the known size of Ndel1 and with Western blotting with other commercially available antibodies (Fig. S1 A). In proliferative keratinocytes cultured in low Ca2+ (0.05 mM), Ndel1 colocalized with γ-tubulin at one of the centrioles, consistent with a previous study in other cell types (Fig. 1, A–C; Lechler and Fuchs, 2007). Once calcium levels are increased (1.2 mM), cadherin-based junctions are able to form, and cells begin a terminal differentiation process. Under these conditions, Ndel1 localized to sites of cell–cell adhesion (Fig. 1 G). This junctional localization was detected with two other antibodies specific for Nde1/Ndel1 (Fig. S1, B and C). Additionally, when GFP-Ndel1 was expressed in cultured keratinocytes, we could detect cells in which GFP-Ndel1 localized to the cell cortex (Fig. S1, D–F). This was a rare event because GFP-Ndel1 aggregated in the majority of cells, which is very similar to what has been reported for ninein-GFP (Delgehyr et al., 2005). However, expression of the amino-terminal 154 amino acids of Ndel1 fused to GFP resulted in a protein that did not aggregate but still localized to cell–cell junctions (Fig. S1 G).
Figure 1.
A subset of centrosomal proteins is recruited to the desmosome. (A–L) Localization of Ndel1 and Lis1 in mouse keratinocytes. (A–C) Anti-Ndel1 (red) and γ-tubulin (green) immunofluorescence staining in WT mouse keratinocytes cultured in low Ca2+. (D–F) Anti-Lis1 (red) and γ-tubulin (green) staining in WT mouse keratinocytes cultured in low Ca2+. A–F are maximum intensity projections. (G) Anti-Ndel1 (green) and anti-DP (red) staining in WT mouse keratinocytes. The boxed region is enlarged on the right to demonstrate the colocalization of Ndel1 and DP. (H) Anti-Lis1 (red) and anti-DP (green) staining in WT mouse keratinocytes. The boxed region is enlarged on the right to demonstrate the colocalization of Lis1 and DP. (I and J) Costaining for Ndel1 (I) or Lis1 (J) in green and β-catenin (β-cat) in red. (K and L) Staining for Ndel1 (K) and Lis1 (L) in mouse keratinocytes grown in high Ca2+ and treated with 10 µM nocodazole (Noc) for 1 h. DNA is labeled with Hoechst (blue) in these images. Bars, 10 µm.
To determine whether Ndel1 colocalized with desmosomes, we costained for DP. With antibodies and with GFP-Ndel1, we saw colocalization of Ndel1 with desmosomes (Figs. 1 G and S1 [F and M]). In contrast, Ndel1 did not significantly colocalize with β-catenin, a component of adherens junctions (Fig. 1 I).
Lis1 is a known binding partner of Ndel1 (Niethammer et al., 2000; Sasaki et al., 2000), but it has not been implicated as a mother centriole–specific protein. We found that in keratinocytes, Lis1 was found either on both centrioles but enriched on one or was only detected on a single centriole (Fig. 1, D–F). Like Ndel1, Lis1 was recruited to the cell cortex upon addition of calcium to the media (Figs. 1 H and S1 I). The antibody used recognized a single band of the expected size (48 kD) in extracts from WT keratinocytes that was lost in Lis1-null keratinocytes (discussed later in this section; Fig. S1 H). In addition, the cortical staining observed with anti-Lis1 antibodies was lost in Lis1-null keratinocytes (Fig. S1, I and J). This localization of Lis1 was seen with another antibody raised to a distinct epitope (Fig. S1 K).
Like Ndel1, Lis1 colocalized with DP at cell junctions (Figs. 1 H and S1 L) but did not significantly colocalize with β-catenin (Fig. 1 J). This suggests that Lis1 and Ndel1 are desmosome-associated proteins in differentiated keratinocytes. Like ninein and CLIP170, the junctional localization of Ndel1 and Lis1 was independent of microtubules. After cells were treated with nocodazole to disrupt microtubules, Ndel1 and Lis1 remained associated with the cell junctions (Fig. 1, K and L). Initial recruitment also does not require microtubules, as nocodazole treatment before a calcium switch does not prevent cortical accumulation of Lis1 and Ndel1 (Fig. S2, P–U). This demonstrates that the desmosomal localization of Ndel1 and Lis1 is independent of any microtubule-binding activity.
To determine the temporal order of recruitment to cell junctions, we performed calcium switch experiments and examined the localization of DP and Ndel1 or Lis1 at early time points after the switch. DP could be detected at some junctions within 1 h and at most junctions within 2 h (Fig. S2, A–I). Ndel1, in contrast, was detected beginning at about 2 h in some cells and thereafter became more robust. Similar results were found with Lis1 (unpublished data). Therefore, the localization of Ndel1 and Lis1 occurs just subsequent to DP recruitment.
Because of their colocalization with desmosomes and their order of appearance at the cortex, we tested whether Lis1 and Ndel1 localization required DP. In cultured DP-null cells grown in high calcium, we did not observe any cortical localization of Lis1 or Ndel1, demonstrating that DP is essential for their recruitment to the membrane (Fig. 2, B and E). Both the initial recruitment and maintenance were affected, as no Lis1 was detected at cell junctions between 2 and 4 h after the calcium switch (Fig. S2 V). In contrast, α-catenin–null keratinocytes showed no defect in cortical localization of Ndel1 or Lis1 (Fig. 2, C and F). These data demonstrate that DP recruits a subset of centrosomal proteins implicated in microtubule anchoring to desmosomes.
Figure 2.
DP-dependent localization of Ndel1 and Lis1 in vitro and in vivo. (A–I) Localization of Ndel1 (A–C), Lis1 (D–F), and CLIP170 (G–I) in either WT keratinocytes, DP-null keratinocytes (DP KO), or α-catenin–null keratinocytes (α-cat KO), as indicated. Note the loss of cortical staining in DP-null cells. Images in A–F are maximum intensity projections of z stacks. (J and K) In vivo localization of Lis1 and Ndel1. WT (J) or DP cKO (K) e17.5 mouse skin was stained with anti-Lis1 (red) and anti–β4 integrin (green denotes the basement membrane separating dermis from epidermis). The boxed regions are magnified below the images to highlight centrosomal staining in dermis and basal cells and cortical staining in suprabasal cells. Note the loss of cortical Lis1 in the DP-null suprabasal cells. (L and M) WT (L) or DP cKO (M) e17.5 mouse skin was stained with anti-Ndel1 (red) and anti–β4 integrin (green). DNA is labeled with Hoechst (blue) in all images. Bars, 10 µm.
Although a role for desmosomes in microtubule organization has only recently been described, the first hint came almost 20 yr ago when the Kreis laboratory showed that the microtubule plus end–binding protein CLIP170 localized to desmosomes (Wacker et al., 1992). Molecular requirements for this localization have not been reported. We confirmed the localization of CLIP170 to desmosomes and further demonstrated that localization was dependent on DP but not α-catenin (Fig. 2, G–I).
To determine whether Lis1 and Ndel1 target to desmosomes in intact tissue, we examined their localization in skin sections. In dermal fibroblasts and basal cells of the epidermis, Lis1 localized to a single punctum, presumably the centrosome (Fig. 2 J). However, in suprabasal cells of the epidermis, Lis1 localized to the cell cortex, where desmosomes are abundant (Fig. 2 J). In DP-null epidermis, Lis1 localization was specifically disrupted in suprabasal cells, where it was diffuse throughout the cytoplasm (Fig. 2 K). Lis1 remained associated with the centrosome in basal cells and in the dermis. This confirms our data in cultured cells that DP is required to recruit Lis1 to the desmosome. The localization pattern of Ndel1 mimicked that of Lis1, though centrosomal staining was poor with this antibody in vivo, and more background staining was noted as well as more cortical staining in basal cells (Fig. 2, L and M).
Collectively, our data demonstrate that at least four proteins that regulate microtubule dynamics and organization are desmosome-associated proteins. Two of these, CLIP170 and Lis1, can directly bind microtubules at the plus ends (Sapir et al., 1997; Diamantopoulos et al., 1999; Perez et al., 1999; Faulkner et al., 2000; Coquelle et al., 2002). Additionally, both ninein and Ndel1 are required for minus end anchoring at the centrosome and perhaps other sites (Mogensen et al., 2000; Dammermann and Merdes, 2002; Guo et al., 2006).
Interactions between Lis1 and Ndel1 and between Lis1 and CLIP170 have been previously identified (Sasaki et al., 2000; Coquelle et al., 2002). To determine whether ninein, Ndel1, Lis1, and CLIP170 physically interact, we performed protein-binding assays using recombinant GST-tagged Ndel1, covalently linked to agarose beads, and WT keratinocyte extracts. Lis1, CLIP170, and ninein were found to be specifically associated with Ndel1 (Fig. S3 A). These data indicate that the centrosomal proteins that associate with the desmosome can biochemically interact. Nevertheless, these proteins exhibited differential turnover at the desmosome, suggesting that they do not form a stable complex (see next paragraph; Fig. S3).
Most desmosomal plaque components are insoluble in protein extracts. We determined the solubility of the desmosome-associated proteins identified here in a harsh buffer (1% Triton X-100 and 0.5% NP-40 with sonication). Whereas DPI is largely insoluble under these conditions, the desmosome-associated proteins are mostly soluble (Fig. S3 B). This is consistent with the observation that CLIP170 is not a core desmosomal plaque protein (Wacker et al., 1992). Gentler treatment and in situ analysis revealed differential dynamics at the cell cortex. A 1-min permeabilization of cultured cells with a lower detergent concentration (0.2% Triton X-100) resulted in almost complete loss of Ndel1 from cell junctions (Fig. S3, G and H). This same treatment had little effect on ninein, Lis1, and DP localization (Fig. S3, C–J). To test whether the cortical turnover rates of some of these components were distinct, we performed FRAP analysis of ninein-GFP and Ndel1(1–154)-GFP. Whereas ninein-GFP recovered slowly and incompletely at cell junctions, the localization domain of Ndel1-GFP recovered quickly and nearly completely (Fig. S3, K and L). The rapid turnover of Ndel1 may explain why it was often detected in a broader cortical region, not always precisely colocalizing with desmosomes (Fig. S1, B and C). These data indicate that the centrosomal proteins are unlikely to form a single stable complex and may play distinct roles at the desmosome.
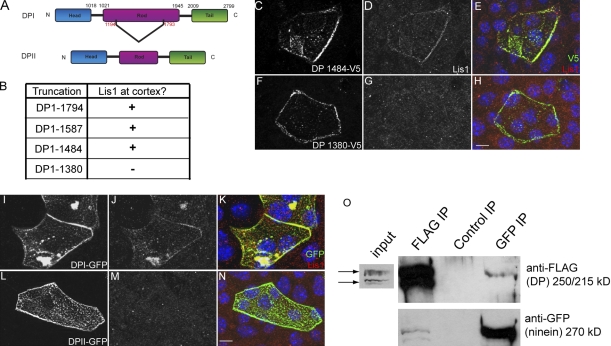
What are the molecular requirements for recruitment of centrosomal proteins to the desmosome? Previous work demonstrated that DP’s intermediate filament-binding activity was unnecessary for ninein recruitment (Lechler and Fuchs, 2007). However, further dissection of the regions required has not been performed. DP has two known isoforms, DPI and DPII, that are generated by alternative splicing (Fig. 3 A). Both contain an amino-terminal domain that targets them to the desmosome and a carboxy-terminal domain that directly interacts with intermediate filaments (Stappenbeck and Green, 1992; Bornslaeger et al., 1996). A central coiled-coil domain present in DPI is largely truncated in DPII. To determine what region of DP is required to recruit centrosomal proteins, we constructed various truncation mutants in the coiled-coil rod domain of DP. After transfecting these constructs into DP-null keratinocytes, we assayed their ability to rescue Lis1 and CLIP170 localization to the desmosome. All of the mutant proteins that we examined were stable and localized to cell–cell junctions. A DP protein truncated at amino acid 1,484 was able to rescue Lis1 and CLIP170 recruitment to the cortex (Figs. 3 [B–E] and S4 [A–C]). However, truncation of DP at amino acid 1,380 resulted in a protein that was unable to rescue recruitment of any centrosomal protein examined (Figs. 3 [F–H] and S4 [D–F]). This demonstrates that a region between amino acids 1,380 and 1,484 of DP’s rod domain is necessary for centrosomal protein recruitment to the cell cortex. We were unable to create a truncation mutant that recruited some of the centrosomal proteins but not others. Therefore, the same region of DP is required to recruit all of the centrosomal proteins. Interestingly, the region between amino acids 1,380 and 1,484 is specific to one isoform of DP, DPI. To verify the isoform-specific function, we examined the ability of full-length GFP-tagged DPI and DPII to recruit centrosomal proteins to the cortex. We confirmed that DPI can rescue centrosomal protein recruitment to the desmosome, whereas DPII cannot (Figs. 3 [I–N] and S4 [G–L]). We further confirmed this by biochemical analysis. Cells were cotransfected with a plasmid to express ninein-GFP and a FLAG-DP construct that generates both DPI and DPII through alternative splicing (Smith and Fuchs, 1998). After immunoprecipitation with an antibody against FLAG, ninein-GFP was detected in the precipitated pool of proteins (Fig. 3 O). Reciprocally, a GFP antibody was used to precipitate ninein and its bound proteins. In this case, we detected only DPI but not DPII in the immunoprecipitate (Fig. 3 O). These data demonstrate an isoform-specific function for DPI. The amounts of coprecipitated proteins were low in both cases; however, this represents the interaction between the small soluble pools of these proteins. It is likely that the interaction is more stable at the desmosome, as the turnover data suggest.
Figure 3.
DP recruits centrosomal proteins to the desmosome in an isoform-specific manner. (A) A diagram of the two isoforms of DP (DPI and DPII) achieved by alternative splicing. Amino acid numbers at splice sites are shown in red, and those marking domain boundaries are shown in black. (B) A table displaying DP truncation mutants. Their ability to recruit Lis1 and CLIP170, as assessed by immunofluorescence staining, is indicated. (C–H) DP knockout (KO) mouse keratinocytes were transfected with V5-tagged truncation mutants, DP 1484 (C–E), and DP 1380 (F–H). (E and H) Merged images of V5-tagged truncation mutants (green) and Lis1 (red). Images are displayed as maximum intensity projections. (I–N) DP knockout mouse keratinocytes were transfected with DPI-GFP (I) or DPII-GFP (L). Immunofluorescence staining of Lis1 in J and M. Merged images are shown in K and N. Images are maximum intensity projections. (O) WT mouse keratinocytes were cotransfected with FLAG-DP and ninein-GFP, and immunoprecipitations (IP) were performed with anti-FLAG, anti-GFP, and normal rabbit sera as a control. Bound proteins were analyzed by Western blotting. Arrows indicate the two isoforms of DP, DPI (top) and DPII (bottom). Bars, 10 µm.
Although we have begun to understand the molecular requirements for centrosomal protein recruitment, the functional importance of these proteins in the epidermis is unknown. To examine this, we took a genetic approach. We isolated and generated lines of Lis1 homozygous floxed (fl/fl) keratinocytes from newborn backskin epidermis. Infection with adenoviral Cre resulted in significant cell death of proliferating keratinocytes. Therefore, we induced differentiation in these cells 12–24 h after Cre infection to avoid any mitotic defects. Under these conditions, we achieved a near-complete loss of Lis1 (see Figs. 6 K and S1 [H–J]). Importantly, these cells did not undergo apoptosis, eliminating the possibility of an analysis of secondary effects.
Figure 6.
Loss of Lis1 results in defects in desmosome stability. (A–J) Staining for desmosomal proteins in WT and Lis1-null keratinocytes. Images are maximum intensity projections and were taken at the same exposure settings and modified in identical ways. Bar, 10 µm. (K) Western blots of whole-cell lysates from WT, DP-null, and Lis1-null keratinocytes for the indicated proteins. Plak, plakoglobin. (L) Western blots of Lis1, epithelial cadherin (E-cad), and α-catenin (α-cat) in WT and Lis1-null cells. (M) Western blots of plakoglobin and Dsg1 from WT cells grown in either high (Hi) or low (Lo) calcium media and Lis1-null cells grown in high calcium media. (N) Levels of Dsg1 were examined from surface proteins that were biotinylated and isolated on avidin-agarose. (O and P) Turnover of desmosomal components after cycloheximide treatment. Levels of plakoglobin (O) and Dsg1 (P) were examined in WT and Lis1-null cells after treatment with cycloheximide for the indicated times. Note that initial levels of both proteins were significantly lower in Lis1-null cells than in WT cells. Error bars are SD. n = 2. (Q) A kymograph analysis of FRAP for DPI-GFP in WT cells (top) and Lis1-null cells (bottom). The bleach point is indicated. (R) Mobile fractions from individual FRAP experiments were plotted. In this box and whisker plot, the boxes represent the 25th and 75th percentiles, whereas the whiskers represent the 10th and 90th percentiles.
Our working model suggests that desmosomes induce the reorganization of the microtubule cytoskeleton to the cell cortex through the recruitment of centrosomal proteins to the desmosome. In culture, microtubules need to be stabilized to reorganize to the cell cortex. In control cells (Lis1 fl/fl;Cre−), the addition of taxol induced the cortical reorganization of microtubules (Fig. 4, A and B). However, upon Lis1 ablation, the microtubules were unable to reorganize to the cell cortex upon taxol addition (Fig. 4 D). This effect was specific for cortical organization, as, without taxol treatment, the cytoplasmic array of microtubules was largely intact in cells lacking Lis1 (Fig. 4 C).
Figure 4.

Lis1 is required for cortical microtubule organization in vitro and in vivo. (A–D) Immunofluorescence staining of β-tubulin in cultured keratinocytes. (A and B) Lis1 fl/fl keratinocytes not treated with Cre adenovirus. (C and D) Lis1 fl/fl keratinocytes infected with Cre adenovirus. (A and C) Control cells have predominantly cytoplasmic microtubules. (B and D) Taxol treatment induces cortical microtubules in control cells (B) but not in cells in which Lis1 has been ablated (D). A–D are maximum intensity projections of z stacks. (E and F) Sections of backskin from K14-ETMB-3GFP transgenic mice that are either WT (E) or Lis1 (F) cKO. Bars, 10 µm.
To determine whether Lis1 is required for microtubule organization in vivo, we generated embryos that lack Lis1 in the epidermis (Lis1 fl/fl;K14-Cre, hereafter referred to as Lis1 conditional knockout [cKO] mice) and that express the fluorescent microtubule-associated protein ensconsin microtubule-binding domain–3GFP (Lechler and Fuchs, 2007). We confirmed the loss of Lis1 in the epidermis by Western blotting (Fig. 5 R). In WT epidermis, microtubules were organized around the cortex of suprabasal cells (Fig. 4 E; Lechler and Fuchs, 2007). However, in Lis1 cKO epidermis, the cortical organization of microtubules was lost, and the microtubules were cytoplasmic (Fig. 4 F). Lis1 is only the second protein identified, after DP, that is required for cortical microtubule organization in the epidermis.
Figure 5.
Lis1-null epidermis exhibits defects in epidermal barrier function and in desmosomes. (A) Barrier assay (X-gal penetration) in Lis1 heterozygous and Lis1 cKO e18.5 embryos. Arrows indicate blisters. Bar, 1 cm. (B) Cornified envelopes from WT or Lis1 cKO mice. Bar, 40 µm. (C–N) Immunofluorescence staining of desmosomal components (as indicated on panels) on sections of backskin from WT and Lis1 cKO e18.5 embryos. The basement membrane is marked by a dashed line. Bar, 10 µm. (O) Quantitation of the cortical/cytoplasmic ratio of fluorescence intensity of DP, plakoglobin, and α-catenin (α-cat). n = 50 from each of two embryos for each genotype. Error bars represent SEM. ***, P < 0.001. (P and Q) Transmission EM images of desmosomes between two spinous layer cells in the backskin of e18.5 embryos. Arrows mark desmosomes. Bar, 0.5 µm. (R) Epidermal lysates from e18.5 WT and Lis1 cKO embryos were blotted with antibodies against the indicated proteins.
Loss of Lis1 in the epidermis resulted in perinatal lethality; no adult Lis1 cKO mice have been recovered. The few live null newborns we were able to recover were severely dehydrated. To test whether this was a result of a barrier defect, we subjected e18.5 (embryonic day 18.5) embryos to an X-gal penetration assay. If the barrier is compromised, X-gal diffuses through the epidermis and forms a blue precipitate in the dermis (Hardman et al., 1998). In WT mice, the barrier formed by the epidermis blocked X-gal from reaching the dermis (Fig. 5 A). However, the Lis1 cKO mouse had severe epidermal barrier defects, as seen by the extensive blue coloring of the skin. Furthermore, this assay highlighted blisters on the surface of the Lis1 cKO epidermis as well as peeling skin (Fig. 5 A, arrows).
Terminal differentiation of the epidermis results in structures called cornified envelopes, which are highly cross-linked networks essential for barrier activity of the epidermis. Preparations of cornified envelopes from Lis1 cKO mice revealed aberrant and irregularly shaped envelopes (Fig. 5 B). This phenotype closely resembled the DP cKO epidermis (Vasioukhin et al., 2001b). We also observed mitotic defects and increased apoptosis in basal epidermal cells, consistent with studies in other tissues (Faulkner et al., 2000; Buttner et al., 2007). However, the underlying cell biological defects we observed can be replicated in cell culture under conditions in which apoptosis does not occur (Figs. 4 [A–D] and 6 [A–J]), strongly arguing against them being secondary defects.
To directly test whether desmosomes were altered in Lis1 cKO epidermis, we performed immunofluorescence analysis of Lis1 cKO embryos. This revealed lower intensity staining of each desmosomal component examined, including desmocollin 1 (Dsc1), desmoglein 1 (Dsg1), desmocollin 2/3 (Dsc2/3), plakoglobin, and DP (Fig. 5, C–L). However, staining for the adherens junction protein α-catenin revealed no defects in its localization at cell junctions (Fig. 5, M and N). To quantitate these differences, we measured the ratios of fluorescence intensity at the cell cortex as compared with the cytoplasm for the nontransmembrane proteins examined. There was a significant decrease in cortical recruitment of both DP and plakoglobin in Lis1 cKO epidermis, whereas the slight decrease in α-catenin was not statistically significant (Fig. 5 O). Ultrastructurally, desmosomes in the spinous layer of Lis1 cKO epidermis were smaller and had less robust attachment to keratin filaments than their WT counterparts (Fig. 5, P and Q). Furthermore, separations between cells were observed, consistent with the blistering that was observed macroscopically (unpublished data).
Next, we examined total levels of desmosomal components in WT and Lis1 cKO epidermis to determine whether the amounts of these proteins were altered (Fig. 5 R). Significant decreases were noted for Dsg1, DPI, and plakoglobin. PKP3 showed little change, as did the adherens junction component α-catenin. These data demonstrate a specific effect on the stability of desmosomal proteins in the Lis1 cKO epidermis.
To understand the mechanism underlying the desmosomal defects in the Lis1 cKO epidermis, we turned to analysis in cultured keratinocytes, which allows for higher resolution imaging and functional studies. Analysis of desmosomal proteins in cultured Lis1-null cells further supported an essential role for Lis1 in maintaining robust desmosomes. Immunofluorescence analysis of Dsg3, Dsc2/3, Dsg1, DP, and plakoglobin revealed substantial (though not complete) loss of cortical staining (Fig. 6, A–J). Biochemical analysis of these cells showed significant decreases in desmosomal components, especially DP and plakoglobin, whereas no changes in adherens junction components were detected (Fig. 6, K and L). Cortical localization of both epithelial cadherin and α-catenin was unchanged in the Lis1-null cells (unpublished data). Interestingly, levels of CLIP170 were decreased in both Lis1- and DP-null cells, whereas levels of Ndel1 were unaffected (Fig. 6 K and not depicted). Therefore, cultured Lis1-null keratinocytes recapitulate the desmosomal defects seen in vivo. Overexpression of plakoglobin could not rescue DP recruitment to cell junctions, suggesting that the low plakoglobin levels are a consequence and not a cause of desmosome dysfunction (unpublished data).
Calcium shift experiments revealed that there was no defect in the initial assembly of desmosomes in Lis1-null cells (Fig. S2, J–O). However, whereas desmosomes continued to form and become more robust in WT cells, they did not do so in Lis1-null cells. These data suggest that the stability of desmosomes and, therefore, desmosomal components, is likely affected. In support of this, the levels of plakoglobin and Dsg1 in the Lis1-null keratinocytes grown in high calcium conditions resembled that of WT cells grown in low calcium conditions (Fig. 6 M). Under low calcium conditions, cadherin components of the desmosomes cannot functionally interact. Upon calcium addition, desmosomes form, and the stability of most desmosomal proteins increases (Penn et al., 1987). Not only were the total levels of Dsg1 reduced in Lis1-null cells, but the plasma membrane pool, as judged by cell surface biotinylation, was also decreased (Fig. 6 N). These biochemical data support the immunofluorescent data, clearly demonstrating that surface pools of this desmosomal cadherin are reduced.
We hypothesized that the decreased levels of desmosomal components were a result of increased turnover of the protein. To measure this, we treated WT and Lis1-null cells with cycloheximide to arrest new protein synthesis and followed the levels of plakoglobin and Dsg1 with time. For both of these proteins, a significantly increased rate of loss was observed in Lis1-null cells as compared with the WT controls (Fig. 6, O and P).
To directly test whether the stable cortical localization of desmosomal proteins is affected in Lis1-null cells, we performed FRAP analysis on DPI-GFP. Although cortical recruitment of DPI-GFP was low in Lis1-null cells, we were able to both bleach and image the recovery of the brightest cortical DPI-GFP puncta. Whereas DPI-GFP is normally very stable in WT cells with a mobile fraction of ∼20%, it became significantly more dynamic in Lis1-null cells (Fig. 6, Q and R). Some cells showed complete recovery of DPI-GFP fluorescence, which was never seen in WT cells. In addition, the mean mobile fraction was ∼40%. This likely underestimates the true value, as we selected the most robust DPI-GFP puncta for analysis in the mutant. We also attempted to examine Dsg3-GFP dynamics. Although this was possible in WT cells, Dsg3-GFP did not significantly accumulate at cell junctions in Lis1-null cells, precluding this analysis (unpublished data). However, this demonstrates that exogenous Dsg3 is unable to rescue the desmosomal phenotype of Lis1-null cells.
In desmosome turnover induced by pathogenic antidesmosomal antibodies (pemphigus), desmosomal cadherins are internalized and move through early endosomes to lysosomes for degradation (Calkins et al., 2006; Delva et al., 2008). Upon chloroquine treatment of Lis1-null cells, we detected desmosomal cadherins in intracellular vesicles that colocalized with lysosomal markers, suggesting that at least a fraction of the internalized cadherins are ultimately degraded in the lysosome (Fig. S5).
Discussion
We have shown that during epidermal differentiation, the desmosome recruits a group of centrosomal and microtubule-associated proteins to the cell cortex. One of these proteins, Lis1, has two functions at the cell cortex. The first is a role in microtubule organization downstream of the desmosome. The second is an unexpected role in desmosome stability.
Although many differentiated cell types undergo microtubule reorganization into noncentrosomal arrays, little is known about the mechanism underlying this change. The work presented here suggests that during epidermal differentiation, a subset of centrosomal proteins is actively recruited to desmosomes to promote cortical microtubule reorganization. This includes ninein and Ndel1, both of which have previously been implicated in microtubule anchoring to the centrosome (Mogensen et al., 2000; Guo et al., 2006). It also includes CLIP170 and Lis1, both of which interact with and stabilize microtubules (Sapir et al., 1997; Diamantopoulos et al., 1999; Perez et al., 1999; Faulkner et al., 2000; Coquelle et al., 2002). We suggest that by recruiting these proteins, desmosomes specify a cortical zone competent for the stabilization and capture of microtubules. This could be mediated, in part, through direct interactions with microtubules but may also involve recruitment of additional factors that can stabilize and/or tether microtubules near the cell cortex.
Loss of ninein from the centrosome has been reported not only in the epidermis but also in neurons, muscle cells, and pillar cells of the inner ear (Mogensen et al., 2000; Bugnard et al., 2005; Ohama and Hayashi, 2009). This suggests that a common mechanism of microtubule reorganization involves specific loss of proteins and functions of the centrosome. Neither the developmental cues nor the signaling pathways that regulate this process are known in mammals. In the Drosophila melanogaster trachea, a noncentrosomal array of microtubules is generated through displacement of microtubule-nucleating proteins from the centrosome to the apical cell cortex (Brodu et al., 2010). Loss of these proteins from the centrosome requires the microtubule-severing protein spastin. It will be interesting to determine whether similar pathways affect the localization of microtubule-anchoring proteins in mammals.
The recruitment of ninein/Ndel1/Lis1/CLIP170 to desmosomes requires the linker protein DP. In DP-null epidermis and keratinocytes, these proteins never get recruited to the cell cortex. Because other desmosomal proteins and the desmosomal plaque remain largely intact in the DP-null epidermis, this strongly implicates DP in the direct recruitment of the centrosomal proteins. Additionally, the recruitment of the centrosomal proteins is dependent on one of the two predominant isoforms of DP, DPI. Although DPI and DPII have structural differences, we believe this to be the first report of a functional difference between these proteins (O’Keefe et al., 1989). It is interesting to speculate that cells may control desmosome function by regulating the splicing of the two isoforms of DP. Although different tissues have different ratios of DPI versus DPII, we do not currently have cellular resolution of the expression patterns and localizations of both of these proteins. However, a single human case involving specific loss of the DPI isoform has been reported and resulted in cardiac and skin defects, leading to lethality (Uzumcu et al., 2006). Whereas part of this phenotype may be a result of decreased levels of total DP in some tissues, the levels of DP in the epidermis appeared normal, yet a phenotype persisted. These data highlight functional relevance for the two DP isoforms in humans.
Although Lis1 function has been studied in detail during neural development, its role in epithelial tissues has not been well characterized. In this study, we have shown that Lis1 translocates from the centrosome to desmosomes upon epidermal differentiation. Lis1 appears to have two distinct functions in differentiated epidermis. First, it is required for the generation of cortical microtubules and is only the second protein (after DP) known to be essential for this process. Whether Lis1’s desmosomal localization or its microtubule-binding activity is necessary for its role in cortical microtubule organization will require the generation of separation of function mutations that can specifically eliminate this activity.
The second and more unexpected finding was that loss of Lis1 results in severe desmosome defects. Lis1-null desmosomes are less robust, have fewer connections to keratin filaments, and undergo faster and more complete turnover. Because of this, loss of Lis1 partially phenocopies the loss of other desmosomal proteins. Human mutations in almost all of the known desmosomal genes have been identified that result in disease, either cutaneous or cardiac or both. Lis1 is unlikely to be included in this group because haploinsufficiency of this gene results in severe brain abnormalities (Hirotsune et al., 1998). However, it will be important to determine whether mutations in ninein/Ndel1/CLIP170 can cause or modulate desmosome-related disease.
Although it is tempting to speculate that the desmosome defects are secondary to microtubule defects, this is not supported by data in the literature or our own observations (Pasdar et al., 1992). Treatment of cells with nocodazole to disrupt microtubules does not significantly affect desmosome stability. In addition, turnover of DP as measured by FRAP does not increase in nocodazole-treated cells (unpublished data). This suggests that Lis1 has another function. Our data demonstrate that Lis1 stabilizes the desmosome at the cortex, preventing its turnover. Similar to desmosomal components in low calcium conditions, in which desmosomal cadherins cannot form productive interactions, the Lis1-null cells show low surface pools of desmosomal cadherins, high turnover of DP, and increased degradation of both transmembrane and plaque components. It may do this in a variety of ways, including directly affecting clustering of components or by regulating the endocytosis of desmosomal components. In other cell types, Lis1–Ndel1 complexes have been implicated in intermediate filament organization and transport, raising the alternative possibility that Lis1 plays a role in coupling the desmosome to the underlying keratin filament network (Nguyen et al., 2004; Shim et al., 2008). In the absence of this, the desmosome may be unstable and quickly turn over.
This work increases the proteomic complexity and functionality of the desmosome and demonstrates additional pathways that control desmosome stability. In addition, it establishes an excellent model in which to study the mechanism and function of noncentrosomal microtubule arrays.
Materials and methods
Cell culture
All keratinocyte cell lines were grown at 37°C and 7.5% CO2 in E low Ca2+ media. DP and α-catenin cKO keratinocytes were isolated and established from backskins of newborn mice of the appropriate genotype (Vasioukhin et al., 2001a,b). Lis1 fl/fl keratinocytes were isolated from backskins of Lis1 fl/fl newborn pups and cultured in E low Ca2+ media. Lis1 fl/fl mice were provided by A. Wynshaw-Boris (University of California, San Francisco, San Francisco, CA). Once cells reached 95% confluency, they were infected with adenoviral Cre-GFP (Vector Development Laboratory, Baylor College of Medicine). 12–24 h after infection, cells were induced to differentiate by increasing the amount of Ca2+ to 1.2 mM. In microtubule organization assays, taxol was added 24 h after induction of differentiation for 1 h at 10 µM. For protein stability experiments, cells were treated with 150 µM cycloheximide (Sigma-Aldrich) for 6–12 h.
Epidermal barrier function assay
e18.5 embryos were immersed in a solution of X-gal as previously described (Hardman et al., 1998), with slight modifications to the X-gal solution (1.3 mM MgCl2, 100 mM NaPO4, 3 mM K3Fe[CN]6, 0.01% sodium deoxycholate, 0.2% NP-40, and 1 mg/ml X-gal).
Immunofluorescence staining
Keratinocytes were grown on glass coverslips in E low Ca2+ media until confluent. Cells were induced to differentiate and form junctions by the addition of calcium to 1.2 mM. After 24 h, cells were fixed in methanol for 2 min at −20°C. For CLIP170 staining, fixation was preceded by a 30-s permeabilization step (50 mM Pipes, 100 mM NaCl, 2 mM MgCl2, 0.5 mM CaCl2, and 0.1% Triton X-100) at 37°C. For Lis1 staining in tissue, slides were fixed in acetone at −20°C for 2 min, and all steps used washes with a Triton X-100 concentration of 1%. For tissues, 8-µm-thick sections were cut and fixed in methanol for 2 min at −20°C. For Ndel1 staining in tissue, an antigen retrieval step was required. Slides were boiled for 5 min in 10 mM sodium citrate, pH 6.0, and then incubated in sodium citrate for 30 min at room temperature. Primary antibodies were incubated overnight at 4°C. For visualization of microtubules in the K14–ensconsin microtubule-binding domain–3GFP transgenic mice, embryos were processed as previously described (Lechler and Fuchs, 2007). In brief, embryos were fixed in a solution containing 80 mM Pipes, pH 6.9, 75 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100, 3% PFA, and 0.5% glutaraldehyde for 30 min at 37°C. Embryos were washed with PBS, treated with sodium borohydride in PBS for 15 min, and then extensively washed with PBS before embedding in optimal cutting temperature. The antibodies used were mouse anti–γ-tubulin (Sigma-Aldrich), rabbit anti-Lis1 (Santa Cruz Biotechnology, Inc. and Abcam), rabbit anti-CLIP170 (Santa Cruz Biotechnology, Inc.), rat anti–β4 integrin (BD), mouse anti–β-tubulin (Sigma-Aldrich), rabbit anti–γ-catenin (Santa Cruz Biotechnology, Inc.), mouse anti-DP1/2 (Millipore), rabbit anti–α-catenin (Sigma-Aldrich), mouse anti-V5 (Invitrogen), guinea pig anti-Dsc1 (a gift from I. King, Medical Research Council, London, England, UK), and rabbit anti-Ndel1 (generated in the laboratory against the carboxy terminus of Ndel1 and affinity purified; ProteinTech Group and Abcam). Images were collected using a microscope (AxioImager Z1; Carl Zeiss) with ApoTome attachment, 63× 1.4 NA Plan Apochromat objective, MRm camera (Axiocam; Carl Zeiss), and AxioVision software (Carl Zeiss). The immersion oil used was Immersol 518F (Carl Zeiss), and the coverslips used (VWR) were no. 1.5 for tissue sections and 12-mm-round no. 1 for cultured cells. Imaging was performed at room temperature. Photoshop (Adobe) was used for postacquisition processing of brightness and contrast. Fluorescence intensity was measured using the Profile tool in the AxioVision software. The two peak pixel values at each side of the cell were used as cortical values, and the mean of the cytoplasmic values was taken. Statistical analysis was performed using a Student’s t test.
FRAP analysis
Mouse keratinocytes were grown on 35-mm glass-bottom culture dishes (no 1.5; MatTek Corporation). In the case of DPI-GFP (a gift from K. Green, Northwestern University, Chicago, IL), ninein-GFP, and Ndel1-GFP FRAP, cells were transfected using TransIT-LT1 transfection reagent (Mirus) and were induced to differentiate 24 h after transfection. In the case of Dsg3-GFP FRAP (a gift from A. Kowalczyk, Emory University, Atlanta, GA), mouse keratinocytes were infected with the adenovirus at 1:100. 24 h after addition of calcium to the media, cells were imaged. Cells were mounted on a temperature-controlled stage for 10–30 min to allow equilibration. Confocal scanning light microscopy was performed using a confocal microscope (LSM 710; Carl Zeiss) with a 63× 1.4 NA oil immersion objective. The pinhole size was 478 µm. All EGFP fusion proteins were excited using the argon 488-nm laser line and emission gated between 493 and 598 nm. FRAP experiments were performed using the regions, bleaching, and time series modules of the ZEN software (Carl Zeiss). A region of interest to be bleached was defined, and 75% laser power at the appropriate wavelength for three iterations was used to bleach signals. After bleaching, images were taken within the same focal plane at regular intervals (0.5–10 s) to monitor recovery. The mean fluorescence intensity of each region was logged for each time point in the ZEN software. For each image, a region was drawn away from a transfected cell and served as a background control. Observation bleaching was not observed, as determined by examining the fluorescence intensity over time of an unbleached region. The percentage of recovery was calculated by normalizing fluorescence intensity to background intensity and then normalizing intensity at each time point to initial intensity. The value of t1/2 was calculated with nonlinear regression without any constraints using SigmaPlot software (SPSS, Inc.). The mobile fraction was determined as mf = Imax − Io/1 − Io as previously described (Shen et al., 2008). Imax is the maximum fluorescence intensity reached after photobleaching, and Io is the initial fluorescence intensity immediately after photobleaching (Shen et al., 2008). Statistical analysis was performed using a Mann–Whitney U test in the SigmaPlot software.
Transmission EM
Backskin from e18.5 embryos was isolated and fixed in a solution of 2% glutaraldehyde, 4% PFA, 1 mM CaCl2, and 0.05 M cacodylate, pH 7.4, for 1 h at room temperature and then overnight at 4°C. Samples were washed in 0.1 M sodium cacodylate buffer containing 7.5% sucrose. Samples were postfixed in 1% osmium tetroxide in 0.15 M sodium cacodylate buffer for 1 h and then washed in two changes of 0.11 M veronal acetate buffer for 15 min each. Samples were placed into en bloc stain (0.5% uranyl acetate in veronal acetate buffer) for 1 h, washed in veronal acetate buffer, and then dehydrated in a series of 70, 95, and 100% ethanol. Samples were placed into propylene oxide for two changes of 5 min each and then placed into a 1:1 mixture of propylene oxide and Epon resin monomer for 1 h. Finally, the 1:1 mixture was replaced with 100% Epon embedding medium for 30 min. After sectioning, samples were imaged with a transmission electron microscope (CM12; Philips) run at 80 kV with a camera (XR60; Advanced Microscopy Techniques). 2Vu software (Advanced Microscopy Techniques) was used for image acquisition.
Cornified envelope preparations
Isolated epidermis was cut into 1-mm pieces and boiled in 10 mM Tris, pH 7.4, 1% β-mercaptoethanol, and 1% SDS. Resulting envelopes were diluted into PBS for microscopic analysis. Images were collected on a CFL microscope (Axiovert 40; Carl Zeiss) with an MRc camera (AxioCam) and AxioVision software.
Biochemical analyses
Epidermal extracts were prepared in 50 mM Hepes, pH 7.4, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, and 1% Triton X-100 with protease inhibitors. For immunoprecipitations, cells were transfected with ninein-GFP and a DP construct that results in production of both DPI-FLAG and DPII-FLAG. Protein G–Sepharose beads (Millipore) were prebound to rabbit anti-GFP (Abcam) or mouse anti-FLAG (Sigma-Aldrich) antibodies. Extracts were incubated with beads for 1 h with rotation at 4°C, washed four times with lysis buffer, and then boiled in Laemmli sample buffer. For pulldowns, full-length GST-tagged mouse NDEL1 was purified from bacterial lysates, and 10 µg was covalently coupled to Affi-Gel 10 (Bio-Rad Laboratories). Epidermal lysate was added, and samples were treated as for immunoprecipitations. For surface protein labeling, cells were treated for 1 h at 4°C with 1 mg/ml sulfo-NHS-SS-biotin (Thermo Fisher Scientific) in PBS with 1 mM CaCl2. After three washes with PBS containing 20 mM Tris, extracts were prepared in radioimmunoprecipitation assay buffer, and biotinylated proteins were isolated on avidin-agarose (Sigma-Aldrich). After four washes with radioimmunoprecipitation assay buffer, bound proteins were analyzed by Western blotting. Dsg3 antibodies were provided by J. Stanley (University of Pennsylvania, Philadelphia, PA). Quantitation of Western blot data was performed with ImageJ gel analysis tools (National Institutes of Health).
Online supplemental material
Fig. S1 includes a characterization of Lis1 and Ndel1 antibodies by Western blot and immunofluorescence analysis. Fig. S2 documents temporal and molecular aspects of Lis1–Ndel1 recruitment to cell junctions. Fig. S3 provides interaction data between centrosomal proteins and demonstrates their differential dynamics at the cell cortex. The DPI-dependent recruitment of CLIP170 is documented in Fig. S4. Fig. S5 shows partial colocalization between Dsg3 and the lysosomal marker LAMP1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104009/DC1.
Acknowledgments
We thank Anthony Wynshaw-Boris for mice, Kathleen Green for constructs and advice, Andrew Kowalczyk for the Dsg3-GFP adenovirus, John Stanley for anti-Dsg3 antibodies, Tim Oliver for assistance with FRAP, Julie Underwood for care of the mice, and Harold Erickson, Michel Bagnat, and members of the Lechler laboratory for comments on the manuscript.
This work was supported by grants from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR055926) and by a Basil O’Connor award from the March of Dimes.
Footnotes
Abbreviations used in this paper:
- cKO
- conditional knockout
- DP
- desmoplakin
- WT
- wild type
References
- Albee A.J., Wiese C. 2008. Xenopus TACC3/maskin is not required for microtubule stability but is required for anchoring microtubules at the centrosome. Mol. Biol. Cell. 19:3347–3356 10.1091/mbc.E07-11-1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D.H., Myers K.A., Mogensen M., Moss D., Baas P.W. 2004. Distribution of the microtubule-related protein ninein in developing neurons. Neuropharmacology. 47:677–683 10.1016/j.neuropharm.2004.07.016 [DOI] [PubMed] [Google Scholar]
- Bartolini F., Gundersen G.G. 2006. Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119:4155–4163 10.1242/jcs.03227 [DOI] [PubMed] [Google Scholar]
- Bornslaeger E.A., Corcoran C.M., Stappenbeck T.S., Green K.J. 1996. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134:985–1001 10.1083/jcb.134.4.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V., Baffet A.D., Le Droguen P.M., Casanova J., Guichet A. 2010. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev. Cell. 18:790–801 10.1016/j.devcel.2010.03.015 [DOI] [PubMed] [Google Scholar]
- Bugnard E., Zaal K.J., Ralston E. 2005. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskeleton. 60:1–13 10.1002/cm.20042 [DOI] [PubMed] [Google Scholar]
- Buttner E.A., Gil-Krzewska A.J., Rajpurohit A.K., Hunter C.P. 2007. Progression from mitotic catastrophe to germ cell death in Caenorhabditis elegans lis-1 mutants requires the spindle checkpoint. Dev. Biol. 305:397–410 10.1016/j.ydbio.2007.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins C.C., Setzer S.V., Jennings J.M., Summers S., Tsunoda K., Amagai M., Kowalczyk A.P. 2006. Desmoglein endocytosis and desmosome disassembly are coordinated responses to pemphigus autoantibodies. J. Biol. Chem. 281:7623–7634 10.1074/jbc.M512447200 [DOI] [PubMed] [Google Scholar]
- Coquelle F.M., Caspi M., Cordelières F.P., Dompierre J.P., Dujardin D.L., Koifman C., Martin P., Hoogenraad C.C., Akhmanova A., Galjart N., et al. 2002. LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089–3102 10.1128/MCB.22.9.3089-3102.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., Bornens M. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Delva E., Jennings J.M., Calkins C.C., Kottke M.D., Faundez V., Kowalczyk A.P. 2008. Pemphigus vulgaris IgG-induced desmoglein-3 endocytosis and desmosomal disassembly are mediated by a clathrin- and dynamin-independent mechanism. J. Biol. Chem. 283:18303–18313 10.1074/jbc.M710046200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantopoulos G.S., Perez F., Goodson H.V., Batelier G., Melki R., Kreis T.E., Rickard J.E. 1999. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144:99–112 10.1083/jcb.144.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner N.E., Dujardin D.L., Tai C.Y., Vaughan K.T., O’Connell C.B., Wang Y., Vallee R.B. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2:784–791 10.1038/35041020 [DOI] [PubMed] [Google Scholar]
- Gallicano G.I., Kouklis P., Bauer C., Yin M., Vasioukhin V., Degenstein L., Fuchs E. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009–2022 10.1083/jcb.143.7.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. 2006. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 17:680–689 10.1091/mbc.E05-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman M.J., Sisi P., Banbury D.N., Byrne C. 1998. Patterned acquisition of skin barrier function during development. Development. 125:1541–1552 [DOI] [PubMed] [Google Scholar]
- Hirotsune S., Fleck M.W., Gambello M.J., Bix G.J., Chen A., Clark G.D., Ledbetter D.H., McBain C.J., Wynshaw-Boris A. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 19:333–339 10.1038/1221 [DOI] [PubMed] [Google Scholar]
- Jonkman M.F., Pasmooij A.M., Pasmans S.G., van den Berg M.P., Ter Horst H.J., Timmer A., Pas H.H. 2005. Loss of desmoplakin tail causes lethal acantholytic epidermolysis bullosa. Am. J. Hum. Genet. 77:653–660 10.1086/496901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. 2007. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 176:147–154 10.1083/jcb.200609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Mushika Y., Ichii T., Takeichi M. 2008. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 135:948–959 10.1016/j.cell.2008.09.040 [DOI] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. 2005. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7:961–968 10.1038/ncb1306 [DOI] [PubMed] [Google Scholar]
- Nguyen M.D., Shu T., Sanada K., Larivière R.C., Tseng H.C., Park S.K., Julien J.P., Tsai L.H. 2004. A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat. Cell Biol. 6:595–608 10.1038/ncb1139 [DOI] [PubMed] [Google Scholar]
- Niethammer M., Smith D.S., Ayala R., Peng J., Ko J., Lee M.S., Morabito M., Tsai L.H. 2000. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 28:697–711 10.1016/S0896-6273(00)00147-1 [DOI] [PubMed] [Google Scholar]
- Ohama Y., Hayashi K. 2009. Relocalization of a microtubule-anchoring protein, ninein, from the centrosome to dendrites during differentiation of mouse neurons. Histochem. Cell Biol. 132:515–524 10.1007/s00418-009-0631-z [DOI] [PubMed] [Google Scholar]
- O’Keefe E.J., Erickson H.P., Bennett V. 1989. Desmoplakin I and desmoplakin II. Purification and characterization. J. Biol. Chem. 264:8310–8318 [PubMed] [Google Scholar]
- Pasdar M., Li Z., Krzeminski K.A. 1992. Desmosome assembly in MDCK epithelial cells does not require the presence of functional microtubules. Cell Motil. Cytoskeleton. 23:201–212 10.1002/cm.970230304 [DOI] [PubMed] [Google Scholar]
- Penn E.J., Burdett I.D., Hobson C., Magee A.I., Rees D.A. 1987. Structure and assembly of desmosome junctions: biosynthesis and turnover of the major desmosome components of Madin-Darby canine kidney cells in low calcium medium. J. Cell Biol. 105:2327–2334 10.1083/jcb.105.5.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Diamantopoulos G.S., Stalder R., Kreis T.E. 1999. CLIP-170 highlights growing microtubule ends in vivo. Cell. 96:517–527 10.1016/S0092-8674(00)80656-X [DOI] [PubMed] [Google Scholar]
- Quintyne N.J., Gill S.R., Eckley D.M., Crego C.L., Compton D.A., Schroer T.A. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321–334 10.1083/jcb.147.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir T., Elbaum M., Reiner O. 1997. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 16:6977–6984 10.1093/emboj/16.23.6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., Hirotsune S. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 28:681–696 10.1016/S0896-6273(00)00146-X [DOI] [PubMed] [Google Scholar]
- Shen L., Weber C.R., Turner J.R. 2008. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J. Cell Biol. 181:683–695 10.1083/jcb.200711165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.Y., Samuels B.A., Wang J., Neumayer G., Belzil C., Ayala R., Shi Y., Shi Y., Tsai L.H., Nguyen M.D. 2008. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J. Biol. Chem. 283:12232–12240 10.1074/jbc.M710200200 [DOI] [PubMed] [Google Scholar]
- Smith E.A., Fuchs E. 1998. Defining the interactions between intermediate filaments and desmosomes. J. Cell Biol. 141:1229–1241 10.1083/jcb.141.5.1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck T.S., Green K.J. 1992. The desmoplakin carboxyl terminus coaligns with and specifically disrupts intermediate filament networks when expressed in cultured cells. J. Cell Biol. 116:1197–1209 10.1083/jcb.116.5.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H.A., Scothern A., McHarg S., Garrod D.R. 2010. Desmosomes: adhesive strength and signalling in health and disease. Biochem. J. 429:419–433 10.1042/BJ20100567 [DOI] [PubMed] [Google Scholar]
- Uzumcu A., Norgett E.E., Dindar A., Uyguner O., Nisli K., Kayserili H., Sahin S.E., Dupont E., Severs N.J., Leigh I.M., et al. 2006. Loss of desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J. Med. Genet. 43:e5 10.1136/jmg.2005.032904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Degenstein L., Wise B., Fuchs E. 2001a. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 104:605–617 10.1016/S0092-8674(01)00246-X [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bowers E., Bauer C., Degenstein L., Fuchs E. 2001b. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085 10.1038/ncb1201-1076 [DOI] [PubMed] [Google Scholar]
- Wacker I.U., Rickard J.E., De Mey J.R., Kreis T.E. 1992. Accumulation of a microtubule-binding protein, pp170, at desmosomal plaques. J. Cell Biol. 117:813–824 10.1083/jcb.117.4.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. 2007. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin. Genet. 72:296–304 10.1111/j.1399-0004.2007.00888.x [DOI] [PubMed] [Google Scholar]