JNK-mediated phosphorylation of the mRNA-decapping protein DCP1a disrupts P body structure, mRNA stability, and gene expression in response to stress and inflammatory stimuli.
Abstract
Cytokines and stress-inducing stimuli signal through c-Jun N-terminal kinase (JNK) using a diverse and only partially defined set of downstream effectors. In this paper, the decapping complex subunit DCP1a was identified as a novel JNK target. JNK phosphorylated DCP1a at residue S315 in vivo and in vitro and coimmunoprecipitated and colocalized with DCP1a in processing bodies (P bodies). Sustained JNK activation by several different inducers led to DCP1a dispersion from P bodies, whereas IL-1 treatment transiently increased P body number. Inhibition of TAK1–JNK signaling also affected the number and size of P bodies and the localization of DCP1a, Xrn1, and Edc4. Transcriptome analysis further identified a central role for DCP1a in IL-1–induced messenger ribonucleic acid (mRNA) expression. Phosphomimetic mutation of S315 stabilized IL-8 but not IκBα mRNA, whereas overexpressed DCP1a blocked IL-8 transcription and suppressed p65 NF-κB nuclear activity. Collectively, these data reveal DCP1a as a multifunctional regulator of mRNA expression and suggest a novel mechanism controlling the subcellular localization of DCP1a in response to stress or inflammatory stimuli.
Introduction
JNKs have been discovered as serine/threonine protein kinases stimulated by cycloheximide (Kyriakis et al., 1994), UV light (Dérijard et al., 1994), or IL-1 (interleukin-1; Kracht et al., 1994). Three JNK genes, which are present only in higher eukaryotes, encode up to 10 homologous isoforms designated “short” JNKs (Mr of 45–48 kD) or “long” JNKs (Mr of 54–57 kD; Gupta et al., 1996). Mice deficient in one JNK gene are viable, whereas double deficiency of JNK1 and JNK2 genes is embryonic lethal (Kuan et al., 1999). In mouse models of disease, JNKs play roles in diabetes, ischemia/reperfusion injury, rheumatoid arthritis, deafness, and tumor progression (Johnson and Nakamura, 2007).
Despite their widespread activation by a plethora of inflammatory or environmental stimuli, including any form of stress, the number of identified JNK substrates or direct downstream effectors that may account for these in vivo phenotypes is still fairly small (Bogoyevitch and Kobe, 2006) and mainly comprises proteins involved in gene transcription (e.g., c-Jun and ATF-2), apoptosis (e.g., bcl2), insulin signaling (IRS-1), or neurodegeneration (e.g., Tau). JNK catalytic activity is stimulated by sequential phosphorylation through kinase cascades involving MAPKKKKs (e.g., germinal center kinase), MAPKKKs (e.g., TAK1 [TGF-β–activated kinase 1] and MEKK1 [mitogen-activated extracellular-regulated kinase kinase kinase 1]), and MAPKKs (e.g., MKK4 and MKK7; Gaestel et al., 2009; Zhong et al., 2009). JNKs can also bind to these activators, their substrates (such as c-Jun), or to scaffolding proteins (such as JIP1–4 [JNK-interacting protein 1–4]), implying that an as yet unknown array of dynamically built signaling complexes recruits JNKs to precisely direct their manifold subcellular functions (Johnson and Nakamura, 2007; Weston and Davis, 2007).
Many of the stressors that activate JNK also affect formation of compositionally related cytoplasmic RNP (ribonucleoprotein) granules, called processing bodies (P bodies) and stress granula (Anderson and Kedersha, 2009; Buchan and Parker, 2009). mRNPs contain messenger RNAs, which are enclosed by a protein coat of factors binding to the m7G (N7-methylguanosine) cap structure elements, to AU (adenine uridine)-rich elements, or to the poly(A) tail. These proteins regulate association with polyribosomes and active translation, or they promote sequential deadenylation (Yamashita et al., 2005), decapping (Sheth and Parker, 2003), and subsequent decay of mRNAs (Cougot et al., 2004; Eulalio et al., 2007a; Buchan and Parker, 2009). P bodies and stress granula share protein components, such as the 5′–3′ exonuclease Xrn1 (Kedersha et al., 2005). P bodies are distinguished by various subunits of the decapping complex, including DCP2 (Beelman et al., 1996) and DCP1a (Lykke-Andersen, 2002; Fenger-Grøn et al., 2005) and are considered as primary sites of mRNA degradation. However, dispersed P body proteins are still fully competent for mRNA decay (Eulalio et al., 2007b). Hence, P bodies may fulfill additional functions, such as transient mRNA storage or sequestration of RNA-metabolizing enzymes (Franks and Lykke-Andersen, 2008).
P bodies become detectable when levels of nontranslatable mRNA increase, and they disassemble upon depletion of cytoplasmic pools of mRNA (Teixeira et al., 2005; Eulalio et al., 2007b). The signals that regulate aggregation or disaggregation of P bodies in mammalian cells are not well understood (Franks and Lykke-Andersen, 2008; Ohn et al., 2008). Recently, phosphorylation of DCP2 by the yeast MAPKKKK Ste20 was shown to affect the decay of distinct mRNAs, providing the first example of signal-dependent regulation of the decapping machinery (Yoon et al., 2010).
In mammalian cells, a TAK1–JNK pathway integrates signals from conditions as divergent as osmotic stress (Huangfu et al., 2006) or inflammatory cytokine signaling (Shim et al., 2005). TAK1, the regulatory subunits TAB1–3 (TAK1-binding protein 1–3), and JNK are core parts of a signaling module that regulates transcription, mRNA stability, and translation of IL-1 target genes (Krause et al., 1998; Holtmann et al., 1999, 2001; Winzen et al., 1999, 2007; Hoffmann et al., 2005; Wolter et al., 2008; Dhamija et al., 2010). TAK1 also activates the transcription factor NF-κB, which cooperates with JNK to activate genes at the level of chromatin (Holtmann et al., 2001; Hoffmann et al., 2005; Thiefes et al., 2005; Wolter et al., 2008).
IL-1–mediated gene expression occurs within 30–60 min and is usually terminated after a few hours. The kinetics and amplitude of mRNA synthesis, degradation, and translation are regulated by multiple mechanisms that operate in parallel within a signaling network, which is still incompletely understood (Hoffmann et al., 2002; Gaestel et al., 2009; Weber et al., 2010).
Here, we report that JNK-mediated phosphorylation of DCP1a at S315 controls subcellular localization of DCP1a as well as P body formation during translational stress but also in response to IL-1. We also demonstrate an additional role of DCP1a in IL-1– and NF-κB–dependent transcriptional regulation.
Results
Ectopic activation of the JNK pathway affects localization and modification of DCP1a
We transiently transfected GFP-DCP1a, which essentially behaves like endogenous DCP1a as a marker for P bodies (Sheth and Parker, 2003; Tritschler et al., 2009). To analyze the cellular localization of GFP-DCP1a under conditions of activated JNK, we cotransfected JNK, MEKK1-BFP, or a TAK1-TAB1 fusion protein (Fig. 1 A; Sakurai et al., 2002). MEKK1-BFP and TAK1-TAB1 strongly activated endogenous JNK as assessed by phosphorylation of the JNK substrate c-Jun, whereas expression of JNK alone had little effect on c-Jun phosphorylation (Fig. 1 B). There was no effect on the overall expression levels of GFP-DCP1a (Fig. 1 B). Cells expressing GFP-DCP1a and MEKK1 or TAK1-TAB1 showed a dramatic dispersion of P bodies to large structures, covering essentially major areas of the entire cytoplasm (Fig. 1 A, lanes 2 and 5). These structures correspond to ubiquitously distributed GFP-DCP1a in the cytoplasm, an effect that was also partially seen with transfected JNK alone (Fig. 1 A, lane 3) but not with a catalytically inactive JNK2 mutant (Fig. 1 A, lane 4). In fact, the JNK mutant colocalized exactly with GFP-DCP1a in P bodies (Fig. 1 A, lanes 4 and 7), implying that loss of enzymatic activity stabilizes potential intracellular interactions of JNK with DCP1a in intact cells. Under conditions of maximal JNK activation (i.e., overexpression of JNK plus TAK1-TAB1 or of MEKK1-BFP), JNK also colocalized with GFP-DCP1a in the cytoplasm as evident from the appearance of overlapping fluorescence signals (Fig. 1 A, lanes 2 and 6). These data indicated that JNK activation is dynamically linked to P body formation and to the association of DCP1a with JNK.
Figure 1.
JNK activators affect localization of DCP1a to P bodies. (A) HEK293IL-1R cells were transfected with vectors encoding GFP-DCP1a together with JNK activators (MEKK1 or a TAK1-TAB1 fusion protein) or with JNK wild type or a catalytically inactive mutant (JNK2K55R). Colocalization of GFP-DCP1a (green) with JNK (red) was analyzed by fluorescence microscopy. The fraction of transfected cells exhibiting a phenotype identical to that shown in the representative image was determined by scoring ≥50 cells from three to four independent transfections. Bar, 10 µm. IF, indirect immunofluorescence. (B) HEK293IL-1R cells were transfected in parallel cultures, and modifications of GFP-DCP1a were analyzed by mobility shifts (labeled 1–3) upon SDS-PAGE and subsequent Western blot analysis. Equal expression and activity of cotransfected constructs were validated by anti-JNK, anti-GFP, anti-TAK1, and antiphospho–c-Jun (P-c-Jun) antibodies. Proteins from the same samples were separated on three gels and transferred to three membranes (M1–M3), which were probed with the indicated antibodies. Ponceau staining or hybridization with antiactin or antitubulin antibodies is shown to indicate equal protein loading. IB, immunoblot; n.s., nonspecifically detected protein band that served as a loading control.
JNK phosphorylates DCP1a at S315
In the experiments shown in Fig. 1 B, JNK activation by MEKK1 or TAK1-TAB1 resulted in strong mobility shifts of DCP1a upon SDS-PAGE. These shifts are indicative of multisite phosphorylation. At least two different phosphorylation states (designated form 2 and form 3) were observed (Fig. 1 B). Form 2 was further converted to form 3 by coexpression of TAK1-TAB1 with wild-type JNK2 but not with JNK2K55R (Fig. 1 B). Using this assay, mutational analysis of a total of 20 candidate proline-directed MAPK phosphorylation sites (Fig. S1 A) revealed S315 as a possible JNK target site in DCP1a because this mutant showed a conversion of form 2 back to form 1 upon TAK1-TAB1 transfection (Fig. S1 B). Moreover, JNK phosphorylated GST-DCP1a directly in vitro (Fig. 2 A). Phosphospecific antibodies against S315 confirmed that DCP1a is phosphorylated by JNK at this site in vitro (Fig. 2 B) as well as in cells overexpressing JNK activators (Fig. 2 C). In contrast, p38 MAPK did not phosphorylate S315 of DCP1a in vitro (unpublished data).
Figure 2.
JNK phosphorylates DCP1a at S315. (A) HEK293IL-1R cells were stimulated for 60 min with 0.5 M sorbitol or were left untreated. JNK was immunoprecipitated from cell extracts, and immune complex protein kinase assays were performed by addition of GST–c-Jun1–135 or GST-DCP1a and radioactive ATP. (top) Reaction mixtures were separated by SDS-PAGE and visualized by autoradiography. Equal loading of GST–c-Jun1–135 or GST-DCP1a substrates was confirmed by Coomassie brilliant blue (CBB) staining of gels. exp., exposure. (B) HEK293IL-1R cells were transfected with HA-JNK2, GFP-DCP1a, or GFP-DCP1aS315A. 24 h later, HA-JNK2–transfected cells were stimulated for 60 min with 0.5 M sorbitol to activate JNK or were left untreated. Lysates were prepared, and JNK or GFP-DCP1a proteins were immunoprecipitated and mixed. In vitro kinase assays were started by addition of ATP and analyzed by immunoblotting for phosphorylation of DCP1a at S315. Proteins in lysates and immunoprecipitates were analyzed by the indicated antibodies. (C) HEK293IL-1R cells were transfected as described in the legend for Fig. 1 B, but GFP-DCP1a constructs were omitted. Phosphorylation of endogenous DCP1a was analyzed by SDS-PAGE/immunoblotting using P(S315)-DCP1a and DCP1a antibodies, respectively. Black lines indicate that intervening lanes have been spliced out. IB, immunoblot; IP, immunoprecipitation.
JNK interacts with DCP1a and mediates DCP1a phosphorylation at S315 in response to cytokines and stressors
The kinetics of JNK activation varies depending on the stimulus. Proinflammatory stimuli like IL-1 cause transient JNK activation, whereas osmotic or translational stress by sorbitol or anisomycin, respectively, causes long-lasting and strong activation of JNK and p38 MAPK (Fig. 3 A). IL-1 induced a more modest and transient increase in DCP1a phosphorylation, whereas sorbitol and anisomycin induced a rapid and quantitative shift of endogenous DCP1a to form 2 as well as phosphorylation at S315 (Fig. 3 A). Inhibition of JNK by SP60025, but not inhibition of p38 MAPK by SB203580, inhibited IL-1– or anisomycin-induced DCP1a phosphorylation by ∼100, whereas sorbitol-induced phosphorylation was inhibited by 30–40% (Fig. 3 B). SP600125 did not strongly affect the mobility shift of DCP1a upon SDS-PAGE, indicating that DCP1a undergoes additional modifications at other amino acids (Fig. 3 B). One of the modified residues is S319, which is also phosphorylated by activated JNK + TAK1-TAB1 (Fig. S1 and not depicted). A specific inhibitor of TAK1, 5Z-7-oxozeaenol (Ninomiya-Tsuji et al., 2003), almost completely prevented both phosphorylation of DCP1a at S315 as well as the mobility shifts upon SDS-PAGE in response to IL-1, anisomycin, and sorbitol (Fig. 3 C). As TAK1 is an upstream activator of several protein kinases, including JNK and p38 MAPK (Shim et al., 2005; Thiefes et al., 2005), this result suggested that DCP1a is subject to multiple modifications exerted by several enzymes.
Figure 3.
JNK interacts with DCP1a and controls phosphorylation of DCP1a at S315 in response to stress stimuli and IL-1. (A) HEK293IL-1R cells were left untreated (−) or were treated for the indicated times with 10 ng/ml IL-1α, 10 µg/ml anisomycin, 0.2% DMSO, or 0.5 M sorbitol. Lysates were immunoblotted, and phosphorylation of DCP1a, JNK, p38 MAPK, and c-Jun was analyzed by the indicated antibodies. (B and C) Cells were left untreated or were treated as indicated for 30 min with solvent (0.05% DMSO) or 20 µM SP600125, 2 µM SB203580, or 1 µM 5Z-7-oxozeaenol to inhibit JNK, p38 MAPK, or TAK1, respectively. Thereafter, cells were treated with IL-1α, anisomycin, or sorbitol for 1 h, and expression and phosphorylation of DCP1a and phosphorylation of p55 JNK were analyzed as in A. In B, proteins from IL-1– and anisomycin-treated samples were separated side by side on three separate gels and transferred to three membranes (M1–M3), which were probed with the indicated antibodies. Loading controls for M1–M3 are shown by antiactin antibodies or Ponceau or Coomassie brilliant blue (CBB) staining of membranes. Proteins from sorbitol-treated samples were loaded twice on the same gel and transferred to the same membrane. One half of the membrane (M4a) was probed with antiphospho-DCP1a (P-DCP1a), DCP1a, and actin antibodies, and the other half (M4b) was probed with antiphospho-JNK (P-JNK) and antiactin antibodies. (D) HEK293IL-1R cells were treated for 1 h with IL-1α, anisomycin, or sorbitol. Then cytosolic (S2) and P body–enriched particular (P2) fractions were prepared as previously described in Teixeira et al. (2005). Immunoprecipitations were performed using anti-JNK or -IgG antibodies and analyzed for coprecipitation of DCP1a. Blots were also probed for p54 JNK. Proteins from the same lysates were separated on two gels and transferred to two membranes (M1 and M2). M1 was hybridized with DCP1a and antiactin antibodies, and M2 was hybridized with anti-JNK and antiactin antibodies. Coomassie brilliant blue stains for M1 and M2 are shown to indicate equal protein loading of S2 and P2 fractions. Black lines indicate that intervening lanes have been spliced out. rel., relative; IB, immunoblot; IP, immunoprecipitation; n.s., nonspecifically detected protein band that served as a loading control.
Here, we focused on JNK, which clearly is a direct S315 kinase. To study the interaction of JNK with DCP1a, we fractionated cell extracts into soluble and particular fractions as described for yeast (Teixeira et al., 2005). The particular fraction (P2) is enriched in P bodies (Fig. S2), whereas the soluble fraction (S2) contains cytosolic DCP1a (Fig. 3 D). DCP1a coprecipitated with endogenous JNK (Fig. 3 D). This interaction was independent of stimulation and occurred in S2 and P2 fractions (Fig. 3 D). We did not find coimmunoprecipitation with Edc4 or Xrn1 (unpublished data). These data indicated that JNKs form a stable complex with DCP1a and interact with specific subunits of the decapping complex.
JNK activation affects localization of DCP1a, Edc4, and Xrn1 to P bodies
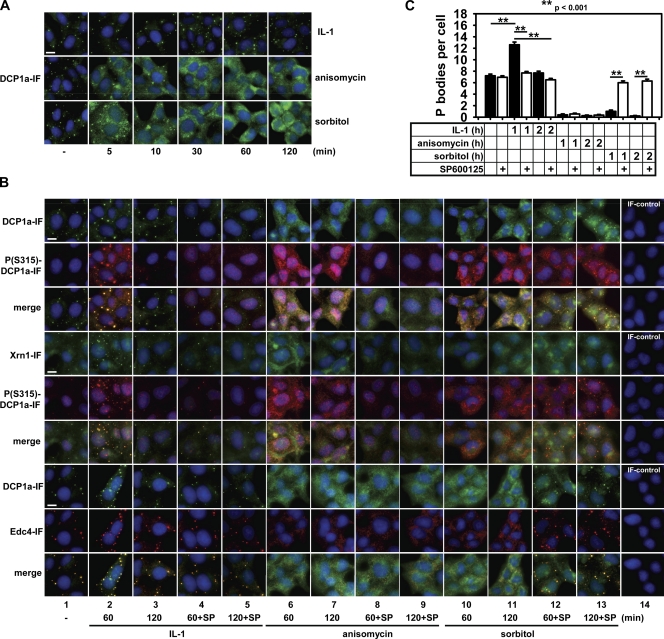
We then studied the localization of P body components under conditions of JNK activation or JNK inhibition in human cells. Sorbitol and anisomycin treatment led to complete disappearance of DCP1a from P bodies and to its redistribution to the cytoplasm within 1–2 h (Fig. 4, A and B), resembling the effects of overexpressed JNK, TAK1-TAB1, or MEKK1 as shown in Fig. 1 A. In contrast, IL-1 treatment transiently increased the number of P bodies by ∼75% after 1 h of stimulation (Fig. 4, A–C). In untreated cells, there was very little DCP1a phosphorylation and localization of DCP1a in P bodies merged with Edc4 and Xrn1 as expected (Fig. 4 B, lane 1). After 1 h of IL-1 stimulation, phosphorylation of DCP1a in the cytoplasm, but much more pronounced in all P bodies, was strongly induced (Fig. 4 B, lane 2). P body–associated Xrn1 colocalized with phosphorylated DCP1a, whereas cytosolic Xrn1 showed fewer overlapping fluorescence signals (Fig. 4 B, lane 2). Upon IL-1 stimulation, Edc4 colocalized with DCP1a and phospho-DCP1a in all P bodies (Fig. 4 B, lane 2). SP600125 strongly inhibited the IL-1–induced increase in P body number as well as DCP1a phosphorylation (Fig. 4 B, lanes 4 and 5). Thus, in the IL-1 pathway, JNK-mediated DCP1a phosphorylation is strictly associated with P body assembly. Anisomycin caused strong DCP1a phosphorylation and disruption of P body structures as judged from the dispersed localization of DCP1a, Xrn1, and Edc4 (Fig. 4 B, lanes 6 and 7). In line with Western blot analyses of total cell extracts (Fig. 3 B), SP600125 blocked anisomycin-induced DCP1a phosphorylation (Fig. 4 B, lanes 8 and 9). However, this did not restore P body structures (Fig. 4 B, lanes 8 and 9). Similar to anisomycin, sorbitol treatment caused strong DCP1a phosphorylation and P body disassembly (Fig. 4 B, lanes 10 and 11). However, in contrast to anisomycin, SP600125 only weakly affected total cellular DCP1a phosphorylation, but it significantly restored the formation of P body–like structures containing Xrn1 and Edc4 (Fig. 4 B, lanes 12 and 13). Anisomycin inhibits translation at the elongation step (Tscherne and Pestka, 1975) similar to cycloheximide, which prevents formation of RNP particles by lowering the cytoplasmic pool of nontranslatable RNA (Cougot et al., 2004). Therefore, these results suggested that in the anisomycin pathway, RNA depletion is likely to override phosphorylation-dependent control by DCP1a, whereas in the IL-1 and sorbitol pathways, JNK controls P body formation through DCP1a phosphorylation. In support for this conclusion, we found that DCP1a phosphorylation was strongly reduced in murine embryonic fibroblasts (MEFs) lacking JNK but not p38 MAPK (Fig. S3 A). Moreover, unlike wild-type or p38-deficient MEFs, cells lacking JNK1/2 were largely devoid of P bodies (Fig. S3 B). These data support a role of JNK, but not p38 MAPK, in P body formation, in line with a lack of SB203580 to affect the sorbitol-induced reduction of P bodies (Fig. S3 C) in human cells.
Figure 4.
JNK-mediated phosphorylation of DCP1a at S315 controls DCP1a localization and P body number. (A) HEK293IL-1R cells were left untreated (−) or were stimulated for the indicated times with 10 ng/ml IL-1α, 10 µg/ml anisomycin, or 0.5 M sorbitol, and DCP1a-containing P bodies (green) were analyzed by fluorescence microscopy. (B) Cells were stimulated for 1 and 2 h as in A. In addition, cells were pretreated for 30 min with 20 µM SP600125 as indicated. Localization of DCP1a, P(S315)-DCP1a, Edc4, and Xrn1 in P bodies was analyzed by double immunofluorescence as indicated. IF-control indicates samples from which first antibodies were omitted. Bars, 10 µm. IF, indirect immunofluorescence. (C) Statistics of P body number per cell under conditions of B. Depicted is the mean P body number per cell ± SEM counted in ≥50 cells taken from two time points. Asterisks indicate significant changes for pairwise comparisons.
Mutation of S315 in DCP1a affects localization but not decapping activity
To establish a direct link between DCP1a phosphorylation, localization, and function, we transiently expressed DCP1a versions carrying inactivating (S315A) or phosphomimetic (S315D) mutants in HEK293IL-1R cells. GFP-DCP2 or a C-terminal truncation mutant of GFP-DCP1a, GFP-DCP1aΔ515–582, was expressed in parallel (Fig. 5). Amino acids 515–582 of DCP1a were recently shown to be required for DCP1a trimerization, interaction with other decapping factors, and decapping enhancer activity (Tritschler et al., 2009). In line with the latter study, immunoprecipitated GFP-DCP2 was active, whereas GFP-DCP1aΔ515–582 had hardly any detectable decapping activity (Fig. 5, left, lanes 2 and 6). Weak endogenous decapping activity was associated with immunoprecipitated GFP-DCP1a, which was, however, independent from mutations of S315 (Fig. 5, left, lanes 3–5). Thus, phosphorylation of DCP1a at S315 is unlikely to affect global cellular decapping activity.
Figure 5.
Phosphorylation at S315 does not affect cellular decapping activity. HEK293IL-1R cells were transiently transfected with expression vectors encoding GFP-DCP2, wild-type GFP-DCP1a, inactivating (GFP-DCP1aS315A), or phosphomimetic (GFP-DCP1aS315D) mutants or a C-terminal deletion mutant of GFP-DCP1a (Δ515–582). 24 h later, cells were lysed, and DCP2 or DCP1a were immunoprecipitated using anti-GFP antibodies or IgG as a control. Three fourths of the immunoprecipitates were incubated with a 32P cap–labeled luciferase mRNA (32P-luc. mRNA). After 30 min at 30°C, reactions were stopped, and reaction mixtures were separated by thin layer chromatography. The luciferase mRNA substrate and the 32P-labeled m7GDP cap structures that were hydrolyzed by exogenous DCP2 or by decapping activity associated with DCP1a immunoprecipitates were visualized by autoradiography. 32P-labeled m7GTP indicates a reaction product that was cleaved unspecifically. The correct size of m7GDP was verified by nonradioactive standards run in parallel (not depicted). Lysates and one fourth of the immunoprecipitates were analyzed for equal expression and immunoprecipitation by anti-GFP antibodies. Shown is one representative out of three experiments. IP, immunoprecipitation.
To reveal a direct link between DCP1a phosphorylation and localization, we generated stable cell lines expressing comparable amounts of inactivating and phosphomimetic mutants of S315 (Fig. 6 A). Compared with wild-type GFP-DCP1a (8.6 ± 0.5 P bodies per cell; Fig. 6 B), mutation of S315 to alanine increased the number (12 ± 0.2 P bodies per cell; Fig. 6 B) and size of P bodies (Fig. 6 C, lane 2), whereas mutation to asparagine strongly reduced P body number (3.4 ± 1.1 P bodies per cell; Fig. 6 B) and led to cytosolic redistribution of GFP-DCP1a (Fig. 6 C, lane 3). In line with the results shown in Fig. 4 B, the distribution of Xrn1 and Edc4 followed the localization of GFP-DCP1a and its mutants, suggesting that S315 affects P body formation (Fig. 6 C, lanes 1–3). JNK was detected throughout the cell, including the nucleus, as expected (Fig. 6 C). However, JNK was also detected in RNP-like structures in which it colocalized with GFP-DCP1a (Fig. 6 C, bottom insets). The S315A mutation stabilized colocalization of JNK with GFP-DCP1a in P body structures (Fig. 6 C). A dispersion of P body structures was also seen with the C-terminal truncation mutant GFP-DCP1aΔ515–582 (Fig. 6, B and C), which was consistant with a recent study (Tritschler et al., 2009). Compared with the GFP-DCP1aS315D mutant, more Edc4- and Xrn1-containing RNPs remained detectable in cells expressing the GFP-DCP1aΔ515–582 mutant (Fig. 6 C). Collectively, the data shown in Fig. 6, therefore, suggest that at the protein level, DCP1a localization is regulated by at least two mechanisms, one involving JNK-mediated phosphorylation at S315 and the other involving oligomerization and interaction of its C terminus with other components of mRNPs, such as Edc4 and DCP2 (Tritschler et al., 2009).
Figure 6.
Phosphorylation at S315 affects number and size of DCP1a-containing P bodies. HEK293IL-1R cells were stably transfected with wild-type GFP-DCP1a (wt), inactivating (GFP-DCP1aS315A), or phosphomimetic (GFP-DCP1aS315D) mutants or a C-terminal deletion mutant of GFP-DCP1a (Δ515–582). (A) All mutants were expressed to similar amounts as assessed by immunoblotting. (B) Quantification of P bodies from stable cell lines described in A. Depicted is the mean GFP-DCP1a–positive P body number per cell ± SEM counted in ≥200 cells taken from two independent stable transfections. P-values indicate significant changes compared with GFP-DCP1a wild-type cells. (C) Colocalization of endogenous Xrn1, (top left, red), Edc4 (top right, red), or JNK (bottom, red) with stably expressed GFP-DCP1a (green) was analyzed by fluorescence microscopy. Boxes and insets show enlarged views of single P bodies that contain JNK. IF-control indicates samples from which the first antibodies were omitted. Bars, 10 µm. IF, indirect immunofluorescence; IB, immunoblotting. Black lines indicate that intervening lanes have been spliced out.
Ectopic expression of DCP1a mutants affect mRNA levels of IL-1–induced genes
The clear differences in subcellular localization of stably transfected GFP-DCP1a mutants suggested that these cells were useful for assessing the relevance of DCP1a phosphorylation for IL-1–mediated gene expression. In two independent experiments, we analyzed genome-wide mRNA expression after 3 h of IL-1 treatment by DNA microarray experiments. Out of 15,713 annotated genes, 10,743 mRNAs were expressed at detectable levels. 42 mRNAs were induced by IL-1 by at least twofold in parental HEK293IL-1R cells in both independent experiments (Fig. S4). Of those, in GFP-DCP1a–expressing cells, 28 genes (67%) and, in GFP-DCP1aS315D– or in GFP-DCP1aΔ515–582–expressing cells, 11 genes (26%) were still inducible by IL-1 by at least 1.5-fold, whereas in the GFP-DCP1aS315A mutant cells, the entire IL-1 gene response was impaired (Fig. S4). In untreated cells, 15–32 genes were up-regulated, and 252–360 genes were down-regulated in the four cell lines, indicating that GFP-DCP1a overexpression exerts minor effects on up-regulation of genes but negatively affects ∼2.3–3.4% of all detectable transcripts. However, there were little differences between the various mutants and wild-type GFP-DCP1a on this set of transcripts (unpublished data).
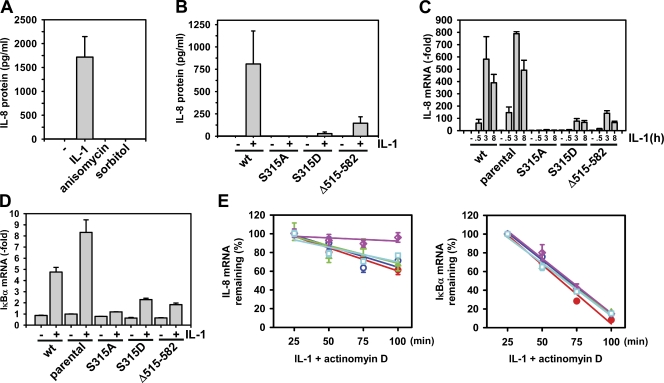
We focused on IL-8, whose transcriptional and posttranscriptional regulation we have investigated previously in great detail (Holtmann et al., 2001; Winzen et al., 2007; Dhamija et al., 2010). As expected, IL-1 induced strong IL-8 secretion in parental HEK293IL-1R cells (Fig. 7 A), whereas the translational inhibitors anisomycin and sorbitol failed to induce IL-8 secretion (Fig. 7 A). Compared with parental cells, ectopic expression of wild-type GFP-DCP1a did not affect IL-8 secretion, whereas expression of either GFP-DCP1aS315D or GFP-DCP1aΔ515–582 strongly suppressed IL-1–induced IL-8 protein secretion. Inducible IL-8 secretion was completely absent in GFP-DCP1aS315A–expressing cells (Fig. 7 B). In line with this result and with the microarray data, GFP-DCP1aS315D or GFP-DCP1aΔ515–582 also strongly suppressed, but not completely abolished, IL-8 mRNA expression (Fig. 7 C). A similar pattern of inhibition by the DCP1a mutants was seen for IκBα mRNA, another IL-1–inducible transcript (Fig. 7 D). To assess the relevance of DCP1a for IL-8 and IκBα mRNA stability, we induced mRNA synthesis by IL-1 treatment for 1 h, and then followed mRNA decay over time after the addition of actinomycin D (Fig. 7 E). About 40–50% of IL-8 mRNA and 90% of IκBα mRNA were degraded after 100 min (Fig. 7 E). IL-8 mRNA decay was almost completely impaired by the GFP-DCP1aS315D mutant but not by the other GFP-DCP1a mutants or wild-type GFP-DCP1a (Fig. 7 E). Moreover, the stabilizing effect of GFP-DCP1aS315D was transcript specific, as none of the GFP-DCP1a versions affected the rapid decay of IκBα (Fig. 7 E). It should be noted that basal mRNA steady-state levels for IκBα and IL-8 were similar in all cell lines (Fig. 7, C and D).
Figure 7.
Phosphomimetic mutants of DCP1a at S315 affect IL-1–induced IL-8 expression and mRNA stability. (A and B) HEK293IL-1R cells (A) or stably transfected cell lines (B) as described in Fig. 6 were left untreated (−) or were treated for 8 h with 10 ng/ml IL-1α, 10 µg/ml anisomycin, or 0.5 M sorbitol. IL-8 protein secreted in the cell culture supernatant was determined by ELISA and normalized for cell number according to total protein content of harvested cells. Shown is the mean IL-8 amount ± SEM from two independent experiments. The minus sign indicates samples from untreated cells. (C) HEK293IL-1R cells (parental) or stable cell lines as described in Fig. 6 were stimulated for the indicated times with 10 ng/ml IL-1α, and IL-8 mRNA expression was determined by quantitative RT-PCR. Shown is the mean fold increase ± SEM compared with unstimulated GFP-DCP1a (wt, wild type)–expressing cells from duplicate measurements of two independent experiments. (D) Cells as in C were stimulated for 1 h, and IκBα mRNA expression was analyzed and quantified as in C. (E) HEK293IL-1R cells (parental, red) or stable cell lines (GFP-DCP1a, blue; GFP-DCP1aS315A, green; GFP-DCP1aS315D, purple; GFP-DCP1aΔ515–582, light blue) were treated for 1 h with 10 ng/ml IL-1α to induce mRNA transcription. Thereafter, 5 µg/ml actinomycin D was added, and total RNA was isolated after the indicated times and analyzed for the expression of IL-8 or IκBα mRNA by quantitative RT-PCR. Shown are relative mRNA levels ± SEM from duplicate measurements of three independent experiments.
Collectively, the results shown in Fig. 5, Fig. 6, and Fig. 7 suggested that phosphorylation of DCP1a at S315 regulates DCP1a localization to P bodies but does not affect total cellular decapping activity. Rather, phosphorylation appears to contribute to mRNA stabilization of selective IL-1 target genes, such as IL-8.
DCP1a suppresses IL-1–induced transcription by negatively regulating p65 NF-κB
Given the strong dominant-negative phenotype of stably expressed GFP-DCP1a mutants on IL-1–induced IL-8 mRNA steady-state levels, we also investigated their role in IL-8 transcription using promoter reporter gene assays. There was an unexpected strong suppression of inducible IL-8 transcription that mirrored the extent of inhibition on IL-8 mRNA levels by the GFP-DCP1aS315A, GFP-DCP1aS315D, or GFP-DCP1aΔ515–582 mutants in the same cell lines (Fig. 8 A). p65 NF-κB is the major transcriptional regulator of IL-1–induced genes, including IL-8 (Hoffmann et al., 2005; Wolter et al., 2008; Weber et al., 2010). To get further insight into the underlying mechanisms of DCP1a-dependent transcriptional inhibition, we fused p65 or DCP1a to the DNA-binding domain of the yeast transcription factor GAL4 to analyze their effects on transcriptional activity in a mammalian one-hybrid system using a luciferase reporter driven by five GAL4 binding sites. Coexpression of GFP-DCP1a wild type or its mutants significantly suppressed GAL4-p65–driven transcription (Fig. 8 B). To assess the relative inhibitory effects of GFP-DCP1a, we normalized the GAL4-p65 activity to GAL4-p65 protein levels in the absence or presence of GFP-DCP1a. Based on this analysis, GFP-DCP1a reduces GAL4-p65 activity by 58–76% (Fig. 8 B). In this assay, a GAL4-DCP1a fusion protein had very low transcriptional activity on its own (0.043 ± 0.0015 relative light units [RLUs]; Fig 8 C, lane 4) compared with background levels (0.00078 ± 0.00037 RLUs; Fig. 8 C, lane 2). This low activity was completely abolished by deleting amino acids 125–377 (unpublished data), confirming previous observations (Bai et al., 2002). In comparison, GAL4-p65 NF-κB was >10,000 times more active when transfected in parallel under identical conditions (54.82 ± 19.17 RLUs; Fig. 8 C, lane 8). The inhibitory effect of GFP-DCP1a, as shown in Fig. 8 B, was enhanced when DCP1a was tethered together with GAL4-p65 to the GAL4 promoter using GAL4-DCP1a fusion proteins (Fig. 8 C). The suppression of p65 activity by GFP-DCP1a or GAL4-DCP1a was independent from S315 but was stronger with GFP-DCP1aΔ515–582 (Fig. 8 B) or GAL4-DCP1aΔ515–582 (Fig. 8 C), respectively, suggesting that it is under negative control of the C-terminal trimerization domain of DCP1a. Inhibition of p65 may involve direct interaction of DCP1a and p65 NF-κB in the nucleus because both proteins could be coimmunoprecipitated using antibodies against p65 or DCP1a. (Fig. 8 D), and we found a small amount of endogenous DCP1a in the nucleus (Fig. 8, E and F). The data shown in Fig. 8 can be reconciled with a transcriptional corepressor function of DCP1a for p65 NF-κB but may also be explained by DCP1a affecting mRNA steady-state levels of other nuclear factors that are required for full p65 NF-κB–dependent transactivation.
Figure 8.
DCP1a inhibits IL-8 transcription and acts as a transcriptional suppressor for p65 NF-κB. (A) Stable cell lines as described in Fig. 6 and Fig. 7 were transiently transfected with the IL-8 promoter luciferase (luc.) and SV40–β-galactosidase (β-gal.) constructs. After 24 h, cells were stimulated for 4 h with 10 µg/ml IL-1α and lysed, and luciferase activity was determined. Shown is the mean luciferase activity ± SEM normalized for β-galactosidase activity from duplicate measurements of two independent experiments. (B) HEK293IL-1R cells were left untransfected or were transfected with 3 µg GFP-DCP1a, empty vector, or a GAL4 expression construct and 1 µg GAL4-p65 in the indicated combinations. 1 µg luciferase vector containing five GAL4 binding sites (pFR-luciferase) and 1 µg SV40–β-galactosidase were cotransfected. 24 h later, cells were lysed, and luciferase activity was determined. Lysates were examined for expression of GAL4 and GFP fusion proteins using anti-GAL4 or anti-GFP antibodies. Blots were probed with antiactin antibodies to confirm equal loading. Shown is the mean luciferase activity (RLU) ± SEM from six independent experiments. Numbers in percentages display relative luciferase activity of GAL4-p65 in the absence or presence of GFP-DCP1a constructs. These values were also normalized to GAL4-p65 expression levels to account for differences in GAL4-p65 amounts as in the particular Western blot shown. (C) HEK293IL-1R cells were transfected in the indicated combinations as in B but with GAL4-DCP1a constructs instead of GFP-DCP1a. Lysates were also examined for equal expression of GAL4 fusion proteins using GAL4 antibodies. A nonspecifically detected protein band served as a loading control. Shown is the mean luciferase activity (RLU) ± SEM from four independent experiments. Numbers in percentages display relative luciferase activity of GAL4-p65 in the absence or presence of GAL4-DCP1a constructs. (D) HEK293IL-1R cells were transfected with expression vectors encoding epitope-tagged p65 NF-κB or GFP-DCP1a. 24 h later, cells were lysed, and immunoprecipitations were performed using anti-p65, anti-DCP1a, or IgG control antibodies in the indicated combinations. Lysates and immunoprecipitates were analyzed by Western blotting for overexpressed p65 and DCP1a proteins as indicated. (E) HEK293IL-1R cells were treated for 1 h with 10 ng/ml IL-1α or left untreated. DCP1a and Edc4 localization was analyzed by double immune fluorescence analysis. The minus sign indicates samples from untreated cells. Bar, 10 µM. (F) Cells were treated as in D for the indicated times with 10 ng/ml IL-1α, and DCP1a and p65 proteins were analyzed in cytosolic and nuclear cell extracts by Western blotting. Black lines indicate that intervening lanes have been spliced out. mIgG, murine IgG; rIgG, rabbit IgG; IB, immunoblot; wt, wild type.
Discussion
JNKs have been implicated in AU-rich element-mediated mRNA stabilization in T cells (Chen et al., 2000), alveolar epithelial cells (Korhonen et al., 2007), and mast cells (Ming et al., 1998), but in general, their role in mRNA decay pathways and in mRNA translation has been elusive. In this study, we found that JNK phosphorylated DCP1a at S315 in intact cells and in vitro, establishing the first example of JNK targeting an RNA-decapping pathway protein directly. The sequence surrounding S315, PLSP, represents a consensus MAPK phosphorylation motif (Bogoyevitch and Kobe, 2006). Experiments using inhibitors for JNK (SP600125), p38 MAPK (SB203580), extracellular-regulated kinase (PD98059; unpublished data), and TAK1 (5Z-7-oxozeaenol) as well as investigations of kinase-deficient embryonic fibroblasts indicated that during translational stress or IL-1 signaling, S315 is specifically targeted by the TAK1–JNK pathway. The TAK1-TAB1 fusion protein and MEKK1 caused mobility shifts and phosphorylation of DCP1a (Fig. 1 B and Fig. 2 C). However, we consider it unlikely that TAK1-TAB1 or MEKK1 directly phosphorylate the PLSP motif within DCP1a even though the TAK1 inhibitor prevents DCP1a modifications (Fig. 3 C). Rather, TAK1-TAB1 and MEKK1 indirectly alter phosphorylation of DCP1a because they activate MAPKs, in particular JNK, via MAPKKs. Our results do not exclude the possibility that in other conditions, additional proline-directed protein kinases may phosphorylate S315 or other residues in DCP1a. In fact, the strong mobility shifts of cellular DCP1a (Fig. 1, Fig. 2, and Fig. 3) and remaining radioactive phosphorylation of GST-DCP1aS315A (not depicted) indicate further modifications of DCP1a. Future studies are required to work out all enzymes and the resulting posttranslational modifications that target DCP1a.
The starting point of this study was the observation that strong upstream activators of JNK promoted drastic changes in P bodies as detected using DCP1a as a marker. Moreover, JNK colocalized and coimmunoprecipitated with endogenous DCP1a. As most evident from the phenotype of a phosphomimetic mutation (S315D), phosphorylation of DCP1a at S315 by JNK regulates the release of DCP1a from P bodies. This was paralleled by disintegration of P bodies as evident from the cytosolic redistribution of two other P body markers, Edc4 and Xrn 1. However, JNKs were also required for P body formation, as JNK inhibitors blocked the IL-1–induced P body increase (Fig. 4), and P bodies were diminished in JNK-deficient cells (Fig. S3). It should be noted that biochemical fractionation (Fig. 3) but also colocalization experiments (Fig. 1 and Fig. 6) suggested that JNKs are permanently associated with DCP1a. In this way, JNKs can regulate multiple aspects of DCP1a function within the cytoplasm, in P bodies or within other cellular compartments by signal-mediated phosphorylation. In conjunction with previous findings (Franks and Lykke-Andersen, 2008), our data now suggest three levels of control of DCP1a localization: (1) the abundance of nontranslatable RNAs in the cytoplasm (Teixeira et al., 2005), (2) C terminus–mediated oligomerization and interaction with other P body components (Fig. 6; Tritschler et al., 2009), and (3) JNK-mediated phosphorylation at S315, which serves as a reversible molecular switch to dynamically control localization of DCP1a and P body formation in different biological situations.
We followed the latter hypothesis, examining conditions representing either strong JNK activation and translational arrest (e.g., anisomycin and sorbitol treatment) or transient JNK activation and highly inducible gene expression (e.g., IL-1 treatment). Sorbitol and anisomycin caused sustained JNK activation as well as sustained DCP1a phosphorylation and disintegration of P bodies as judged by the cytosolic redistribution of Edc4 and Xrn1. This mechanism may be operative for those mRNAs whose decay is supposed to be accelerated in the cytoplasmic pool of nontranslated mRNAs during stress.
In contrast, transient JNK activation in response to IL-1 was paralleled by enhanced DCP1a phosphorylation and by an increase in Edc4- and Xrn1-positive P bodies. This mechanism may serve to selectively sequester mRNAs from the RNA decay machinery in a gene-specific manner. We propose that some IL-1–induced mRNAs are directed to P bodies to become rapidly degraded after initial induction as exemplified for IκBα. Others are retained in the cytoplasm to delay P body–mediated decay. These mRNAs are more stable and are expressed and translated for longer times as exemplified for IL-8. Support of this interpretation is derived from the observation that a phosphomimetic mutant of DCP1a stabilized IL-8 mRNA but not IκBα mRNA. These data point to a novel role of P bodies in selective termination of proinflammatory cytokine action. However, full proof of this hypothesis will require the development of methods to separately determine mRNA species and decapping activities in P bodies and within the cytoplasm, respectively.
Microarray analyses revealed that mutants of DCP1a that interfere with phosphorylation-mediated shuttling (S315D and S315A) or with its trimerization (Δ515–582) exerted a strong dominant-negative effect on virtually all IL-1 response genes (Fig. S4). The suppression of steady-state mRNA levels of all of these genes and the inhibition of an IL-8 promoter construct suggested that DCP1a played an unexpected role in transcriptional regulation of IL-1 response genes. We found that a minor fraction of DCP1a was localized in the nucleus (Fig. 8, E and F), whereas other decapping proteins, such as Edc4, were localized exclusively in the cytoplasm or in P bodies (Fig. 8 E). DCP1a interacted with p65 NF-κB and suppressed p65-mediated transcription (Fig. 8, B–D). All these results point to DCP1a as a novel multifunctional regulator of gene expression in cytokine signaling.
This interpretation is in line with previous observations suggesting that DCP1a can have both transcriptional and posttranscriptional functions. In yeast, DCP1a was identified as a regulatory subunit of the decapping enzyme DCP2 (Beelman et al., 1996; Dunckley and Parker, 1999). In metazoan cells, it requires more complex interactions with other proteins (Lsm1-7, Edc3, Edc4 [hedls], and DDX6/RCK) to enhance decapping (Fenger-Grøn et al., 2005; Bail and Kiledjian, 2006; Franks and Lykke-Andersen, 2008; Buchan and Parker, 2009). The phosphorylation site S315 is not part of a region that has been previously found to participate in decapping or in binding to DCP2 or to other subunits of the decapping complex (Franks and Lykke-Andersen, 2008). Accordingly, mutation of S315 did not affect decapping activity associated with DCP1a immunoprecipitates (Fig. 5).
In an independent study, DCP1a was identified as a SMAD4-interacting protein (called SMIF) that acted as a transcriptional coactivator in the TGF-β pathway (Bai et al., 2002; Callebaut, 2002). Recently, DCP1a was also found to interact with PNRC2 (proline-rich nuclear receptor coregulatory protein 2) in P bodies as part of the nonsense-mediated decay pathway (Cho et al., 2009). PNRC2 and its homologue PNRC1 also act as transcriptional coactivators of various steroid receptors (Zhou et al., 2000; Zhou and Chen, 2001). Neither SMADs nor PNRC1/2 has so far been implicated in IL-1–mediated nuclear signaling (Weber et al., 2010). Most likely, therefore, DCP1a has separate roles in TGF-β, nuclear hormone, and IL-1 signaling that warrant further investigations.
In summary, our study reveals two hitherto unrecognized aspects of DCP1a regulation: first, phosphorylation-mediated control of subcellular localization and second, nuclear suppression of NF-κB target genes. Signal-regulated localization of DCP1a has evolved late, as JNKs are not found in lower eukaryotes, such as yeast, and the sequence motif PLSP is only found in primates and rodents (Fig. 9). This pathway may specifically serve to direct DCP1a to transcriptional and posttranscriptional functions in response to physiological stimuli.
Figure 9.
Scheme of DCP1a as a multifunctional downstream effector of the JNK pathway. In addition to its known role in AP-1–mediated transcriptional control through the phosphorylation of c-Jun, JNK binds to and colocalizes with DCP1a. JNK activation results in phosphorylation-dependent shuttling of DCP1a between P bodies and the cytoplasm to modulate the translational output of stress- or inflammation-induced genes. DCP1a also affects IL-1–induced mRNA transcription by affecting p65 NF-κB–mediated gene expression. The gray shading indicates phosphorylated JNK substrates. (inset) Conservation of the JNK phosphorylation motif surrounding S315 (shaded in gray) of human DCP1a. For details see Discussion. P, phosphorylated.
Materials and methods
Cells
HEK293 cells stably expressing the IL-1 receptor (HEK293IL-1R; Thiefes et al., 2006), JNK1/2, and p38 MAPK-deficient MEF lines have been previously described (Thiefes et al., 2005; Wolter et al., 2008) and were gifts from E. Wagner (Cancer Cell Biology Program, Spanish National Cancer Center, Madrid, Spain) and A. Nebreda (Institute for Research in Biomedicine Barcelona, Barcelona, Spain), respectively. Cells were cultured in Dulbecco’s modified Eagle’s medium complemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Materials
Antibodies against the following proteins or peptides were used: actin (JLA20; EMD) and DCP1a (H00055802-M06; Abnova); MYC (9E10), GFP (clones 7.1 and 13.1), and HA (12CA5; all obtained from Roche); P(T180/Y182)-p38 MAPK (36–850; Invitrogen); TAB1 (sc-13956), TAK1 (sc-7162), JNK (D-2 and sc-7345), tubulin (TU-02; sc-8035), Xrn1 (C-1; sc-165985), and p65 NF-κB (C-20; sc-372; all obtained from Santa Cruz Biotechnology, Inc.); P(Ser73)-c-Jun (9164), JNK (9252), P(T183/Y185)-JNK (9251), and Edc4 (25485; all obtained from Cell Signaling Technology); and rabbit and mouse IgG (Santa Cruz Biotechnology, Inc.). Other antibodies were as follows: long JNK2 (SAK9 raised against DSSLDASTGPLEGCR; human with C-terminal 15 residues), short JNK3 (SAK10 raised against GVVKGQPSPSAQVQQ; human with C-terminal 15 residues), JNK (Ab681; chicken antibody raised against GST-JNK3), and p38 MAPK (SAK7 raised against ISFVPPPLDQEEMES; rat p38α with C-terminal 15 residues). All were gifts from J. Saklatvala (The Kennedy Institute of Rheumatology, Imperial College London, London, England, UK; Krause et al., 1998; Finch et al., 2001). Purified polyclonal antibodies that recognize specific phosphorylation at S315 in human DCP1a were raised against the following peptides: TYTIPL-(phospho)S-PVLSPT (Eurogentec) or NEKHAPTYTIPL-(phospho)-S-PVLSPTLPAEAPTAQV (PickCell Laboratories). Polyclonal rabbit antibodies against GFP or GAL4 were gifts from E. Izaurralde (Max Planck Institute for Developmental Biology, Tübingen, Germany) or L. Schmitz (Justus Liebig University Giessen, Giessen, Germany), respectively. HRP-coupled secondary antibodies were goat anti–mouse IgG and goat anti–rabbit IgG obtained from Dako, rabbit anti–chicken IgG (A-9046) obtained from Sigma-Aldrich, and TrueBlot anti–mouse or –rabbit IgG obtained from eBioscience.
The expression plasmid for GST–c-Jun (amino acids 1–135) was a gift from J.R. Woodgett (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada). Expression of GST fusion proteins was induced in the Escherichia coli BL21 strain by 0.5–1 mM IPTG for 4 h at 37°C, and proteins were purified from bacterial extracts by glutathione–Sepharose beads, eluted with 20 mM glutathione, dialysed against 20 mM Tris, pH 7.4, 100 mM NaCl, and 2 mM DTT, and stored at −20°C in 50% glycerol according to standard methods.
Protein A– or G–Sepharose were purchased from GE Healthcare. Human recombinant IL-1α was a gift from J. Saklatvala and was expressed in E. Coli and purified by sequential column chromatography as described previously (Krause et al., 1998; Wolter et al., 2008). SP600125 and 5Z-7-oxozeaenol were obtained from Tocris Bioscience, and SB203580 was purchased from EMD. Anisomycin, sorbitol, and all other reagents were purchased from Sigma-Aldrich or Thermo Fisher Scientific and were of analytical grade or better.
Plasmids, transfections, stable cell lines, and reporter gene assays
The following plasmids have been described previously: pUHC13-3 IL-8 promoter luciferase (Holtmann et al., 1999), peVHA-JNK2, peVHA-JNK2K55R (Krause et al., 1998), pQBI-MEKK1Δ998–2743-BFP (Thiefes et al., 2006), pGAL4-p65, pBSPACΔP (Thiefes et al., 2005), and pEGFP-p65 (Buss et al., 2004). pEGFP-DCP1a, pEGFP-DCP2, and pEGFP-DCP1αΔ515–582 were cloned by RT-PCR, pCDNA3.1-TAK1kdTAB1ad was obtained from H. Sakurai (Institute of Natural Medicine, University of Toyama, Sugitani, Toyama, Japan), pFA-DCP1a and pFA-DCP1AΔ125–377 were obtained from J. Duyster and S. Kuhn (Laboratory of Leukemogenesis, Technical University of Munich, Munich, Germany; Bai et al., 2002), pEBB-HA-p65 was obtained from L. Schmitz, pSV–β-galactosidase and pGL2-control were purchased from Promega, pEGFP-C1 was purchased from Takara Bio Inc., and pFC2–DNA-binding domain and pFR-luciferase were purchased from Agilent Technologies. The following plasmids were constructed by standard cloning techniques: pEGFP-DCP1aS60/62A, pEGFP-DCP1aS140/142A, pEGFP-DCP1aS179/180A, pEGFP-DCP1aT289/290A, pEGFP-DCP1aS315A, pEGFP-DCP1aS315D, pEGFP-DCP1aS319A, pEGFP-DCP1aS334A, pEGFP-DCP1aT347/348A, pEGFP-DCP1aS353A, pEGFP-DCP1aT401A, pEGFP-DCP1aS422A, pEGFP-DCP1aS522/523/525A, pEGFP-DCP1aT531A, pDEST15-DCP1a, pDEST15-DCP1aS315A, pFA-DCP1aS315A, pFA-DCP1aS315D, and pFA-DCP1Δ515–582.
Unless indicated otherwise, cells were seeded at 2–5 × 105 for 6-well plates, 4.5 × 106 for T75 (75 cm2) flasks, and 9.8 × 106 for T175 (175 cm2) flasks and transfected with 5–7.5, 20–40, or 50–100 µg plasmid DNA, respectively. Transfections were performed with the CaPO4 method as previously described (Holtmann et al., 2001). Stable cell lines of HEK293IL-1R cells were generated by transfecting 5 µg pEGFP-DCP1a constructs (Fig. 5) and 1 µg pBSPACΔP. Cells were selected and maintained in1 µg/ml puromycin. 50–65% of all cells in stably transfected cell pools expressed GFP-DCP1a constructs.
Cell lysis, luciferase, and β-galactosidase assays for determination of reporter gene activities have been previously described (Holtmann et al., 2001; Thiefes et al., 2005). In brief, cells were lysed in ice-cold potassium phosphate buffer (8.5 mM KH2PO4, 91.5 mM K2HPO4, pH 7.8, 0.2% Triton X-100, 1 µg/ml pepstatin, 10 µg/ml leupeptin, and 1 mM PMSF) for 15 min on ice. Lysates were cleared for 15 min at 10,000 g at 4°C. 20 µl lysate was mixed with 100 µl luciferase buffer (20 mM tricin, 1.07 mM (MgCO3)4Mg(OH)2 × 5H2O, 2.67 mM MgSO4, 33.3 mM DTT, 0.53 mM ATP, 30 mg coenzyme A, and 472 µM d-luciferine), and chemiluminescence was recorded for 10 s in a 96-well luminometer (Fluoroscan Ascent FL; Laboratory systems). β-Galactosidase activity was measured with reagents from Takara Bio Inc.
Immunostaining and fluorescence microscopy
HEK293IL-1R cells were seeded for 24 h in µ-Slides VI (ibidi). After washing, cells were fixed with 4% paraformaldehyde in HBSS (PAA Laboratories) for 5 min, blocked with 10% normal donkey serum (Dianova) for 30 min, and incubated with primary and secondary antibodies diluted in HBSS containing 0.1% saponin (Sigma-Aldrich) for 2 h at room temperature. Cy3-conjugated (1:200; Millipore) and FITC-conjugated (1:100; Sigma-Aldrich) secondary antibodies were used. For controls, primary antibodies were omitted. Nuclei were stained with Hoechst 33342 (Invitrogen).
Image acquisition and processing
Immunofluorescence analyses were performed on an inverse microscope (DMIRE2; Leica) equipped with a monochrome digital camera (DFX350 FX R2; Leica). Fluorescence image acquisition was performed at room temperature using a 40-fold objective lens (40×/0.6 NA dry; HCX PL Fluotar; Leica) and appropriate filter cubes (DAPI: excitation 359 nm and emission 461 nm; FITC: excitation 490 nm and emission 520 nm; Cy3: excitation 550 nm and emission 570 nm; GFP: excitation 489 nm and emission 508 nm). Standard image processing was performed with the FW4000 software (version 1.2.1; Leica) using contrast, deconvolution, and merge applications (DAPI: γ reduction = 0.90, 2D deconvolution [haze removal 70%, smoothing 4, and spot removal 60], and 70% merged; FITC: γ reduction = 0.70, local contrast enhancement = 30%, and 90% merged; Cy3: γ reduction = 0.90, local contrast enhancement = 30%, and 100% merged). In Fig.6 C (bottom), additional image processing was preformed by using the Deblur software (version 2.3.2; Leica) with 25% background equalization and removing of negative intensities. Subsequently, 2D blind deconvolution was performed with the following settings: all channels, low noise smoothing, 20 total iterations, point spread function correction factor 2, and super resolution factor 1.
In vitro kinase assays
For immune complex protein kinase assays, whole-cell extracts (0.85–2 mg protein) were diluted in 500 µl ice-cold immunoprecipitation buffer (20 mM Tris-HCl, pH 7.4, 154 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100, and 2 mM DTT). Samples were incubated for 2–3 h with 2–4 µl antibodies against JNK followed by the addition of 30 µl protein A– or protein G–Sepharose beads and incubation for 1–2 h at 4°C. Beads were spun down, washed 3× in 1 ml immunoprecipitation buffer, and resuspended in 10 µl of the same buffer. Then, 2–3 µg recombinant protein substrates (GST, GST–c-Jun1–135, or GST–DCP1-α) in 10 µl H2O and 10 µl kinase buffer (150 mM Tris-HCl, pH 7.4, 30 mM MgCl2, 15 µM ATP, and 8 µCi γ-[32P]ATP) was added. After 30 min at room temperature, the SDS sample buffer was added, and proteins were eluted from the beads by boiling for 5 min. After centrifugation at 16,000 g for 5 min, supernatants of both types of kinase assays were separated by 7.5–10% SDS-PAGE. Phosphorylated proteins were visualized by autoradiography.
For nonradioactive immune complex protein kinase assays (Fig. 2 B), whole-cell extracts from separate cell cultures transfected with plasmids encoding protein kinases or substrates were prepared as described in the previous paragraph. 4–5 mg lysate proteins were incubated with antibodies against JNK (SAK9; 3 µl) or GFP (anti-GFP; 2 µg) precoupled to 30 µl protein G–Sepharose. Immunoprecipitations were performed in 500 µl immunoprecipitation buffer for 2.5 h at 4°C. Immunopurified kinase and substrate samples were washed twice with 1 ml immunoprecipitation buffer, combined, washed again, and resuspended in 20 µl immunoprecipitation buffer. 20 µl H2O and 20 µl kinase buffer (150 mM Tris-HCl, pH 7.4, 30 mM MgCl2, and 135 µM ATP) were added. After 30 min at room temperature, proteins were eluted from the beads by boiling for 10 min as described in the previous paragraph. Proteins were separated on SDS-PAGE and detected by immunoblotting.
Cell lysis and immunoprecipitations from soluble and particular fractions
For whole-cell extracts, cells were lysed in Triton X-100 cell lysis buffer (10 mM Tris, pH 7.05, 30 mM NaPPi, 50 mM NaCl, 1% Triton X-100, 2 mM Na3VO4, 50 mM NaF, 20 mM β-glycerophosphate, and freshly added 0.5 mM PMSF, 0.5 µg/ml leupeptin, 0.5 µg/ml pepstatin, and 1 µg/ml microcystin) as previously described (Holtmann et al., 1999). Cell lysates were subjected to SDS-PAGE on 6–10% gels, and immunoblotting was performed as described in the Immunoblotting and ELISA section.
Nuclear and cytosolic extracts were prepared as described previously (Holtmann et al., 1999). In brief, cells were suspended and pelleted in buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.3 mM Na3VO4, freshly added 200 µM leupeptin, 10 µM E64, 300 µM PMSF, 0.5 µg/ml pepstatin, 5 mM DTT, 400 nM okadaic acid, and 20 mM β-glycerophosphate). The pellet was resuspended in buffer A containing 0.1% NP-40. After centrifugation at 10,000 g for 5 min at 4°C, supernatants were taken as cytosolic extracts. Pellets were resuspended in buffer B (20 mM Hepes, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.3 mM Na3VO4, 20 mM β-glycerophosphate, 200 µM leupeptin, 10 µM E64, 300 µM PMSF, 0.5 µg/ml pepstatin, 5 mM DTT, and 400 nM okadaic acid). After 1 h on ice, nuclear extracts were cleared at 10,000 g for 5 min at 4°C, and supernatants were collected. Protein concentration of cell extracts was determined by the Bradford method, and samples were stored at −80°C.
Soluble (S2) and particular (P2) fractions (Fig. 3 D) were prepared as previously described (Teixeira et al., 2005). In brief, cells from one T175 (175 cm2) flask were washed in ice-cold PBS, pelleted, and lysed in 1 ml lysis buffer (50 mM Tris, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 1 mM β-mercaptoethanol, proteinase inhibitor mix [Roche], 0.4 U/µl RNase inhibitor [RiboLock; Fermentas], and 1 µM microcystin). Samples were kept for 15 min on ice, vortexed three times, and then centrifuged for 2 min at 2,000 g at 4°C. The supernatant was recovered and centrifuged for 10 min at 10,000 g at 4°C. The supernatant (S2) was taken as cytosolic extract, and the pellet (P2) was taken as a P body–enriched particular fraction. P2 was diluted in 1 ml immunoprecipitation buffer (50 mM Hepes, pH 7.4, 50 mM NaCl, 1% Tween 20, 2.5 mM EGTA, 1 mM EDTA, 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM PMSF, protease inhibitor mix, 1 mM DTT, and 1 µM microcystin) and sonified three times for 20 s on ice. 370 µg S2 proteins were adjusted to 150 µl with lysis buffer. Both S2 and P2 containing equal amounts of proteins were diluted into 500 µl immunoprecipitation buffer and were precleared with 15 µl TrueBlot anti–rabbit IgG immunoprecipitation beads (00–8800). The supernatant was incubated with 1 µg rabbit IgG or with anti-JNK antibodies (2.5 µl SAK9 + 2.5 µl SAK10) precoupled to 15 µl IgG beads. Samples were end-to-end rotated overnight at 4°C, and beads were washed three times in immunoprecipitation buffer including 450 mM NaCl. Proteins were eluted by boiling in SDS-PAGE sample buffer, separated by SDS-PAGE, immunoblotted, and detected by the indicated primary antibodies followed by TrueBlot HRP-conjugated anti–rabbit IgG (18-8816) or anti–mouse IgG (18-8817).
Coimmunoprecipitations
For coimmunoprecipitations of overexpressed GFP-p65, HA-p65, and GFP-DCP1 (Fig. 8 D), cells were lysed in 50 mM Hepes, pH 7.4, 50 mM NaCl, 1% Tween 20, 2.5 mM EGTA, 1 mM EDTA, 1 mM NaF, 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM PMSF, protease inhibitor mix, and 1 mM DTT, sonified five times for 30 s on ice in a sonicator (Biorupter; Diagenode) at the high setting with 30-s intervals, and centrifuged at 100,000 g for 20 min at 4°C. 1 mg of the supernatant in 400 µl lysis buffer was precleared with 25 µl TrueBlot anti–rabbit or anti–mouse Ig immunoprecipitation beads (00-8811). Then, 1 µg anti-DCP1, anti-p65, or IgG antibodies precoupled overnight to 15 µl TrueBlot Ig immunoprecipitation beads was added for 4 h at 4°C. Thereafter, beads were washed three times in 1 ml lysis buffer. Proteins were eluted by boiling in SDS-PAGE sample buffer, and immunoblotted proteins were detected by the indicated primary antibodies and TrueBlot HRP-conjugated anti-IgG antibodies.
Immunoblotting and ELISA
Immunoblotting was performed essentially as previously described (Hoffmann et al., 2005). Proteins were separated on 6–10% SDS-PAGE and electrophoretically transferred to polyvinylidene fluoride membranes (Millipore). After blocking with 2 or 5% dried milk in Tris-HCl–buffered saline/0.05% Tween (TBST) for 1 h, membranes were incubated for 12–24 h with primary antibodies, washed in TBST, and incubated for 1–2 h with the peroxidase-coupled secondary antibody. Proteins were detected by using ECL systems from Thermo Fisher Scientific, Millipore, or GE Healthcare. IL-8 secretion was measured by the IL-8 ELISA development kit (DuoSet; R&D Systems).
RT-PCR
1 µg total RNA was prepared by a minispin kit (NucleoSpin RNA II; Macherey-Nagel) and transcribed into cDNA using Moloney murine leukemia virus reverse transcription (Fermentas) in a total volume of 20 µl. 2 µl of this reaction mixture was used to amplify cDNAs using assays on demand (Applied Biosystems) for IL-8 (Hs00174103_m1), IκBα (Hs00153283_m1), and β-actin (Hs99999903_m1) on a real-time PCR instrument (ABI 7500). The cycle threshold (ct) value for each individual PCR product was calculated by the instrument’s software, and ct values obtained for IL-8 or IκBα were normalized by subtracting the ct values obtained for β-actin. The resulting Δct values were then used to calculate relative changes of mRNA expression as ratio (R) of mRNA expression of stimulated/unstimulated cells according to the equation R = 2−(Δct[stimulated] − Δct[unstimulated]).
Decapping assays
Decapping assays were performed essentially as previous described in Tritschler et al. (2009). In brief, a 176-bp luciferase cDNA template was amplified by PCR from pGL2-control (Promega) using the primers sense, 5′-GCGTAATACGACTCACTATAGGGAGAATGGAACCGCTGGAGAGCA-3′,and antisense, 5′-TCTGTGATTTGTATTCAGCC-3′. mRNA was transcribed in vitro using the MEGAscript T7 kit (Invitrogen), purified by the mirVana kit (Invitrogen) using the total RNA protocol, and eluted in 100 µl H2O. 2 µg mRNA was heated at 65°C for 10 min and then capped in a reaction mixture containing 2 µl of 10× ScriptCap capping buffer, 2 µl (20 µCi) α-[32P]GTP, 1 µl of 2-mM S-adenosylmethionine, 0.5 µl ScriptGuard RNase inhibitor (40 U/µl), and 1 µl (10 U) script capping enzyme in a total volume of 20 µl using the capping system kit (ScriptCap m7G; Epicentre). After 60 min at 37°C, mRNA was purified by the mirVana kit as described previously in this paper. For the experiment shown in Fig. 5, one T175 (175 cm2) flask of HEK293IL-1R cells was transfected with 50 µg GFP-DCP2 or GFP-DCP1a expression vectors and 30 µg pCDNA3 using the calcium phosphate method. After 24 h, cells were lysed in 1 ml NET buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, and 10% glycerol plus freshly added protease inhibitors). After 15 min on ice, lysates were cleared at 18,000 g for 15 min at 4°C. GFP-tagged proteins were immunoprecipitated from 3 mg protein in 1 ml NET buffer for 2 h at 4°C using 1 µg anti-GFP antibodies precoupled to 25 µl protein G–Sepharose slurry. Beads were washed 3× in 1 ml NET buffer, 1× in 1 ml NET buffer 2 (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Triton X-100), and 1× in 0.975 ml decapping buffer (50 mM, Tris-HCl, pH 7.9, 30 mM ammonium sulfate, and 1 mM MgCl2). One fourth of the material was used for Western blotting. The remaining three fourths of the immunoprecipitate was resuspended in 1 vol decapping buffer including 0.1 mM cap structure analogue G(5′)ppp(5′)G RNA (New England Biolabs, Inc.) and 0.4 U/µl RNase inhibitor. 5 µl of this reaction was mixed with 50 ng 32P-labeled luciferase mRNA in a total volume of 10 µl decapping buffer and rotated horizontally at 1,400 rpm. After 30 min at 30°C, the reaction was stopped by addition of 50 mM EDTA, and 5 µl was separated on polyethylenimine thin layer chromatography plates (Merck) using 0.75 M LiCl as a solvent. 25 µg m7GDP and m7GTP (Sigma-Aldrich) was used as a standard.
Microarray-based mRNA expression analysis
The Whole Human Genome Oligo Microarray (Agilent Technologies) used in this study contains 45,015 oligonucleotide probes covering the entire human transcriptome. Synthesis of Cy3-labeled cRNA was performed with the Quick Amp Labeling kit, one color (Agilent Technologies) according to the manufacturer’s recommendations. cRNA fragmentation, hybridization, and washing steps were also carried out exactly as recommended for the One-Color Microarray-Based Gene Expression Analysis Protocol V5.7 (Agilent Technologies). Slides were scanned on a microarray scanner (pixel resolution of 5 µm and bit depth of 20; Microarray Scanner G2565CA; Agilent Technologies). Data extraction was performed with the Feature Extraction Software V10.7.3.1 by using the recommended default extraction protocol file GE1_107_Sep09.xml.
Processed intensity values of the green channel (gProcessedSignal or gPS) were normalized by global linear scaling: all gPS values of one sample were multiplied by an array-specific scaling factor. This scaling factor was calculated by dividing a reference 75th percentile value (set as 1,500 for the whole series) by the 75th percentile value of the particular microarray (Array i in the formula shown in this paragraph). Accordingly, normalized gPS values for all samples (microarray datasets) were calculated by the following formula: normalized gPSArray i = gPSArray i × (75th PercentileReference Array/75th PercentileArray i).
A lower intensity threshold was defined as 1% of the reference 75th Percentile value (threshold = 15). All of those normalized gPS values that fell below this intensity border were substituted by the respective surrogate value of 15. Calculation of ratio values of relative gene expression and data filtering was performed using Excel macros (Microsoft).
Statistics
Statistics were calculated by the Mann–Whitney rank test or paired t test using Sigma Plot12 (Systat Software, Inc.).
Online supplemental material
Fig. S1 shows mapping of JNK-dependent phosphorylation sites in DCP1a. Fig. S2 shows microscopic analysis of P body–enriched (P2) fractions. Fig. S3 shows DCP1a phosphorylation and P body formation in JNK-deficient MEFs. Fig. S4 shows impaired IL-1 response in HEK293IL-1R cells upon overexpression of DCP1a constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006089/DC1. Multicolor images shown in Fig. 1 A, Fig. 4 B, and Fig. 6 C can be viewed via the JCB DataViewer at http://jcb-dataviewer.rupress.org/.
Acknowledgments
We thank Erwin Wagner and Angel Nebreda for the gift of JNK- and p38-deficient MEFs, respectively. We thank J. Duyster and S. Kuhn for pFA-SMIF constructs and J. Saklatvala for recombinant human IL-1α and antibodies.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Kr1143/5-3, Kr1143/7-1, SFB566/Z02, and TRR81/B2 to M. Kracht). Work from M. Kracht was further supported by the Excellence Cluster Cardio-Pulmonary System and the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz/Universities of Giessen and Marburg Lung Center program.
Footnotes
Abbreviations used in this paper:
- ct
- cycle threshold
- MEF
- murine embryonic fibroblast
- RLU
- relative light unit
References
- Anderson P., Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Bai R.Y., Koester C., Ouyang T., Hahn S.A., Hammerschmidt M., Peschel C., Duyster J. 2002. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFbeta signalling. Nat. Cell Biol. 4:181–190 10.1038/ncb753 [DOI] [PubMed] [Google Scholar]
- Bail S., Kiledjian M. 2006. More than 1 + 2 in mRNA decapping. Nat. Struct. Mol. Biol. 13:7–9 10.1038/nsmb0106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A., Stevens A., Caponigro G., LaGrandeur T.E., Hatfield L., Fortner D.M., Parker R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 382:642–646 10.1038/382642a0 [DOI] [PubMed] [Google Scholar]
- Bogoyevitch M.A., Kobe B. 2006. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol. Mol. Biol. Rev. 70:1061–1095 10.1128/MMBR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss H., Dörrie A., Schmitz M.L., Hoffmann E., Resch K., Kracht M. 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-kappaB at serine 536 is mediated by multiple protein kinases including IkappaB kinase (IKK)-alpha, IKKbeta, IKKepsilon, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279:55633–55643 10.1074/jbc.M409825200 [DOI] [PubMed] [Google Scholar]
- Callebaut I. 2002. An EVH1/WH1 domain as a key actor in TGFbeta signalling. FEBS Lett. 519:178–180 10.1016/S0014-5793(02)02751-5 [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Andersen J.S., Gaietta G., Jürchott K., Royer H.D., Mann M., Karin M. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14:1236–1248 [PMC free article] [PubMed] [Google Scholar]
- Cho H., Kim K.M., Kim Y.K. 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell. 33:75–86 10.1016/j.molcel.2008.11.022 [DOI] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Séraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dérijard B., Hibi M., Wu I.H., Barrett T., Su B., Deng T., Karin M., Davis R.J. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 76:1025–1037 10.1016/0092-8674(94)90380-8 [DOI] [PubMed] [Google Scholar]
- Dhamija S., Doerrie A., Winzen R., Dittrich-Breiholz O., Taghipour A., Kuehne N., Kracht M., Holtmann H. 2010. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IκBζ mRNA. J. Biol. Chem. 285:29165–29178 10.1074/jbc.M110.146365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Parker R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411–5422 10.1093/emboj/18.19.5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. 2007a. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. 2007b. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27:3970–3981 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 20:905–915 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Finch A., Davis W., Carter W.G., Saklatvala J. 2001. Analysis of mitogen-activated protein kinase pathways used by interleukin 1 in tissues in vivo: activation of hepatic c-Jun N-terminal kinases 1 and 2, and mitogen-activated protein kinase kinases 4 and 7. Biochem. J. 353:275–281 10.1042/0264-6021:3530275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell. 32:605–615 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M., Kotlyarov A., Kracht M. 2009. Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug Discov. 8:480–499 10.1038/nrd2829 [DOI] [PubMed] [Google Scholar]
- Gupta S., Barrett T., Whitmarsh A.J., Cavanagh J., Sluss H.K., Dérijard B., Davis R.J. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760–2770 [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847–855 [PubMed] [Google Scholar]
- Hoffmann E., Thiefes A., Buhrow D., Dittrich-Breiholz O., Schneider H., Resch K., Kracht M. 2005. MEK1-dependent delayed expression of Fos-related antigen-1 counteracts c-Fos and p65 NF-kappaB-mediated interleukin-8 transcription in response to cytokines or growth factors. J. Biol. Chem. 280:9706–9718 10.1074/jbc.M407071200 [DOI] [PubMed] [Google Scholar]
- Holtmann H., Winzen R., Holland P., Eickemeier S., Hoffmann E., Wallach D., Malinin N.L., Cooper J.A., Resch K., Kracht M. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmann H., Enninga J., Kalble S., Thiefes A., Dorrie A., Broemer M., Winzen R., Wilhelm A., Ninomiya-Tsuji J., Matsumoto K., et al. 2001. The MAPK kinase kinase TAK1 plays a central role in coupling the interleukin-1 receptor to both transcriptional and RNA-targeted mechanisms of gene regulation. J. Biol. Chem. 276:3508–3516 10.1074/jbc.M004376200 [DOI] [PubMed] [Google Scholar]
- Huangfu W.C., Omori E., Akira S., Matsumoto K., Ninomiya-Tsuji J. 2006. Osmotic stress activates the TAK1-JNK pathway while blocking TAK1-mediated NF-kappaB activation: TAO2 regulates TAK1 pathways. J. Biol. Chem. 281:28802–28810 10.1074/jbc.M603627200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.L., Nakamura K. 2007. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim. Biophys. Acta. 1773:1341–1348 10.1016/j.bbamcr.2006.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen R., Linker K., Pautz A., Förstermann U., Moilanen E., Kleinert H. 2007. Post-transcriptional regulation of human inducible nitric-oxide synthase expression by the Jun N-terminal kinase. Mol. Pharmacol. 71:1427–1434 10.1124/mol.106.033449 [DOI] [PubMed] [Google Scholar]
- Kracht M., Truong O., Totty N.F., Shiroo M., Saklatvala J. 1994. Interleukin 1 α activates two forms of p54 α mitogen-activated protein kinase in rabbit liver. J. Exp. Med. 180:2017–2025 10.1084/jem.180.6.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause A., Holtmann H., Eickemeier S., Winzen R., Szamel M., Resch K., Saklatvala J., Kracht M. 1998. Stress-activated protein kinase/Jun N-terminal kinase is required for interleukin (IL)-1-induced IL-6 and IL-8 gene expression in the human epidermal carcinoma cell line KB. J. Biol. Chem. 273:23681–23689 10.1074/jbc.273.37.23681 [DOI] [PubMed] [Google Scholar]
- Kuan C.Y., Yang D.D., Samanta Roy D.R., Davis R.J., Rakic P., Flavell R.A. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 22:667–676 10.1016/S0896-6273(00)80727-8 [DOI] [PubMed] [Google Scholar]
- Kyriakis J.M., Banerjee P., Nikolakaki E., Dai T., Rubie E.A., Ahmad M.F., Avruch J., Woodgett J.R. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 369:156–160 10.1038/369156a0 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114–8121 10.1128/MCB.22.23.8114-8121.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming X.F., Kaiser M., Moroni C. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039–6048 10.1093/emboj/17.20.6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. 2003. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278:18485–18490 10.1074/jbc.M207453200 [DOI] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10:1224–1231 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Nishi A., Sato N., Mizukami J., Miyoshi H., Sugita T. 2002. TAK1-TAB1 fusion protein: a novel constitutively active mitogen-activated protein kinase kinase kinase that stimulates AP-1 and NF-kappaB signaling pathways. Biochem. Biophys. Res. Commun. 297:1277–1281 10.1016/S0006-291X(02)02379-3 [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.H., Xiao C., Paschal A.E., Bailey S.T., Rao P., Hayden M.S., Lee K.Y., Bussey C., Steckel M., Tanaka N., et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19:2668–2681 10.1101/gad.1360605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiefes A., Wolter S., Mushinski J.F., Hoffmann E., Dittrich-Breiholz O., Graue N., Dörrie A., Schneider H., Wirth D., Luckow B., et al. 2005. Simultaneous blockade of NFkappaB, JNK, and p38 MAPK by a kinase-inactive mutant of the protein kinase TAK1 sensitizes cells to apoptosis and affects a distinct spectrum of tumor necrosis factor [corrected] target genes. J. Biol. Chem. 280:27728–27741 10.1074/jbc.M411657200 [DOI] [PubMed] [Google Scholar]
- Thiefes A., Wolf A., Doerrie A., Grassl G.A., Matsumoto K., Autenrieth I., Bohn E., Sakurai H., Niedenthal R., Resch K., Kracht M. 2006. The Yersinia enterocolitica effector YopP inhibits host cell signalling by inactivating the protein kinase TAK1 in the IL-1 signalling pathway. EMBO Rep. 7:838–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F., Braun J.E., Motz C., Igreja C., Haas G., Truffault V., Izaurralde E., Weichenrieder O. 2009. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc. Natl. Acad. Sci. USA. 106:21591–21596 10.1073/pnas.0909871106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne J.S., Pestka S. 1975. Inhibition of protein synthesis in intact HeLa cells. Antimicrob. Agents Chemother. 8:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Wasiliew P., Kracht M. 2010. Interleukin-1 (IL-1) pathway. Sci. Signal. 3:cm1 10.1126/scisignal.3105cm1 [DOI] [PubMed] [Google Scholar]
- Weston C.R., Davis R.J. 2007. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19:142–149 10.1016/j.ceb.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C.Y., Shyu A.B., Müller M., Gaestel M., Resch K., Holtmann H. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969–4980 10.1093/emboj/18.18.4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen R., Thakur B.K., Dittrich-Breiholz O., Shah M., Redich N., Dhamija S., Kracht M., Holtmann H. 2007. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell. Biol. 27:8388–8400 10.1128/MCB.01493-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter S., Doerrie A., Weber A., Schneider H., Hoffmann E., von der Ohe J., Bakiri L., Wagner E.F., Resch K., Kracht M. 2008. c-Jun controls histone modifications, NF-kappaB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol. Cell. Biol. 28:4407–4423 10.1128/MCB.00535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054–1063 10.1038/nsmb1016 [DOI] [PubMed] [Google Scholar]
- Yoon J.H., Choi E.J., Parker R. 2010. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189:813–827 10.1083/jcb.200912019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Gavrilescu L.C., Molnár A., Murray L., Garafalo S., Kehrl J.H., Simon A.R., Van Etten R.A., Kyriakis J.M. 2009. GCK is essential to systemic inflammation and pattern recognition receptor signaling to JNK and p38. Proc. Natl. Acad. Sci. USA. 106:4372–4377 10.1073/pnas.0812642106 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou D., Chen S. 2001. PNRC2 is a 16 kDa coactivator that interacts with nuclear receptors through an SH3-binding motif. Nucleic Acids Res. 29:2003–2011 10.1093/nar/29.10.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Quach K.M., Yang C., Lee S.Y., Pohajdak B., Chen S. 2000. PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors including orphan receptors SF1 (steroidogenic factor 1) and ERRalpha1 (estrogen related receptor alpha-1). Mol. Endocrinol. 14:986–998 10.1210/me.14.7.986 [DOI] [PubMed] [Google Scholar]