RanBP2 captures RanGTP–importin-β complexes at cytoplasmic fibrils to ensure adequate classical NLS–mediated protein import and cell viability.
Abstract
RanBP2/Nup358, the major component of the cytoplasmic filaments of the nuclear pore complex (NPC), is essential for mouse embryogenesis and is implicated in both macromolecular transport and mitosis, but its specific molecular functions are unknown. Using RanBP2 conditional knockout mouse embryonic fibroblasts and a series of mutant constructs, we show that transport, rather than mitotic, functions of RanBP2 are required for cell viability. Cre-mediated RanBP2 inactivation caused cell death with defects in M9- and classical nuclear localization signal (cNLS)–mediated protein import, nuclear export signal–mediated protein export, and messenger ribonucleic acid export but no apparent mitotic failure. A short N-terminal RanBP2 fragment harboring the NPC-binding domain, three phenylalanine-glycine motifs, and one Ran-binding domain (RBD) corrected all transport defects and restored viability. Mutation of the RBD within this fragment caused lethality and perturbed binding to Ran guanosine triphosphate (GTP)–importin-β, accumulation of importin-β at nuclear pores, and cNLS-mediated protein import. These data suggest that a critical function of RanBP2 is to capture recycling RanGTP–importin-β complexes at cytoplasmic fibrils to allow for adequate cNLS-mediated cargo import.
Introduction
Traffic of macromolecules between the cytosol and the nucleus occurs through nuclear pore complexes (NPCs). Each NPC is composed of multiple copies of ∼30 different proteins called nucleoporins, which form a central transport channel that perforates the nuclear envelope, eight filaments that protrude from the cytoplasmic face of the pore, and eight filaments that converge into a basket structure at the nuclear side of the pore (Strambio-De-Castillia et al., 2010). Nucleoporins with phenylalanine-glycine (FG) repeats line the central channel, where they create a permeability barrier for larger macromolecules and contribute to transport receptor–mediated traffic through the NPC (Terry and Wente, 2009). Transport receptors bind to NLSs or nuclear export signals (NESs) in macromolecules to be transported and modulate cargo translocation across the NPC via sequential FG nucleoporin interactions. Most transport receptors belong to a family of related proteins, which in humans consists of >21 members, including the protein import receptors importin-β and transportin 1 and the protein export receptor Crm1 (Chook and Suel, 2010).
RanGTPase regulates the ability of nuclear transport factors to bind and release cargo (Wente and Rout, 2010). Import receptor–cargo complexes reaching the nuclear face of the NPC bind RanGTP, resulting in cargo release. On the other hand, binding of export receptors to RanGTP promotes cargo loading rather than release. As export complexes arrive at the cytoplasmic face of the pore, hydrolysis of RanGTP to RanGDP triggers cargo release into the cytoplasm. RanGTP hydrolysis in the cytosol is activated by the RanGTPase-activating protein RanGAP1 and is facilitated by binding to RanBP1. In the nucleus, Ran is maintained in the GTP-bound form by the guanine nucleotide exchange factor RCC1.
Despite great progress in identifying the components and principles of the nucleocytoplasmic transport machinery, in-depth mechanistic understanding of individual components of this system is often difficult to obtain because of the dynamic nature of macromolecular transport and the intricacy of the NPC (Terry and Wente, 2009). Adding to the complexity is that several nucleoporins are not only implicated in nucleocytoplasmic transport in interphase but also in the segregation of chromosomes during mitosis (Wozniak et al., 2010). One of these proteins is RanBP2 (or Nup358), which is the major nucleoporin component of the cytoplasmic filaments of the NPC (Walther et al., 2002). In addition to FG repeats, RanBP2 has various non-FG domains that are implicated in cargo transport (Wu et al., 1995; Yokoyama et al., 1995). For instance, RanBP2 has four Ran-binding domains (RBDs) and a small ubiquitin-like modifier (SUMO) E3 ligase domain that binds SUMO-modified RanGAP1, which have been proposed to stimulate dissociation of RanGTP-exportin-cargo and RanGTP-recycling import receptor complexes exiting the central channel (Mahajan et al., 1997; Matunis et al., 1998), thereby presumably facilitating both nuclear export and nuclear import (Bernad et al., 2004; Engelsma et al., 2004; Hutten and Kehlenbach, 2006; Hutten et al., 2008, 2009). In addition to binding SUMO-RanGAP1, the SUMO E3 ligase domain has been proposed to mediate cargo sumoylation at the cytoplasmic face of the NPC (Pichler et al., 2002; Reverter and Lima, 2005). Positioned in the central portion of RanBP2 are zinc finger domains that provide a binding platform for Crm1 (Singh et al., 1999) and that are thought to play a role in its transport back into the nucleus (Bernad et al., 2004).
When the nuclear envelope disintegrates and NPCs disassemble at the start of prometaphase, RanBP2-SUMO1–RanGAP1-Ubc9 subcomplexes disperse into the mitotic cytosol. In certain human cell lines, these subcomplexes accumulate at plus ends of spindle microtubules (MTs) and, in a Crm1-dependent fashion, at unattached kinetochores (Dasso, 2006). In HeLa and RGG cells, depletion of RanBP2 causes various mitotic abnormalities, including formation of multipolar spindles, chromosome misalignment, and mislocalization of several kinetochore-associated proteins (Salina et al., 2003; Joseph et al., 2004). Mouse embryonic fibroblasts (MEFs) from mice with low amounts of RanBP2 form chromatin bridges in anaphase, resulting in aneuploidy (Dawlaty et al., 2008). This led to the discovery that RanBP2 sumoylates TopII-α, thereby targeting this decatenating enzyme to inner centromeres to resolve sister chromosomes and prevent bridging.
Although RanBP2 has been implicated in diverse functions related to both transport and mitosis, it is unknown how RanBP2 performs its diverse tasks and how critical each function is for cellular integrity. Here, we have addressed these central questions using RanBP2 conditional knockout MEFs and a series of RanBP2 domain mutants.
Results
RanBP2 is essential for cell viability
Inactivation of RanBP2 causes early embryonic lethality (Aslanukov et al., 2006; Dawlaty et al., 2008), but whether RanBP2 is essential for viability at the cellular level has not been established. To examine this, we generated RanBP2 conditional knockout MEFs by intercrossing RanBP2+/F mice. We created these mice by interbreeding our RanBP2 hypomorphic mice (Dawlaty et al., 2008) with Flp recombinase transgenic mice (Fig. 1 A). Independent RanBP2F/F MEF lines were immortalized by expression of SV40 large T antigen and then transduced with pTSIN-Cre lentivirus to establish RanBP2−/− MEFs. Progressive cell death started on day 5 after transduction, resulting in a complete loss of viable cells between days 8 and 10 (Fig. 1 B). Western blot analysis of MEF extracts prepared at various days after lentiviral transduction showed that RanBP2 protein was undetectable by day 6 (Fig. 1 C). Consistent with this, RanBP2F/F MEFs at day 6 after Cre expression, designated as RanBP2−/− (D6) MEFs, showed no detectable nuclear rim staining for RanBP2 (Fig. 1 D). SUMO1-conjugated RanGAP1, whose association with the NPC is known to be dependent on RanBP2 (Matunis et al., 1998), was also undetectable (Fig. 1, E and F). SUMO1-RanGAP1 levels at the nuclear envelope were strongly decreased in RanBP2−/− (D6) MEFs (Fig. 1 G), strengthening the notion that SUMO1-RanGAP1 requires RanBP2 for protection against SUMO-isopeptidase activity (Zhang et al., 2002; Zhu et al., 2009). Furthermore, declining SUMO1-RanGAP1 levels were accompanied by corresponding increases in nonconjugated RanGAP1 levels. Together, these data show that RanBP2 is essential for cell viability.
Figure 1.
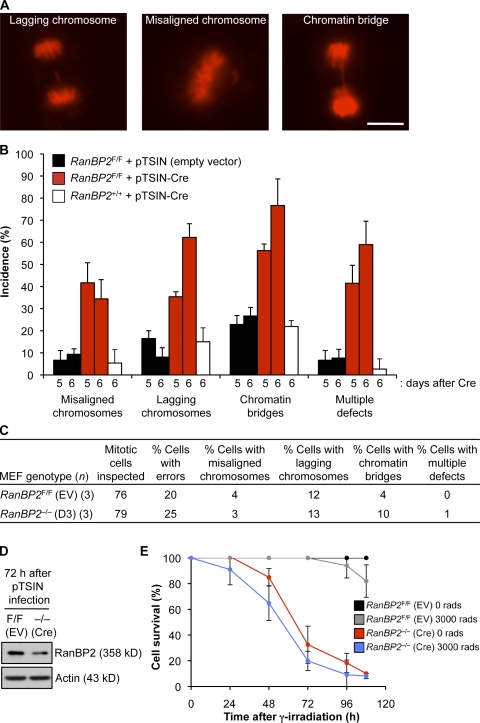
Generation of MEFs lacking RanBP2. (A) An overview of the knockout strategy. Relevant portions of the RanBP2 hypomorphic allele (H), the floxed allele (F), and the null allele (−) are shown. (B) Images of RanBP2F/F MEF cultures taken 8 d after infection with empty or Cre-containing lentivirus. (C) Western blots of MEF extracts prepared at the indicated times after infection with Cre lentivirus. (D) Images illustrating that NPCs of RanBP2−/− (D6) MEFs lack RanBP2. (E) Images showing that RanGAP1 is unable to localize to nuclear pores in RanBP2−/− (D6) MEFs. (F) Images showing that SUMO1 fails to decorate the nuclear envelope in RanBP2−/− (D6) MEFs. (G) Western blots of MEF extracts of RanBP2F/F and RanBP2−/− (D6) MEFs probed for RanGAP1. Actin served as a loading control. Bars: (B) 500 µm; (D–F) 5 µm.
RanBP2-null cells exhibit chromosome missegregation but not mitotic failure
RanBP2 hypomorphic MEFs that express low amounts of RanBP2 protein undergo frequent chromosome missegregation in the absence of overt transport-related defects (Dawlaty et al., 2008). This led us to speculate that MEFs without RanBP2 might die in mitosis as a result of severe segregation errors, a phenomenon referred to as mitotic catastrophe (Vitale et al., 2011). To explore this possibility, chromosome segregation and cell fate of RanBP2F/F MEFs at days 5–7 after Cre infection were monitored using time-lapse microscopy. To visualize chromosomes, RanBP2F/F MEFs were transduced with a lentivirus expressing H2B–monomeric RFP (mRFP; Malureanu et al., 2009) before RanBP2 inactivation. Chromosome misalignment, chromosome lagging, and chromatin bridging were observed at high incidence at day 5 (Fig. 2, A and B). Chromosome lagging and chromosome bridging rates were even higher on day 6. In addition, more cells were experiencing multiple types of segregation errors (Fig. 2 B). All RanBP2−/− MEFs exhibited chromosome missegregation on day 6, mostly involving small numbers of chromosomes. On day 7, we were unable to identify any mitotic cells in our cultures. RanBP2F/F MEFs receiving empty virus showed the same types of segregation errors as their Cre-treated counterparts but at much lower rates and without progression.
Figure 2.
High chromosome missegregation rates but no mitotic catastrophe in MEFs lacking RanBP2. (A) Representative chromosome segregation errors in RanBP2−/− (D6) MEFs. Images are from live H2B-mRFP–expressing cells. Bar, 10 µm. (B) Analysis of chromosome segregation errors at days 5 and 6 after infecting RanBP2F/F MEFs with empty or Cre-containing lentivirus. RanBP2+/+ MEFs were used to control for chromosome missegregation that might result from expression of Cre recombinase (Loonstra et al., 2001). Error bars represent SEM. Three independent lines were analyzed per genotype/per treatment (z25 cells per line, z75 total per genotype/per treatment). (C) Chromosome segregation errors at 72 h after infecting RanBP2F/F MEFs with empty or Cre-containing lentivirus. EV, empty vector. (D) Western blot analysis of MEF extracts of RanBP2F/F and RanBP2−/− (D3) MEFs probed for RanBP2. Actin served as a loading control. (E) Survival curves of RanBP2-null and control MEFs with and without γ irradiation. RanBP2F/F MEFs were infected with empty or Cre-containing lentivirus and selected with puromycin for 24 h. Cells were then seeded in dishes and either not irradiated or exposed to 3,000 rads of γ irradiation at 72 h after infection (t = 0 in the graph). Cells were counted at 24-h intervals. n = 5 lines per genotype. Error bars represent SD.
Although the incidence of chromosome missegregation was 100% for RanBP2−/− (D6) MEFs, cells never died during mitosis. Furthermore, RanBP2−/− (D6) MEFs that we continued to monitor for 12 h after exit from mitosis rarely died during this period (unpublished data), suggesting that mitotic errors may not be the primary cause of death of RanBP2−/− MEFs. We reasoned that if mitotic errors were indeed not driving cell death, mitotically inactivated and cycling RanBP2−/− MEFs should die at similar rates. To test this, we arrested RanBP2−/− MEFs at G2/M with a low dose of γ irradiation 72 h after Cre transduction (day 3). At this time, RanBP2 protein levels are still relatively high, and 75% of cells still segregate their chromosomes accurately (vs. 80% of RanBP2F/F control cells; Fig. 2, C and D). However, even though most of the irradiated RanBP2−/− (D3) cells had no preexisting chromosome segregation errors, mitotic inactivation did not improve cell survival (Fig. 2 E).
Multiple nuclear import and export pathways are RanBP2 dependent
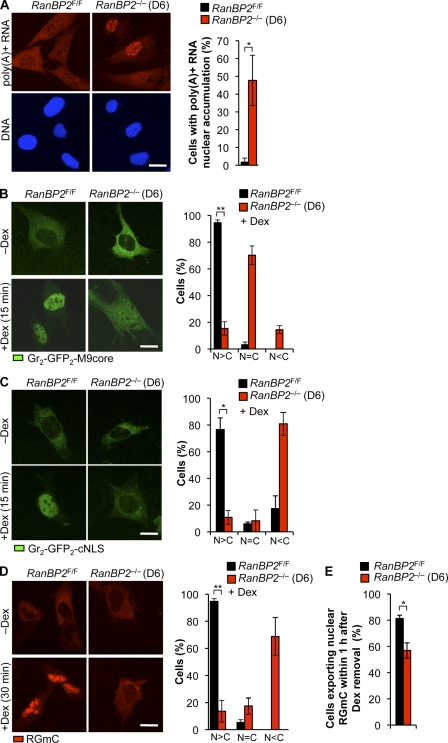
Earlier work in Drosophila melanogaster Schneider (S2) cells showed that depletion of RanBP2 results in nuclear accumulation of polyadenylated gene transcripts, presumably by perturbing efficient recycling of the mRNA export receptor NXF1 to the nucleus after cargo release at the NPC (Forler et al., 2004). However, RanBP2 depletion in HeLa cells by RNAi had little or no impact on mRNA export (Hutten and Kehlenbach, 2006). The cause of the observed discrepancy is currently unclear, although it has been proposed that it might be caused by the lack of RanBP1 in Drosophila (Hutten and Kehlenbach, 2006). To determine the impact of RanBP2 ablation on MEFs, we analyzed the subcellular distribution of poly(A)+ RNA in RanBP2−/− (D6) and RanBP2F/F MEFs by FISH with an oligo(dT)50 probe. As expected, in control MEFs, poly(A)+ RNA levels were high in the cytoplasm and relatively low in the nucleus (Fig. 3 A). In contrast, nuclear accumulation of poly(A)+ RNA was observed in nearly half of the RanBP2−/− (D6) MEFs. Poly(A)+ RNA levels in the cytoplasmic compartment of these cells was clearly reduced but never undetectable. The remaining cells had a staining pattern that was not markedly different from control cells. Immunostaining experiments revealed that RanBP2−/− (D6) MEFs had normal Nxf1 localization (unpublished data), implying that, unlike in RanBP2-depleted S2 cells, impairment of mRNA export does not correlate with aberrant release of the mRNA export receptor Nxf1 into the cytoplasm.
Figure 3.
Multiple transport pathways are perturbed in RanBP2-null MEFs. (A) Measurement of poly(A)+ RNA export from the nucleus (n = 3 MEF lines per genotype; >50 cells per line). (B) Measurement of protein import using the Gr2-GFP2-M9 reporter. Three independently generated cell lines were used (RanBP2F/F, n = 168; RanBP2−/− [D6], n = 97). (C) Measurement of protein import using the Gr2-GFP2-cNLS reporter. Three independently generated cell lines were used (RanBP2F/F, n = 167; RanBP2−/− [D6], n = 157). (D and E) Combined measurement of NLS protein import (D) and NES export (E) using RGmC. Three independently generated cell lines were used. (D) RanBP2F/F, n = 188; RanBP2−/− [D6], n = 173. (E) RanBP2F/F, n = 305; RanBP2−/− [D6], n = 332. (A–D) *, P < 0.01; **, P < 0.001 (two-tailed unpaired t test). Error bars indicate SD. C, cytoplasm; N, nucleus. Bars, 10 µm.
Contrasting results have also been obtained on the role of RanBP2 in protein import. For instance, in vitro import assays on RanBP2-deficient nuclei generated from immunodepleted Xenopus laevis egg extracts revealed efficient classical NLS (cNLS)– and M9-mediated protein import, even as nuclear pores embedded in these nuclei lacked cytoplasmic fibrils (Walther et al., 2002). However, more recent knockdown experiments in HeLa cells revealed that cNLS- and M9-mediated protein import are both less efficient when RanBP2 levels are reduced (Hutten et al., 2008, 2009). To thoroughly investigate the requirement for RanBP2 in protein import, RanBP2−/− (D6) and RanBP2F/F MEFs were subjected to various kinds of protein transport assays.
First, we measured M9-mediated protein import using the dexamethasone (Dex)-inducible GR2-GFP2-M9 reporter (Hutten et al., 2009). In the absence of Dex, this reporter is localized exclusively in the cytoplasm of both RanBP2−/− (D6) and RanBP2F/F MEFs. Upon addition of Dex, most RanBP2F/F MEFs accumulated GR2-GFP2-M9 into the nucleus within 15 min (Fig. 3 B). In contrast, a relatively low proportion of RanBP2−/− (D6) MEFs did so, with most cells showing near equal GR2-GFP2-M9 levels throughout the cell, reflecting impaired cargo import. A similar approach was used to study cNLS-mediated protein import using the GR2-GFP2-cNLS reporter (Hutten et al., 2008). Again, whereas most RanBP2F/F MEFs efficiently imported this cargo into the nucleus within 15 min after Dex treatment, RanBP2−/− (D6) MEFs failed to do so, with most cells exhibiting considerably lower nuclear than cytoplasmic fluorescence (Figs. 3 C and S1 A). A relatively small proportion of RanBP2−/− (D6) cells retained the ability to import cargo. Although these cells had no detectable RanBP2 at the nuclear rim (unpublished data), the potential presence of subdetectable RanBP2 levels that still might stimulate nuclear import cannot be excluded. Importantly, nearly identical results were obtained using a Dex-inducible mCherry-labeled chimeric HIV-1–Rev protein (Rev-Gr-mCherry [RGmC]) as the reporter (Fig. 3 D; Love et al., 1998). The NES of HIV-1–Rev allowed us to use the RGmC reporter for analysis of Crm1-mediated nuclear export in RanBP2-null cells. RanBP2−/− (D6) MEFs showed a slight, but statistically significant, decrease in the ability to export RGmC from the nucleus within 1 h after removal of Dex (Fig. 3 E). This result is in accord with earlier studies documenting that RanBP2 knockdown causes a mild Crm1-mediated nuclear export defect (Bernad et al., 2004; Hutten and Kehlenbach, 2006). RCC1, which generates the RanGTP necessary for assembly of nuclear export complexes, was properly localized to the nuclear compartment (Fig. S1 B). In summary, these data demonstrate that RanBP2 loss substantially reduces the efficiency with which various transport pathways operate and raise the possibility that cell death in the absence of RanBP2 is caused by the combination of defects in cellular processes and pathways that require rapid nucleocytoplasmic exchange of macromolecules.
C-terminal domains of RanBP2 are not essential for cell growth and survival
To dissect the critical RanBP2 domains and to provide insight into the in vivo significance of discrete domains within the complex modular architecture of this giant nucleoporin, we wanted to stably introduce mutant RanBP2 expression constructs into RanBP2F/F MEFs, inactivate endogenous RanBP2 with pTSIN-Cre lentivirus, and monitor cells for growth and survival. Stable expression of DNA sequences encoding a GFP-tagged human RanBP2-1–3224 fusion protein (Fig. 4, A and B) confirmed that full-length RanBP2 is able to rescue cell survival in the absence of endogenously expressed RanBP2 (Fig. 4, C–F).
Figure 4.
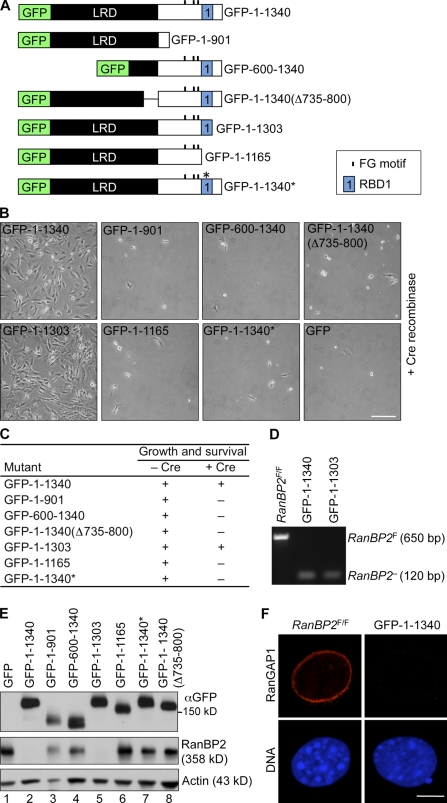
The RanBP2 SUMO E3 ligase domain is not essential for cell viability. (A) An overview of wild-type and mutant RanBP2 proteins. The LRD includes the NPC-targeting domain. CY, cyclophilin homology domain; E3, SUMO E3 ligase domain; KBD, kinesin-binding domain; ZnFs, zinc finger motifs. (B) Fluorescent images of RanBP2−/− (D6) MEFs containing the indicated GFP-tagged RanBP2 mutants. Bar, 5 µm. (C) Images of RanBP2−/− (D8) MEFs containing the indicated expression constructs. Bar, 200 µm. (D) Viability of RanBP2F/F MEFs expressing the indicated RanBP2 proteins in the presence or absence of endogenous RanBP2. (E) PCR genotyping of RanBP2−/− MEFs expressing the indicated RanBP2 proteins showing complete absence of RanBP2F alleles. (F) Western blot analysis of MEFs carrying the indicated GFP-tagged RanBP2 expression constructs. Actin was used as a loading control. MEFs in the first three lanes were Cre treated and lacked endogenous RanBP2, whereas MEFs in the last two lanes were not treated with Cre (nonsurviving mutants). The antibody against RanBP2 did not detect GFP-1–1889 and GFP-Δ2562–2802 proteins because these mutants lack the antibody epitope (Dawlaty et al., 2008). Asterisks mark endogenous RanBP2 protein.
Next, we created four RanBP2 mutants (Fig. 4 A): GFP-RanBP2-Δ2562–2802 (lacking the SUMO E3 ligase domain), HA-RanBP2-2553–2838 (consisting of the SUMO E3 ligase domain), GFP-RanBP2-1–1889 (N terminus), and GFP-RanBP2-1930–3224 (C terminus). As expected, only mutants containing the NPC-binding domain localized to the nuclear rim (Fig. 4 B and not depicted). Of the four mutants, only GFP-RanBP2-Δ2562–2802 and GFP-RanBP2-1–1889 maintained cell growth and survival upon inactivation of endogenous RanBP2 expression (Fig. 4, C and D). PCR analysis confirmed that the floxed alleles of the surviving cells had been converted into null alleles (Fig. 4 E). Western blot analysis using anti-GFP antibody validated that each mutant was accurately expressed (Fig. 4 F; Dawlaty et al., 2008). Furthermore, probing of these same blots with anti-RanBP2 antibody demonstrated that MEFs expressing HA-RanBP2-Δ2553–2802 and GFP-RanBP2-1–1889 lacked endogenous RanBP2 protein and that our GFP-tagged proteins were expressed at near endogenous levels. From these experiments, we conclude that the C-terminal RanBP2 domains are not essential for cell viability.
A relatively short N-terminal fragment of RanBP2 is sufficient for cell viability
To identify the essential domains within the RanBP2 N terminus, we first generated GFP-RanBP2-1–1340, which lacks the zinc finger domain cluster (Fig. 5 A). As shown in Fig. 5 (B–E), this mutant was capable of rescuing cell growth and survival of RanBP2−/− MEFs. Then, three additional truncation mutants were designed, focusing on the role of the leucine-rich domain (LRD; GFP-RanBP2-600–1340 and GFP-RanBP2-1–1340(Δ735–800)) and the region containing the three residual FG repeats and RBD1 (GFP-RanBP2-1–901). However, none of these mutants was able to rescue cell growth and survival in the absence of endogenous RanBP2 (Fig. 5, B and C), even though they were properly expressed (Fig. 5 E). GFP-RanBP2-600–1340 did not accumulate at the nuclear envelope (Fig. S2 A), indicating that NPC binding is essential.
Figure 5.
The N terminus of RanBP2 is sufficient for cell growth and survival. (A) An overview of mutant RanBP2 proteins. 1–1340*, 1–1340 containing a W1211R and K1212M double mutation in RBD1 designed to disrupt Ran binding and potentiation of RanGAP1. (B) Images of RanBP2−/− (D8) MEFs containing the indicated expression constructs. Bar, 200 µm. (C) Viability of RanBP2F/F MEFs expressing the indicated RanBP2 proteins in the presence or absence of endogenous RanBP2. (D) PCR analysis of DNA isolated from viable RanBP2−/− (D6) MEFs expressing the indicated RanBP2 proteins. (E) A Western blot of MEFs carrying the indicated GFP-tagged RanBP2 expression constructs. Actin was used as a loading control. MEFs in lanes 2 and 5 were Cre treated (lacking endogenous RanBP2), whereas MEFs in the remaining lanes were untreated. (F) RanBP2−/− MEFs expressing GFP-RanBP2-1–1340 and control MEFs stained for RanGAP1. Bar, 5 µm.
To specifically determine the requirement for RBD1, we generated the following three mutants: GFP-RanBP2-1–1303, which is truncated up to RBD1; GFP-RanBP2-1–1165, which completely lacks RBD1; and GFP-RanBP2-1–1340*, containing two point mutations in RBD1 designed to perturb Ran binding and potentiation of RanGAP1 activity (Petersen et al., 2000). Of these, only GFP-RanBP2-1–1303 was able to provide cell viability, indicating that RanGTP binding is essential. Thus, the critical RanBP2 functions are concentrated in a relatively short N-terminal segment of the protein. We confirmed that this fragment cannot recruit RanGAP1 to the cytoplasmic face of the NPC (Fig. 5 F) and cannot stabilize SUMO1-RanGAP1 (Fig. S2 B).
The N terminus and the SUMO E3 ligase domain of RanBP2 have mitotic functions
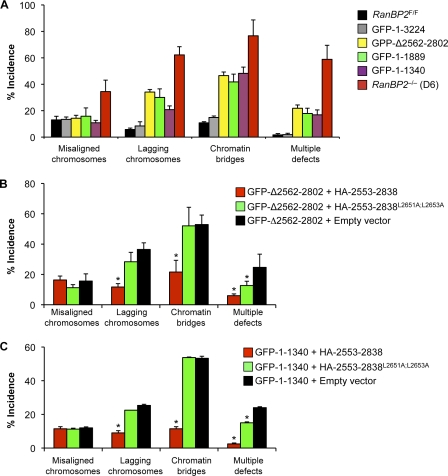
To determine the RanBP2 domain requirements for accurate chromosome segregation, chromosome movements of RanBP2−/− MEFs expressing RanBP2 mutants that rescue cell growth and survival were followed by live-cell microscopy. Importantly, full-length RanBP2 fully restored the chromosome misalignment, lagging, and bridging errors resulting from complete loss of RanBP2 (Fig. 6 A). Although GFP-RanBP2-Δ2562–2802, the mutant specifically lacking the SUMO E3 ligase domain of RanBP2, fully corrected chromosome alignment errors, it only partially corrected chromosome lagging and chromatin bridging. Surprisingly, GFP-RanBP2-1–1889 and GFP-RanBP2-1–1340 had similar corrective potentials as GFP-RanBP2-Δ2562–2802, indicating that the SUMO E3 ligase domain is the only C-terminal domain with mitotic relevance.
Figure 6.
The RanBP2 SUMO E3 ligase domain acts autonomously in mitosis. (A) An analysis of segregation errors in RanBP2−/− (D6) MEFs and RanBP2−/− MEFs expressing the indicated RanBP2 mutants (n = 3 MEF lines per mutant; ∼25 cells per line). Error bars indicate SEM. All mutants are P < 0.05 versus RanBP2−/− (D6) MEFs (χ2 test). (B) An incidence of chromosome segregation errors in GFP-RanBP2-Δ2562–2802 RanBP2−/− MEFs expressing wild-type or mutant SUMO E3 ligase domain (n = 3 MEF lines evaluated for each mutant combination; ∼20–25 cells per line). (C) An incidence as described in B but for GFP-RanBP2-1–1340 RanBP2−/− MEFs. (B and C) *, P < 0.05 versus GFP-RanBP2-Δ2562–2802 RanBP2−/− MEFs carrying empty expression vector (χ2 test). Error bars indicate SEM.
To provide further support for the aforementioned conclusion and to determine whether the SUMO E3 ligase domain can perform mitotic functions as an independent functional unit in the complete absence of endogenous RanBP2, we expressed the SUMO E3 ligase domain in RanBP2−/− MEFs that already express either GFP-RanBP2-Δ2562–2802 or GFP-RanBP2-1–1340 and measured the accuracy of chromosome segregation. Empty vector and mutant SUMO E3 ligase domain (RanBP2-2553–2838L2651A;L2653A), which lacked Ubc9 interaction and SUMO E3 ligase activity (Dawlaty et al., 2008), were used as controls in these experiments. As shown in Fig. 6 (B and C), combined expression of functional SUMO E3 ligase domain and either GFP-RanBP2-Δ2562–2802 or RanBP2-1–1340 corrected both chromosome lagging and chromatin bridging errors to rates seen in RanBP2F/F MEFs expressing normal amounts of endogenous RanBP2. No such corrective effect was observed in empty vector or HA-RanBP2-2553–2838L2651A;L2653A–transduced cells. These data demonstrate that the RanBP2 SUMO E3 ligase domain is the only C-terminal domain that regulates accurate chromosome segregation and suggest that this domain is functionally autonomous. Given the strong corrective effect of the SUMO E3 ligase domain on chromosome missegregation, we wondered whether its expression in RanBP2−/− MEFs containing GFP-RanBP2-1–1165 might restore growth and survival, but this was not the case (Fig. S3).
The mitotic phenotype of RanBP2−/− (D6) MEFs is different from that of RanBP2−/− MEFs expressing GFP-RanBP2-1–1340 in two ways. First, RanBP2−/− MEFs have increased rates of chromosome misalignment, and GFP-RanBP2-1–1340 RanBP2−/− MEFs do not. Second, although both RanBP2−/− MEFs and GFP-RanBP2-1–1340 RanBP2−/− MEFs have increased rates of chromosome lagging and chromatin bridging, the increases are much more profound in RanBP2−/− MEFs. These findings suggest that the RanBP2 N terminus also contributes to the chromosome segregation process. Previous studies of HeLa cells have documented that depletion of RanBP2 by RNAi perturbs mitotic spindle formation and loading of certain mitotic checkpoint proteins onto kinetochores in early mitosis (Salina et al., 2003; Joseph et al., 2004; Arnaoutov et al., 2005). However, no such mitotic defects were detectable in RanBP2−/− (D6) MEFs (Fig. S1 C and not depicted). Furthermore, RanBP2 itself is found at kinetochores in a subset of mammalian cell lines, including HeLa cells (Joseph et al., 2004; Arnaoutov and Dasso, 2005; Arnaoutov et al., 2005), but neither endogenous RanBP2 nor GFP-RanBP2-1–1340 was localized to kinetochores in mitotic MEFs (unpublished data). Together, these data suggest that the RanBP2 N terminus may exert its effect on chromosome segregation through an unknown mitotic function or indirectly through a nucleocytoplasmic transport function.
Ran binding ability of RanBP2 is required for adequate cNLS protein import
Next, we used our collection of RanBP2 mutants to investigate the domain requirements for each of the transport pathways that are defective in the absence of endogenous RanBP2. Restoration of cell viability by GFP-RanBP2-1–1340 coincided with near complete correction of the bulk mRNA export defect of RanBP2−/− (D6) MEFs (Fig. 7 A). However, strong corrective effects were also established by GFP-RanBP2-1–1340* and GFP-RanBP2-1–1165, two mutants that were unable to restore cell viability, suggesting that efficient mRNA export does not depend on RanBP2’s Ran binding ability. These data also imply that rescue of nuclear mRNA export is not sufficient to restore viability in the absence of endogenous RanBP2. In contrast, GFP-RanBP2-600–1340 established no corrective effect, indicating that the LRD is essential for mRNA export. Furthermore, GFP-RanBP2-1–901 had only a slight corrective effect, suggesting the involvement of FG repeats in efficient mRNA export.
Figure 7.
cNLS-mediated import selectively requires Ran binding ability. (A) An analysis of poly(A)+ RNA distribution in RanBP2−/− (D6) MEFs expressing the indicated RanBP2 mutants (n = 3 MEF lines per mutant). 1–1340*, 1–1340 containing a W1211R and K1212M double mutation in RBD1 designed to disrupt Ran binding and potentiation of RanGAP1. (B) Measurement of protein import using the Gr2-GFP2-cNLS reporter. C, cytoplasm; N, nucleus. (C) Measurement of protein import using the NES-GFP2-cNLS reporter. (D) In vitro import of FITC-labeled cNLS-BSA (at 50 mM importin-β). (A–C) Error bars indicate SD. (B and C) *, P < 0.01; **, P < 0.001 (two-tailed unpaired t test). n = 3 MEF lines per mutant genotype; ∼50 cells per line. Bars,10 µm.
Restoration of cell growth and survival by GFP-RanBP2-1–1340 also concurred with near complete correction of the GR2-GFP2-M9 nuclear import defect (Fig. S4 A). Because fluorescence from GFP-RanBP2-1–1340 and other GFP-tagged RanBP2 mutants was very low relative to that of GR2-GFP2-M9, it did not complicate the analysis (Fig. S5 A). Similar results were obtained for RanBP2−/− (D6) MEFs expressing GFP-RanBP2-1–1340* and GFP-RanBP2-1–1165, implying that cell death is not related to M9-mediated protein import and that RBD1 is not essential for this type of protein import. In vitro import assays on RanBP2−/− MEFs expressing the aforementioned mutants yielded similar results (Figs. S4 [B and C] and S5 B), confirming that M9-mediated import is RBD1 independent. In contrast, GFP-RanBP2-600–1340 and GFP-RanBP2-1–901 had no corrective effects, indicating that efficient M9-mediated transport is dependent on the LRD and the presence of the region with several FG repeats.
GFP-RanBP2-1–1340 also restored efficient import of GR2-GFP2-cNLS reporter protein (Fig. 7 B). In contrast, GFP-RanBP2-1–1340* and GFP-RanBP2-1–1165 did not, implying that RBD1 is required for efficient cNLS-mediated protein import. The validity of these findings was confirmed using the shuttling reporter protein NES-GFP2-cNLS (Fig. 7 C). Furthermore, in in vitro import assays with FITC-labeled recombinant cNLS-BSA as a substrate, efficient cargo import occurred in RanBP2−/− MEFs expressing GFP-RanBP2-1–3224 or GFP-RanBP2-1–1340 but not in those expressing GFP-RanBP2-1–1340 versions that cannot bind Ran (Figs. 7 D and S5 C). Thus, multiple independent assays indicate that RanBP2 Ran binding ability is critical for efficient cNLS-mediated protein import. We found that RanBP2 Ran binding ability is also essential for efficient in vitro import of Snail (Fig. S5 D), a protein that binds directly to importin-β and does not require importin-α (Yamasaki et al., 2005). Crm1-mediated protein export, in contrast, does not seem to be dependent on RanBP2 Ran binding ability (Fig. S5 E).
RBD1 of RanBP2 retains importin-β at nuclear pores
Ran binding by RanBP2 is unlikely to play a direct role in nuclear import of cNLS protein importin-α/β transport complexes because these complexes lack Ran. However, recycling of importin-α and -β to the cytoplasm after cargo release in the nucleus is Ran dependent (Wente and Rout, 2010). Immunostaining experiments for importin-α and -β revealed that neither transport factor was mislocalized in RanBP2−/− (D6) MEFs and RanBP2−/− (D6) MEFs expressing GFP-RanBP2-1–1340* (unpublished data). However, using a mild fixation procedure that allows for the removal of soluble protein fractions before staining, we observed a dramatic decline in importin-β levels at the nuclear rim in RanBP2−/− (D6) MEFs and RanBP2−/− (D6) MEFs expressing RanBP2 mutants without an (functional) RBD (Fig. 8 A). Western blot analysis demonstrated that this decline was not caused by a decrease in overall importin-β levels (Fig. S5 F). These data suggested that Ran-dependent docking of importin-β to the RBD of RanBP2 at the cytoplasmic face of the NPC might be essential for efficient cNLS-mediated nuclear protein import. Data from two additional experiments support this model. First, RanBP2−/− (D6) MEFs expressing GFP-RanBP2-1–1340* or GFP-RanBP2-1–1165 showed a strong increase in nuclear import of cNLS-BSA-FITC cargo when in vitro import assays were conducted at a twofold higher importin-β concentration (Fig. 8 B). Second, in a blot overlay assay (Delphin et al., 1997), GFP-RanBP2-1–1340 interacted with importin-β in the presence but not in the absence of RanGTP, whereas GFP-RanBP2-1–1340* or GFP-RanBP2-1–1165 was defective in binding importin-β both in the presence and absence of RanGTP (Fig. 8 C).
Figure 8.
Importin-β accumulation at the NPC is dependent on RanBP2 RBD1. (A) Images of RanBP2−/− (D6) MEFs expressing the indicated RanBP2 mutants immunostained for importin-β. Nuclei were visualized with Hoechst. 1–1340*, 1–1340 containing a W1211R and K1212M double mutation in RBD1 designed to disrupt Ran binding and potentiation of RanGAP1. (B) In vitro import of FITC-labeled NLS-BSA into RanBP2−/− (D6) MEFs expressing the indicated RanBP2 mutants. Import assays were conducted at a twofold higher importin concentration as described in Fig. 7 D. (C) Blot overlay assay measuring binding of importin-β to various N-terminal RanBP2 fragments in the absence or presence of RanGTP. (D) Images of digitonin-permeabilized RanBP2−/− MEFs expressing the indicated RanBP2 mutants stained for Crm1. Nuclei were visualized with Hoechst staining. (E) Quantification of Crm1 signal at the nuclear rim. Mean values of 10 cells per line are presented. Error bars represent SEM. **, P < 0.001 versus RanBP2−/− cells expressing GFP-1–3224 (two-tailed unpaired t test). Bars, 10 µm.
Crm1 is known to accumulate at the cytoplasmic face of the NPC in a RanBP2-dependent fashion, presumably to allow for efficient recycling of Crm1 to the nucleus after cargo release (Engelsma et al., 2004). However, RanBP2 mutants that fully restored the NES-mediated export defect of RanBP2−/− MEFs showed weak nuclear rim staining for Crm1 (Fig. 8, D and E), implying that accumulation of Crm1 at the cytoplasmic face of the pore is not a critical step in the NES transport pathway. The aforementioned analysis indicated that the RanBP2 domain responsible for accumulation of Crm1 at cytoplasmic filaments is located in the C-terminal half of the protein. RanBP2−/− MEFs expressing GFP-RanBP2-Δ2562–2802 showed normal Crm1 staining (Fig. 8, D and E), suggesting that Crm1 binding to RanBP2 filaments does not involve the SUMO E3 ligase domain.
Discussion
In this study, we have used a RanBP2 conditional knockout approach in MEFs to dissect the critical cellular functions of RanBP2. We show that RanBP2 is essential for viability at the cellular level and provides compelling evidence to suggest that cell death is associated with transport defects rather than mitotic failure. Rescue experiments in which we expressed various RanBP2 mutant constructs before the inactivation of endogenous RanBP2 not only revealed the RanBP2 domains critical for cell growth and survival but also provided mechanistic insight into the dependence of various major transport pathways on these domains.
A link between RanBP2 and mRNA export in mammalian cells
A hitherto unrecognized role of mammalian RanBP2 in mRNA export was uncovered by our experiments. Previously, such a role had only been demonstrated in flies (Forler et al., 2004). There, RanBP2 serves as a major docking site for the mRNA transport receptor Nxf1. In binding to Nxf1, RanBP2 prevents Nxf1 diffusion into the cytoplasm after cargo release, allowing for efficient recycling of this transport receptor. Nxf1 is not mislocalized to the cytoplasm in RanBP2−/− MEFs, implying that the mechanisms by which RanBP2 controls mRNA export in insect and mouse cells may have diverged. The basis for this divergence may involve a differential requirement for RBDs of RanBP2. Insect cells are thought to be highly dependent on these domains for cargo release into the cytoplasm because RanBP1, the functional equivalent of RanBP2 RBDs, is lacking (Villa Braslavsky et al., 2000). Although RanBP2−/− MEFs and RanBP2 knockdown Drosophila cells show robust nuclear accumulation of poly(A)+ RNA (Forler et al., 2004), in both cases there is residual cytoplasmic poly(A)+ staining, indicating that RanBP2 is a facilitator rather than an indispensible component of this pathway. In addition to Nxf1, mRNPs contain various shuttling binding proteins (Strambio-De-Castillia et al., 2010), whose potential mislocalization in RanBP2−/− MEFs might be indirectly implicated in nuclear accumulation of poly(A)+ RNA.
New insights into key actions of RanBP2 from trimmed filaments
RanBP2 is thought to provide a platform where RanGTP-exportin-cargo and RanGTP–importin complexes exiting at the cytoplasmic side of the NPC dock to disassemble via RanBP1- and RanGAP1-stimulated GTP hydrolysis on RanGTP (Yaseen and Blobel, 1999). In this model, GTP hydrolysis is not only stimulated by soluble RanBP1 and RanGAP1 but also by RanBP2-bound RanGAP1 and RBDs with RanBP1-like properties (Mahajan et al., 1997; Matunis et al., 1998; Villa Braslavsky et al., 2000). Furthermore, RanBP2 is thought to mediate efficient exportin recycling to the nucleus and efficient reloading of cargo onto newly exported importins (Bernad et al., 2004; Hutten and Kehlenbach, 2006; Hutten et al., 2008).
Here, we have critically tested these and other presumed RanBP2 functions in vivo using RanBP2−/− MEF lines expressing various RanBP2 mutants. The absence of endogenous RanBP2 is key here because, as a result of the catalytic nature of the proposed RanBP2 functions, small amounts of protein that may remain with RNAi approaches could easily mask certain RanBP2 functional deficiencies. Particularly insightful was a short N-terminal RanBP2 fragment that restored cell viability and corrected all transport defects of RanBP2-null cells, despite only containing the NPC-anchoring domain, three FG repeats, and RBD1. However, when RBD1 was mutated, cNLS protein import rates dropped substantially, and cells were unable to sustain growth and survival. The finding that M9-mediated protein import, mRNA export, and NES-mediated export were largely unaffected indicates a selective rather than a generic requirement of transport pathways for RanGTP binding sites at the cytoplasmic face of the NPC. When the sole RBD is mutated or depleted, RanGTP–importin-β binding was perturbed, and importin-β failed to accumulate at nuclear pores, suggesting that a critical function of this RanBP2 domain is to concentrate importin-β at the cytoplasmic face of the pore. The most plausible scenario is that in the absence of RBD1, RanGTP–importin-β complexes recycling from the nuclear compartment fail to dock at the cytoplasmic face of the pore and diffuse into the cytoplasm. The drop in importin-β at the cytoplasmic face might impair the rate at which unloaded importin-β molecules can reload and start a new import cycle. Consistent with this interpretation is our finding that the importin-α/β–mediated protein import defect observed in the absence of RBD1 is largely corrected when the importin-β concentration in in vitro import assays is raised.
RanBP2−/− MEFs expressing RanBP2-1–1340 are viable and have quite normal NES protein export rates, even though they fail to accumulate Crm1 at the cytoplasmic face. This, together with the finding that RanBP2−/− MEFs expressing RanBP2-1–1340 show a normal Crm1 distribution pattern, suggests that the docking of Crm1 to RanBP2 is not critical for efficient cargo unloading and/or Crm1 recycling. The RanBP2 domains that mediate Crm1 accumulation at pores reside in the C-terminal half of the protein; our data suggest that the SUMO E3 ligase domain is not implicated. Interestingly, we find that transportin-mediated protein import and Nxf1-mediated mRNA export rely on the same RanBP2 domains. Because transportin recycles Nxf1 back into the nucleus after mRNA is discharged at the cytoplasmic side of the pore, it is tempting to speculate that impaired mRNA export is a result of inadequate transport receptor recycling rather than inefficient release of mRNA cargo at the cytoplasmic side of the pore. As mentioned, we did not observe an increase in the steady-state levels of Nxf1 in the cytoplasmic compartment, which, at the surface, argues against this possibility. However, if mRNA release and Nxf1 recycling were coupled events, the distribution of Nxf1 would be expected to remain unchanged.
An RNAi study in HeLa cells has suggested that transportin becomes rate limiting for efficient M9 protein import when RanBP2 levels are low (Hutten et al., 2009), but how RanBP2 facilitates M9 protein import is unknown. Our structure–function analysis demonstrates that the RanBP2 N terminus can largely sustain efficient M9-mediated export in the absence of RBD1 but not when the three FG motifs or the NPC-anchoring domain is deleted. The fact that RBD1 is not required indicates that RanBP2 does not exert its effect by stimulating release of recycled transportin at the cytoplasmic face of the pore. Perhaps RanBP2 FG motifs play a role in retaining transportin at the NPC after GTP hydrolysis to allow for efficient loading of M9-containing cargo. We were unable to test this potential mechanism because antibodies detecting mouse transportin are not available.
RanBP2 in mitosis
Four observations presented here suggest that the chromosome segregation errors resulting from loss of RanBP2 are unlikely to cause cell death. First, similar cell death kinetics were observed in cycling and mitotically inactivated RanBP2-null MEFs. Importantly, at the time of mitotic inactivation, most RanBP2-null MEFs accurately separated their chromosomes. Second, unlike cells lacking mitotic checkpoint proteins such as Mad2 and BubR1 (Dobles et al., 2000; Malureanu et al., 2009), RanBP2-null MEFs did not die by mitotic catastrophe, a type of cell death that occurs in mitosis after a failed chromosome segregation attempt (Castedo et al., 2004). Third, chromosome missegregation events in RanBP2-null MEFs typically involved relatively small numbers of chromosomes and rarely resulted in cell death within 12 h after completing cell division. Fourth, the RanBP2-1–1340 fragment repairs transport and viability but still has frequent mitotic errors.
Using RanBP2 hypomorphic MEFs that have ∼20% of normal RanBP2 levels, we previously identified a key role for the RanBP2 SUMO E3 ligase domain in chromosome segregation (Dawlaty et al., 2008). Our structure–function analysis demonstrates that it is the sole domain in the C-terminal two thirds of the protein implicated in mitosis. Our finding that RanBP2-null cells show near complete correction of accurate chromosome segregation upon coexpression of the SUMO E3 ligase domain and RanBP2-1–1340 indicates that the E3 ligase domain can perform its mitotic functions in an autonomous fashion. Mutation of the SUMO E3 ligase domain precludes such correction, revealing that it exerts its corrective effect through its ability to sumoylate TopII-α and potentially other mitotic regulators (Dawlaty et al., 2008) or through regulation of the mitotic functions of RanGTPase via RanGAP1 (Arnaoutov and Dasso, 2005).
Our finding that the N-terminal RanBP2 fragment that mediates cell survival fully restores proper chromosome misalignment in RanBP2-null MEFs suggested that this fragment might have an independent role in mitosis. Interestingly, in certain human cell lines, RanBP2 accumulates at kinetochores in a Crm1-dependent fashion, presumably to stabilize kinetochore–MT attachments (Joseph et al., 2004; Arnaoutov et al., 2005). However, two observations suggest that the N terminus of RanBP2 does not exert its corrective effect on chromosome–MT attachment through this mechanism in MEFs. First, the N-terminal fragment lacks Crm1 binding ability. Second, neither the N terminus of RanBP2 nor endogenous RanBP2 accumulates at kinetochores in MEFs. Perhaps the N terminus acts to stabilize MT–kinetochore attachments in a Crm1-independent fashion. The LRD has been suggested to bind and regulate the dynamics of MTs (Joseph and Dasso, 2008). Alternatively, the N terminus may regulate the activities of RanGTPase in MT–chromosome attachment by stimulating RanGTP hydrolysis of cargo complexes containing spindle assembly factors (Dasso, 2002). Finally, RanBP2 depletion in cancer cell lines interferes with kinetochore targeting of proteins implicated in mitotic checkpoint signaling and proper MT–kinetochore attachment. However, no such defect was observed in MEFs lacking RanBP2.
In conclusion, our data suggest that docking of recycling importin-β–RanGTP complexes to RanBP2 RBDs at the cytoplasmic fibrils of NPCs is necessary to achieve importin-β–mediated nuclear import rates high enough to sustain cell growth and survival. Whether cell death resulting from loss of this docking ability involves inadequate nuclear import of a broad spectrum of nuclear proteins or just a small subset of perhaps highly dynamic shuttling proteins remains to be determined.
Materials and methods
Generation and culture of RanBP2 conditional knockout MEFs
We created heterozygous RanBP2 conditional knockout mice (RanBP2+/F mice) by interbreeding RanBP2 hypomorphic mice (RanBP2H/H mice) with FLPeR transgenic mice purchased from The Jackson Laboratory. MEFs were subsequently generated from 13.5-d-old embryos of RanBP2+/F females mated with RanBP2+/F males. Three independent RanBP2F/F MEF lines were obtained and immortalized by transduction with SV40 large T antigen–containing murine stem cell virus retrovirus. Primers used for the detection of RanBP2F and RanBP2− alleles have been previously described (Dawlaty et al., 2008). An inverted microscope (TMS; Nikon) with a 10× Ph1 dual lens objective was used to acquire images of MEF cultures.
Western blot analysis, indirect immunofluorescence, and confocal microscopy
Western blot analysis (Kasper et al., 1999) and indirect immunofluorescence (Kasper et al., 1999; Taylor et al., 2001) were performed as previously described. Standard fixations for immunostainings were performed with 3% PFA for 15 min at RT. For nuclear pore localization studies, cells were fixed with 1% PFA for 15 min at RT (mild fixation). For kinetochore localization studies, cells were fixed with 1% PFA for 5 min at RT. Fixation was followed by permeabilization in 0.2% Triton X-100 for 10 min. For staining of Crm1 at the cytoplasmic face of the NPC, cells were permeabilized with 0.4 µg/ml digitonin for 5 min and then fixed with 3% PFA for 15 min at RT (no Triton X-100 permeabilization). A laser-scanning microscope (LSM 510 v3.2SP2; Carl Zeiss) with Axiovert 100M (Carl Zeiss) and c-Apochromat 100 or 63× objectives was used to analyze immunostained cells and acquire images. Quantification of fluorescence was performed on confocal images using ImageJ software (National Institutes of Health). Fluorescence intensity at the nuclear rim was measured at four points per cell and averaged. Fluorescence intensity inside the nucleus and in the intercellular background was subtracted from the mean nuclear rim value.
Antibodies
The following antibodies were used: rabbit anti–hRanBP2-2500–3224 (Dawlaty et al., 2008) and mouse anti-RanBP2 (D-4; Santa Cruz Biotechnology, Inc.; Fig. 1, E and F); rabbit anti-RanGAP1 (provided by M. Dasso, National Institutes of Health, Bethesda, MD); goat anti-RanGAP1 (N-19; Santa Cruz Biotechnology, Inc.); mouse anti-GFP (B-2; Santa Cruz Biotechnology, Inc.); rabbit anti-hCrm1 (provided by M. Fornerod, Erasmus University, Rotterdam, Netherlands); mouse anti-MAb414 (Covance); rabbit anti–CENP-E (provided by D. Cleveland, Ludwig Institute for Cancer Research, La Jolla, CA); rabbit anti-Bub1 (Jeganathan et al., 2007); rabbit anti-Mad2 (Malureanu et al., 2010); goat antiimportin-β (C-19; Santa Cruz Biotechnology, Inc.); rabbit anti-SUMO1 (Cell Signaling Technology); mouse anti-SUMO2/3 (Zhang et al., 2008); mouse anti–β actin (A5441; Sigma-Aldrich); and goat anti-RCC1 (N-19; Santa Cruz Biotechnology, Inc.).
Live-cell imaging
Analyses of chromosome missegregation and mitotic timing by live-cell imaging were performed as previously described (Jeganathan et al., 2007; Malureanu et al., 2009). In brief, MEFs were transduced with a lentivirus encoding an mRFP-tagged H2B to allow for visualization of chromosomes by fluorescence microscopy. Transduced cells were seeded onto 35-mm glass-bottomed culture dishes. Approximately 24 h later, chromosome movements of MEFs progressing through an unchallenged mitosis were followed at interframe intervals of 3 min using an Axio Observer Z1 system equipped with CO2 Module S, TempModule S, Heating Unit XL S, a Pln Apo 63×/1.4 oil differential interference contrast III objective, AxioCam MRm camera, and AxioVision 4.6 software (Carl Zeiss). PowerPoint software (Microsoft) for Macintosh was used for image processing, and Prism software (version 4.0a; GraphPad) for Macintosh was used for statistical analysis. Three independent clones per genotype were used unless otherwise noted.
Plasmids
pEGFP-C2 full-length human RanBP2 was previously described (Joseph and Dasso, 2008). All RanBP2 mutants were constructed from pEGFP-C2 full-length human RanBP2 using standard cloning procedures and verified by DNA sequencing. RanBP2-1–1340* contains W1211R and K1212M mutations in RBD1 of RanBP2. These residues correspond to W67R and K68M mutations in murine RanBP1, which have been shown to disrupt the RanBP1’s ability to bind Ran and potentiate RanGAP1 activity (Petersen et al., 2000). For stable expression of wild-type and mutant RanBP2 cDNAs, we used Tol2-based transposition (Balciunas et al., 2006) using pKTol2C-hygromycin and pKC-Tol2 (provided by S. Ekker, Mayo Clinic, Rochester, MN). pKTol2C-hygromycin was generated from pKTol2C-EGFP as follows: EGFP was removed by XhoI–BglII digestion and replaced with an adaptor with multiple cloning sites to create pKTol2C–multiple cloning site. Herpes simplex virus–thymidine kinase promoter–hygromycin resistance gene–herpes simplex virus thymidine kinase pA cassette was PCR amplified with primers 5′-TTATTTACCGGTCTGCTTCATCCCCGTGGC-3′ and 5′-TTATTTGCTAGCTTGCGCCAGAAATCCGCG-3′ using pJTI Fast (Invitrogen) as a template. AgeI–NheI-digested PCR product was inserted into AgeI–NheI-opened pKTol2C-MCS to generate pKTol2C-hygromycin. HA-tagged RanBP2-2553–2838 and RanBP2-2553–2838L2561A;L2563A were previously described (Dawlaty et al., 2008). HA-RanBP2-2553–2838 and HA-RanBP2-2553–2838L2561A;L2563A were cloned into the MluI site of the retroviral vector pMSCV-bsd. Procedures for the production of retroviral vectors were performed as previously described (Kasper et al., 1999; Malureanu et al., 2009).
Transfection and viral transduction
To create cell lines stably expressing RanBP2 or RanBP2 mutants, pKTol2Chyg-EGFP-RanBP2 vectors were cotransfected with pKC-Tol2 into immortalized RanBP2F/F MEFs using Amaxa Nucleofector II (Lonza). Nucleofections were performed on 2 × 106 cells in MEF Nucleofector Solution 2 (VPD-1005; Lonza). After 24 h, nucleofected cells were selected in 300 µg/ml hygromycin B (Roche) for 7 d before experiments. For inactivation of endogenous RanBP2, RanBP2F/F MEFs or RanBP2F/F MEFs expressing various RanBP2 mutants were transduced with pTSINpuro-Cre lentivirus and cultured in DME/10% FCS with 3 µg/ml puromycin (InvivoGen) after 24 h. Puromycin was maintained in culture medium throughout the experiments.
Detection of polyadenylated RNA and nuclear transport assays
FISH for the detection of polyadenylated RNA was performed as previously described (Bastos et al., 1996). In brief, cells were fixed with 3% PFA for 30 min at RT. After five washes in PBS, cells were permeabilized in PBS/0.5% Triton X-100 for 5 min at RT. Cells were rinsed five times in PBS, prehybridized for 60 min at 37°C in hybridization mix without probe, and then hybridized overnight at 37°C. The hybridization mix consisted of 2× SSC containing Denhardt’s solution (Sigma-Aldrich), 1 mg/ml yeast tRNA (Invitrogen), 10% dextran sulfate, 25% formamide, 0.5 mg/ml salmon sperm DNA (Invitrogen), and 50 µg/ml biotin–oligo dT(50) probe. After hybridization, the cells were washed twice in 2× SSC for 15 min at RT, once in 1× SSC for 15 min at RT, and once in 0.5× SSC for 15 min at 37°C. Immediately upon completion of the last wash, cells were fixed with 3% PFA for 5 min at RT. After three washes in PBS, cells were incubated for 30 min at RT with A594-streptavidin (Invitrogen) diluted into PBS/0.2% Triton X-100. After three washes in PBS/0.2% Triton X-100/Hoeschst and two final washes in PBS, cells were mounted in Vectashield (Vector Laboratories) and analyzed by confocal microscopy.
For in vivo transport studies, we used the following expression constructs: Gr2-GFP2-cNLS (Hutten et al., 2008), Gr2-GFP2-M9 (Hutten et al., 2009), and NES-GFP2-cNLS (Hutten et al., 2008). RGmC was generated by inserting XhoI-digested Rev-GR fragment from pXRGG (Love et al., 1998) into the XhoI site engineered after the start codon of mCherry in pTSINmCherry lentiviral vector (a gift from E. Poeschla, Mayo Clinic, Rochester, MN).
In vivo import studies were performed as previously described (Hutten et al., 2009). In brief, NES-GFP2-cNLS, Gr2-GFP2-cNLS, Gr2-GFP2-M9, or RGmC was transfected into MEFs using Lipofectamine 2000 (Invitrogen). The next day, cells expressing the latter three constructs were incubated in the presence (5 µM) or absence of Dex for 15 min and then fixed with 2% PFA for 8 min at RT. Cells expressing NES-GFP2-cNLS were fixed immediately. The number of cells with cytoplasmic accumulation of fluorescent protein (N < C), nuclear accumulation of fluorescent protein (N > C), or even distribution of fluorescent protein between the nucleus and the cytoplasm (N = C) was scored (Bernad et al., 2004; Hutten and Kehlenbach, 2006). Three independent MEF lines were evaluated per genotype (∼30 cells per line).
For in vivo export, cells transfected with RGmC were treated with 5 µM Dex for 2 h at 37°C, washed three times with PBS, and released in DME/10% FCS for 1 h. Cells were then fixed with 2% PFA for 8 min at RT. The percentages of cells with N ≥ C after 2 h in Dex and 1 h after removal of Dex were determined. The percentages of cells exporting RGmC from the nucleus were calculated from the total pool of cells with nuclear localization after 2 h of Dex treatment to take into account the preexisting import defect of RGmC in RanBP2 mutant cells.
In vitro nuclear import analysis was essentially performed as previously described (Hutten et al., 2009). In brief, MEFs grown on glass slides were permeabilized with 40 µg/ml digitonin (EMD) in transport buffer for 8 min on ice. For standard cNLS-mediated protein import, cells were incubated with 500 nM importin-α, 50 nM importin-β, 900 nM Ran, ATP regeneration system (1 mM ATP, 5 mM creatine phosphate, and 50 µg/ml creatine kinase), and 15.6 µg/ml cNLS-BSA-FITC in transport buffer for 10 min at RT and fixed with 3% PFA on ice. For M9-mediated protein import, cells were incubated in 200 nM transportin, 900 nM Ran, ATP regeneration system, and 40 µg/ml M9-BSA-FITC (Wu et al., 2001). In vitro import of GST-Snail-GFP (provided by Y. Yoneda, Osaka University, Osaka, Japan) was performed as previously described (Yamasaki et al., 2005).
Blot overlay assays
GFP-RanBP2-1–1340, GFP-RanBP2-1–1340*, and GFP-RanBP2-1–1165 were immunoprecipitated from MEF lines stably expressing these proteins as previously described (Kasper et al., 1999). The precipitated proteins were separated by SDS-PAGE and transferred to Immobilon-P membrane. Overlay binding assays were then performed essentially as previously described (Delphin et al., 1997). In brief, membranes with renatured proteins were incubated for 30 min at RT with 20 nM recombinant His–importin-β in the presence of 40 nM RanGTP or RanGDP or in the absence of Ran. Blots were then washed with 0.1% Tween 20 and 4 mM DTT in PBS and probed for His–importin-β using mouse anti–penta-His (QIAGEN). RanGTP and RanGDP were prepared by incubating 1 µM Ran with 1 mM GTP or GDP in 10 mM EDTA, 2 mM ATP, 4 mM DTT, and 50 mM Hepes, pH 7.4, for 30 min at 30°C. Reactions were stopped by adding MgCl2 to the final concentration of 15 mM at 4°C.
Online supplemental material
Fig. S1 shows that the subcellular localization of RCC1 and mitotic checkpoint proteins is normal in RanBP2-null MEFs. Fig. S2 shows that SUMO1 modification of RanBP1 requires the SUMO E3 ligase domain of RanBP2. Fig. S3 shows that RanBP2 SUMO E3 ligase activity cannot rescue cell death of GFP-RanBP2-1–1165 RanBP2−/− MEFs. Fig. S4 shows that M9-mediated protein import is not dependent on RBD1 of RanBP2. Fig. S5 contains results of various transport-related experiments performed on RanBP2 mutant MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201102018/DC1.
Acknowledgments
We thank S. Ekker, M. Dasso, D. Cleveland, Y. Yoneda, and E. Poeschla for providing critical reagents and Drs. P. Galardy, R. Ricke, and D. Baker for comments on the manuscript.
This work was supported by the National Institutes of Health grant RO1-CA077262.
Footnotes
Abbreviations used in this paper:
- cNLS
- classical NLS
- Dex
- dexamethasone
- LRD
- leucine-rich domain
- MEF
- mouse embryonic fibroblast
- mRFP
- monomeric RFP
- MT
- microtubule
- NES
- nuclear export signal
- NPC
- nuclear pore complex
- RBD
- Ran-binding domain
- RGmC
- Rev-Gr-mCherry
- SUMO
- small ubiquitin-like modifier
References
- Arnaoutov A., Dasso M. 2005. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 4:1161–1165 10.4161/cc.4.9.1979 [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. 2005. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 7:626–632 10.1038/ncb1263 [DOI] [PubMed] [Google Scholar]
- Aslanukov A., Bhowmick R., Guruju M., Oswald J., Raz D., Bush R.A., Sieving P.A., Lu X., Bock C.B., Ferreira P.A. 2006. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2:e177 10.1371/journal.pgen.0020177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Wangensteen K.J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P.B., Largaespada D.A., McIvor R.S., Ekker S.C. 2006. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2:e169 10.1371/journal.pgen.0020169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R., Lin A., Enarson M., Burke B. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141–1156 10.1083/jcb.134.5.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., van der Velde H., Fornerod M., Pickersgill H. 2004. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24:2373–2384 10.1128/MCB.24.6.2373-2384.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M., Perfettini J.L., Roumier T., Andreau K., Medema R., Kroemer G. 2004. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 23:2825–2837 10.1038/sj.onc.1207528 [DOI] [PubMed] [Google Scholar]
- Chook Y.M., Suel K.E. 2010. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta. 10.1016/j.bbamcr.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. 2002. The Ran GTPase: theme and variations. Curr. Biol. 12:R502–R508 10.1016/S0960-9822(02)00970-3 [DOI] [PubMed] [Google Scholar]
- Dasso M. 2006. Ran at kinetochores. Biochem. Soc. Trans. 34:711–715 10.1042/BST0340711 [DOI] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C., Guan T., Melchior F., Gerace L. 1997. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell. 8:2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M.L., Benezra R., Sorger P.K. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 101:635–645 10.1016/S0092-8674(00)80875-2 [DOI] [PubMed] [Google Scholar]
- Engelsma D., Bernad R., Calafat J., Fornerod M. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23:3643–3652 10.1038/sj.emboj.7600370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D., Rabut G., Ciccarelli F.D., Herold A., Köcher T., Niggeweg R., Bork P., Ellenberg J., Izaurralde E. 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol. 24:1155–1167 10.1128/MCB.24.3.1155-1167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Kehlenbach R.H. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26:6772–6785 10.1128/MCB.00342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Flotho A., Melchior F., Kehlenbach R.H. 2008. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol. Biol. Cell. 19:2300–2310 10.1091/mbc.E07-12-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Wälde S., Spillner C., Hauber J., Kehlenbach R.H. 2009. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell Sci. 122:1100–1110 10.1242/jcs.040154 [DOI] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Dasso M. 2008. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 582:190–196 10.1016/j.febslet.2007.11.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Liu S.T., Jablonski S.A., Yen T.J., Dasso M. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617 10.1016/j.cub.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Kasper L.H., Brindle P.K., Schnabel C.A., Pritchard C.E., Cleary M.L., van Deursen J.M. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra A., Vooijs M., Beverloo H.B., Allak B.A., van Drunen E., Kanaar R., Berns A., Jonkers J. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 98:9209–9214 10.1073/pnas.161269798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D.C., Sweitzer T.D., Hanover J.A. 1998. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. USA. 95:10608–10613 10.1073/pnas.95.18.10608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 88:97–107 10.1016/S0092-8674(00)81862-0 [DOI] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L., Jeganathan K.B., Jin F., Baker D.J., van Ree J.H., Gullon O., Chen Z., Henley J.R., van Deursen J.M. 2010. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J. Cell Biol. 191:313–329 10.1083/jcb.201003090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Wu J., Blobel G. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499–509 10.1083/jcb.140.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Orem N., Trueheart J., Thorner J.W., Macara I.G. 2000. Random mutagenesis and functional analysis of the Ran-binding protein, RanBP1. J. Biol. Chem. 275:4081–4091 10.1074/jbc.275.6.4081 [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Reverter D., Lima C.D. 2005. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 435:687–692 10.1038/nature03588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Enarson P., Rattner J.B., Burke B. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001 10.1083/jcb.200304080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.B., Patel H.H., Roepman R., Schick D., Ferreira P.A. 1999. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J. Biol. Chem. 274:37370–37378 10.1074/jbc.274.52.37370 [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395 [DOI] [PubMed] [Google Scholar]
- Terry L.J., Wente S.R. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell. 8:1814–1827 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa Braslavsky C.I., Nowak C., Görlich D., Wittinghofer A., Kuhlmann J. 2000. Different structural and kinetic requirements for the interaction of Ran with the Ran-binding domains from RanBP2 and importin-beta. Biochemistry. 39:11629–11639 10.1021/bi001010f [DOI] [PubMed] [Google Scholar]
- Vitale I., Galluzzi L., Castedo M., Kroemer G. 2011. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12:385–392 10.1038/nrm3115 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Pickersgill H.S., Cordes V.C., Goldberg M.W., Allen T.D., Mattaj I.W., Fornerod M. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63–77 10.1083/jcb.200202088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R., Burke B., Doye V. 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell. Mol. Life Sci. 67:2215–2230 10.1007/s00018-010-0325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Matunis M.J., Kraemer D., Blobel G., Coutavas E. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209–14213 10.1074/jbc.270.23.14209 [DOI] [PubMed] [Google Scholar]
- Wu X., Kasper L.H., Mantcheva R.T., Mantchev G.T., Springett M.J., van Deursen J.M. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA. 98:3191–3196 10.1073/pnas.051631598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Sekimoto T., Ohkubo T., Douchi T., Nagata Y., Ozawa M., Yoneda Y. 2005. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 10:455–464 10.1111/j.1365-2443.2005.00850.x [DOI] [PubMed] [Google Scholar]
- Yaseen N.R., Blobel G. 1999. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin nup358. J. Biol. Chem. 274:26493–26502 10.1074/jbc.274.37.26493 [DOI] [PubMed] [Google Scholar]
- Yokoyama N., Hayashi N., Seki T., Panté N., Ohba T., Nishii K., Kuma K., Hayashida T., Miyata T., Aebi U., et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature. 376:184–188 10.1038/376184a0 [DOI] [PubMed] [Google Scholar]
- Zhang H., Saitoh H., Matunis M.J. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498–6508 10.1128/MCB.22.18.6498-6508.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.D., Goeres J., Zhang H., Yen T.J., Porter A.C., Matunis M.J. 2008. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell. 29:729–741 10.1016/j.molcel.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Goeres J., Sixt K.M., Békés M., Zhang X.D., Salvesen G.S., Matunis M.J. 2009. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell. 33:570–580 10.1016/j.molcel.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]