Abstract
In budding yeast, commitment to cell division corresponds to activating the positive feedback loop of G1 cyclins controlled by the transcription factors SBF and MBF. This pair of transcription factors has over 200 targets, implying that cell cycle commitment coincides with genome-wide changes in transcription. Here, we find that genes within this regulon have a well-defined distribution of transcriptional activation times. Combinatorial use of SBF and MBF results in a logical OR function for gene expression and partially explains activation timing. Activation of G1 cyclin expression precedes the activation of the bulk of the G1/S regulon ensuring that commitment to cell division occurs before large-scale changes in transcription. Furthermore, we find similar positive feedback-first regulation in the yeasts S. bayanus and S. cerevisiae, as well as human cells. The widespread use of the feedback-first motif in eukaryotic cell cycle control, implemented by non-orthologous proteins, suggests its frequent deployment at cellular transitions.
INTRODUCTION
Order may be produced in a sequence of biochemical events through feedback control mechanisms or substrate-specific chemical kinetics. In the cell cycle, regulatory checkpoints ensure the proper order of many essential events through feedback control. DNA replication must be finished and damage repaired before mitosis, while anaphase is initiated only after complete spindle assembly (Morgan 2007). Checkpoints use designated regulatory molecules to restrain cell cycle progression until a set of criteria are satisfied (Hartwell and Weinert 1989). However, order without checkpoint control is observed in Xenopus embryos as cell cycle events are entrained by oscillations in cyclin dependent kinase (CDK) activity. Furthermore, addition of CDK substrates to Xenopus egg extracts in different stages of mitosis revealed that the order of substrate phosphorylation is independent of cell cycle phase(Georgi, Stukenberg et al. 2002). Thus, temporal order of phosphorylation in mitosis is likely the result of substrate-specific kinetics. Here, we investigate the integration of chemical kinetics and feedback control at the Start transition in budding yeast.
Start marks the point of commitment to the mitotic cell cycle, which is located between cell division and DNA replication (Hartwell, Culotti et al. 1974). Prior to Start, cells integrate internal (e.g., cell size) and external (e.g., mating pheromone) signals to make an all-or-none decision to divide. Beyond Start, cells are committed to divide regardless of changes in extracellular signals. In another article in this issue, we show that passage through Start corresponds precisely to the activation of the G1 cyclin positive feedback loop (Doncic, Fettig and Skotheim 2011). Thus, Start is a member of a growing list of cellular and developmental transitions driven by positive feedback (Pomerening, Sontag et al. 2003; Xiong and Ferrell 2003; Holt, Krutchinsky et al. 2008; Justman, Serber et al. 2009; Lopez-Aviles, Kapuy et al. 2009).
Positive feedback at Start is initiated by the G1 cyclin, Cln3 in complex with the cyclin dependent kinase Cdc28 (Figure 1A). The primary target of Cln3 is the transcriptional inhibitor Whi5, whose inactivation is rate-limiting for the expression of the G1/S regulon (Costanzo, Nishikawa et al. 2004; de Bruin, McDonald et al. 2004). Cln3-Cdc28 phosphorylates and initiates Whi5 inactivation, which allows some transcription of two additional G1 cyclins, CLN1 and CLN2 (Tyers, Tokiwa et al. 1993). The downstream G1 cyclins then complete the positive feedback loop through the inactivation and nuclear exclusion of Whi5 and the full activation of the transcription factors SBF (Swi4-Swi6) and MBF (Mbp1-Swi6) (Andrews and Herskowitz 1989; Nasmyth and Dirick 1991; Koch, Moll et al. 1993; de Bruin, McDonald et al. 2004; Skotheim, Di Talia et al. 2008).
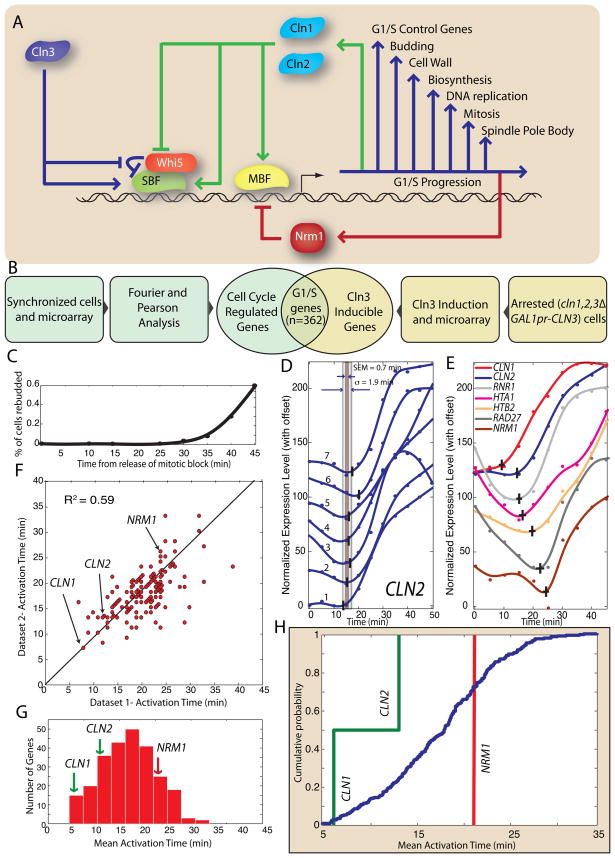
Figure 1. Positive feedback precedes genome-wide change in transcription at G1/S in S. cerevisiae.
(A) Schematic diagram of the G1/S transition. (B) The G1/S regulon is defined as the intersection of the set of cell cycle regulated genes with the set of Cln3-inducible genes. (C) Synchrony of cdc20Δ GALLpr-CDC20 metaphase block-release from Di Talia et al (2009). (D) An algorithm is applied to a smoothing-spline fit to detect activation of CLN2 transcription in 7 mitotic block-release datasets (See Figures S1-3 for algorithm description; specific genotypes of Datasets 1–7 described in Figure S1D). The standard deviation σ and the standard error of the mean (SEM) is calculated for each gene. (E) 7 genes in the G1/S regulon are activated at different times; data shown from a single dataset. The vertical and horizontal bars indicate the activation time and its SEM respectively. (F) Gene activation time correlation between two of the 7 datasets (R2 = 0.59; see supplementary information for additional correlations). Histogram (G) and corresponding cumulative distribution (H) of mean activation times for the 7 mitotic block-release datasets. CLN1 and CLN2, two genes responsible for positive feedback, are among the earliest-activated genes. NRM1, a negative regulator of MBF, is activated later.
Surprisingly, the transcription factors at the center of the positive feedback loop, SBF and MBF, are also responsible for the transcription of over 200 additional genes (Ferrezuelo, Colomina et al. 2010). Indeed, cell cycle commitment appears to coincide with the coordinated transcriptional activation of approximately 5% of all genes (Spellman, Sherlock et al. 1998). Although Whi5 phosphorylation is rate-limiting for activation of positive feedback, it is also likely to be rate-limiting for the transcription of all SBF regulated genes due to the direct Whi5-SBF interaction (de Bruin, McDonald et al. 2004). The concurrent activation of the related heterodimeric transcription factor MBF also requires CDK activity, possibly through phosphorylation of the shared component Swi6 (Wijnen, Landman et al. 2002). Thus, given the integrated nature of the regulatory circuit and the ability of the upstream cyclin Cln3 to activate SBF- and MBF-dependent transcription in cln1Δ cln2Δ cells (Dirick, Bohm et al. 1995; Stuart and Wittenberg 1995), it is unclear if genome-wide changes in transcription occur after commitment to division.
Although G1/S transcription is largely regulated by SBF and MBF, single-cell studies have revealed significant differences in transcriptional activation of the 3 regulon members CLN2, RAD27 and RFA1 (Skotheim, Di Talia et al. 2008). A rapid, feedback-driven increase in CDK activity drives the coherent and nearly simultaneous induction of these three genes in WT cells. However, significant differences in transcriptional activation timing are revealed in cln1Δ cln2Δ cells lacking positive feedback. CLN2 is induced earlier than two other regulon members, which suggests a model in which full regulon expression would only occur after feedback loop activation to avoid detrimental transcription in cases where the cell does not commit to the mitotic cell cycle. Therefore, we hypothesized that the G1 cyclins CLN1 and CLN2, involved in positive feedback, would be activated earlier than other genes in the G1/S regulon to ensure that commitment precedes the genome-wide change in transcription.
In this study, we observed that the two SBF/MBF-regulated G1 cyclins, namely CLN1 and CLN2, are among the earliest activated genes of the G1/S regulon, which supports the hypothesis that genome-wide changes in transcription occur after a cell is committed to division. By comparing sets of genes regulated by SBF, MBF, or by both factors together, we found that both transcriptional activation and inactivation can be approximated as logical OR functions. Furthermore, CLN1 and CLN2 remain among the earliest activated cell cycle regulated genes in the related yeast, S. bayanus, which has significantly diverged gene expression(Tirosh, Weinberger et al. 2006; Guan, Dunham et al. 2010). A similar analysis of human tissue culture cells revealed that functionally analogous feedback loop components E2F1, Skp2, and the cyclins E1 and E2 (Blagosklonny and Pardee 2002; Yung, Walker et al. 2007) are among the earliest activated cell cycle regulated targets of the E2F family of transcription factors. Taken together, our results demonstrate that feedback-first regulation, which places genome-wide changes in transcription downstream of positive feedback-dependent cell cycle commitment, is a common feature of G1/S control across eukaryotes.
RESULTS
Defining the G1/S regulon
To test our model that induction of positive feedback and concomitant cell cycle commitment precedes large-scale transcriptional change, we first need to accurately define the G1/S regulon. We are interested in the set of genes whose transcription is initiated due to increasing cyclin activity rather than upstream cyclin-independent processes (MacKay, Mai et al. 2001; Di Talia, Wang et al. 2009). The set of cell cycle regulated genes was defined as the 800 genes with the largest amplitude mRNA concentration oscillation through the cell cycle (Spellman, Sherlock et al. 1998). To identify the set of G1 cyclin regulated genes, we relied on a second experiment by Spellman et al (1998), which identified a set of genes responding to exogenous Cln3 induction in G1 arrested cln1Δ cln2Δ cln3Δ cells. We took the top 413 as the set of G1 cyclin inducible genes. The intersection of these two sets defines the 362-gene regulon (Figure 1B; Table S1).
Automated detection of gene activation
Next, we developed an algorithm to determine the time at which a specific gene is induced during the cell cycle. We analyzed 7 previously published microarray time-course datasets with 5-minute temporal resolution (Di Talia, Wang et al. 2009). All experiments were performed on cdc20Δ GALLpr-CDC20 cells that were synchronized by mitotic arrest. Cells were released by switching to media containing galactose resulting in CDC20 expression and a synchronous first cell cycle (Figure 1C).
Although manually identifying activation points of cell cycle regulated genes is not difficult, we developed an automated algorithm to both avoid potential bias and increase throughput. Our algorithm is robust to noisy data, which can produce incorrect estimates for the activation time. We normalized all the time series and assumed that the time scale for changing transcript concentration is greater than 10 minutes. We therefore remove data points associated with large concentration changes on shorter timescales. Data points further than 20% of the dynamic range of the time series (maximum – minimum) from adjacent points are removed. We discarded time series with two or more removed data points. The mRNA level is then estimated using smoothing-splines. We selected the point where the 1st derivative first reaches 10% of its maximum. The smoothing parameter is optimized to minimize variation in biological replicates and the 1st derivative method is shown to be superior in estimating activation times relative to other methods (Figure S1A–C).
Figure 1D shows the activation times for 7 independent CLN2 expression profiles and their standard deviation and standard error of the mean. Because we have multiple time-courses, our error in estimating the activation time is low, e.g., for CLN2 we find the activation time to be 13 minutes after galactose addition with a standard deviation of 1.9 minutes and a standard error of the mean of 0.7 min. For genes within the G1/S regulon, we find that the average standard deviation is 4.7 min and the average standard error of the mean is 2.1 min. Despite regulation by the same transcription factors, the activation times of G1/S regulon members has a defined distribution (mean = 17.2 minutes; standard deviation = 5.9 minutes; Figure 1E–H, S1D; Table S2).
To test our model that feedback activation precedes regulon induction, we averaged the activation times from all 7 datasets for each gene (Figure 1G–H). These results were consistent with induction times measured in rtPCR time-courses (see Figure S1E). The positive feedback genes CLN1 and CLN2 are activated significantly earlier than the bulk of the G1/S regulon. Indeed, within error, CLN1 is the earliest activated gene, 5 minutes earlier than CLN2, suggesting a different temporal role even though these two genes are generally thought to be functionally redundant. However, it has been shown that CLN1, but not CLN2, transcription affects cell size (Flick, Chapman-Shimshoni et al. 1998), which our data suggests is due to timing. We note that for the feedback-first model to work it is sufficient to express either G1 cyclin, not necessarily both, prior to the majority of the regulon. Thus, we see that induction of the G1 cyclin positive feedback loop, which coincides with cell cycle commitment, precedes large-scale changes in the transcriptional program.
Interestingly, NRM1, the negative feedback element responsible for inactivating MBF regulated genes (de Bruin, Kalashnikova et al. 2006), is activated 15 min later than CLN1 (Figure 1G–H) even though both genes are MBF targets (Ferrezuelo, Colomina et al. 2010). Thus, distinct temporal regulation allows positive feedback sufficient time for regulon transcription prior to NRM1-dependent inactivation.
Delayed positive feedback does not rescue cln1Δ cln2Δ cells
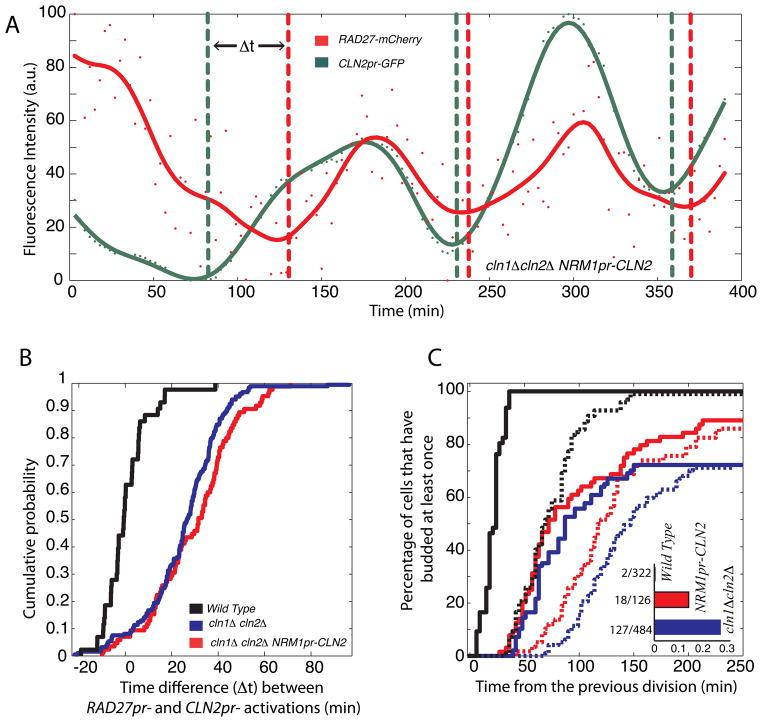
To examine the functional consequences of feedback timing, we integrated a CLN2 allele regulated by the NRM1 promoter into a cln1Δ cln2Δ cell containing MET3pr-CLN2, CLN2pr-GFPpest and RAD27-mCherry. Cells were grown overnight on media lacking methionine (MET3pr-CLN2 on) prior to switching to media containing methionine (MET3pr-CLN2 off) for single-cell analysis of one cell cycle (Skotheim, Di Talia et al. 2008). Cells exhibited similarly incoherent gene expression (time between CLN2pr and RAD27pr induction) and cell size defect as cln1Δ cln2Δ cells (Figure 2A–B; S2). However, the fitness defect was partially reduced (Figure 2C). This indicates the importance of running the positive feedback loop from an early activated promoter.
Figure 2. Phenotypic consequences of delayed positive feedback.
(A) Time course of incoherent RAD27-mCherry and CLN2pr-GFP expression in a single cln1Δ cln2Δ NRM1pr-CLN2 cell. (B) Time difference between CLN2pr and RAD27pr induction measured as in Skotheim et al (2008); cells not showing significant induction of either promoter were omitted from the analysis. (C) A cumulative plot for the first bud emergence measured from cell division. Solid and dashed lines correspond to mother and daughter cells respectively. Inset shows fraction of G1-arrested cells.
Feedback-first regulation is robust to changes in carbon source and synchronization method
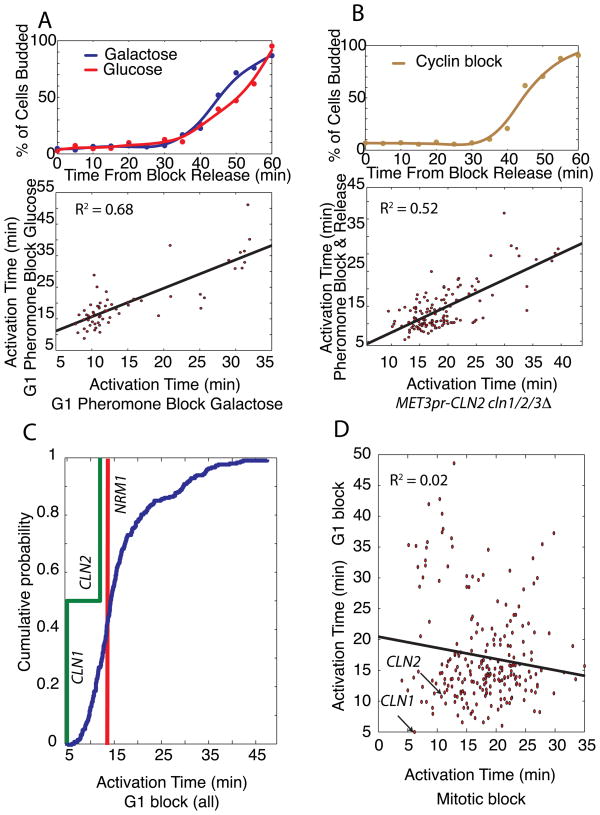
To further test our feedback-first model, we examined the effects of varying carbon source and synchronization method, which are both known to affect gene expression (Flick, Chapman-Shimshoni et al. 1998; Levy, Ihmels et al. 2007; Brauer, Huttenhower et al. 2008). We performed a micro-array time course after synchronizing cells with mating pheromone in media with either glucose or galactose. Carbon source does not have a large effect as differences in activation times were similar to experimental replicates (Figure 3A).
Figure 3. Synchronization phase, but not carbon source or synchronization method, affects gene activation timing.
(A) Bud-index measurements and gene activation time correlation for G1 pheromone block-release time-course microarray experiments with glucose or galactose carbon sources. (B) Bud index for G1 block-release using cln1Δ cln2Δ cln3Δ MET3pr-CLN2 cells and correlation of gene activation times for pheromone and G1 cyclin block-release experiments. (C) Significant correlation between the 3 G1 block-release datasets allows them to be pooled together to produce a histogram of activation times for the G1/S regulon again demonstrating feedback-first regulation. (D) Activation times from G1 and mitotic block-release experiments are not correlated.
To analyze the effect of synchronization method, we examined cells lacking endogenous G1 cyclins (cln1Δ cln2Δ cln3Δ) but containing an integrated MET3pr-CLN2 construct (see methods). Cells were arrested in G1 before being transferred to media with a low level of methionine to activate exogenously controlled CLN2 transcription at physiological levels. We then compared activation times between the cyclin blocked and the pheromone blocked cells (Figure 3B). Our three G1 block-release experiments varying carbon source and synchronization method produced similar timing profiles.
We examined the distribution of activation times pooled from the 3 separate G1 block experiments (Figure 3C). Although transcriptional order is affected by the arrest phase (Figure 3D, S3), CLN1 is activated at the first possible time-point (5 minutes after release) in agreement with the feedback-first model.
Gene activation is correlated in freely cycling cells and mitotic block-release experiments
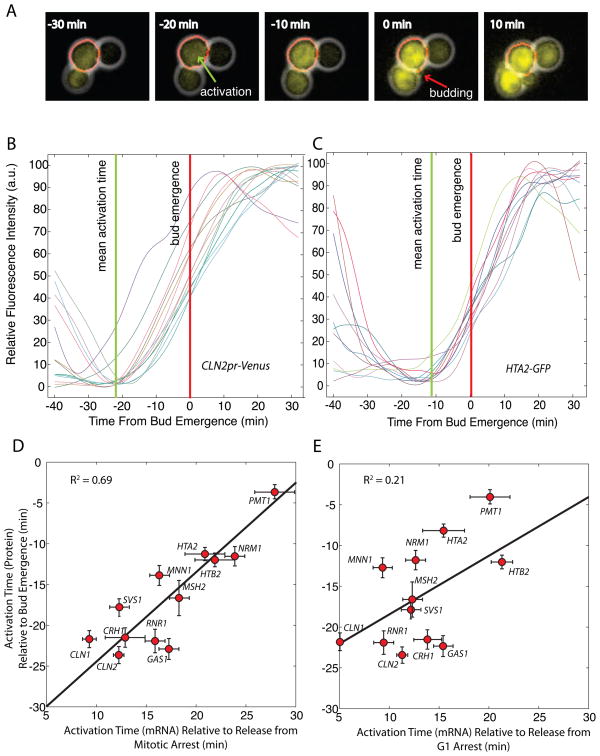
Since transcriptional order changes with the arrest phase, we decided to investigate which block is more similar to the free-running cell cycle using time-lapse fluorescence microscopy (Skotheim, Di Talia et al. 2008). We analyzed protein accumulation in 10 strains expressing C-terminal GFP fusion proteins from the endogenous loci (Ghaemmaghami, Huh et al. 2003), and two strains containing an integrated CLN1 or CLN2 promoter driving the expression of a destabilized VenusPEST (Mateus and Avery 2000). We selected this group of strains to span the distribution of activation times. Automated cell segmentation allows us to analyze the fluorescent intensity change in single-cells through the cell cycle (figure 4A). We detected activation timing relative to bud emergence (Figure 4B–C; Table S3). We found that the mean single-cell activation times in the unperturbed cell cycle correlated more with the mitotic block experiments (R2 = 0.72; Figure 4D) than the G1 block experiments (R2 = 0.21; Figure 4E). This result also implies that the order of mRNA transcription is largely reflected in protein accumulation. Thus, the mitotic block experiments are more representative of freely cycling cells.
Figure 4. Gene activation is correlated in the free running cell cycle and mitotic block-release experiments.
(A) Composite phase and fluorescence images of CLN2pr-VenusPEST cells. Venus yellow fluorescent protein contains a destabilizing PEST sequence. The red contour denotes the cell boundary detected by automatic segmentation. Gene activation time calculated from fluorescence intensity time courses aligned at bud emergence for (B) CLN2pr-VenusPEST and (C) HTA2-GFP cells. Gene activation times ± SEM for 10 strains containing GFP-fused proteins and 2 strains containing promoter-Venus constructs expressed at the endogenous locus correlated with mean activation times from microarray time-courses for cells synchronized at mitosis (D) or G1 (E).
Since transcription activation times change with the phase of the block used, we decided to analyze previously published cell cycle synchronized microarray time courses (Spellman, Sherlock et al. 1998; Pramila, Wu et al. 2006; Orlando, Lin et al. 2008). Although quantitative comparisons of individual genes are difficult due to either poor temporal resolution or lack of experimental replicates, we are able to detect correlations of genes within the G1/S regulon. We found that G1 blocks, including elutriation, correlate with our G1 block data (see Table S4). Interestingly, the cdc15ts data from Spellman et al (1998) correlates with our G1 block experiments rather than the mitotic block experiments even though this is an anaphase block indicating that an event occurring in cells blocked downstream of Cdc20 may be responsible for differences in gene activation timing. We note that release from G1 arrest and free cycling are both likely to be physiologically relevant.
SBF- and MBF-dependent activation is a logical OR gate
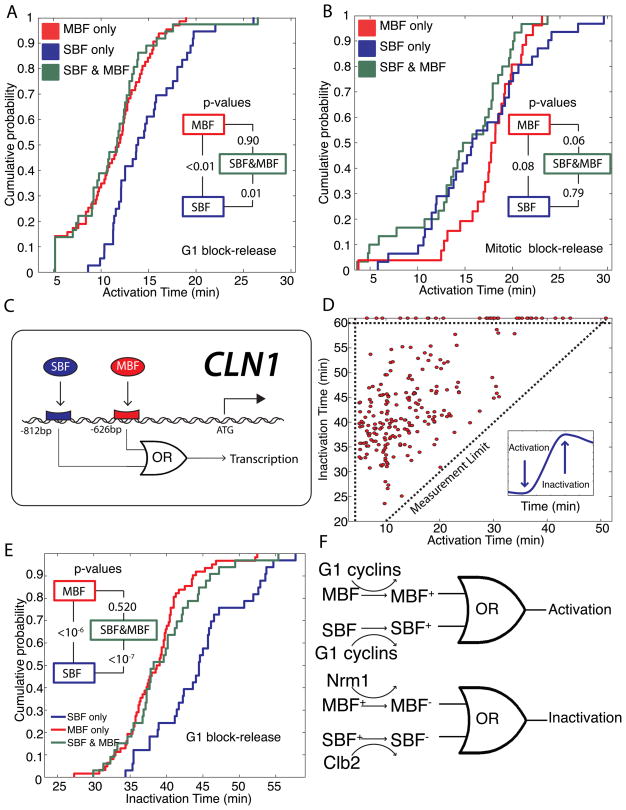
We hypothesized that the observed differences in gene activation time in different blocks might be due to differential regulation of specific transcription factors. The majority of genes in what we defined as the G1/S regulon are regulated by the transcription factors SBF and MBF (Ferrezuelo, Colomina et al. 2010). For our analysis, we divided the activation times of the G1/S genes into three categories: 136 SBF-only targets, 63 MBF-only targets, and 36 dual-regulated SBF and MBF targets.
Since combinatorial use of transcription factors may yield differential activation timing, we analyzed the activation times of the SBF only, MBF only, and dual-regulated genes. For our G1 arrest data, we find that MBF-only targets are activated earlier than SBF-only targets (p <0.01). Furthermore, the distribution of the dual regulated targets is more similar to the earlier-activated MBF-only targets (p = 0.90) than the more tardy SBF-only targets (p = 0.01; Figure 5A).
Figure 5. SBF and MBF dual-regulated promoters act as logical OR gates in response to activation and inactivation signals.
Cumulative probability of activation times for SBF-only, MBF-only and SBF/MBF dual-regulated targets are plotted for (A) G1 block-release and (B) mitotic block-release experiments. Inset shows p-values comparing each pair of distributions. (C) Schematic showing logical regulation of the early-activated CLN1 promoter denoting SBF and MBF consensus binding sites. (D) Inactivation time for each gene, where the 1st derivative is zero and the 2nd derivative is negative (inset), is uncorrelated with activation for G1 block-release experiments. Points above the horizontal dotted line represent genes peaking later than 60 min. (E) Cumulative probability of inactivation for SBF-only, MBF-only and SBF/MBF dual targets for G1 block-release experiments. Inset shows p-values comparing each pair of distributions. (F) The transcriptional activation and inactivation can be modeled as a logical OR gate. For dual-regulated genes, activating either SBF or MBF suffices for activation, while inactivating MBF suffices for inactivation. Different colors denote different possible states of a transcription factor.
In the mitotic block-release, the SBF-only targets are activated earlier than the MBF-only targets (p = 0.08). This is the opposite order than in the G1-block experiments and consistent with the lack of correlation between activation times of individual G1/S regulon members (Figure 3D). Furthermore, we find that the common targets are much more likely to follow the SBF-only distribution (p = 0.79) than the MBF-only distribution (p = 0.06; Figure 5B). We note that the SBF distribution is broader so that the late-activated SBF genes are activated later than the late-activated MBF genes. However, the late-activated dual-regulated genes now appear to follow MBF.
Taken together, our results from the two different types of experiments suggest that the dual-regulated targets are activated by the earliest active transcription factor. In the G1 block experiments, the co-regulated genes are activated by MBF, while in the mitotic block experiments the co-regulated genes are activated by SBF. This implies that transcriptional activation is functioning as a logical OR gate, where either an active SBF or an active MBF is sufficient to activate transcription.
Logical inactivation
Our results analyzing transcriptional activation encouraged us to perform a similar analysis on transcriptional inactivation, which we estimate as the time of the peak transcript level (Figure 5C). The peak time is defined to be the point where the 1st derivative of the smoothed data is zero and the 2nd derivative is negative. We then implemented an algorithm for unbiased peak detection and analyzed our G1 block-release data. Inactivation is not well correlated with activation (Figure 5D).
Next, we decided to analyze inactivation in light of our SBF-only, MBF-only and dual regulated gene lists. Whereas mitotic cyclins are responsible for SBF inactivation(Amon, Tyers et al. 1993), MBF inactivation is performed by Nrm1 possibly through a direct interaction (de Bruin, Kalashnikova et al. 2006). In nrm1Δ cells, mitotic cyclins are capable of inactivating MBF-regulated genes; however, inactivation is delayed about 10 minutes relative to WT (de Bruin, Kalashnikova et al. 2006). This suggests that mitotic cyclin-dependent inactivation occurs later than Nrm1-dependent inactivation and that we should expect to see MBF-only targets inactivated earlier than SBF-only targets. Consistent with previous results (Ferrezuelo, Colomina et al. 2010), we find that MBF-only targets are inactivated earlier than SBF-only targets (p <10−7; Figures 5E, S4). The distribution of inactivation times for the dual regulated genes was much more similar to the MBF-only genes (p=0.52) than the SBF-only genes (p < 10−7). Inactivation of MBF is sufficient to turn off gene expression regardless of the presence of an active SBF transcription factor. Thus, both activation and inactivation may be represented by logical OR gates (Figure 5F).
Feedback-first regulation in the budding yeast S. bayanus
We found that S. cerevisiae activates positive feedback and commits to another round of cell division before making large-scale changes to its transcriptional program. This temporal organization of the G1/S regulon may be an efficient way to ensure that cell cycle associated genes are only transcribed after a cell has decided to divide. If feedback-first regulation increases fitness then we should expect to see it conserved in divergent evolutionary lineages.
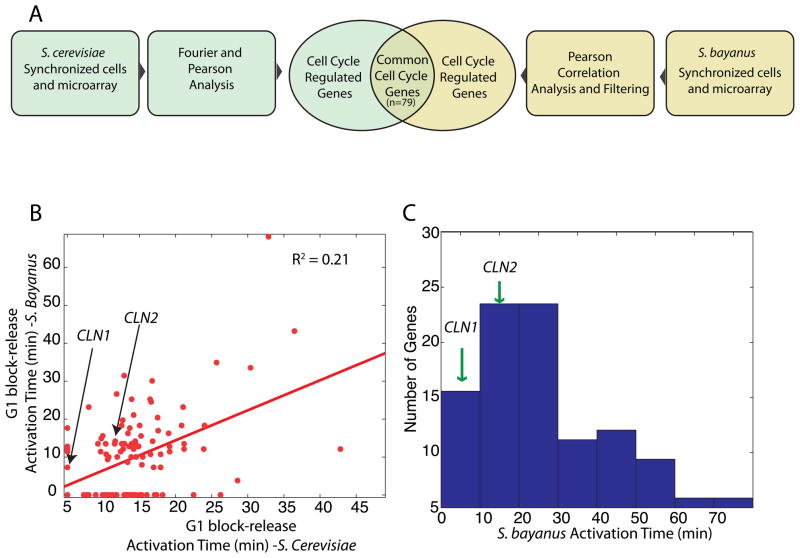
To examine the conservation of feedback-first regulation, we analyzed a closely related yeast Saccharomyces bayanus, for which cell cycle synchronized microarray data was available. Compared to S. cerevisiae, S. bayanus has 67% local similarity of intergenic regions indicating significant divergence of gene regulation (Cliften, Sudarsanam et al. 2003). Gene orthologs are easily identified by sequence and the S. bayanus genes are conveniently annotated using the S. cerevisiae nomenclature (Cliften, Sudarsanam et al. 2003). Indeed, studies on the evolution of gene expression among sensu stricto yeast species revealed substantial differences (Tirosh, Weinberger et al. 2006; Guan, Dunham et al. 2010).
We analyzed the S. bayanus time-course microarray dataset from the GEO database (GSE16544). Cells were synchronized in G1 using mating pheromone and samples were taken every 10 minutes for 300 minutes following release(Guan, Dunham et al. 2010). To define a set of genes that are cell cycle regulated, we calculated the cross-correlation coefficients with two known cell cycle regulated genes, CLN2 and the G2 gene KIN2. We sorted the genes based on their cross-correlation scores and selected the 714 genes that were in the top 1000 of both cross-correlations. To eliminate spurious profiles, we considered only genes showing multiple well-defined oscillations.
We analyzed the correlation of cell cycle regulated gene expression in the two budding yeasts. Of the 800 and the 223 well-defined cell cycle regulated genes in S. cerevisiae and S. bayanus respectively, only 79 were cell cycle regulated in both species (Figure 6A). Furthermore, the activation times of the common cell cycle regulated genes is weakly correlated (R2 = 0.22; Figure 6B). Our observation of significant changes in transcriptional activation timing through the cell cycle is consistent with the emerging picture of significantly diverged transcription across the sensu stricto (Tirosh, Weinberger et al. 2006).
Figure 6. Feedback-first regulation is conserved in the budding yeast S. bayanus.
Activation times are analyzed for all cell cycle regulated genes in a S. bayanus pheromone block-release microarray time-course (Guan, Dunham et al. 2010). (A) Intersection of cell cycle regulated genes in both budding yeasts. (B) Weak correlation between gene activation times in S. bayanus and S. cerevisiae for G1 block-release experiments. (C) Histogram of activation times of the cell cycle regulated genes in S. bayanus indicates that the G1 cyclins responsible for positive feedback, CLN1 and CLN2, are among the early-activated genes.
To test for the conservation of feedback-first regulation, we analyzed the distribution of first activation times (<80 min). The activation times for CLN1 and CLN2 was calculated to be 6 and 15 minutes respectively. Thus, the G1 cyclins are among the earliest activated genes in the S. bayanus cell cycle, which indicates conservation of feedback-first regulation (Figure 6C).
Temporal analysis of E2F-dependent transcription in human cells
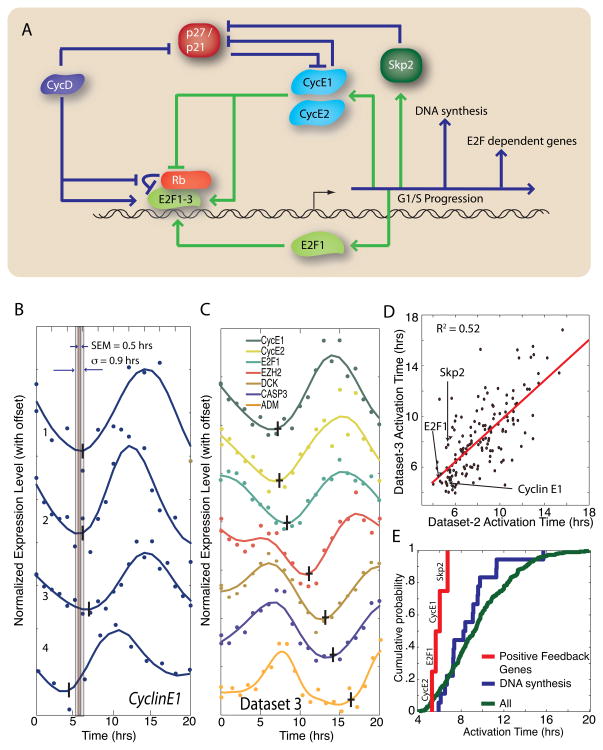
Our finding that two yeasts engage positive feedback prior to full regulon activation suggests that this regulatory motif is widespread. Thus, we chose to examine a mammalian system. Although many of the components of the genetic network regulating the G1/S transition in mammals do not have well-defined orthologs in yeast, both networks contain multiple positive feedback elements indicating similar network topology (Figure 7A). There is a functional analogy between the cyclin D-E2F-Rb-cyclinE and the Cln3-SBF/MBF-Whi5-Cln1/2 pathways. Furthermore, both budding yeasts and mammals regulate commitment to cell division in response to multiple internal and external signals at the G1/S transition (Planas-Silva and Weinberg 1997; Blagosklonny and Pardee 2002; Yao, Lee et al. 2008).
Figure 7. Feedback-first regulation is conserved in human cells.
(A) Schematic diagram of G1/S regulation in human cells. (B) Cyclin E1 activation is consistent in 4 different cell cycle synchronized microarray experiments from Whitfield et al. (2002). The standard deviation σ and the standard error of the mean (SEM) is calculated for each gene. (C) 7 genes regulated by the E2F family of transcription factors are activated at different times; data shown from a single dataset. The vertical and horizontal bars indicate the mean activation time and the SEM respectively. (D) Gene activation time correlation between 2 datasets (R2 = 0.52). (E) Cumulative distribution of mean activation times for cell cycle regulated E2F-targets. Genes responsible for positive feedback at the G1/S transition, including the cyclins E1 and E2 the transcription factor E2F1, and the SCF component Skp2, are transcribed earlier than other E2F-targets (p<0.01) and earlier than the set of E2F targets specifically involved in DNA replication (p=0.03). This demonstrates the conservation of feedback-first regulation in eukaryotes.
Mammalian G1 progression is initiated by mitogen-dependent activation of cyclin D–CDK4/6 complexes, which phosphorylate and partially inactivate the transcriptional inhibitor Rb (Blagosklonny and Pardee 2002). This allows for the activation of transcription by three members of the E2F family (E2F1-3) of transcription factors. Included in the set of targeted genes are cyclin E1 and cyclin E2, which complex with CDK2 to phosphorylate Rb and thereby complete a positive feedback loop (Bracken, Ciro et al. 2004). Additionally, E2F1-3 activate transcription of E2F1, which may form an additional transcriptional positive feedback loop(Johnson, Ohtani et al. 1994). The SCF component Skp2, responsible for the specific degradation of the CDK inhibitor p27, is also an E2F target(Yung, Walker et al. 2007). Therefore, multiple potential positive feedback loops may act during the mammalian G1/S transition. If our feedback-first model applies to mammalian cell cycle control, we expect to see feedback loop components transcribed before other E2F targets.
To test our hypothesis that the positive feedback elements are transcribed early, we first need to define a set of cell cycle regulated E2F targets (Markey, Angus et al. 2002; Ren, Cam et al. 2002). Therefore, we compiled a list of 315 cell cycle regulated E2F targets from two previous studies (Muller, Bracken et al. 2001; Whitfield, Sherlock et al. 2002; Xu, Bieda et al. 2007).
We analyzed gene activation timing in human HeLa cells for 4 cell cycle synchronized microarray time courses (Whitfield, Sherlock et al. 2002). We see consistent activation of individual genes across the datasets. For example, cyclin E1 is activated at 5.9 ± 0.5 hours on average with a standard deviation of 1.1 hours (Figure 7B). Our analysis identifies distinct activation times for E2F regulated genes (Figure 7C). In three experiments, cells were synchronized with a double thymidine block, while in one experiment, cells were synchronized with a thymidine block followed by a nocodazole block (Whitfield, Sherlock et al. 2002). However, we see no difference in relative activation timing due to the two different synchronization methods as all four data sets yield comparable results (Figure 7D). We observe that genes responsible for positive feedback (cyclin E1, cyclin E2, Skp2 and E2F1) are among the first transcribed at the G1/S transition consistent with our feedback-first model (Figure 7E).
DISCUSSION
We showed that genome-wide transcription is restricted until positive feedback commits a cell to division. This regulatory organization was previously unclear because transcription of the G1 cyclins and the rest of the G1/S regulon are both dependent on the same transcription factors and appear concurrent when analyzed with clustering-based algorithms. In contrast to clustering methods, both our activation detection algorithm and parametric algorithms preserve the dynamic information required for our analysis (Chechik, Oh et al. 2008).
Towards the mechanistic basis of transcription order
We observed considerable variation in gene activation timing among genes regulated by a specific transcription factor. In budding yeast, we showed that a significant amount of this variation is due to combinatorial use of the transcription factors SBF and MBF resulting in logical OR gates for both transcriptional activation and inactivation. Thus, the genes regulated by both SBF and MBF transcription factors are activated early in mitotic block-release experiments, where SBF is activated before MBF, and in G1 block-release experiments, where MBF is activated before SBF. This may be functionally relevant as the earliest activated G1 cyclin CLN1 is regulated by both factors (Flick, Chapman-Shimshoni et al. 1998; Ferrezuelo, Colomina et al. 2010), which may ensure feedback-first regulation in a variety of physiological contexts.
Future work will aim at explaining the molecular basis for the significant temporal variation in G1/S transcription unexplained by the combinatorial use of SBF and MBF. One possibility is that differential transcription timing may arise through the combinatorial use of additional transcription factors (Kato, Hata et al. 2004). In such a model, intermediate times might be produced by regulating a promoter with a late-activated and an early-activated transcription factor. An example of this type of regulation is that the Fkh2-regulated genes show different activation times at G2/M depending on Yox1-coregulation (Darieva, Clancy et al. 2010). This model suggests that the late activated SBF targets might also be regulated by a late-activated transcription factor such as Fkh2. A large number of transcription factors might therefore account for the variation in gene activation time.
A second possibility is that promoter-specific rate-constants underlie gene activation kinetics. This could arise through promoter-specific transcription factor and nucleosome arrangements or TATA-box sequence (Lam, Steger et al. 2008; Chechik and Koller 2009; Bai, Charvin et al. 2010; Mogno, Vallania et al. 2010). Thus, in response to a single input such as CDK activity, the organization of kinetic parameters can result in differential activation timing(Shen-Orr, Milo et al. 2002). We note that these two classes of mechanisms are not mutually exclusive and likely cooperate to tune gene expression.
Temporal separation of positive and negative feedback loops
An interesting feature of the G1/S regulon is that both positive (CLN1,2) and negative (NRM1) feedback elements are regulated under the same CDK-dependent transcription factors. Activating both feedbacks at the same time would be much like stepping on the brake and gas pedals simultaneously to detrimental effect (Figure 2). To avoid this outcome, promoter specific kinetics may allow temporal separation of positive and negative feedback loops. A similar process was found to regulate mitotic entry in Xenopus egg extracts(Georgi, Stukenberg et al. 2002). Wee1 and Cdc25 phosphorylation by CDK1, which is associated with positive feedback at mitotic entry, occurs before the phosphorylation of other CDK1 targets including the APC component Cdc27. Thus, both feedback-first regulation and the temporal separation of positive and negative feedback loops may be enacted through the evolution of differential rate constants.
Feedback-first regulation ensures commitment to cell division prior to large-scale changes in gene expression
Transcribing genes when they are needed may increase efficiency by avoiding unnecessary protein synthesis. The subunits of the E. coli flagella were found to be synthesized in the order that they are needed for assembly (Kalir, McClure et al. 2001). Fine temporal control of transcription during amino acid synthesis ensured that enzymes were made in the order they were needed (Zaslaver, Mayo et al. 2004). In the cell cycle context, ribonucleotide reductase is transcribed just before S phase(Elledge and Davis 1990), and histones are transcribed during S phase to be assembled with newly replicated DNA (Borun, Gabrielli et al. 1975; Hereford, Osley et al. 1981). Taken together, these studies indicate that fine temporal order of events may provide a fitness advantage.
A transcriptional oscillation with specific temporal order occurs through the cell division cycle in both prokaryotes and eukaryotes. This extensive oscillation entails ~10–20% of all Caulobacter and budding yeast genes (Cho, Campbell et al. 1998; Spellman, Sherlock et al. 1998; Laub, McAdams et al. 2000). However, a comparative analysis of the yeasts S. pombe and S. cerevisiae revealed that temporal regulation of most orthologous genes is not well conserved (Rustici, Mata et al. 2004; Oliva, Rosebrock et al. 2005; Peng, Karuturi et al. 2005). Indeed, we report here that the order of gene activation at the G1/S transition in S. cerevisiae depends on the synchronization phase (Figure 3D). Further comparison of the two yeasts with human cell lines and the plant Arabidopsis revealed that only 5 orthologs are cell cycle regulated in all 4 species (Jensen, Jensen et al. 2006). However, different protein subunits of the same complex were often found to have cell cycle regulated transcription in different species, suggesting conserved transcriptional control of the complex rather than the individual subunits (Jensen, Jensen et al. 2006). Thus, although periodic transcription of individual genes varies, there may still be conserved regulatory features.
Here, we identify such a conserved regulatory feature of the eukaryotic cell cycle. We find that commitment via positive feedback precedes large-scale transcriptional activation at the G1/S transition. Our study was able to identify feedback-first regulation because we employ an algorithm to analyze activation and inactivation separately. We revealed feedback-first regulation in the yeasts, S. cerevisiae and S. bayanus as well as in human cells. The conservation of feedback-first regulation leads us to anticipate its widespread use in cellular and developmental transitions.
EXPERIMENTAL PROCEDURES
Microarray experiments and analysis
Mitotic block-release analysis was based on data collected in (Di Talia, Wang et al. 2009). The sequential order of activations is highly consistent between datasets indicating a defined temporal regulation even though the genotypes for the 7 time-courses were not identical (Figure 1F, Figure S1D; Table S2).
G1 block-release experiments were performed at 30°C. Cells were harvested immediately after inoculation and then every 5 minutes thereafter. Microarray hybridization was performed at the Stony Brook Microarray Facility. For pheromone block experiments, bar1Δ cells were grown in log-phase in either SCD (%2) or SCG (3%) before being arrested for 135 minutes in 95nM α-factor. Cells were then washed and inoculated into pheromone-free media. The cln-block experiment was performed using cln1Δ cln2Δ cln3Δ MET3pr-CLN2 cells grown to early log-phase in SCD - met (media without methionine; exogenous CLN2 on), then 0.2g/L met was added for 120 minutes to arrest cells in early G1 (CLN2 off). Cells were then washed and inoculated into 4mg/L met (CLN2 partially on) to provide the amount of CLN2 expression resulting in budding kinetics similar to WT cells released from a pheromone block.
There was some ambiguity in identifying the gene activation time for CLN1 in the S. bayanus data set because either the 2nd or 3rd data point for CLN1 was likely an outlier. Therefore, we averaged the activation times for the dataset after having removed either the 2nd or 3rd data point.
Time-Lapse Fluorescence Microscopy
Wide-field fluorescence and phase-contrast images were captured every 3 minutes for 6 hours from cells prepared as previously described (Bean, Siggia et al. 2006). Cells were automatically segmented and the mean fluorescence intensity was measured. Bud emergence was identified manually using phase images.
Supplementary Material
Acknowledgments
We thank Chris Aakre, Amanda Amodeo, Lucy Bai, Fred Cross, Stefano Di Talia, Bruce Futcher, Amy Gladfelter and Eric Siggia for insightful discussions and comments on the manuscript. The Burroughs Wellcome Fund, the NIH (GM092925) and the NSF (CAREER award #1054025) supported this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: GSM738421
References
- Amon A, Tyers M, et al. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74(6):993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57(1):21–9. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- Bai L, Charvin G, et al. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18(4):544–55. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, et al. Coherence and timing of cell cycle Start examined at single-cell resolution. Mol Cell. 2006;21(1):3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1(2):103–10. [PubMed] [Google Scholar]
- Borun TW, Gabrielli F, et al. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975;4(1):59–67. doi: 10.1016/0092-8674(75)90134-8. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, et al. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19(1):352–67. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, Koller D. Timing of gene expression responses to environmental changes. Journal of Computational Biology. 2009;16(2):279–290. doi: 10.1089/cmb.2008.13TT. [DOI] [PubMed] [Google Scholar]
- Chechik G, Oh E, et al. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nature biotechnology. 2008;26(11):1251–1259. doi: 10.1038/nbt.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, et al. A Genome-Wide Transcriptional Analysis of the Mitotic Cell Cycle. Molecular cell. 1998;2(1):65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301(5629):71–6. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117(7):899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Darieva Z, Clancy A, et al. A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol Cell. 2010;38(1):29–40. doi: 10.1016/j.molcel.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23(4):483–96. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, et al. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117(7):887–98. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Di Talia S, Wang H, et al. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 2009;7(10):e1000221. doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Bohm T, et al. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. Embo J. 1995;14(19):4803–13. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4(5):740–51. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, et al. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biology. 2010;11:6. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, et al. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 2010;11(6):R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K, Chapman-Shimshoni D, et al. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol Cell Biol. 1998;18(5):2492–501. doi: 10.1128/mcb.18.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi AB, Stukenberg PT, et al. Timing of events in mitosis. Curr Biol. 2002;12(2):105–14. doi: 10.1016/s0960-9822(01)00662-5. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham M, et al. Systematic planning of genome-scale experiments in poorly studied species. PLoS Comput Biol. 2010;6(3):e1000698. doi: 10.1371/journal.pcbi.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, et al. Genetic control of the cell division cycle in yeast. Science. 1974;183(120):46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hereford LM, Osley MA, et al. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, et al. Positive feedback sharpens the anaphase switch. Nature. 2008;454(7202):353–7. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Jensen TS, et al. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443(7111):594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, et al. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8(13):1514–25. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Justman QA, Serber Z, et al. Tuning the activation threshold of a kinase network by nested feedback loops. Science. 2009;324(5926):509–12. doi: 10.1126/science.1169498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalir S, McClure J, et al. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292(5524):2080–3. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- Kato M, Hata N, et al. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004;5(8):R56. doi: 10.1186/gb-2004-5-8-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, et al. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261(5128):1551–7. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, et al. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453(7192):246–50. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, et al. Global Analysis of the Genetic Network Controlling a Bacterial Cell Cycle. Science. 2000;290(5499):2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Levy S, Ihmels J, et al. Strategy of Transcription Regulation in the Budding Yeast. PLoS ONE. 2007;2(2):e250. doi: 10.1371/journal.pone.0000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Aviles S, Kapuy O, et al. Irreversibility of mitotic exit is the consequence of systems-level feedback. Nature. 2009;459(7246):592–5. doi: 10.1038/nature07984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay VL, Mai B, et al. Early cell cycle box-mediated transcription of CLN3 and SWI4 contributes to the proper timing of the G(1)-to-S transition in budding yeast. Mol Cell Biol. 2001;21(13):4140–8. doi: 10.1128/MCB.21.13.4140-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey MP, Angus SP, et al. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62(22):6587–97. [PubMed] [Google Scholar]
- Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16(14):1313–23. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Mogno I, Vallania F, et al. TATA is a modular component of synthetic promoters. Genome Res. 2010;20(10):1391–7. doi: 10.1101/gr.106732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. The cell cycle: principles of control. London Sunderland, MA: New Science Press; Sinauer Associates; 2007. [Google Scholar]
- Muller H, Bracken AP, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15(3):267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66(5):995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Oliva A, Rosebrock A, et al. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3(7):e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, et al. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453(7197):944–7. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Karuturi RK, et al. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell. 2005;16(3):1026–42. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Silva MD, Weinberg RA. The restriction point and control of cell proliferation. Curr Opin Cell Biol. 1997;9(6):768–72. doi: 10.1016/s0955-0674(97)80076-2. [DOI] [PubMed] [Google Scholar]
- Pomerening JR, Sontag ED, et al. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5(4):346–51. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- Pramila T, Wu W, et al. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20(16):2266–78. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–56. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, Mata J, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36(8):809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, et al. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics. 2002;31(1):64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, et al. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454(7202):291–6. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9(22):2780–94. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, et al. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38(7):830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, et al. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. Embo J. 1993;12(5):1955–68. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Molecular Biology of the Cell. 2002;13(6):1977. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, et al. Identification of Genes Periodically Expressed in the Human Cell Cycle and Their Expression in Tumors. Molecular Biology of the Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Landman A, et al. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol Cell Biol. 2002;22(12):4402–18. doi: 10.1128/MCB.22.12.4402-4418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426(6965):460–5. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, et al. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17(11):1550–61. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Lee TJ, et al. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10(4):476–82. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- Yung Y, Walker J, et al. A Skp2 autoinduction loop and restriction point control. The Journal of cell biology. 2007;178(5):741. doi: 10.1083/jcb.200703034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung Y, Walker JL, et al. A Skp2 autoinduction loop and restriction point control. J Cell Biol. 2007;178(5):741–7. doi: 10.1083/jcb.200703034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, et al. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36(5):486–91. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.