Summary
Two papers in this issue of Molecular Cell, Doncic et al. (2011) and Eser et al. (2011), present some satisfyingly simple ideas for the organization of the complex network that controls cell cycle progression and cell fate specification in budding yeast.
The default fate for a newly born budding yeast cell is to proceed through the cell cycle. Up to a certain point during the G1-phase of the cell cycle, termed Start, this fate is not inevitable; the cell has the option of mating rather than growing and dividing. But once Start has been passed, the cell is committed to doing a round of DNA replication and mitosis before it can once again consider mating. Much is known about the traversal of Start, based on decades of genetic and biochemical analysis of the pathways that regulate cell cycle progression in S. cerevisiae. Dozens of genes and proteins have been identified and organized into complex regulatory circuits. Still, we are left with the question of what exactly happens when Start is traversed. What does it mean for this regulatory circuit, built out of reversible elements, to irreversibly commit to a particular course? And of the hundreds of genes whose expression changes once Start has been passed, is there any logic to the question of which gene is expressed when? Two papers from the Skotheim group published in this issue of Molecular Cell (Doncic et al., 2011; Eser et al., 2011) provide interesting new insights into these issues.
Whi5 Translocation Defines Start
To address the first question, Doncic et al. (2011) made use of a live-cell video microscopy approach. The main probe for the status of the Start trigger was a fluorescent version of the Whi5 protein, a yeast transcriptional inhibitor that plays a role analogous to that of Rb in animal cells (Costanzo et al., 2004; de Bruin et al., 2004). At about the time Start occurs, Whi5 exits the nucleus, and thus loses its ability to repress G1/S transcription. The authors treated yeast cells at different times after mitosis with mating pheromone to see if the cells had passed Start yet—if a cell goes directly into the mating program, it is pre-Start, whereas if it carries out another cell cycle before going into the mating program, it is post-Start. Simultaneously, they determined how much the nuclear concentration of Whi5 had dropped from its post-mitotic peak. This allowed them to determine how tight the connection was between Whi5 redistribution and Start.
The results are remarkably clear. If mating pheromone is applied to a cell when more than 50% of the Whi5 is still in the nucleus, the cell will almost always directly arrest; it is pre-Start. If mating pheromone is applied when more than 50% of the nuclear Whi5 is gone, the cell will almost always carry out one more mitotic cycle before arresting; it is post-Start. This correlation holds up in both mothers and daughters, and in cells treated with both low and high concentrations of pheromone. Two other plausible surrogates of Start (time after mitosis and cell size) do a poorer job of predicting whether a cell will arrest directly when treated with phermomone. Thus, the metric that the cell uses to decide whether or not to proceed through the cell cycle is the nuclear Whi5 concentration.
Three Interlinked Positive/Double-Negative Feedback Loops Constitute a Modular Trigger for Start
The next question is what type of system regulates the nuclear concentration of Whi5: the answer is a small network of positive and double-negative feedback loops. For example, Whi5 inhibits CDK1 activation by inhibiting CLN1 and CLN2 transcription, and, conversely, CDK1 inhibits Whi5 function by promoting its exit from the nucleus. Thus the Whi5/Cln1,2-CDK1 system constitutes a double negative feedback loop, which, under the right circumstances, can function as a bistable trigger. Moreover, the Whi5/Cln1,2-CDK1 system is one of several interlinked positive or double-negative feedback loops (Figure 1). Active Cln1,2-CDK1 promotes more Cln1/2 transcription through the transcription factors MBF and SBF—a positive feedback loop—and active Cln1,2-CDK1 promotes the inactivation of its inhibitor Far1—a double negative feedback loop. In principle, these three interlinked loops could function together as a single bistable trigger that operates more robustly than any of the loops individually would (Ferrell, 2008). If so, then mutations that make any individual loop harder to flip from its pre-Start state to its post-Start state (for example, overexpression of Far1 or deletion of Cln1 and Cln2) should make it so that more Whi5 must be exported from the nucleus to get the cell through Start.
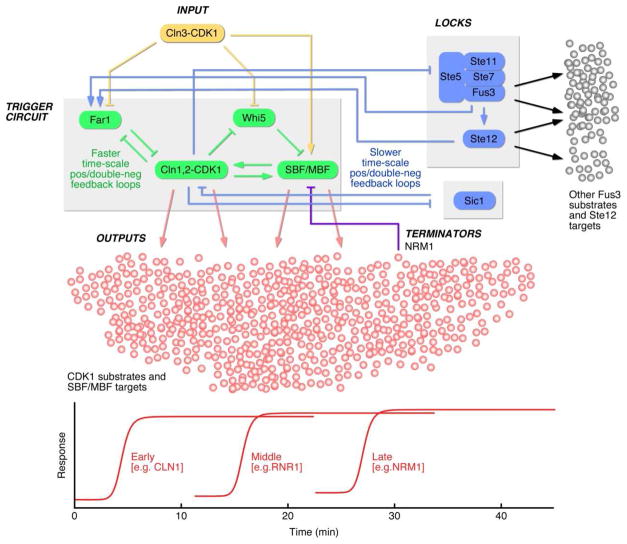
Figure 1. Schematic View of the Regulation of Start.
The key to cell cycle commitment appears to be a system of interlinked, rapid positive feedback and double-negative feedback loops—the trigger circuit. The input to the trigger circuit is Cln3-CDK1, which probably regulates multiple components of the trigger (Far1, Whi5, SBF/MBF) in a feed-forward arrangement. Slower double-negative feedback loops involving Sic1 and Ste5 may serve as locks, helping to maintain the trigger in its post-Start state and to suppress the mating response. The output of the trigger circuit also includes hundreds of CDK1 substrates and SBF/MBF-regulated genes, which are regulated in a complex temporal program. SBF/MBF targets that are themselves components are turned on early ensuring commitment to cell division precedes significant changes in the cellular transcription program; targets that execute the G1/S program tend to be turned on later; and targets that reverse the trigger’s activation tend to be turned on later as well.
Positive/Double-Negative Feedback as a Recurring Theme
Fifty years ago, Jacob and Monod hypothesized that the control systems that regulate eukaryotic cell differentiation might be double-negative feedback loops that established and maintained stable patterns of gene expression (Monod and Jacob, 1961). The details of the Start trigger are a bit different from what Jacob and Monod were originally thinking—the feedback loops here include not only double-negative loops but also positive feedback loops; it is not just patterns of gene expression but also patterns of protein phosphorylation that are critical to the system; and the biology here is not cell differentiation, but rather cell cycle commitment. Nevertheless, the ideas put forward by Jacob and Monod and expanded upon by theoretical biologists over the last several decades have held up extremely well. It seems that when nature needs to convert reversible, continuously graded inputs into irreversible, switch-like biological outputs, the solution often involves bistability, positive feedback, and double-negative feedback.
From Triggers to Locks: Suppressing the Mating Response after Start
There are at least two more double negative feedback loops present in the Cln1,2-CDK1 system. The first involves Sic1, a stoichiometric inhibitor of CDK1 (Sic1 inhibits CDK1) whose destruction is brought about by CDK1 phosphorylation (CDK1 inhibits Sic1) (Nash et al., 2001). Here, Doncic et al. (2011) show that Sic1 destruction occurs post-Start, and so is not likely to be a part of the trigger. Another double-negative loop connects CDK1 to the mating pheromone response pathway. Cln1,2-CDK1 phosphorylates and inactivates the MAPK scaffold Ste5, which is required for Fus3 (MAPK) activation; Fus3 phosphorylates Far1 and also activates the transcription factor Ste12; Ste12 turns on Far1 transcription; and Far1 stoichiometrically inactivates CDK1. Like the Sic1 loop, this loop appears to operate on a slower time-scale than the Start trigger, and when it is compromised, the result is a post-Start state where some aspects of the mating pathway are aberrantly expressed, rather than a shift in the Whi5 threshold. Thus the transition through Start involves interlinked positive/double-negative feedback loops operating on two different time-scales. The quick feedback loops function as a trigger for cell cycle progression; the slow loops then help lock the system into an unambiguous, non-mating state (Figure 1). Previous work has demonstrated that this type of trigger-and-lock arrangement offers important performance advantages, and may be a recurring motif in cellular regulation (Brandman et al., 2005).
Feedback First
Once Cln1,2-CDK1 has been activated and has turned on the transcription factors SBF and MBF, hundreds of target genes get induced (Ferrezuelo et al. 2010). To dissect this complicated regulatory program, Eser and colleagues (Eser et al., 2011), begin by analyzing microarray data for 362 G1/S genes in S. cerevisiae cells released from an M-phase (CDC20) arrest. They find that the transcripts appear in a defined temporal order, with the trigger components CLN1 and CLN2 being transcribed early, before genes like ribonucleotide reductase whose products carry out the nuts-and-bolts business of S and M phase. Thus, the first priority of the G1/S regulon is to get itself completely turned on.
Next, the regulatory system (Figure 1) focuses on the execution of the G1/S program (Eser et al., 2011), activating genes that are important for specific cell biological changes. Thus genes with some gene ontology terms (e.g. “budding”) tend to appear before genes with others (e.g.”DNA replication” or “mitosis”). The negative feedback component NRM1, which helps turn off MBF-regulated genes, is transcribed late, which again seems intuitively sensible; the regulon should not turn itself off until the G1/S program has been largely carried out. The order of the G1/S genes is a bit different in cells released from a G1-block rather than a mitotic block, but CLN1 is still among the earliest. The same is true for the closely-related yeast S. bayanus, and, more remarkably, for mammalian cells in culture. Analysis of previously published data on HeLa cells released from either a double thymidine (G1/S) block or a thymidine/nocodazole (M-phase) block, shows that cyclin E1, cyclin E2, E2F1 and Skp2—four genes thought to be critical for positive and double-negative feedback at the onset of S-phase—are among the first-transcribed genes. This suggests that this feedback-first strategy is a recurring theme in the organization of complex cellular regulatory programs.
These findings (Eser et al., 2011) harken back to a study published by Georgi and co-workers a decade ago on the timing of mitotic phosphorylations in Xenopus egg extracts (Georgi et al., 2002). Like the G1/S regulators examined here, the master regulator of mitosis, cyclin B-CDK1 is thought to control hundreds of target proteins. Georgi et al. examined the timing of the mitotic phosphorylation of various putative CDK targets, and found that two substrates important for positive/double-negative feedback (Cdc25C and Wee1) were regulated early, whereas one substrate important for terminating mitosis (the APC component Cdc27) was regulated late.
Twenty-five years ago, one of the most thrilling discoveries in cell biology was the realization that the genes and proteins that regulate cell cycle progression in fungi also regulate cell cycle progression in clams, frogs and humans. Now with we have examples of different cell cycle transitions (G1/S regulation vs. mitosis) in different organisms (S. cerevisiae vs. Xenopus laevis) being orchestrated by non-homologous proteins (e.g. Whi5 has no obvious orthologs outside of fungi). Yet the processes share common systems-level organizational principles, like feedback-first. Thus the field has another exciting convergence.
Strategies to Establish Timing
The next question is how the timing of gene activation within the G1/S regulon is established. Part of the answer arises out of the heirarchical nature of the control system: a small number of transcription factors (SBF and MBF, which can be regarded here as network hubs) regulate the transcription of a large number of genes (the G1/S regulon), so there is bound to be some coordination between the individual transcriptional events. Beyond this level of coordination, Eser et al. show that part of the answer has to do with whether an individual gene is regulated by SBF, by MBF, or by both. In the CDC20 (mitotic) block/release experiments, SBF and SBF/MBF targets are transcribed earlier than MBF-only targets; in G1 block/release experiments MBF and SBF/targets are transcribed earlier than SBF-only target. Taken together, these experiments indicate that the transcriptional activation of a gene regulated by both factors can be represented as a logical OR function: activation of one of the two promoter bound factors suffices for activation. However, this does not explain the fine-tuning of gene activation. Several mechanisms could explain why some SBF targets are transcribed earlier than others: (1) an affinity mechanism, where a rising concentration of active SBF activates high affinity targets first and low affinity targets later; (2) a kinetic mechanism, where active transcriptional complexes take longer to assemble on late genes than on early genes; or (3) a combinatorial mechanism, where other transcription factors contribute to some the transcription of some but not all of the SBF targets.
Note that precisely analogous mechanisms could also account for the differences in timing of mitotic phosphorylations in Xenopus extracts, and the experiments of Georgi et al. are most consistent with hypothesis #2—a kinetic mechanism where it takes longer for some substrates to be phosphorylated than others. There are numerous analogies between the regulation of proteins by phosphorylation and the regulation of genes by transcription factors. For example, both often involve multiple poorly-conserved, rapidly evolving sites (Moses et al., 2007). It will be of interest to determine whether similar systems-level strategies are used to organize regulatory programs in both gene expression networks and phosphorylation networks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;***:****–****. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;***:****–****. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr Feedback regulation of opposing enzymes generates robust, all-or-none bistable responses. Curr Biol. 2008;18:R244–245. doi: 10.1016/j.cub.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Futcher B, Aldea M. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 2010;11:R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi AB, Stukenberg PT, Kirschner MW. Timing of events in mitosis. Curr Biol. 2002;12:105–114. doi: 10.1016/s0960-9822(01)00662-5. [DOI] [PubMed] [Google Scholar]
- Monod J, Jacob F. General conclusions: teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harbor Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- Moses AM, Heriche JK, Durbin R. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 2007;8:R23. doi: 10.1186/gb-2007-8-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multi-site phosphorylation of a CDK inhibitor sets a threshold for the onset of S-phase. Nature (London) 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]