Abstract
This study uses scanning acoustic microscopy (SAM) ultrasonic profilometry to determine acceptable vs. failed tissue engineered oral mucosa. Specifically, ex vivo-produced oral mucosal equivalents (EVPOMEs) under normal or thermally stressed culture conditions were scanned with the SAM operator blinded to the culture conditions. As seeded cells proliferate, they fill in and smooth out the surface irregularities; they then stratify and produce a keratinized protective upper layer. Some of these transformations could alter backscatter of ultrasonic signals and in the case of the thermally stressed cells, produce backscatter similar to an unseeded device. If non-invasive ultrasonic monitoring could be developed, then tissue cultivation could be adjusted to measure biological variations in the stratified surface. To create an EVPOME device, oral mucosa keratinocytes were seeded onto acellular cadaveric dermis. Two sets of EVPO-MEs were cultured: one at physiological temperature 37 °C and the other at 43 °C. The specimens were imaged with SAM consisting of a single-element transducer: 61 MHz center frequency, 32 MHz bandwidth, 1.52 f#. Profilometry for the stressed and unseeded specimens showed higher surface irregularities compared to unstressed specimens. Elevated thermal stress retards cellular differentiation, increasing root mean square values; these results show that SAM can potentially monitor cell/tissue development.
Keywords: Ultrasound, Non-invasive assessment, Tissue engineering, Profilometry
INTRODUCTION
The development and use of an ex vivo-produced oral mucosal equivalent (EVPOME)—an engineered human oral mucosa developed for reconstructive intraoral grafting procedures—has demonstrated its clinical efficacy in intra-oral surgical grafts.9 EVPOME has shown to be an effective soft tissue replacement in cases where patients’ oral mucosa was absent or damaged due to disease, injury, or other pathological conditions.9–11 Previous studies have shown the efficacy of using scanning acoustic microscopy (SAM) to image and assess the differentiation of these tissues.18,19 In this study, we tested the ability of SAM to differentiate between seeded EVPOME devices which have undergone periods of elevated thermal incubation (stressed) to those which were incubated continuously at physiological temperature (unstressed).
Background on EVPOME
The oral mucosa’s histology resembles that of skin: as new cells are formed on the basal lamina, the more matured cells migrate toward the apex, undergoing apoptosis and keratinization as they migrate.10 This same process occurs in the EVPOME tissues, making it an excellent tissue for repair and replacement of damaged or diseased soft tissue.
We earlier examined and compared the morphologies of AlloDerm®—a commercially available acellular human cadaver dermis (the tissue goes through a cell removal process while retaining biochemical and structural components.), and EVPOME at different stages of development using SAM.20 By first comparing the morphologies of the oral mucosal tissues—both natural and engineered, we can better understand the physical compositions when it comes to testing for similarities in the tissues’ physical properties. We can then quantify the cellular constituents among each of the tissue types and correlate how each contributes to the physical behavior of their respective tissues. This would assist us in establishing criteria for standardization of manufacture of the engineered tissues as well as criteria for their uses in reconstructive surgery based on their mechanical properties.
Background on SAM
SAM is an effective tool to study both the morphology and non-linear elastic characteristics of natural and engineered oral mucosal tissues. The advantages of using SAM over conventional optical and electron microscopy include being able to image the cells and tissues without doing any preparations which could potentially kill or alter the tissues; this provides a more accurate representation of the tissues’ natural properties. We have also shown that SAM can provide evidence as to the degree of differentiation which the cells are undergoing without altering their physical conditions.11,14,15,19 In addition, ultrasound imaging systems can indirectly estimate and image elastic properties of tissue using ultrasonic measurements of externally or internally induced tissue motion.1
Other imaging methods which do not require preparing cell/tissue cultures include atomic force microscopy (AFM).13 Although AFM can provide imaging of the cultures, there are several disadvantages associated with this imaging method. These include the AFM cantilever systems deforming or tearing apart living cells and other biological materials and the force being too great to image cells with sufficient resolution.13,17
We are currently using SAM to examine and correlate any changes in the radiofrequency (RF) data to the EVPOME (and mucosal cells in general) undergoing differentiation, apoptosis, and keratinization. This will allow us to better understand the physiological processes of these cells as they undergo stratification on the AlloDerm® over time.
Profilometry
This is a standard method used to measure a surface’s profile, in order to quantify its roughness: the number of surface irregularities of the overlying basement membrane (AlloDerm® without cells) or of the stratified epithelium (EVPOME). The SAM is utilized to check for surface roughness and correlate this to the growth and development of the keratinized layer on the surface: the smoother the surface (as determined by the reflectivity of the SAM), the higher the keratinized surface as the cells differentiate and stratify when migrating up to the surface. Surface profilometry will also provide evidence as to the degree of differentiation which the cells are undergoing without chemically affecting their properties.
MATERIALS AND METHODS
Tissue Preparation—EVPOME
The protocol for harvesting human oral mucosal tissue was approved by a University of Michigan Internal Review Board. All individuals signed informed consent before the tissue samples were procured. A keratinized oral mucosa sample was taken from an out-patient at the University of Michigan Oral and Maxillofacial Surgery Clinic. The methods for preparation have been described in greater detail previously.4,5 In brief, oral mucosa keratinocytes were enzymatically dissociated from the tissue sample, and a primary cell culture was established and propagated in a chemically defined, serum- and xenogeneic products-free culture medium, with calcium concentration of 1.2 mM. The AlloDerm® was soaked in 5 µg/cm2 human type IV collagen at 4 °C overnight prior to seeding cells to assist the adherence of cells, then approximately 1.5 × 105 cells/cm2 of oral keratinocytes were seeded onto the type IV collagen pre-soaked AlloDerm®. The composites of the keratinocytes and the AlloDerm® were then cultured, in the submerged condition, for 4 days to form a continuous epithelial monolayer. After 4 days, the equivalents were raised to an air–liquid interface to encourage epithelial stratification and cultured for another 7 days, resulting in a fully differentiated, well-stratified epithelial layer on the AlloDerm®. At day 11 post-seeding, EVPOME samples were collected for SAM imaging.
On day 9 post-seeding, one set of EVPOME devices was incubated at 43 °C for 24 h, then switched back to 37 °C for another 24 h (thermally stressed). Another set of EVPOMEs was kept at 37 °C up through day 11 post-seeding (non-stressed). The total seeding times for both conditions were equal. Three specimens were used for each study set: an unseeded AlloDerm®, an unstressed control, and a thermally stressed EVPOME.
SAM Apparatus and Imaging
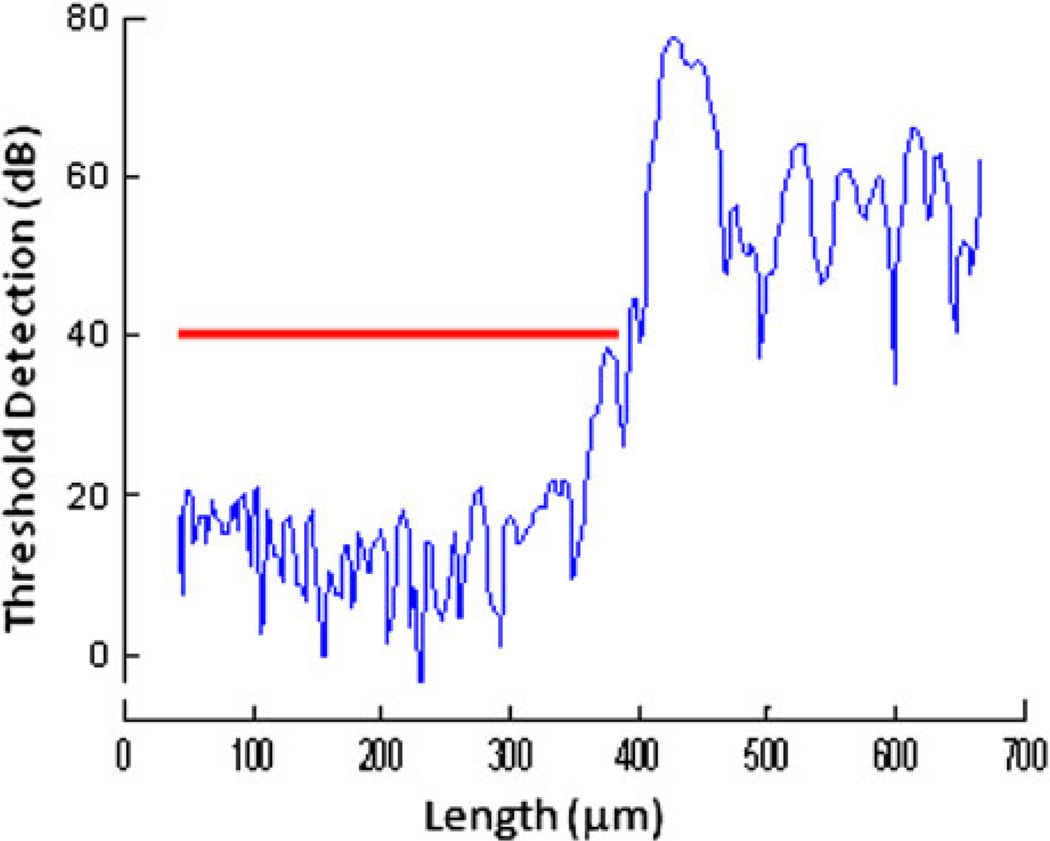
Details of the scanning and ultrasound system are similar to those described elsewhere.3,4,6 EVPOME samples were immersed in deionized water and imaged with a single element fixed focus transducer, producing ultrasonic B-scans. The transducer has an approximate frequency of 61 MHz, the element is 3 mm in diameter and focused to a depth of 4.1 mm, giving an f/number of approximately 1.4. The transducer was fastened to an optical mount and the angular position was adjusted until the ultrasonic beam was normal to the deflecting plate. We scanned the surfaces of the EVPOME, showing the acoustic signal between the interface of the sample and water on the sample’s apical side. DC stepper motors accurately positioned the transducer above the specimen. B-scan images were obtained by stepping the transducer element laterally across the desired region. At each position, the transducer fired and an RF A-line was recorded. After repeated firings at one position, the transducer moved to the next, where the image was constructed from A-lines acquired at all lateral positions. Because low f/number, single element transducers have a short depth of field, a composite B-scan image was generated from multiple scans at different heights. Ultrasonic images were created using confocal image reconstruction. The spacing was determined by the step sizes along each of the axes: 15 µm scanning step size in both the transverse and horizontal directions; a lateral resolution (Rlat) of 37 µm; an axial resolution (Rax) of 24 µm; a depth of field of 223 µm. Z-axis was sampled at 300 mega samples per second. Rax is the resolution in the direction of propagation and is determined by the length of the ultrasound pulse propagating in the tissue; Rlat is the resolution orthogonal to the propagation direction of the ultrasound wave. The tissue surface was determined by thresholding the magnitude of the signal at the first axial incidence of a value 20–30 dB safely above noise; a typical A-line is illustrated in Fig. 1. Four sets of scans were performed, each set containing a thermally stressed and unstressed specimen. An unseeded AlloDerm® specimen was also scanned as a control. All specimens were imaged for approximately 30 min, thus minimizing the risk for contamination to occur.
FIGURE 1.
Plot diagram of the SAM, showing the detection of the device surface (red line) after setting the threshold detection safely above the signal of the water.
Surface profilometry was determined by first finding the instance of threshold value, fitting, and subtracting the planar surface, then calculating root mean squared (RMS) height.5 The RMS value was computed over the surface and therefore a function of x and y. Because the surface height is z, its function is z(x,y). It has a two-dimensional sum and is divided by nx and ny inside the square root:
| (1) |
where n is number of x and y samples.
Histology
EVPOMEs (both the stressed and unstressed samples) were fixed with 10% formalin, embedded in paraffin, cut in 5 µm sections, stained with hematoxylin and eosin. The histology of the EVPOME samples were then examined under a Nikon Ti-U inverted microscope (Nikon Optical, Tokyo, Japan) using bright-field light.
Statistical Analyses
Both one-way analyses of variance (ANOVA) and linear regression analyses were performed on the surface characteristics for both categories of EVPOME cultures. The RMS values with their standard deviation values were compared between the stressed and unstressed EVPOME specimens and to an unseeded AlloDerm® control specimen.
RESULTS
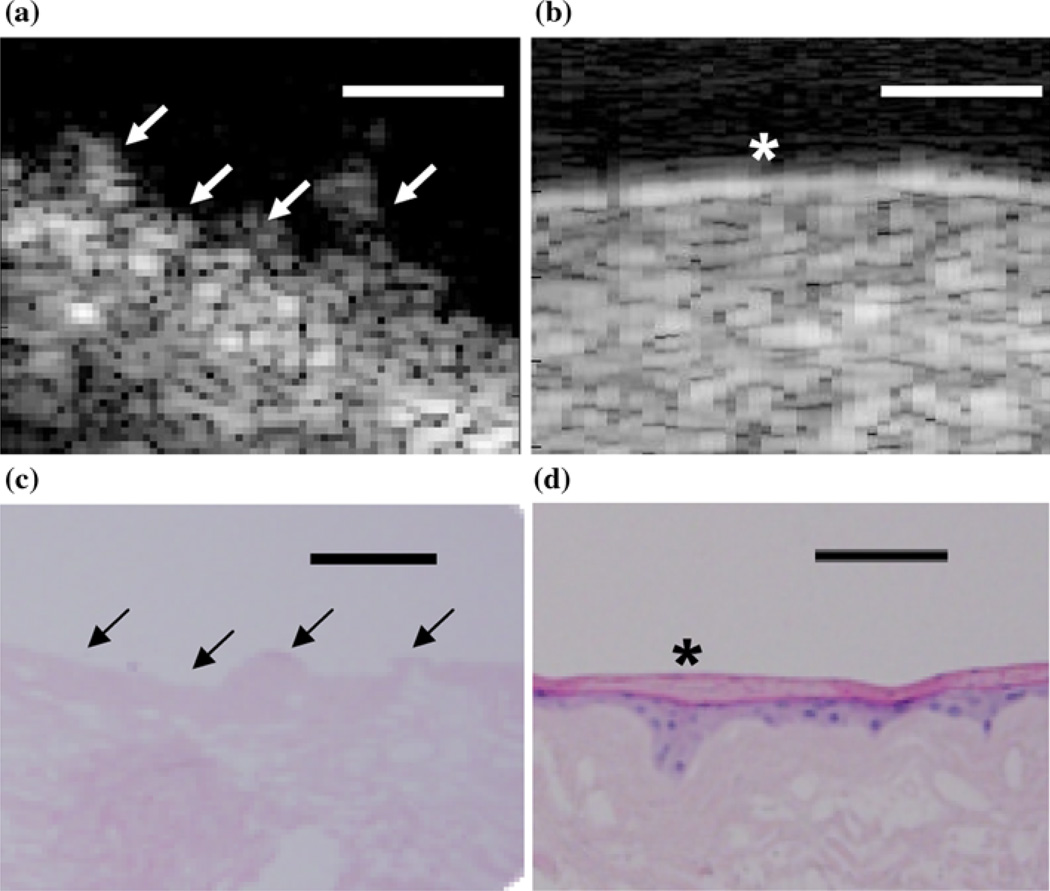
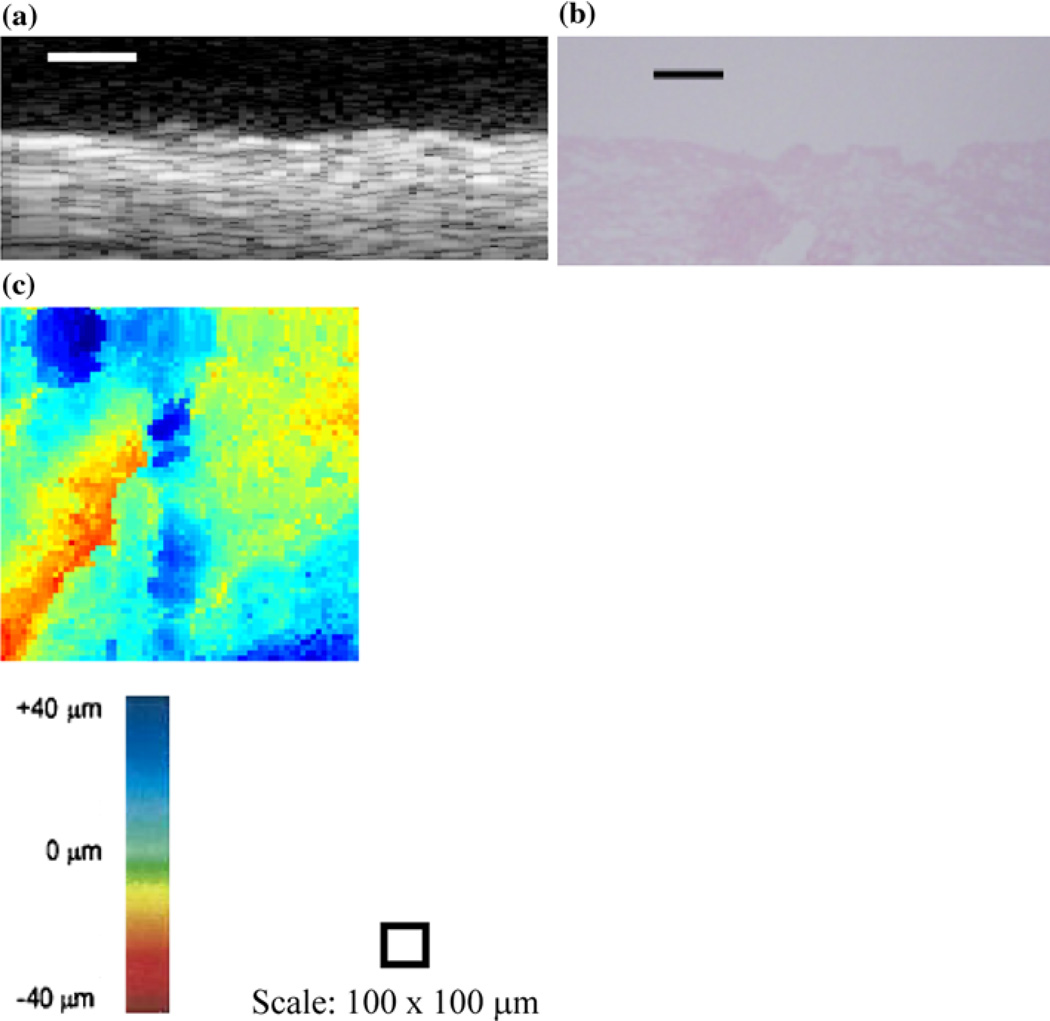
For the thermally stressed devices, the apical sides showed similar surface irregularities to those of the 2D B-scans of unseeded AlloDerm® devices (Fig. 2a). Comparing the B-scans taken of EVPOME at 11 days post-seeding (Fig. 2b), there is a distinct reflection off of the surface which is not found in the unseeded AlloDerm®; a result of the seeded keratinocytes differentiating.18,19 The histology images of these same devices—AlloDerm® and EVPOME at 11 days post-seeding (Figs. 2c and 2d, respectively) verify this finding as the keratinized layer in the latter image shows a smooth surface, with less surface irregularities compared to the former image.18
FIGURE 2.
2D B-scan images of the untreated AlloDerm® device (a) and EVPOME at 11 days post-seeding (b). Surface irregularities (arrows) are clearly present on the AlloDerm® compared to the EVPOME; the latter shows a brighter surface reflection (asterisk), the result of the differentiating keratinocytes. The corresponding histology images for the untreated AlloDerm® device (c). The EVPOME at 11 days post-seeding (d) also show a high number of surface irregularities (arrows) on the former compared to the keratinized layer (asterisk) on the EVPOME surface. Scale bars represent 100 µm.
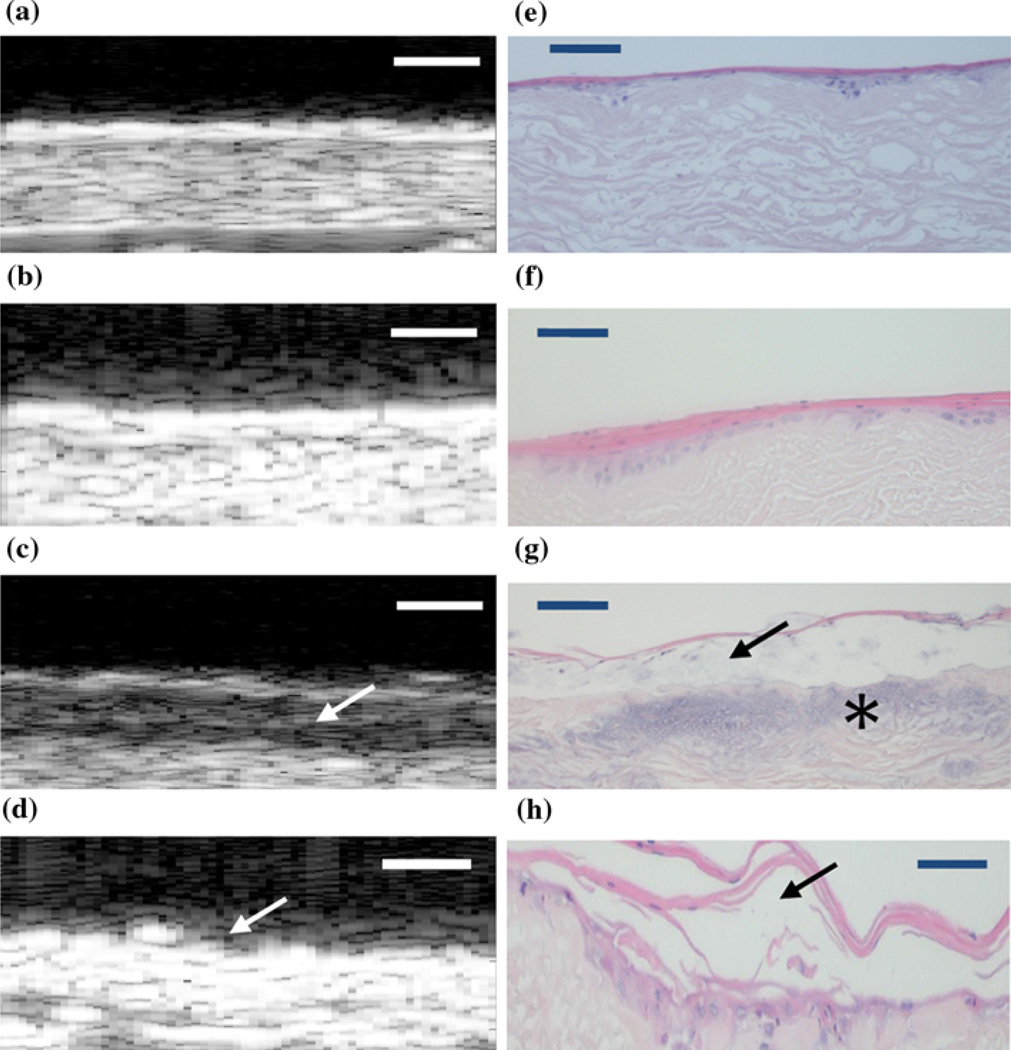
The SAM 2D B-scans showed higher reflectivity on the surfaces for the non-stressed devices (Figs. 3a and 3b) than those of the stressed (Figs. 3c and 3d). This is typical to the results we had obtained from our previous study to examine the morphological differences between the AlloDerm® device and EVPOME at different stages of post-seeding.18 This was confirmed by examining the device sections using standard optical microscopy to compare non-stressed (Figs. 3e and 3f) and stressed (Figs. 3g and 3h) devices. Further, the scans for the stressed devices displayed a darkened region between two more brightly reflected regions (Fig. 3c); an indication that the keratin layer had partitioned from the surface of the device. We validated this by comparing the histological images of the same device (Fig. 3g), which confirmed our findings. For one of the thermally stressed devices the irregular surface of the AlloDerm® device is visible, but no keratinized layer; the likely result of the keratin completely sloughing off from the surface (Fig. 3d). The histology from this latter device showed the keratin layer more fractionated and partitioning from the surface to a greater degree than other stressed devices (3 h); this validates our claim of the keratin sloughing from the device.
FIGURE 3.
2D B-scan composites of the EVPOME devices for both the thermally unstressed (a–b) and stressed (c–d) groups. The corresponding histology micrographs of both unstressed (e–f) and stressed devices (g–h) compare the conditions of the keratinized surfaces for both imaging modalities. Comparing (c) with (g), there is a clear partitioning (arrows) between the keratinized layer and the device due to the heat stress; evident in the darkened region of the B-scan image (arrow). For (d), the irregular surface of the AlloDerm® device is visible (arrow), but no keratinized layer; the likely result of the keratin layer sloughing off from the device surface. For (g), the blue staining on the device (asterisk) is a result of excessive hematoxylin used in their preparation and not a result of the thermal treatment. Device surfaces are at the top of each image. Scale bars represent 100 µm.
Filtering any existing artifacts in the RMS processing showed uniformity within the non-stressed devices. This again validated the findings from our previous studies: the space-filling properties of the keratinocytes on the devices.18,19
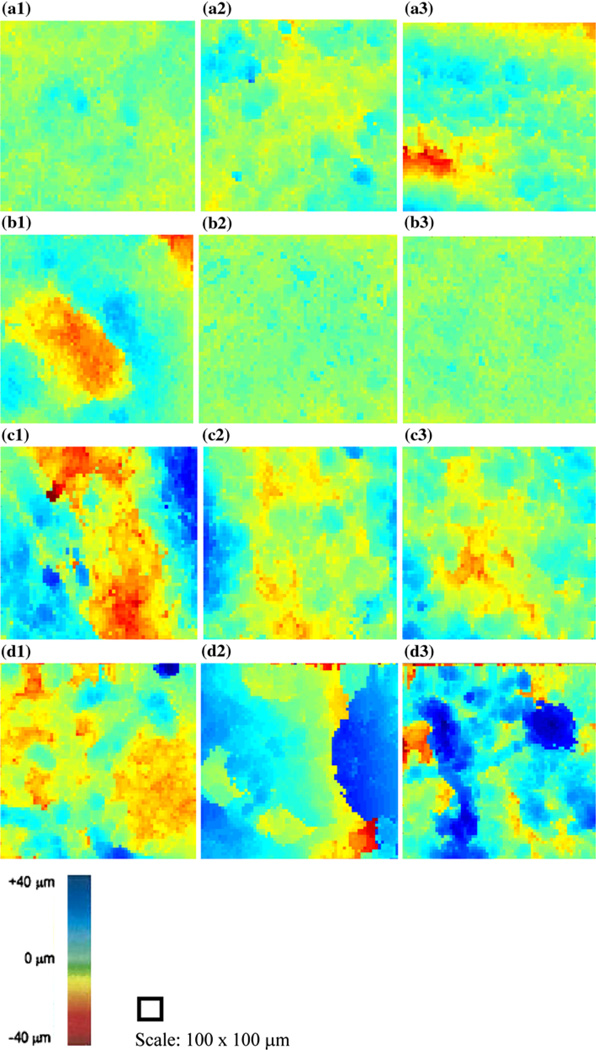
The surface profilometry images for all scans performed on each device displayed higher degrees of variation for the stressed devices compared to the non-stressed ones (Fig. 4). The images displaying red and blue in certain regions show greater variations in the surfaces; this is particularly the case for the thermally stressed specimens. The classification is based on fitting the planar surface of each representative sample and subtracting the difference. Therefore, the more changes in colors represent greater irregularities in the surface of the specimen. The unstressed devices’ scans showed minimal variation in the RMS height values; either low or high (Figs. 4a and 4b). By contrast, there are greater changes in these RMS values in the scans for the stressed devices (Figs. 4c and 4d). There was some variation found in the unstressed device scans; a result of the way in which the device was mounted to the bottom of the water tank (we also took this into account for when examining the stressed device scans). This compares with the 2D B-scan, histology, and C-scan profilometry images produced by scanning the unseeded AlloDerm® control (Figs. 5a–5c, respectively).
FIGURE 4.
RMS profilometry images for each scan performed on the thermally non-stressed (a–b) and stressed (c–d) devices. For the latter, there is higher variation in the surface features—presumably from the thermal stress fractionating the keratinized layer. With the non-stressed devices, there is little variation in the surface profiles, characteristic of the uniformity of the keratin layer on their surfaces.
FIGURE 5.
SAM 2D B-scan (a), histology micrograph (b), and C-scan profile (c) of an unseeded AlloDerm® device. Scale bars represent 100 µm.
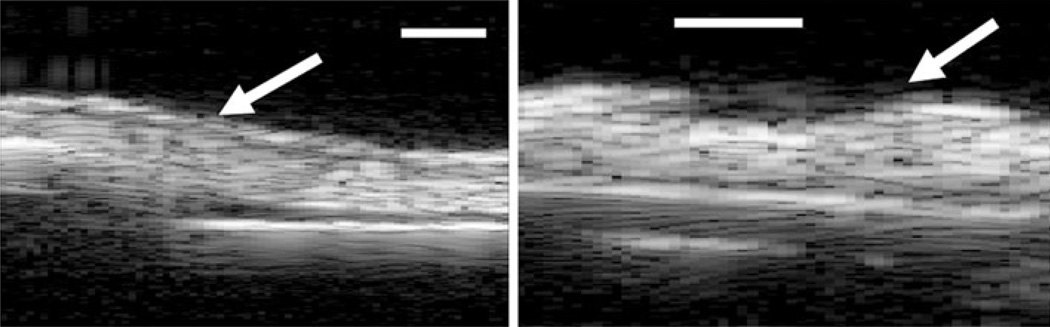
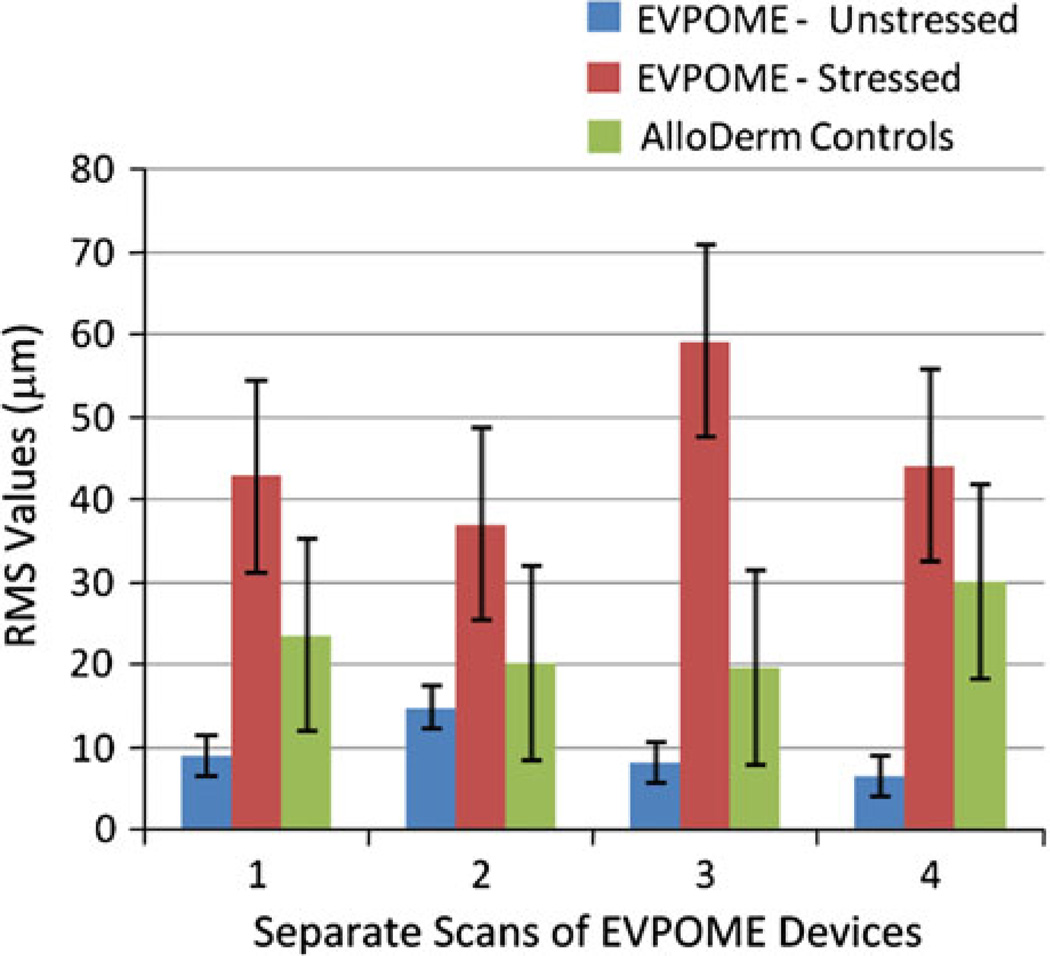
Examining the 2D B-mode images found irregularities caused by the tissue with respect to the placement of the transducer which caused the increased RMS heights. For the non-stressed device scans, there was one scan from each sample which produced relatively higher RMS results than the others. This is noticeable in both the SAM surface profilometry images (Figs. 4a3 and 4b1) and in the 2D B-scans (Fig. 6) of the devices. Consequently, the standard deviations were comparatively greater for the non-stressed devices than the stressed group, while the mean values remained lower for the non-stressed group. The compendium of RMS values between the thermally stressed and unstressed EVPOME devices shows significant values: the RMS heights for the stressed devices are four to five times greater than the unstressed devices, with the unseeded AlloDerm® control mean displaying RMS value approximately 2.5 times greater than the unseeded EVPOMEs (Fig. 7).
FIGURE 6.
2D B-scans of non-stressed devices showing surface irregularities (arrows) in these two selected scan areas of the devices—due to positioning the devices. This resulted in higher (outlier) RMS values when processing the SAM scans and consequently, higher variations. Scale bars are 100 µm.
FIGURE 7.
Comparative RMS heights for EVPOME devices, both unstressed, stressed, and the unseeded AlloDerm®. Values provided are from each separate set of SAM scans.
DISCUSSION
Ultrasonic profilometry does not detect cells on the thermally stressed EVPOME filling in the irregularities on the surfaces compared to the unstressed EVPOME. The decrease in the RMS values for the unstressed device is evidence of the proliferation/differentiation by the seeded cells on the EVPOME surface; in contrast the thermally stressed cells show little to no activity. The latter case indicates SAM’s ability to measure physiological activity by measuring the degree of ultrasonic scatter. These initial results show that ultrasonic characterization may have the potential to monitor EVPOME development during its manufacturing process: SAM could be utilized in a sterile environment such as a laminar hood and its applicability to monitor in vitro cell and tissue growth over specified time periods (i.e., 24 h) exists. Comparing similarities on SAM 2D B-mode images of the unstressed EVPOME to earlier studies’ images of EVPOME at 11 days post-seeding10,11 provided the best evidence to select the correct devices.
The two outliers in the non-stressed devices are likely the result of their mounting prior to performing the scan and not any error in the SAM detection and imaging. Nevertheless, they present a serious challenge toward properly imaging and accurately identifying the structural characteristics of the devices and any other subsequent cells, tissues, and other biological devices examined using SAM: in the specific case of the sample being observed, there was a significant slope caused by inadvertently positioning the sample in the manner described. A similar case involving an optical micrograph would be having the tissue specimen slightly thicker on one end, which could lead to the image being slightly out of focus on one side. Although we are able to correct for this error through filtering and background subtraction and analyses, improving methods for mounting the devices during the SAM scans must take effect. This is to minimize any potential errors in calculating RMS heights caused by the devices buckling or tilting.
Although SAM identified differences in cell and tissue development on the 11-day EVPOME, we cannot conclude that the devices are devoid of surface irregularities, regardless of the amount of keratinization present: neither acoustic nor optical microscopy can show that the EVPOME are consistently uniform across their entire surfaces. This was verified by the aforementioned outliers seen in the SAM scans of both thermally unstressed devices.
This study provides strong supplemental evidence to demonstrate SAM as a method to objectively quantify cell and tissue development and differentiation. In addition, it provides insight on mature EVPOME’s characteristics when subjected to an elevated temperature. Subsequent studies will further examine these differentiation characteristics through continuous ultrasound monitoring of in vitro EVPOME devices under aseptic conditions.
Future considerations will examine the use of apparent integrated backscatter (AIB) to further demonstrate and quantify cell development using SAM. AIB is a measure of the frequency-averaged (integrated) backscattered power contained in some portion of a backscattered ultrasonic signal; it has been used extensively to study soft tissues.2,5,12,21
ACKNOWLEDGMENTS
This study was supported through the National Institutes of Health (NIH) Regenerative Sciences Training Grant Number 5T90DK070071 and NIH Grant Numbers R21EY018727, R01 DE13417, and NIH center core (P30) Grant, EY007003. National Institutes of Health, Bethesda, MD. We gratefully acknowledge the NIH Resource Center for Medical Ultrasonic Transducer Technology at the University of Southern California (Los Angeles, CA) for designing and building the high-frequency transducer used in this study.
REFERENCES
- 1.Bridal SL, Fornes P, Bruneval P, Berger G. Parametric (integrated backscatter and attenuation) images constructed using backscattered radio frequency signals (25–56 MHz) from human aortae in vitro. Ultrasound Med. Biol. 1997;23:215–229. doi: 10.1016/s0301-5629(96)00201-3. [DOI] [PubMed] [Google Scholar]
- 2.Cohen RD, Mottley JG, Miller JG, Kurnik PB, Sobel BE. Detection of ischemic myocardium in vivo through the chest wall by quantitative ultrasonic tissue characterization. Am. J. Cardiol. 1982;50:838–843. doi: 10.1016/0002-9149(82)91242-5. [DOI] [PubMed] [Google Scholar]
- 3.Cohn NA, Emelianov SY, Lubinski MA, O’Donnell M. An elasticity microscope. Part I: methods. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 1997;44:1304–1319. [Google Scholar]
- 4.Cohn NA, Emelianov SY, O’Donnell M. An elasticity microscope. Part II: experimental results. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 1997;44:1320–1331. [Google Scholar]
- 5.Endo T, Finger WJ, Kanehira M, Utterodt A, Komatsu M. Surface texture and roughness of polished nanofill and nanohybrid resin composites. Dent. Mater. J. 2010;29(2):213–223. doi: 10.4012/dmj.2009-019. [DOI] [PubMed] [Google Scholar]
- 6.Holland MR, Hall CS, Lewis SH, Handley SM, Finch-Johnston AE, D’Sa AP, Perez JE, Miller JG. Comparison of integrated backscatter values obtained with acoustic densitometry with values derived from spectral analysis of digitized signals from a clinical imaging system. J. Am. Soc. Echocardiogr. 1997;10(5):511–517. doi: 10.1016/s0894-7317(97)70004-x. [DOI] [PubMed] [Google Scholar]
- 7.Hollman KW, Emelianov SY, Neiss JH, Jotyan G, Spooner GJR, Juhasz T, Kurtz RM, O’Donnell M. Strain imaging of corneal tissue with an ultrasound elasticity microscope. Cornea. 2002;21:68–73. doi: 10.1097/00003226-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Insana MF, Hall TJ. Parametric ultrasound imaging from backscatter coefficient measurements—image-formation and interpretation. Ultrason. Imaging. 1990;12:245–267. doi: 10.1177/016173469001200402. [DOI] [PubMed] [Google Scholar]
- 9.Izumi K, Feinberg SE, Iida A, Yoshizawa M. Intraoral grafting of an ex vivo produced oral mucosa equivalent: a preliminary report. Int. J. Oral Maxillofac. Surg. 2003;32:188–197. doi: 10.1054/ijom.2002.0365. [DOI] [PubMed] [Google Scholar]
- 10.Izumi K, Song J, Feinberg SE. Development of a tissue-engineered human oral mucosa: from the bench to the bed side. Cells Tissues Organs. 2004;176:134–152. doi: 10.1159/000075034. [DOI] [PubMed] [Google Scholar]
- 11.Izumi K, Terashi H, Marcelo CL, Feinberg SE. Development and characterization of a tissue-engineered human oral mucosa equivalent produced in a serum-free culture system. J. Dent. Res. 2000;79:798–805. doi: 10.1177/00220345000790030301. [DOI] [PubMed] [Google Scholar]
- 12.Kolios MC, Czarnota GJ, Lee M, Hunt JW, Sherar MD. Ultrasonic spectral parameter characterization of apoptosis. Ultrasound Med. Biol. 2002;28:589–597. doi: 10.1016/s0301-5629(02)00492-1. [DOI] [PubMed] [Google Scholar]
- 13.Marchant RE, Kang I, Sit PS, Zhou Y, Todd BA, Eppell SJ, Lee I. Molecular views and measurements of hemostatic processes using atomic force microscopy. Curr. Protein Pept. Sci. 2002;3(3):249–274. doi: 10.2174/1389203023380611. [DOI] [PubMed] [Google Scholar]
- 14.Matsuyama T, St. Goar FG, Tye TL, Oppenheim G, Schnittger I, Popp RL. Ultrasonic tissue characterization of human hypertrophied hearts in vivo with cardiac cycle-dependent variation in integrated backscatter. Circulation. 1989;80:925–934. doi: 10.1161/01.cir.80.4.925. [DOI] [PubMed] [Google Scholar]
- 15.Olerud JE, Obrien WD, Riedererhenderson MA, Steiger DL, Debel JR, Odland GF. Correlation of tissue constituents with the acoustic properties of skin and wound. Ultrasound Med. Biol. 1999;16:55–64. doi: 10.1016/0301-5629(90)90086-r. [DOI] [PubMed] [Google Scholar]
- 16.Taggart LR, Baddour RE, Giles A, Czarnota GJ, Kolios MC. Ultrasonic characterization of whole cells and isolated nuclei. Ultrasound Med. Biol. 2007;33:389–401. doi: 10.1016/j.ultrasmedbio.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 17.Verstraeten VL, Lammerding J. Experimental techniques for study of chromatin mechanics in intact nuclei and living cells. Chromosome Res. 2008;16(3):499–510. doi: 10.1007/s10577-008-1232-8. [DOI] [PubMed] [Google Scholar]
- 18.Wickline SA, Thomas LJ, Miller JG, Sobel BE, Perez JE. The dependence of myocardial ultrasonic integrated backscatter on contractile performance. Circulation. 1985;72:183–192. doi: 10.1161/01.cir.72.1.183. [DOI] [PubMed] [Google Scholar]
- 19.Winterroth F, Hollman KW, Izumi K, Feinberg SE, Fowlkes JB, Hollister SJ. Examination and comparisons of EVPOME/AlloDerm® compositions with human mucosal tissues using scanning acoustic microscopy; Society for Biomaterials 2009 Annual Meeting and Exposition; 2009. [Google Scholar]
- 20.Winterroth F, Kuo S, Izumi K, Feinberg SE, Hollister SJ, Fowlkes JB, Hollman KW. High-resolution ultrasonic monitoring of cellular differentiation in ex vivo oral mucosal equivalent (EVPOME) tissue culturing; Proceedings of the IEEE Bioultrasonics; 2009. [Google Scholar]
- 21.Zuber M, Gerber K, Erne P. Myocardial tissue characterization in heart failure by real-time integrated backscatter. Eur. J. Ultrasound. 1999;9:135–143. doi: 10.1016/s0929-8266(99)00019-1. [DOI] [PubMed] [Google Scholar]