Abstract
Protein ubiquitylation is a key process in the regulation of many cellular processes. The balance between the activity of ubiquitin ligases and that of proteases controls the level of ubiquitylation. In response to extracellular stimuli, stress-activated protein kinases (SAPK) modulate gene expression to maximize cell survival. In yeast, the Hog1 SAPK has a key role in reprogramming the gene expression pattern required for cell survival upon osmostress. Here, we show that the Ubp3 ubiquitin protease is a target for the Hog1 SAPK to modulate gene expression. ubp3 mutant cells are defective in expression of osmoresponsive genes. Hog1 interacts with and phosphorylates Ubp3 at serine 695, which is essential to determine the extent of transcriptional activation in response to osmostress. Furthermore, Ubp3 is recruited to osmoresponsive genes to modulate transcriptional initiation as well as elongation. Therefore, Ubp3 activity responds to external stimuli and is required for transcriptional activation upon osmostress.
Keywords: gene expression, Hog1, osmostress, SAPK, Ubp3
Introduction
Ubiquitylation is a reversible post-translational modification with essential roles in various signal transduction cascades and determines protein stability (Amerik and Hochstrasser, 2004; Komander et al, 2009; Reyes-Turcu et al, 2009). The balance of ubiquitin ligases and ubiquitin proteases determines the ubiquitylation state of a protein. To date, several ubiquitin-specific proteases (UBPs) have been identified in the budding yeast Saccharomyces cerevisiae. The large number and the diversity of these enzymes suggest that each might have a specific function in regulating biological functions. Indeed, de-ubiquitylating enzymes are implicated in many processes, including free ubiquitin generation and ubiquitin removal from substrate fragments after proteasomal degradation. Also, they are responsible for the reversal of ubiquitylation to prevent degradation or to modify substrate activity (reviewed by Amerik and Hochstrasser, 2004; Soboleva and Baker, 2004; Komander et al, 2009). Ubp3, the yeast homologue of human USP10, was originally isolated by Baker et al (1992) on the basis of the ability to cleave a ubiquitin-β-galactosidase fusion protein. Ubp3 requires Bre5 as a cofactor to form an active de-ubiquitylation symmetric heterotetrameric complex (Li et al, 2005, 2007). Ubp3 has the ability to remove ubiquitin chains from target proteins before the ubiquitylated target binding to the proteasome and thus can negatively regulate proteasome-mediated degradation (Brew and Huffaker, 2002; Cohen et al, 2003a, 2003b; Baxter et al, 2005; Kraft et al, 2008; Ossareh-Nazari et al, 2010). Ubp3 has also been implicated in transcription since it interacts with Sir4 (Moazed and Johnson, 1996) as well as the general transcription factor TFIID (Auty et al, 2004). Recently, it was shown that Ubp3 acts as a transcriptional co-activator of Gal4 and Gcn4 by protecting protein Tbp1/Spt15 from degradation (Chew et al, 2010). In addition, Ubp3 acts during transcriptional elongation by reversing RNA Pol II ubiquitylation (Kvint et al, 2008).
p38-related stress-activated protein kinase (SAPK) Hog1, which is activated in response to osmostress, has a pivotal role for reprogramming the gene expression capacity of the cell (reviewed by de Nadal and Posas, 2010; Martinez-Montanes et al, 2010; Weake and Workman, 2010). Hog1 regulates different steps of the transcription process. At initiation, it modulates several unrelated transcription factors, each of them responsible for controlling the expression of a subset of osmoresponsive genes directly or in collaboration with other factors (Capaldi et al, 2008; Ni et al, 2009). Active Hog1 interacts with and directly phosphorylates some of these transcription factors, which alters their level of activity (Proft et al, 2001; de Nadal et al, 2003). Hog1 also associates with DNA at stress-responsive promoters via such specific transcription factors (Alepuz et al, 2001; Pascual-Ahuir et al, 2006; Pokholok et al, 2006; Proft et al, 2006) and functions as a platform to recruit general transcription factors, chromatin modifying activities and RNA Pol II (Proft and Struhl, 2002; Alepuz et al, 2003; de Nadal et al, 2004; Zapater et al, 2007). During elongation, Hog1 SAPK serves as a selective transcriptional elongation factor for stress-responsive genes (Pascual-Ahuir et al, 2006; Pokholok et al, 2006; Proft et al, 2006). In addition, Hog1 recruits the RSC chromatin remodelling complex at the stress-responsive genes to modify nucleosome organization (Mas et al, 2009). Here, we report crosstalk between phosphorylation and ubiquitin signalling networks. We demonstrate that Hog1 SAPK interacts with Ubp3 upon osmotic stress and regulates its de-ubiquitylase activity to determine the extent of transcription outcome. Therefore, Ubp3 and the control of its activity by the SAPK is an important factor in the modulation of gene expression in response to osmostress.
Results
Ubp3 is required for full transcriptional response upon osmostress
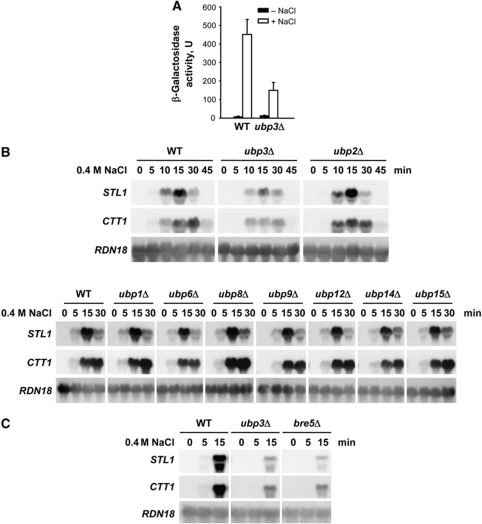
Cells rapidly change their pattern of gene expression upon osmostress. To uncover novel activities required for maximal gene expression in response to osmostress, we undertook an exhaustive genome-wide genetic screen searching for mutations that rendered cells with defective expression of an osmoresponsive gene reporter at high osmolarity. A high-throughput screen was performed by robotically pinning an ordered array of ∼800 haploid yeast deletion mutants related to transcriptional processes and containing an integrated osmostress-responsive gene reporter construct (STL1∷LacZ) onto YPD and YPD 0.4 M NaCl X-gal plates and scoring their ability to induce gene expression in response to stress (β-galactosidase production at 0.4 M NaCl). One of the mutants identified corresponded to the Ubp3 ubiquitin protease and induction of the STL1∷LacZ reporter upon stress was impaired in cells deficient in UBP3 (Figure 1A).
Figure 1.
The Ubp3–Bre5 de-ubiquitylase complex is essential for the transcriptional response upon osmostress. (A) Activation of the STL1∷LacZ reporter upon stress is impaired in cells deficient in UBP3. WT and ubp3 mutant strains carrying a multicopy STL1∷LacZ reporter construct were assayed for β-galactosidase activity (expressed in nmol/min per mg) in cells grown to mid-log phase before (filled bars) or after (open bars) a brief osmotic stress (0.4 M NaCl for 30 min). The results are presented as the mean±s.d. of three independent experiments. (B) Impaired gene expression in a ubp3 mutant strain upon osmostress. WT and the indicated mutant strains were grown to mid-log phase in rich medium and then subjected to osmotic stress (0.4 M NaCl) for the indicated length of time. Total RNA was assayed by northern blot for transcript levels of STL1, CTT1 and RDN18 (as a loading control). (C) Osmostress-impaired gene expression of cells lacking BRE5. Northern blot of WT, ubp3 and bre5 strains was done as described for (B).
We assessed whether expression of osmoresponsive genes was affected in ubp3 cells upon stress to determine the importance of Ubp3 in the regulation of gene expression. Expression of osmoresponsive genes such as STL1, CTT1 and ALD3 was reduced in a ubp3 strain as compared with the wild type (WT) (Figure 1B and data not shown). S. cerevisiae contains several UBPs and we followed gene expression in yeast strains lacking individual ubiquitin proteases to determine their role in gene activation in response to osmostress. Specific deletion of other ubiquitin proteases did not have any particular effect on gene expression upon stress (Figure 1B; Supplementary Figure S1A). These results define a specific role for Ubp3 de-ubiquitylase in modulating gene expression upon osmostress. Albeit having altered gene expression, ubp3 mutant cells were not osmosensitive (Supplementary Figure S1B). Ubp3 forms a complex with the accessory factor Bre5 (see Introduction). Expression of STL1 and CTT1 was also strongly reduced by deletion of BRE5 (Figure 1C), suggesting that Ubp3 and Bre5 function together in transcriptional activation in response to osmostress.
Ubp3 interacts with Hog1 in an osmotic stress-dependent manner
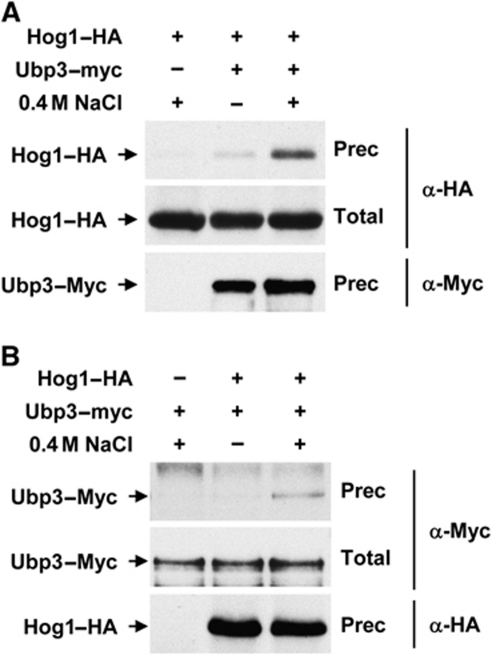
Hog1 interacts with a number of substrates to control gene expression. The possibility that Hog1 and Ubp3 physically interact was addressed by testing whether Hog1 was immunoprecipitated with Ubp3 from cell extracts. Yeast cells expressing Myc-tagged Ubp3 and HA-tagged Hog1 expressed from their genomic locus were subjected to osmostress and Ubp3 was immunoprecipitated using specific monoclonal antibodies against the Myc epitope. As shown in Figure 2A, Ubp3 was able to coprecipitate Hog1 in a stress-dependent manner. Similarly, Hog1–HA was precipitated with Ubp3 in response to osmostress (Figure 2B). Moreover, in vitro experiments using purified proteins from Escherichia coli showed that Ubp3 binds directly to Hog1 (Supplementary Figure S7A). Thus, the data show that these proteins interact upon osmostress, which provides biochemical evidence for the relationship between Ubp3 ubiquitin protease and the SAPK.
Figure 2.
In vivo binding of Hog1 and Ubp3. Hog1–HA and Ubp3–Myc-tagged proteins were expressed from the WT locus. Samples were taken before (−) or 10 min after (+) the addition of 0.4 M NaCl. (A) Ubp3–Myc was immunoprecipitated by monoclonal antibodies against Myc and the Ubp3–Myc (bottom) and Hog1–HA (top) proteins were detected by western blotting against Myc and HA epitopes, respectively. Total represents 2.5% of total input protein (middle). (B) Hog1–HA was immunoprecipitated by monoclonal antibodies against HA and proteins were detected as described for (A).
Ubp3 associates with osmostress-responsive genes upon stress
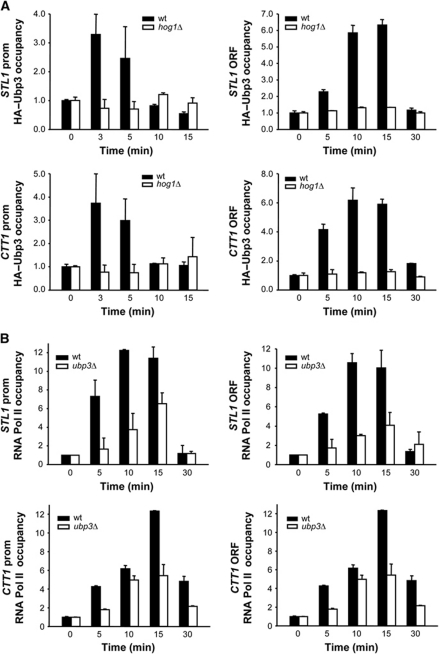
Hog1 associates with stress-responsive gene promoters and throughout the entire transcribed region of target genes in response to stress (Alepuz et al, 2001; Pokholok et al, 2006; Proft et al, 2006). We asked whether Ubp3 was recruited to osmoresponsive genes upon stress. We used chromatin immunoprecipitation (ChIP) to follow the binding of Ubp3 to promoters and ORFs of osmoresponsive genes (such as STL1 and CTT1) before and after the addition of NaCl. Chromatin from WT and from hog1 cells, both expressing integrated tagged Ubp3, was immunoprecipitated and quantified by real-time PCR. As shown in Figure 3A, Ubp3 associated specifically with osmoresponsive promoters and ORFs only in response to osmostress. However, the binding of Ubp3 to stress-responsive promoters was weaker than that observed at ORFs. Furthermore, no binding of Ubp3 was observed in a hog1 strain upon stress, indicating that recruitment of Ubp3 to chromatin in response to osmostress is dependent on Hog1 (Figure 3A). It is noteworthy that binding of Hog1 to stress-responsive promoters was not affected in a ubp3 strain (see Supplementary Figure S2). Therefore, Hog1 is required for Ubp3 recruitment at stress-responsive genes upon osmostress.
Figure 3.
In response to osmostress, Ubp3 binds to stress-dependent genes through the Hog1 SAPK and it is required for recruitment of RNA Pol II. (A) Hog1 mediates the recruitment of Ubp3 to stress-responsive genes in response to osmotic shock. WT (filled bars) and hog1 (open bars) strains carrying HA-tagged Ubp3 were grown to mid-log phase and subjected to osmostress (0.4 M NaCl for the indicated length of time). Proteins were immunoprecipitated with anti-HA monoclonal antibodies and binding to the promoter (left-hand panels) and ORF (right-hand panels) regions of STL1 and CTT1 loci was analysed. The real-time PCR results are shown as the fold induction of treated relative to non-treated (time zero) samples normalized to a telomere internal control. Data are the mean and s.d. of three independent experiments. (B) Ubp3 is required for proper RNA Pol II occupancy at stress-responsive genes. WT (filled bars) and ubp3 (open bars) strains were analysed by ChIP as described for (A) using anti-Rpb1 antibody (8WG16, Covance).
We used ChIP to assess the presence of RNA Pol II (Rpb1) at the promoter and coding region of stress-responsive genes in WT and ubp3 cells in response to osmostress. RNA Pol II was recruited at the promoters and ORFs of STL1 and CTT1 only in response to osmostress. It is noteworthy that recruitment of RNA Pol II upon osmostress was strongly reduced in ubp3 cells (Figure 3B). These results are consistent with the fact that in a ubp3 strain, CTT1 and STL1 gene expression is reduced in response to stress (Figure 1B). Taken together, these data provide evidence that Ubp3 is involved directly in the regulation of transcription of osmoresponsive genes.
Phosphorylation of Ubp3 by the Hog1 SAPK is required for the transcriptional response upon osmostress
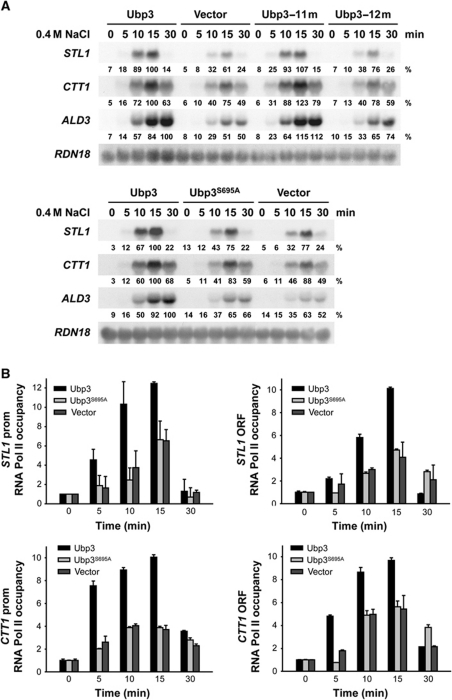
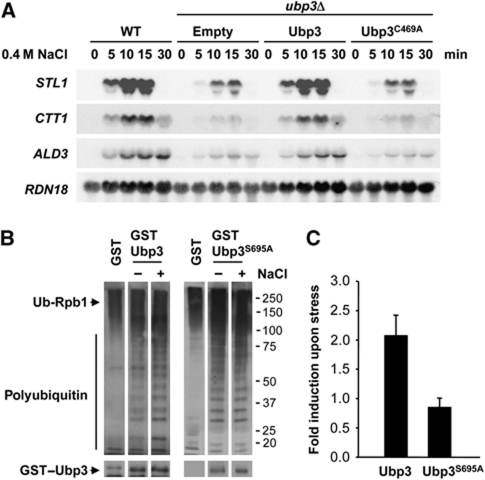
To determine if Hog1 was controlling Ubp3 function, we assessed whether Ubp3 phosphorylation was crucial for transcriptional activation upon stress. There are 12 sequences corresponding to the consensus phosphorylation site for SAPKs (Ser-Pro or Thr-Pro) in the Ubp3 protein. Initially, we created a single-copy plasmid carrying a mutant version of Ubp3 replacing all 12 putative Ser and Thr sites by SAPKs to Ala (Ubp3-12m), transformed in a ubp3 strain and assessed transcriptional activation in vivo. As shown in Figure 4A (upper panel), cells expressing Ubp3-12m were unable to restore STL1, CTT1 or ALD3 expression upon stress, when compared with the WT, indicating that phosphorylation of Ubp3 might be important for Ubp3 function in the regulation of gene expression. We reverted each of the 12 mutations to the WT to identify the specific Ubp3-phosphorylation site relevant to stress transcriptional activation. Strikingly, when a mutant carrying 11 mutated sites and containing only Ser695 (Ubp3-11m) was expressed, the transcriptional expression pattern was virtually indistinguishable from that of the WT (Figure 4A, upper panel), suggesting that the essential phosphorylation site in Ubp3 is Ser695. We then created and expressed in ubp3 cells a single point mutant at Ser695 (Ubp3S695A). This mutant displayed reduced expression of osmoresponsive genes upon osmostress similar to that of ubp3 cells (Figure 4A, lower panel). Correspondingly, cells carrying the Ubp3S695A mutation showed reduced binding of RNA Pol II to STL1 and CTT1, promoters and coding regions, compared with the WT (Figure 4B). Of note, Ubp3S695A associated with osmoresponsive promoters upon stress similar to WT. Ubp3S695A binding at coding regions was slightly reduced if compared with WT, possibly due to the reduced amount of RNA Pol II (Supplementary Figure S3). It is worth mentioning that the protein levels of WT and mutated versions of Ubp3 were expressed equally in yeast cells (see Supplementary Figure S4). The drug 6-azauracil (6-AU) reduces the cellular pools of nucleotides and is frequently used as a tool to study the genetics of transcript elongation (Mason and Struhl, 2005). Strains lacking UBP3 or expressing a version of Ubp3 with impaired catalytic activity (Ubp3C469A) are sensitive to 6-AU (Kvint et al, 2008). Expression of the unphosphorylatable allele Ubp3S695A was able to rescue the 6-AU-sensitive phenotype of a ubp3 strain (Supplementary Figure S5), indicating the functionality of the Ubp3S695A allele. Together, our data suggest that the phosphorylation of Ser695 is crucial for mediation of gene expression upon osmostress.
Figure 4.
Phosphorylation modulates Ubp3-mediated transcriptional activation in vivo. (A) Impaired gene expression in an unphosphorylatable Ubp3 version upon osmostress. The ubp3 yeast strain was transformed with the indicated plasmids, grown to mid-log phase and subjected to osmotic stress (0.4 M NaCl) for the indicated length of time. Total RNA was assayed by northern blot for transcript levels of STL1, CTT1, ALD3 and RDN18 (as a loading control). Quantification data came from the same original blot and it was normalized to the loading control. The values of maximum gene expression of the WT strain were used as 100% reference. (B) The phosphorylation site of Ubp3 is required for proper RNA Pol II occupancy at stress-responsive genes. The ubp3 strain was transformed with an empty vector as a control (grey bars), a WT Ubp3 (dark bars) and the unphosphorylatable Ubp3S695A proteins (light grey bars). Cells were subjected to osmostress (0.4 M NaCl) for the indicated length of time and binding of Rpb1 to the promoter (left-hand panels) and ORF (right-hand panels) regions of STL1 and CTT1 was analysed by ChIP as described for Figure 3B.
Ubp3 is phosphorylated upon osmostress in a Hog1-dependent manner
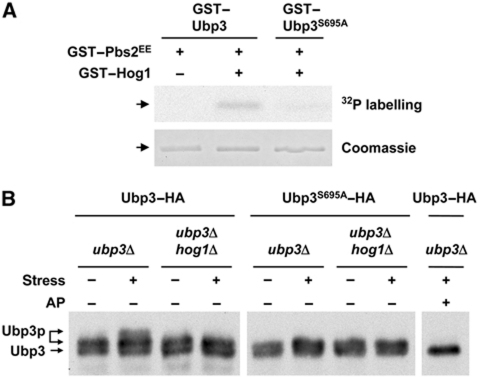
We used in vitro and in vivo phosphorylation assays to confirm that the Hog1 SAPK directly phosphorylated Ubp3 Ser695. Initially, we asked whether Hog1 phosphorylated Ubp3 directly. In vitro kinase assays were done with E. coli purified proteins in which Hog1 was activated in the presence of a constitutive MAPKK allele (Pbs2EE) (Proft et al, 2001). As shown in Figure 5A, GST-fused Ubp3 expressed from E. coli was phosphorylated only when it was incubated with activated Hog1, indicating that Ubp3 is a direct substrate for the Hog1 SAPK. Notably, phosphorylation of Ubp3 was reduced significantly in the Ubp3S695A mutant when compared with the WT.
Figure 5.
Ubp3 is phosphorylated by the Hog1 SAPK at Ser695 in vitro and in vivo. (A) In vitro phosphorylation of Ubp3 by Hog1. Recombinant GST-fused proteins were purified from E. coli. Hog1 and the constitutively activated Pbs2EE allele were incubated (when indicated) in kinase buffer containing ATP. Ubp3 or Ubp3S695A was then added in the presence of radioactive ATP. Phosphorylated proteins were resolved by SDS–PAGE and detected by autoradiography (upper panel). GST-tagged Ubp3 proteins were detected by staining with Coomassie brilliant blue (lower panel). (B) In vivo phosphorylation of Ubp3 upon osmotic stress depends on Hog1. Ubp3–HA and Ubp3S695A–HA were expressed in a low-copy vector in ubp3 and ubp3 hog1 cells. Samples were taken in mid-log phase and fixed in 20% TCA for SDS–PAGE and immunoblotting with monoclonal HA-specific antibodies (12CA5). Cells were subjected or not subjected to brief osmotic shock (0.4 M NaCl for 10 min) and treated or not treated with alkaline phosphatase (AP).
Hog1 was rapidly phosphorylated and activated when the yeast cells were exposed to osmostress. To determine whether Ubp3 Ser695 was also phosphorylated in vivo by Hog1 in response to osmostress, fully functional Ubp3–HA and Ubp3S695A–HA-tagged proteins were transformed in ubp3 and ubp3 hog1 strains and probed using specific monoclonal antibodies against the HA epitope. As shown in Figure 5B, the mobility pattern of Ubp3 in SDS–polyacrylamide gel electrophoresis (PAGE) was altered in WT cells subjected to osmostress. Protein extracts were treated with alkaline phosphatase and the mobility shift was abolished, confirming that the shift in electrophoretic mobility was the result of phosphorylation. Strikingly, mutation of Ser695 abolished the mobility shift because of the phosphorylation of Ubp3 in response to osmostress. Furthermore, phosphorylation of Ubp3 in response to osmotic stress was completely dependent on Hog1 because Ubp3–HA expressed in hog1 cells did not undergo the mobility shift observed for WT cells. It is noteworthy that Ubp3 was partially phosphorylated under basal conditions, although this phosphorylation did not depend on stress or Hog1. Taken together, these results show that in vivo, Ubp3 Ser695 is the specific phosphorylation site for Hog1 upon osmotic shock and that this modification depends on the Hog1 SAPK.
Ubp3 regulates transcription initiation and elongation in stress-responsive genes
Ubp3 is involved in the regulation of transcriptional initiation and elongation by controlling the ubiquitylation of different substrates (Kvint et al, 2008; Chew et al, 2010). We found that Ubp3 is recruited to promoters and ORFs of osmoresponsive genes (Figure 4A), suggesting that this ubiquitin protease might control initiation as well as elongation upon stress.
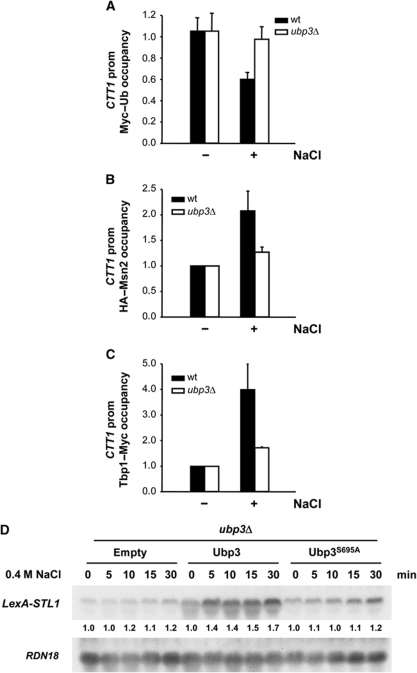
To assess the role of Ubp3 at initiation upon osmostress, we used ChIP from WT and ubp3 cells expressing a single-copy Myc-tagged ubiquitin and monitored the ubiquitylation state of proteins recruited to the CTT1 promoter upon osmostress. As shown in Figure 6A, the level of ubiquitylation was reduced upon osmostress in WT cells, whereas no change was observed in ubp3 cells. Thus, the level of ubiquitylation of proteins at the osmoresponsive promoters was altered upon osmostress in a Ubp3-dependent manner. Several unrelated transcription factors that can act independently or in combination are responsible for controlling the transcriptional initiation of osmoresponsive genes (Capaldi et al, 2008; Ni et al, 2009). Among them, the activators Msn2 and Msn4 and the Tbp1/Spt15 proteins are controlled by protein ubiquitylation (Chi et al, 2001; Chew et al, 2010). We followed the occupancy of HA-tagged Msn2 and Tbp1/Spt15 to the CTT1 promoter and found that occupancy in osmoresponsive promoters was reduced in the absence of UBP3 (Figure 6B and C), indicating that Ubp3 modulates the presence of key elements for transcription initiation at promoters upon stress. This is consistent with reduced recruitment of RNA Pol II in ubp3 cells (Figure 3B). Together, these results show that Ubp3 has a role in transcription initiation in response to stress.
Figure 6.
Ubp3 regulates transcriptional initiation and elongation of stress-responsive genes. (A) Ubp3 affects the ubiquitylation state of the recruited proteins to osmoresponsive promoters. WT (filled bars) and ubp3 (open bars) strains expressing Myc-tagged ubiquitin were grown to the mid-log phase and subjected (+) or not subjected (−) to osmostress (0.4 M NaCl for 10 min). Proteins were immunoprecipitated with anti-Myc monoclonal antibodies and binding to the promoter region of CTT1 loci was analysed. For simplicity of presentation, the level of ubiquitin binding normalized to a telomere internal control in the absence of stress was set to 1, and the level of stressed cells was expressed relative to that. The data represent the mean and s.d. of three independent experiments. (B) Msn2 occupancy of osmostress promoters depends on Ubp3. WT (filled bars) and ubp3 (open bars) strains expressing HA-tagged Msn2 under the control of the ADH1 promoter were grown to mid-log phase and subjected (+) or not subjected (−) to osmostress (0.4 M NaCl for 5 min.). Proteins were immunoprecipitated with anti-HA monoclonal antibodies and binding to the promoter region of CTT1 loci was analysed. The results are shown as the fold induction of treated relative to non-treated (time zero) samples normalized to a telomere internal control. Data represent the mean and s.d. of three independent experiments. (C) Tbp1/Spt15 occupancy to osmostress promoters is reduced in ubp3 mutant cells. WT (filled bars) and ubp3 (open bars) strains expressing Myc-tagged Tbp1/Spt15 were grown and stressed as described for (B). Proteins were immunoprecipitated with anti-Myc monoclonal antibodies and binding to the promoter region of CTT1 loci was analysed. The results are shown as in (B). (D) Ubp3 is required for STL1 mRNA production when expressed by the LexA-VP16 activator. ubp3 cells lacking endogenous STL1 and expressing an empty vector, a single copy of WT Ubp3 or the unphosphorylatable mutant Ubp3S695A were transformed with a vector carrying STL1 under the LexA promoter and with a plasmid containing the LexA-binding domain fused to the VP16 transcriptional activator. Cells were treated with 0.4 M NaCl for the indicated length of time and transcript levels of STL1 and RDN18 (as loading control) were measured. Quantification data came from the same original blot for each strain and referenced to that at 0 time point.
It is known that Ubp3 has a role in transcriptional elongation by directly controlling the ubiquitylation levels of the RNA Pol II subunit 1 (Rpb1) (McCullock et al, 2006; Bilsland et al, 2007; Kvint et al, 2008). To test whether Ubp3 also affected elongation upon osmostress, we uncoupled the processes of transcriptional initiation and elongation by driving the expression of an osmo-inducible gene with the LexA-VP16 activator (Proft et al, 2006). Specifically, we measured the RNA expression level of the STL1 osmoresponsive gene, which is driven by the LexA promoter (LexA-STL1) and the LexA-VP16 activator in the presence or in the absence of osmotic stress. In such system, the STL1 transcript levels in response to stress can be attributed exclusively to an effect on elongation and not to changes in initiation. As shown in Figure 6D, the expression of STL1 mRNA was induced in response to osmostress. It is noteworthy that the induction of mRNA accumulation was impaired in a ubp3 mutant strain, while the unphosphorylatable allele Ubp3S695A was unable to restore STL1 expression in contrast to the WT. The use of a constitutive activator such as VP16 explains the observed basal transcription in the absence of stress. Expression of the non-responsive stress gene LexA-ADH1 was not affected in ubp3 cells (Supplementary Figure S6A). Moreover, we have assessed RNA Pol II association through the ORF of the LexA-STL1 gene in ubp3 cells carrying a plasmid containing the WT Ubp3 or the Ubp3S695A allele. Cells expressing Ubp3S695A showed reduced RNA Pol II at STL1 coding region upon osmostress, compared with WT (Supplementary Figure S6B). Together, these results show that the phosphorylation of Ubp3 by Hog1 is crucial during transcriptional elongation and that the Ubp3 de-ubiquitylase has a role in transcription elongation in response to stress.
Ubp3 activity is regulated via phosphorylation by Hog1 SAPK
The observation that Ser695 in Ubp3 is required to control transcriptional initiation and elongation suggested that the ubiquitin protease activity might be regulated directly by phosphorylation. First, we assessed whether the function of Ubp3 in osmostress requires its de-ubiquitin protease activity. We created a point mutant allele in Cys469 (Cys469 to Ala469), which has been shown to abolish its catalytic activity (Cohen et al, 2003a). Cells expressing the catalytically impaired Ubp3 mutant (Ubp3C469A) did not restore gene expression upon osmostress, whereas cells carrying a single-copy plasmid containing the WT Ubp3 rescued expression of the osmoresponsive genes in a ubp3 strain (Figure 7A). It is noteworthy that the protein levels of WT and catalytically impaired Ubp3 proteins were expressed equally in yeast cells (see Supplementary Figure S4). These results indicate that the de-ubiquitylating activity of Ubp3 is essential for its function in the Hog1-mediated transcription response.
Figure 7.
Ubp3 de-ubiquitylase activity is required for adaptation to osmostress. (A) The catalytic activity of Ubp3 is essential for osmostress gene activation. ubp3 mutant cells were transformed with a centromeric plasmid containing the WT Ubp3, a catalytically inactive allele (Ubp3C469A) or an empty vector as a control, grown to mid-log phase and subjected to osmostress (0.4 M NaCl) for the indicated length of time. Total RNA was assayed by northern blot analysis for transcript levels of STL1, CTT1, ALD3 and RDN18 as a loading control. (B) Ubp3 phosphorylated by Hog1 in response to osmostress is more active in vitro. Highly purified RNA Pol II, ubiquitylated using pure ubiquitylation factors, was de-ubiquitylated with purified WT or S695A mutant Ubp3 fractions isolated from control or osmostressed yeast. The fractions were separated by SDS–PAGE and western blotted with anti-ubiquitin and anti-GST antibodies. (C) Quantification of the results in (B), adjusting for loading of GST–Ubp3. The results are shown as the fold induction of treated relative to non-treated samples normalized to GST–Ubp3 protein levels. Error bars indicate the s.d. among experiments.
We analysed Ubp3 activity on a ubiquitylated RNA Pol II substrate to assess whether the activity of Ubp3 was modulated by Hog1 phosphorylation. RNA Pol II was purified using an Rpb3-TAP-tagged strain and ubiquitylated in vitro by pure ubiquitylation factors Uba1, Ubc5 and Rsp5 (described by Somesh et al, 2005). Polyubiquitylated RNA Pol II was then incubated with purified GST-tagged Ubp3 from yeast cells treated or not treated with NaCl and samples were probed with an anti-ubiquitin antibody. RNA Pol II was de-ubiquitylated efficiently by Ubp3 from yeast as measured by the accumulation of shorter ubiquitin chains (Figure 7B). Of note, a moderate but reproducible two-fold increase of RNA Pol II de-ubiquitylation was observed when Ubp3 was purified from osmostressed cells (Figure 7C). The higher rate of Rpb1 de-ubiquitylation in response to stress suggested that Ubp3 was more active upon Hog1 phosphorylation. We used similar assays expressing the yeast Ubp3 mutant allele unable to be phosphorylated by Hog1 (GST–Ubp3S695A) to confirm this possibility. As shown in Figure 7B, the activity of Ubp3S695A did not change in response to osmostress. Of note, the activity of WT and Ubp3S695A proteins under basal conditions was very similar. Taken together, phosphorylation of Ser695 in Ubp3 by Hog1 is crucial to induce Ubp3 activity upon osmostress.
Discussion
SAPKs are key elements for intracellular signalling networks that respond and adapt to extracellular changes. Upon osmostress, the p38-related SAPK Hog1 has a deep impact on different aspects of cellular physiology, such as cell cycle (Clotet and Posas, 2007; Yaakov et al, 2009), cytoskeleton reorganization, ion homoeostasis and metabolic adjustments (Hohmann, 2002; Hohmann et al, 2007), as well as major effects on gene expression (de Nadal and Posas, 2010; Martinez-Montanes et al, 2010; Weake and Workman, 2010). Here, we provide evidence that the ubiquitin protease Ubp3 is a novel target of the Hog1 SAPK for modulating gene expression in response to extracellular input. First, Ubp3 copurifies with Hog1 in an osmotic stress-dependent manner. Second, Hog1 phosphorylates Ubp3 at serine 695 upon osmotic stress. Third, this phosphorylation is essential for the transcriptional response to osmostress because the non-phosphorylated mutant shows impaired osmoresponsive gene expression and reduced binding of RNA Pol II.
The balance of ubiquitylation is crucial to the modulation of many activities. There are several examples of control of E3 ubiquitin ligases; however, little is known about the regulation of ubiquitin proteases in response to external stimuli. Here, we show that Ubp3 phosphorylated by the Hog1 SAPK upon stress alters its activity. Ubp3p is also phosphorylated under basal conditions in alternative phosphorylation sites than Hog1, suggesting that Ubp3 can be targeted by alternative kinases in different growth conditions. Actually, analysis by mass spectrometry of purified Ubp3 from yeast cells identified several Ubp3 phosphorylation sites that were not osmo-regulated. How Ubp3 de-ubiquitylase activity is regulated is a major question. Ubp3 function is modulated by binding cofactor Bre5 (Li et al, 2005, 2007). Because the phosphorylatable Ubp3 serine 695 is far away from its binding domain with Bre5 in the tertiary structure, it is unlikely that the interaction of Ubp3 with Bre5 is altered by phosphorylation. An alternative scenario is that binding between Ubp3 and its substrates is altered upon phosphorylation of Ubp3. Again, this is an unlikely scenario because the affinity between RNA Pol II, a well-known Ubp3 substrate, and the de-ubiquitylase is not affected by osmostress or by the presence of the SAPK (Supplementary Figure S7). Interestingly, our data suggest that Ubp3 activity might be up-regulated by phosphorylation that occurs on the catalytic domain of Ubp3 and the enzyme, which is able to de-ubiquitylate ubiquitylated RNA Pol II in vitro, does it more efficiently when phosphorylated by the Hog1 SAPK. Correspondingly, the activity of the non-phosphorylated Ubp3S695A mutant does not change in response to osmostress. In addition, the complementation of the 6-AU sensitivity of the ubp3 mutant by Ubp3S695A shows that this allele is able to perform some of the functions of Ubp3 under normal growth conditions. Thus, these results suggest that Ubp3 activity is increased by Hog1 phosphorylation in response to stress.
Ubiquitin proteases are implicated in multiple processes, including the reversal of ubiquitylation, which functions to prevent degradation or to modify substrate activity. In this study, we found that Ubp3 is implied in regulating both initiation and elongation of transcription in response to stress. Ubp3 reverses the ubiquitylation of Tbp1/Spt15 during transcriptional activation (Chew et al, 2010), which opens the possibility that Ubp3 recruited by the Hog1 SAPK protects promoter-bound Tbp1/Spt15. Indeed, Tbp1/Spt15 occupancy at stress-responsive promoters is reduced in the absence of UBP3. Alternatively, the transcription factor Msn2, which acts downstream of Hog1, is known to be ubiquitylated (Chi et al, 2001). It is noteworthy that binding of Msn2 in osmoresponsive promoters is reduced in ubp3 cells. Thus, it seems likely that the basic machinery involved in transcription initiation upon osmostress is altered in UBP3-deficient cells. Correspondingly, there is less expression of a reporter construct that contains only a stress-responsive promoter fused to LacZ and there is a strong reduction of the amount of RNA Pol II recruited at stress-responsive genes upon osmostress. Binding of Ubp3 occurs in both promoters and ORFs of stress-activated genes, indicating a role of this ubiquitin protease in transcriptional initiation and in elongation in response to osmostress. Ubp3 has a role in rescuing RNA Pol II from degradation by reversing the ubiquitylation of RNA Pol II (Kvint et al, 2008). We show that Ubp3 is important for induction of stress-activated genes at coding regions, suggesting that Ubp3 also targets elongating RNA Pol II upon osmostress. Thus, although we cannot exclude other targets of Ubp3 on osmostress-mediated transcriptional activation, Ubp3 clearly has a prominent role in this process. The Hog1 SAPK interacts and travels with elongating RNA Pol II through the coding regions of osmoresponsive genes (Proft et al, 2001). Because Hog1 associates specifically with ORF of osmoresponsive genes in response to stress, it seems likely that the SAPK itself dictates the specificity of targeting RNA Pol II and, presumably also Ubp3, to osmoresponsive ORFs. Together, our data suggest that the control of Ubp3 activity by the HOG pathway in response to extracellular stimuli controls the balance of ubiquitylated proteins and this is a key factor in determining the outcome of gene expression in response to external stimuli.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strain BY4741 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) and its derivatives YEN257 (UBP3∷NatMX), YEN262 (UBP3∷NatMX HOG1∷kanMX4), YCS168 (UBP5∷kanMX4) and YCS48 (UBP2∷kanMX4) were obtained in this work. Genomic disruptions were made by long flanking homology PCR-based gene disruption. Deletion strains and strain Rpb3-TAP were obtained from the EUROSCARF collection. The tagging strains obtained in this study are YEN255 (UBP3–6HA∷HIS3), YEN256 (UBP3–6HA∷HIS3 HOG1∷kanMX4), YCS9 (HA–UBP3), YCS175 (HA–UBP3S695A), YCS41 (HA–UBP3 HOG1∷kanMX4), YMZ33 (HOG1–6HA∷HIS3), YCS33 (UBP3–18Myc∷HIS3 HOG1–6HA∷kanMX4), YCS22 (UBP3–18Myc∷HIS3), YEN254 (HOG1–HA∷HIS3 UBP3∷kanMX4), YCS98 (TBP1–18Myc∷HIS) and YCS101 (UBP3∷kanMX4 TBP1–18Myc∷HIS). Tagging of genomic ORFs with HA or Myc epitopes was done with a PCR-based strategy.
The plasmids used in this study were pGEX4T-Hog1 and pGEX4T-Pbs2EE (PBS2 with Ser514-Glu and Thr518-Glu mutations), which are described by Bilsland-Marchesan et al (2000). UBP3 (PEN253), UBP3-S695A (UBP3 with Ser695-Ala mutation) (PCS81) and RSP5 (PCS31) were cloned into pGEX4T. UBP3 (PEN298), UBP3-C469A (UBP3 with Cys469-Ala mutation) (PEN301), UBP3-S695A (PCS71), UBP3-11mut (UBP3 with S20A, T25A, S74A, S146A, S171A, S339A, S341A, S343A, T376A, S400A and S646A mutations) (PCS68) and UBP3-12mut (UBP3 with S20A, T25A, S74A, S146A, S171A, S339A, S341A, S343A, T376A, S400A, S646A and S695A mutations) (PCS69) were cloned into pRS415. For HA tagging, UBP3 (PCS70) and UBP3-S695A (PCS75) were cloned into the yeast vector pRS415-HAx6∷HIS3. UBP3 (PEN285) and UBP3-S695A (PCS84) were fused to glutathione S-transferase (GST) under the TEF1 promoter (pRS426TEG1). YCpLac111 PADH1MSN2–3HA was a gift from Dr C Schuller (Gorner et al, 1998). PADH1MSN2–3HA was cloned into pRS416 (PCS95). CEN-based vector PTEF19Myc-Ubiquitin; LEU2+ (PGR140) was a gift from Dr M Peter. The STL1∷LacZ fusion was constructed by insertion of the −825/+4 nucleotides of the STL1 gene into the multicopy plasmid YEp358R containing the reporter PEN9. The LexA-binding domain fused to the VP16 transcriptional activator and the LexA-STL1 (PGM45) and LexA-ADH1 (PGM66) vectors are described by Proft et al (2006).
Screening and β-galactosidase assays
An ordered array of 800 MATa viable haploid yeast gene deletion mutants obtained from EUROSCARF (Saccharomyces Gene Deletion Project) was crossed with MATα STL1∷LacZ and replica pinned in duplicate onto YPD and YPD with 0.4 M NaCl, 5-bromo-4-chloro-3-indolyl-β,D-galactopyranoside (X-gal) plates. The screen was done with an automated system in which yeast cells are transferred using a Biomek FX robot and the 384 floating-pin replicator (Biomek FX HDR 384-Pin plate) as described (Tong et al, 2001). The screen was done twice and the plates were incubated at 30°C for 2 days before scoring. X-gal (solid medium contains selective medium buffered with 2-(N-morpholino) ethanesulfonic acid (Mes)) at pH 7 plus 0.1 mg/ml X-gal. For liquid β-galactosidase assays, exponentially growing cells were subjected to osmotic stress (0.4 M NaCl for 30 min), permeabilized by treatment with ethanol/toluene and β-galactosidase measured as described (Proft et al, 2001).
Northern blot analysis
Yeast strains were grown to mid-log phase in rich medium or in minimal medium for plasmid selection at an absorbance at 660 nm (A660) of 0.6–0.9 and then subjected to osmotic shock (0.4 M NaCl) for the length of time indicated. Total RNA and expression of specific genes were probed using radiolabelled PCR fragments containing the entire ORF of STL1 (1.7 kbp), CTT1 (1.7 kbp), ALD3 (1.5 kbp) and the non-coding exon of RDN18 (1.8 kbp). Signals were quantified with a Fujifilm BAS-5000 phosphorimager and autoradiographs were obtained on Kodak Biomax XAR films (Sigma-Aldrich).
In vitro kinase assays
The GST fusion proteins encoding the WT or the Ubp3 mutant, Hog1 and Pbs2EE were expressed in E. coli DH5α and purified using glutathione-Sepharose beads (GE Healthcare) in STET buffer (10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA pH 8.0, 5% Triton X-100, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, 2 μg/ml leupeptin, 2 g/ml of pepstatin). A 1-μg sample of Hog1 was activated with 0.5 μg of Pbs2EE in the presence of kinase buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM DTT) and 50 μM ATP. After 20 min at 30°C, 2 μg of WT or mutated Ubp3 protein was added to the previous mixture together with [γ-32P]ATP (0.1 μCi/μl) and incubated for 15 min at 30°C. The reaction was terminated by the addition of 5 × SDS loading buffer. Labelled proteins were resolved by SDS/PAGE, stained, dried and detected by autoradiography.
In vitro coprecipitation assay
GST or GST–Ubp3 was expressed in E. coli DH5α cells and purified using Glutathione-Sepharose beads (GE Healthcare) as described above. His-tagged Hog1 was expressed in E. coli BL21 strain and 150 μg of total extract was incubated with TALON metal affinity resin (Clontech) in binding buffer (20 mM Tris pH 8, 150 mM NaCl plus anti-proteases inhibitors) for 1 h. A total of 0.5 μg of GST-purified proteins was then added for 3 h. Beads were extensively washed (50 mM Tris pH 8, 150 mM NaCl, 0.1% Triton X-100) and samples were boiled with 2 × SDS loading buffer. Bound proteins were analysed by western blotting using anti-GST (GE Healthcare) and anti-Hog1 (yC-20, Santa Cruz Biotechnology) antibodies.
In vivo Ubp3 phosphorylation assays
ubp3 or ubp3 hog1 cells were transformed with HA-tagged WT Ubp3 protein or the indicated mutant form and grown in minimal medium for plasmid selection. Cells at the mid-log phase were subjected to osmotic shock (0.4 M NaCl for 5 min) and 1 ml of culture was fixed with 25% trichloroacetic acid (TCA), pelleted and washed with 100 μl of 12.5% TCA. Following the addition of glass beads, pellets were lysed by vortex mixing for 5 min, washed in 1 ml of iced acetone and dried at 55°C for 15 min. Protein precipitates were resuspended in 15 μl of 1% SDS, 100 mM Tris–HCl pH 8.0, 1 mM EDTA. Finally, 35 μl of AP buffer (100 mM Tris–HCl pH 9.5, 1 mM EDTA) was added to all samples and, when indicated, 0.2 μl of alkaline phosphatase (20 U/μl; Roche) for 30 min at 37°C. Samples were separated by SDS–PAGE and Ubp3 protein was detected by immunoblotting using monoclonal anti-HA 12CA5.
In vivo coprecipitation assay
Ubp3 and/or Hog1-tagged cells in mid-log phase (50 ml) were treated with 0.4 M NaCl for 15 min or left untreated and then collected by brief centrifugation at 4°C. Pellets were harvested with glass beads in the FastPrep-24 (Qbiogene, 30 s at speed 5) in lysis buffer A (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 15 mM EDTA, 15 mM EGTA, 2 mM DTT, 0.1% Triton X-100, 1 mM PMSF, 1 mM benzamidine, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 25 mM β-glycerophosphate, 1 mM sodium pyrophosphate, 10 mM sodium fluoride, 100 μM sodium orthovanadate), and lysates were clarified by centrifugation and quantified by the Bradford assay (Bio-Rad Laboratories). As a control, 25 μg of whole-cell extract was blotted with anti-HA 12CA5 or anti-Myc 9E10 to check the expression levels of the tagged proteins (total, shown in Figure 2). Alternatively, 1 mg of cleared supernatant was subjected to immunoprecipitation with anti-Rpb1 (8WG16, Covance), anti-HA 12CA5 or anti-Myc 9E10 overnight at 4°C. Antibodies were recovered with protein G and were resolved by SDS–PAGE.
ChIP assays
ChIP was performed as described (Zapater et al, 2007). Yeast cultures were grown to early log phase (A660 0.6–0.9) before samples of the culture were exposed to osmotic stress (0.4 M NaCl) for the length of time specified in the legends of Figures 3 and 4. For crosslinking, yeast cells were treated with 1% formaldehyde for 20 min at room temperature. Antibodies used in this study were mouse polyclonal anti-Rpb1 (8WG16, Covance), monoclonal anti-Myc 9E10 and anti-HA 12CA5. Quantitative PCR analysis of stress genes and constitutively expressed genes used the following primers with locations indicated by the distance from the respective ATG initiation codon: STL1 promoter, −682/−582; STL1 ORF, +1475/+1575; CTT1 promoter, −432/−302 (−275/+125 for TBP1 binding); CTT1 ORF, +736/+836; and TEL (telomeric region on the right arm of chromosome VI). Experiments were done on three independent chromatin preparations and quantitative PCR analysis was done in real time using an Applied Biosystems 7700 sequence detector. Immunoprecipitation efficiency was calculated in triplicate by dividing the amount of PCR product in the immunoprecipitated sample by that in the TEL sequence control. The binding data are presented as fold induction with respect to the non-treated condition.
Recombinant protein purification
The ubp3 strain was transformed with the pRS426TEG1 Ubp3 or Ubp3-S695A mutant and the mid-log phase culture (2 l) was subjected or not to a brief osmotic shock (0.4 M NaCl for 15 min). GST pull-down experiments were done using glutathione-Sepharose 4B (GE Healthcare) in buffer A. Bound protein was eluted by 10 mM reduced glutathione (GSH), 50 mM Tris–HCl pH 9.5, 2 mM DTT. Eluted proteins were concentrated (Amicon Ultra centrifugal filters, 30 kDa cutoff membrane; Millipore) and resuspended in ubiquitination buffer (25 mM Tris pH 8, 125 mM NaCl, 2 mM MgCl2, 50 μM DTT). The TAP-tagged Rpb3 strain was used for the purification of RNA Pol II as described (Puig et al, 2001).
De-ubiquitylation of Rpb1 in vitro
For de-ubiquitylation reactions in vitro, 250–300 ng of RNA Pol II was incubated with 40 ng of Uba1 (Boston Biochem), 40 ng of Ubc5H5c (Boston Biochem), 90 ng of GST–Rsp5, 2 μg of ubiquitin (Boston Biochem), 2 mM ATP and 40 ng of purified Ubp3 proteins in ubiquitylation buffer for 90 min at 30°C. Reactions were stopped by the addition of SDS–PAGE loading buffer. The samples were fractionated in BisTris 4–12% polyacrylamide gel or acetate 7% polyacrylamide gel (NuPAGE Invitrogen) then blotted and probed with an anti-ubiquitin antibody (Sigma) and an anti-GST antibody (Sigma).
Supplementary Material
Acknowledgments
We are grateful for technical assistance from L Subirana and S Obejas. We thank Dr Jesper Q Svejstrup, Dr Gustav Ammerer and Dr Bernat Crosas for helpful comments. MN is the recipient of an FIS (Spanish Government) fellowship and FP is the recipient of an ICREA Acadèmia (Generalitat de Catalunya). This work was supported by Fundación Marcelino Botín (FMB) and grants from the Spanish Ministry of Science and Innovation (BFU2008-00530 to EN and BIO2009-07762 to FP) and the FP7 (UNICELLSYS) framework programme.
Author contributions: CS, MN, CK, FP and EN performed the experiments and analysed the results. CS, MN, CK, MP, EN and FP participated in the experimental design. CS, FP, MP and EN wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F (2003) Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J 22: 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G (2001) Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7: 767–777 [DOI] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207 [DOI] [PubMed] [Google Scholar]
- Auty R, Steen H, Myers LC, Persinger J, Bartholomew B, Gygi SP, Buratowski S (2004) Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J Biol Chem 279: 49973–49981 [DOI] [PubMed] [Google Scholar]
- Baker RT, Tobias JW, Varshavsky A (1992) Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem 267: 23364–23375 [PubMed] [Google Scholar]
- Baxter BK, Abeliovich H, Zhang X, Stirling AG, Burlingame AL, Goldfarb DS (2005) Atg19p ubiquitination and the cytoplasm to vacuole trafficking pathway in yeast. J Biol Chem 280: 39067–39076 [DOI] [PubMed] [Google Scholar]
- Bilsland E, Hult M, Bell SD, Sunnerhagen P, Downs JA (2007) The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair (Amst) 6: 1471–1484 [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F (2000) Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol 20: 3887–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew CT, Huffaker TC (2002) The yeast ubiquitin protease, Ubp3p, promotes protein stability. Genetics 162: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK (2008) Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet 40: 1300–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew BS, Siew WL, Xiao B, Lehming N (2010) Transcriptional activation requires protection of the TATA-binding protein Tbp1 by the ubiquitin-specific protease Ubp3. Biochem J 431: 391–399 [DOI] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ (2001) Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J, Posas F (2007) Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol 428: 63–76 [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C (2003a) Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5: 661–667 [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Dargemont C (2003b) Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J Biol Chem 278: 51989–51992 [DOI] [PubMed] [Google Scholar]
- de Nadal E, Casadome L, Posas F (2003) Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23: 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F (2010) Multilayered control of gene expression by stress-activated protein kinases. EMBO J 29: 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374 [DOI] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C (1998) Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 12: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B (2007) Yeast osmoregulation. Methods Enzymol 428: 29–45 [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2008) Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell 30: 498–506 [DOI] [PubMed] [Google Scholar]
- Li K, Ossareh-Nazari B, Liu X, Dargemont C, Marmorstein R (2007) Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J Mol Biol 372: 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zhao K, Ossareh-Nazari B, Da G, Dargemont C, Marmorstein R (2005) Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor. J Biol Chem 280: 29176–29185 [DOI] [PubMed] [Google Scholar]
- Martinez-Montanes F, Pascual-Ahuir A, Proft M (2010) Toward a genomic view of the gene expression program regulated by osmostress in yeast. OMICS 14: 619–627 [DOI] [PubMed] [Google Scholar]
- Mas G, de NE, Dechant R, Rodriguez de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F (2009) Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J 28: 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- McCullock S, Kinard T, McCullough L, Formosa T (2006) blm3-1 is an allele of UBP3, a ubiquitin protease that appears to act during transcription of damaged DNA. J Mol Biol 363: 660–672 [DOI] [PubMed] [Google Scholar]
- Moazed D, Johnson D (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86: 667–677 [DOI] [PubMed] [Google Scholar]
- Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, Gerstein MB, Snyder M (2009) Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev 23: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van DA, Dargemont C (2010) Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11: 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Struhl K, Proft M (2006) Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods 40: 272–278 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313: 533–536 [DOI] [PubMed] [Google Scholar]
- Proft M, Mas G, de NE, Vendrell A, Noriega N, Struhl K, Posas F (2006) The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23: 241–250 [DOI] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de NE, Arino J, Serrano R, Posas F (2001) Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K (2002) Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva TA, Baker RT (2004) Deubiquitinating enzymes: their functions and substrate specificity. Curr Protein Pept Sci 5: 191–200 [DOI] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2010) Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11: 426–437 [DOI] [PubMed] [Google Scholar]
- Yaakov G, Duch A, Garcia-Rubio M, Clotet J, Jimenez J, Aguilera A, Posas F (2009) The stress-activated protein kinase Hog1 mediates S phase delay in response to osmostress. Mol Biol Cell 20: 3572–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E (2007) Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27: 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.