Abstract
Piwi proteins function in an RNAi-like pathway that silences transposons. Piwi-associated RNAs, also known as piRNAs, act as a guide to identify Piwi targets. The tudor domain-containing protein Tdrd1 has been linked to this pathway but its function has thus far remained unclear. We show that zebrafish Tdrd1 is required for efficient Piwi-pathway activity and proper nuage formation. Furthermore, we find that Tdrd1 binds both zebrafish Piwi proteins, Ziwi and Zili, and reveals sequence specificity in the interaction between Tdrd1 tudor domains and symmetrically dimethylated arginines (sDMAs) in Zili. Finally, we show that Tdrd1 complexes contain piRNAs and RNA molecules that are longer than piRNAs. We name these longer transcripts Tdrd1-associated transcripts (TATs). TATs likely represent cleaved Piwi pathway targets and may serve as piRNA biogenesis intermediates. Altogether, our data suggest that Tdrd1 acts as a molecular scaffold for Piwi proteins, bound through specific tudor domain–sDMA interactions, piRNAs and piRNA targets.
Keywords: piRNA, piRNA biogenesis, Piwi, Tdrd1, zebrafish
Introduction
In gonads of animals, the activity of a specialized RNAi-like pathway is essential for reproduction. This pathway is characterized by a subclass of the Argonaute protein family known as the Piwi subfamily (Carmell et al, 2002), which is required for efficient transposon silencing, germ-cell differentiation and meiosis (Cox et al, 1998, 2000; Aravin et al, 2001; Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004; Brennecke et al, 2007; Carmell et al, 2007; Houwing et al, 2007, 2008; Ghildiyal and Zamore, 2009). The Piwi pathway operates in distinct subcellular compartments, with nuage—an electron dense, germ cell-specific perinuclear structure, often associated with mitochondria—functioning as a major site of Piwi protein activity (Harris and Macdonald, 2001; Lim and Kai, 2007; Pane et al, 2007; Aravin et al, 2009; Li et al, 2009; Lim et al, 2009; Malone et al, 2009; Shoji et al, 2009; Patil and Kai, 2010). Nuage is known to associate with clusters of nuclear pores in Caenorhabditis elegans (Pitt et al, 2000; Updike et al, 2011), suggesting that nuage may have a function in the trafficking of molecules between the nucleus and the cytoplasm. Interestingly, nuage-like structures named Yb bodies have been described in Drosophila somatic nurse cells surrounding the oocyte (Szakmary et al, 2009). These Yb bodies have been implicated in Piwi-pathway activity as well, in particular, in the loading of Drosophila Piwi with piRNA and its subsequent transition into the nucleus (Olivieri et al, 2010; Saito et al, 2010; Qi et al, 2011).
Small RNAs named piRNAs, originally named rasiRNAs in Drosophila, guide Piwi proteins to their targets (Aravin et al, 2006; Girard et al, 2006; Lau et al, 2006; Saito et al, 2006). Two types of piRNAs have been identified: primary and secondary (Li et al, 2009; Malone et al, 2009). Primary piRNAs are characterized by uracil at their 5′-end. They are likely derived from long single-stranded transcripts through cleavage by an unknown nuclease, after which the 5′-end of the 3′-cleavage product is bound by a Piwi protein followed by 3′-end processing (Vagin et al, 2006; Horwich et al, 2007; Houwing et al, 2007; Kirino and Mourelatos, 2007; Saito et al, 2007). Primary piRNAs can trigger the generation of secondary piRNAs through cleavage of single-stranded transcripts by the Piwi proteins themselves (Brennecke et al, 2007; Gunawardane et al, 2007; Li et al, 2009). This step defines the 5′-end of a novel piRNA, which is then again presumably followed by 3′-end trimming. Consequently, secondary piRNAs have opposite polarity compared with primary piRNAs and are characterized by adenosine at position 10. In turn, if complementary target RNA molecules are present, secondary piRNAs may trigger generation of additional primary-like piRNAs. Because of the reciprocal interactions involved, this mode of piRNA biogenesis has been dubbed ‘ping-pong’ (Brennecke et al, 2007).
Additional components of these mechanisms have been identified. Many of these interactors (Tdrd proteins) contain so-called tudor domains (Chen et al, 2009; Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009; Vasileva et al, 2009; Wang et al, 2009; Kirino et al, 2009a; Patil and Kai, 2010; Liu et al, 2011; Yabuta et al, 2011). It was shown for tudor domains of the p100 and Tdrd3 proteins that they can bind peptides containing symmetrically dimethylated arginine (sDMA) (Cote and Richard, 2005) and subsequent structural studies have revealed the physical basis behind these interactions (Friberg et al, 2009; Liu et al, 2010a, 2010b). Indeed, sDMAs have been identified in the N-terminal tails of Piwi proteins and in other Piwi-pathway components like Vasa (Kirino et al, 2010), and these mediate interaction with tudor domain-containing proteins (Chen et al, 2009; Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009; Kirino et al, 2009a). Interestingly, multiple sDMA sites in Piwi proteins have been described and many Tdrd proteins contain multiple tudor domains that potentially can bind multiple peptides simultaneously (Liu et al, 2010a). It is thus far, however, not clear whether tudor–Piwi interactions display sequence specificity.
Many tudor domain-containing proteins contain other domains as well. For example, Tdrd1 contains an additional MYND domain. This is a zinc-binding domain that is most likely involved in protein–protein interactions (Matthews et al, 2009). Other tudor domain-containing proteins contain additional domains like, for example, RNA helicase (Tdrd9) or RNA-binding domains (TdrKH). The molecular functions of these domains in the context of the Piwi pathway are currently not known.
Mutants in tdrd genes have been described in both mice and Drosophila. In mice, mutants lacking the tudor domain-containing proteins Tdrd1, Tdrd5, Tdrd6 and Tdrd9 result in male sterility (Chuma et al, 2006; Shoji et al, 2009; Vasileva et al, 2009; Wang et al, 2009; Yabuta et al, 2011). In addition, relatively weak effects on piRNA production in tdrd1 and tdrd9 mutants have been reported (Reuter et al, 2009; Shoji et al, 2009; Vagin et al, 2009), demonstrating a functional role of these proteins in the Piwi pathway. In Drosophila, mutations in Piwi pathway-associated tdrd genes have defects in fertility, transposon silencing and subcellular localization of Piwi-pathway components, again revealing functional relevance of these Piwi-interacting proteins (Aravin et al, 2001; Kennerdell et al, 2002; Nishida et al, 2009; Kirino et al, 2009b; Patil and Kai, 2010; Liu et al, 2011). Nishida et al (2009) reported on long RNA molecules associated with the Drosophila multi-tudor domain protein Tud and suggested that these may be piRNA precursors. However, a detailed molecular analysis supporting this notion was not provided. Taken together, the molecular functions of Tdrd proteins in Piwi pathways have not yet been described.
Zebrafish germ cells express two Piwi proteins, Ziwi and Zili (Houwing et al, 2007, 2008). Ziwi predominantly binds primary(-like) piRNAs, which are antisense with respect to targeted transposons, whereas Zili mostly associates with sense, secondary piRNAs. Both Piwi proteins are required for normal germ-cell development. Here, we demonstrate that zebrafish Tdrd1 is an interactor of both Ziwi and Zili. It is required for nuage formation and tdrd1 mutation results in mild delocalization of Zili. Other Piwi pathway-related defects such as transposon desilencing and germ-cell defects are observed as well in tdrd1 mutants, consistent with results obtained in mice. We then go on to reveal sequence specificity in the tudor domain–Zili interaction and identify arginines at specific sites in the N-terminal tail of Zili that, when symmetrically dimethylated, bind Tdrd1. Finally, we describe Tdrd1-associated RNA molecules that display all the characteristics of piRNA biogenesis intermediates. Taken together, we demonstrate that sequence specificity is a relevant factor in assessing Tdrd–Piwi interactions and that Tdrd1 is found together with multiple RNA and protein components of the Piwi pathway. This, for the first time, provides direct support for the idea that Tdrd proteins may function as molecular scaffolds in this pathway.
Results
Zili-interacting proteins
We performed mass spectrometry on Zili immunoprecipitates (IPs; Supplementary Table S1). Apart from Zili, we mainly enriched for tudor domain-containing proteins, in line with previously published findings regarding Piwi protein interactors (Chen et al, 2009; Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009; Wang et al, 2009; Kirino et al, 2009b; Siomi et al, 2010). We focused our analysis on Tdrd1, a strongly conserved protein with four tudor domains and an N-terminal MYND domain that has been implicated in nuage formation, germ-cell differentiation and Piwi-pathway activity in mice (Chuma et al, 2006; Hosokawa et al, 2007; Reuter et al, 2009; Vagin et al, 2009; Wang et al, 2009). We raised an antibody against zebrafish Tdrd1 that recognizes a gonad-specific protein of roughly the expected molecular weight of 130 kDa (Figure 1A). Immunostainings using this antibody reveal germ cell-specific signals in wild-type but not in tdrd1 mutant animals (see below). Mass spectrometry on Tdrd1 IPs identified both Ziwi and Zili but also Vasa and the tudor domain-containing proteins Tdrd4 and Tdrd5 (Supplementary Table S2).
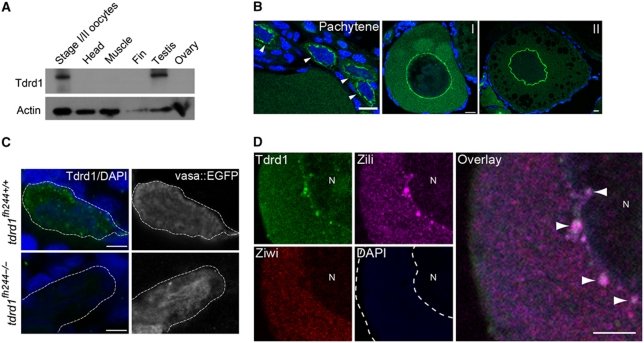
Figure 1.
Tdrd1 expression in zebrafish. (A) Western blot analysis of Tdrd1 in diverse tissues. (B) Immunohistochemistry on ovary. Arrowheads indicate pachytene stage oogonia. Scale bars are 10μm. I, II: oocyte stage. (C) Immunohistochemistry on PGCs in 4 days post fertilization (dpf) embryos. Vas::EGFP expression marks PGCs. Green: Tdrd1. Blue: DAPI. Scale bars are 10 μm. Dashed line outlines a single PGC. (D) Co-staining of Ziwi, Zili and Tdrd1 in stage I oocyte. N: nucleus. Arrowheads: sites of colocalization. Scale bar is 5 μm. Green: Tdrd1. Red: Ziwi. Magenta: Zili.
Tdrd1 expression
Using in situ hybridization, RT–PCR, immunohistochemistry and western blotting analysis (Figure 1A; Supplementary Figure S1A–C), we can detect Tdrd1 expression in testis and immature oocytes. On western blot, the Tdrd1 signal in total ovary, mostly consisting of large, mature oocytes is often not detectable (Figure 1A), most likely due to the fact that maturing oocytes accumulate high amounts of yolk proteins, diluting the Tdrd1 signal. Consistent with this, we only detect strong Tdrd1 signals in oocytes of stage II and younger in immunohistochemistry experiments (Figure 1B). During development, Tdrd1 protein starts to be detectable in the primordial germ cells (PGCs) at 4 days post fertilization (4dpf; Figure 1C; Supplementary Figure S1D). At this stage, Tdrd1 is cytoplasmic, displaying fine granular distribution (Figure 1B) distinct from the nuclear localization of Zili at 4dpf (Houwing et al, 2008). At 5dpf, Tdrd1 starts to assemble into larger perinuclear granules (Supplementary Figure S1D), much like we have described for Zili (Houwing et al, 2008). We do not detect strong Tdrd1 staining within nuclei of PGCs (Supplementary Figure S2). At later time points, Tdrd1 and Zili display very similar subcellular localization, most prominently in nuage-like structures in early stages of both oocyte and sperm development (Figure 1B and D; Supplementary Figure S1C). Using immunogold electron microscopy (EM), we confirmed that indeed both Zili and Tdrd1 localize to electron-dense structures (Supplementary Figures S1E and S3A). This localization profile closely resembles that described for mouse Tdrd1 (Chuma et al, 2006; Aravin et al, 2009).
Loss of Tdrd1 affects nuage and Zili localization
We obtained a tdrd1 mutant allele, fh224, harbouring a premature stop codon truncating Tdrd1 between the MYND and the first tudor domain. Tdrd1 immunohistochemistry on tdrd1 mutant cells shows no detectable staining (Figure 1C), indicating that fh224 represents a strong loss-of-function allele, perhaps a null allele. Tdrd1 homozygous mutants are viable but lose their germ cells during development (Figure 2A; Supplementary Figure S3C), similar to the ziwi and zili loss-of-function phenotypes (Houwing et al, 2007, 2008). However, in contrast to ziwi and zili mutants, tdrd1 mutants contain differentiating germ cells at 3 weeks of age (Figure 2A; Supplementary Figure S3C). This indicates that the tdrd1 mutant phenotype is relatively weak. The zebrafish genome (version Zv9) only encodes one tdrd1 gene making redundancy through gene duplication unlikely. Although we cannot exclude the possibility that other Tdrd proteins can partially substitute for Tdrd1, we prefer the interpretation that Tdrd1 function is not absolutely required for Piwi-pathway activity but rather potentiates it.
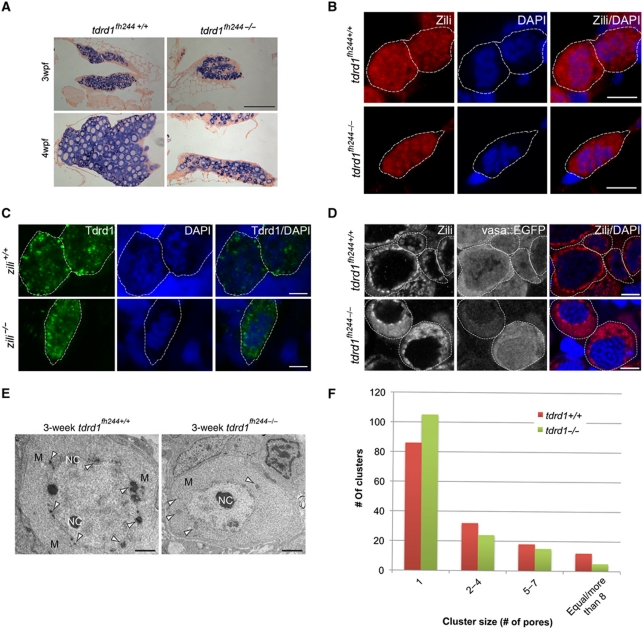
Figure 2.
tdrd1 mutant analysis. (A) Vasa ISH (purple) on gonads from wild-type and tdrd1 mutant females at 3 and 4 weeks post fertilization (wpf). Scale bar: 100 μm. (B) Zili immunohistochemistry (red) in wild-type and tdrd1 PGCs (5dpf). Blue: DAPI. Dashed line outlines individual PGCs. (C) Tdrd1 immunohistochemistry (green) in wild-type and zili mutant PGCs (5dpf). Blue: DAPI. Dashed line outlines individual PGCs. (D) Zili staining on wild-type and tdrd1 oocytes at 3wpf. Scale bars B–D: 5 μm. (E) Transmission electron microscopy on 3wpf ovaries from wild-type and tdrd1 mutant animals. Mitochondria (M), nuage (arrowheads) and nucleoli (NC) are labelled. Scale bar: 1 μm. (F) Cluster sizes of nuclear pores were determined from TEM images of wild-type (n=31 oocytes; 406 pores) and tdrd1 mutant (n=15 oocytes; 313 pores) ovaries at 3wpf. Y axis shows how often each cluster size was found. X axis shows cluster size (P<0.001; χ2-test).
Next, we determined the subcellular localization of Tdrd1 and Zili in each other's absence. At 5 days of development, Zili remains nuclear in tdrd1 mutant PGCs (Figure 2B). Reversely, Tdrd1 remains present in fine cytoplasmic granules in the absence of Zili (Figure 2C). At later stages, when most Zili is found in nuage, we observe that Zili immunostaining becomes less focused around the nucleus and spreads into the cytoplasm in tdrd1 mutants (Figure 2D; Supplementary Figure S3D and E). However, perinuclear Zili-positive granules are still present. To follow-up on this, we also used transmission EM to analyse nuage in wild-type and tdrd1 mutant oocytes in more detail. Generally, tdrd1 mutant oocytes contain only few nuage-like patches compared with wild type, and these patches are much less electron dense (Figure 2E; Supplementary Figure S4). These structures are no longer tightly associated with mitochondria but their overall association with the nuclear envelope is less affected (Supplementary Figure S5). Immunogold labelling showed that Zili is no longer detectable in the structures that persist in the absence of Tdrd1 (Supplementary Figure S3B). This implies that the perinuclear localization of Piwi proteins observed in immunofluorescence experiments does not reflect nuage as defined by EM and that other, non-Tdrd1-mediated mechanisms are in place to steer Piwi proteins towards perinuclear regions.
We noted in the TEM experiments that nuage in oocytes associates with clusters of nuclear pores. This resembles findings in C. elegans, where P-granules have been found associated with nuclear pore clusters (Pitt et al, 2000; Updike et al, 2011). Interestingly, in tdrd1 mutant oocytes we find significantly more solitary pores and nuclear pore aggregates tend to consist of fewer pores (Figure 2F), suggesting that Tdrd1 is a factor driving the clustering of nuclear pores in zebrafish oocytes.
Tdrd1 affects piRNA populations
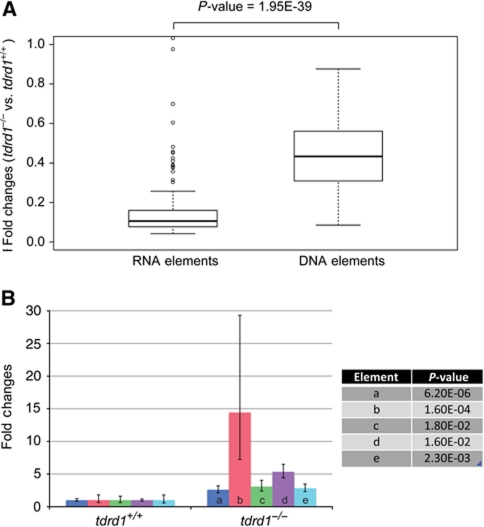
Mouse Tdrd1 has been described to have effects on ping-pong efficiency (Vagin et al, 2009) and to filter RNA molecules entering the ping-pong cycle (Reuter et al, 2009). We probed the effect of zebrafish Tdrd1 on piRNAs to see if these effects can be seen in the zebrafish. We made small RNA libraries from 3-week-old wild-type and mutant gonads, a stage in which tdrd1 mutant gonad morphology is relatively normal, and analysed these using deep sequencing (Supplementary Table S3). First, assuming miRNA levels remain unchanged, a mild decrease in piRNA abundance was observed (Supplementary Figure S6A). In contrast to what has been described in mice (Reuter et al, 2009), we do not observe an increase in gene-derived or other RNA species-derived piRNAs (Supplementary Table S3). We then looked at transposon-derived piRNAs specifically, and noted an especially strong effect of tdrd1 mutation on retrotransposon piRNAs (Figure 3A). This may derive from the fact that retrotransposons, while representing a minority of all transposable elements in the zebrafish genome, produce most piRNAs (Houwing et al, 2007), indicating that the majority of Piwi activity is directed against retrotransposons. For some elements, we observed a change in strand bias of the piRNAs (Supplementary Figure S6B), and in those cases the class that is mostly bound by Ziwi (Houwing et al, 2008) was reduced. Next, we quantified transcript levels of five individual transposons using qRT–PCR on RNA isolated from FACS purified germ cells. This revealed that loss of Tdrd1 results in a mild upregulation of transcripts from all elements tested (Figure 3B). Interestingly, the element with the strongest reduction in piRNA levels (I-1) also shows the strongest increase in transcript levels (Supplementary Figure S6C). Finally, we analysed Piwi-cleavage activity by looking at the so-called ping-pong signature, the characteristic signal of 10-nucleotide sense–antisense 5′-overlaps between individual piRNAs. We observed a very low ping-pong signature in tdrd1 mutants (Supplementary Figure S6D). However, when we correct for the amount of overlapping reads in total, a wild-type signature is evident (Supplementary Figure S6E). Taken together, these data suggest that in absence of Tdrd1, Piwi proteins can still cleave their targets, but do so less frequently.
Figure 3.
Effect of tdrd1 mutation on transposons and piRNAs. (A) Boxplot displaying the effects of tdrd1 mutation on piRNA levels derived from transposable elements that transpose either via an RNA or a DNA intermediate. Ratios are based on all transposon-derived reads that were normalized between the wild-type and tdrd1 libraries. (B) qRT–PCR analysis on five individual transposon types, using RNA from FACS isolated germ cells from wild-type and tdrd1 mutant gonads. a–e: EnSpmN1 (DNA or class II), I-1 (RNA or class I), GypsyDR2 (RNA), Ngaro1 (RNA), Polinton1 (DNA). P-values (two-tailed t-test) relate to the differences between wild-type and mutant samples for each element.
Tdrd1–Zili binding characteristics
Co-IP experiments using zebrafish Tdrd1-specific antibodies in testis lysate confirmed that Tdrd1 binds, directly or indirectly, to Zili (Figure 4A). In mouse, Tdrd1 has been suggested to bind exclusively to Mili (Reuter et al, 2009). However, other studies suggested Tdrd1 is not so specific, binding Mili, Miwi and Miwi2 (Chen et al, 2009; Vagin et al, 2009). In zebrafish, Tdrd1 clearly interacts with both Zili and Ziwi (Figure 4A), although little Tdrd1 is detected in Ziwi IPs. The latter may be due to Tdrd1 shielding the antibody epitope on Ziwi or to an abundance of non-Tdrd1-bound Ziwi.
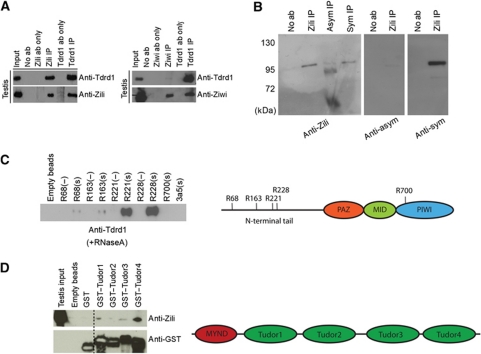
Figure 4.
Specific arginine methylation on Zili mediates Tdrd1 binding. (A) Western blots on IP material from testis extracts using antibodies directed against Ziwi, Zili and Tdrd1. (B) IP-western blot analysis on testis extracts using antibodies recognizing Zili, and symmetrically and asymmetrically dimethylated arginines. (C) Pull-down experiments using biotinylated peptides spanning Zili residues R68, R163, R221, R228 and R700 (indicated in the schematic on the right). A histone H3-derived peptide containing sDMA (3a5) was also used as a control. Pull-down sample was analysed through western blotting using Tdrd1 antibodies. (−): unmethylated arginine; (s): sDMA. (D) Tdrd1 tudor domains (as indicated in the schematic on the right) were individually expressed as GST-fusion proteins and used as baits in testis extracts. Bound material was probed for Zili and GST using western blotting.
Binding of tudor domains to Piwi proteins has been shown to require sDMA residues in the N-terminal tails of the Piwi proteins (Nishida et al, 2009; Reuter et al, 2009; Vagin et al, 2009; Kirino et al, 2009a). Using arginine methylation-specific antibodies, we found that also Zili contains this type of arginine modification (Figure 4B). Using mass spectrometry on immunopurified Zili protein, we identified four methylation sites in the N-terminal region of Zili (R68, 163, 221 and 228) (Supplementary Figure S7 and data not shown). We tested whether these four sites can mediate Tdrd1 binding by performing pull-down experiments in testis extracts, using synthetic biotinylated peptides as bait. We found that Tdrd1 binds most strongly to peptides containing dimethylated R221 or R228 (Figure 4C). Peptides with unmethylated arginines (Figure 4C) and of unrelated sequence but containing sDMA (Figure 4C) do not interact with Tdrd1. These interactions are RNA independent as they are performed in the presence of RNaseA. We also tested which of the Tdrd1 tudor domains can bind Zili through GST-pull-down experiments using testis extracts. This revealed that the most C-terminal tudor domain of Tdrd1 most strongly associates with Zili (Figure 4D). This differs from previously published observations on Tdrd1–Mili interactions in a heterologous setting (293T cells). These implicated the N-terminal half of Tdrd1 in Mili binding (Wang et al, 2009).
Tdrd1-associated piRNAs
Next, we analysed the RNA molecules associated with Tdrd1 in zebrafish testis. As can be expected, we found RNA species that resemble mature piRNAs in Tdrd1 IPs (Figure 5A). We size selected these small RNAs and sequenced them. As comparison we sequenced testis-derived piRNAs from Ziwi and Zili IPs and piRNAs from total testis extracts. The observed length profiles from Ziwi and Zili IPs correlate well with our previously published results (Houwing et al, 2008). Interestingly, the Tdrd1-bound piRNA length profile is intermediate between the Ziwi and Zili profiles (Figure 5B), suggesting roughly equimolar amounts of Ziwi- and Zili-bound piRNAs in Tdrd1 IPs. We further analysed transposon-derived piRNA sequences in terms of their strand biases. As described before (Houwing et al, 2008) for many repeats (287 out of 866) Ziwi and Zili enrich for antisense and sense piRNAs, respectively, but we also detect a set of transposons in which this bias is inverted (97 out of 866). The Tdrd1-bound piRNAs from these two sets of transposons (producing 50% of all transposon piRNAs) display intermediate strand biases, consistent with a mix of Ziwi- and Zili-bound piRNAs (Figure 5C) in Tdrd1 IPs.
Figure 5.
Tdrd1-associated piRNAs. (A) Radiolabelled RNA from the indicated IPs was separated on a denaturing gel, followed by autoradiography. Bracket indicates piRNAs. The solid bar indicates Tdrd1-associated transcripts (TATs). Arrows indicate rRNA species. (B) Length distributions of piRNAs from Ziwi, Zili and Tdrd1 IPs. (C) Heatmap showing strand biases of piRNAs from individual transposons in the indicated libraries. Map is relative from left to right and is sorted on strand ratios of Tdrd1-associated piRNAs. Low Z-score (red) indicates relative enrichment for antisense reads. (D) Scatterplot displaying the coverage of different transposon types (open circles) by piRNAs from a Zili IP (X axis) and a Tdrd1 IP (Y axis). Correlation coefficient R=0.93.
One potential function for Tdrd1 could be to select specific transposon transcripts for entry into the piRNA amplification cycle. Such a function has been proposed for mouse Tdrd1 (Reuter et al, 2009). We, therefore, checked whether the piRNAs retrieved by Tdrd1 IP display the same transposon profile as we find in the total Ziwi and Zili IPs (Figure 5D). This revealed a very strong correlation between piRNA obtained from both Tdrd1 and Ziwi IPs and Tdrd1 and Zili IPs, strongly suggesting that Tdrd1 does not have a significant role in discrimination between different transposon substrates.
Tdrd1-associated transcripts
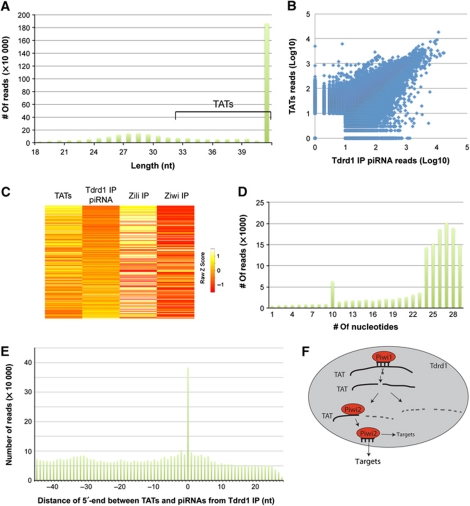
We also cloned RNA species from Tdrd1 IP samples without size selection or enzymatic pre-treatments. Interestingly, this resulted in a majority of reads that were significantly longer than mature piRNAs, even longer than 46 nucleotides, our sequencing limit (Figure 6A). We named these Tdrd1-associated transcripts (TATs). To probe their length distribution further, we subcloned the library and fully sequenced the inserts of a random set of 235 clones (Supplementary Table S4). This revealed that TATs display a wide size distribution, ranging from mature piRNA length up to over 100 bases long. This experiment also revealed that the more discrete RNA fragments visible in Figure 5A are derived from rRNA molecules and most likely represent aspecific background. TATs likely lack 2′-OMe modifications since we cloned almost exclusively TATs from the Tdrd1 IP while piRNAs are present at apparently higher levels (Figure 5A). Since we do not treat the RNA with either phosphatases or kinases, the majority of TATs likely contain 5′P and 3′OH termini. Strikingly, we find that loci producing TATs tend to also produce piRNAs, and abundances of both RNA types correlate well (Figure 6B; Supplementary Figure S8A). Overall, TATs, like the Tdrd1-associated piRNAs, display a roughly 1:1 ratio of sense versus antisense polarities (Supplementary Figure S8B). Interestingly, at the level of individual transposons we find that the strand ratios of Tdrd1-associated piRNAs anticorrelate with the strand ratios of TATs (Figure 6C). In other words, whenever the piRNA population tends to be of antisense polarity the TAT population displays a tendency for sense polarity and vice versa. This suggests a guide–target relationship between these two RNA populations, with piRNAs being the guides and TATs being the targets.
Figure 6.
Tdrd1-associated transcripts (TATs). (A) Length distribution of repeat-derived sequences isolated from a Tdrd1 IP. Significant tailing of the piRNA peak indicates the presence of RNA species longer than mature piRNAs. We name these TATs. TATs of 46 bases and longer are used in further analysis. (B) Scatterplot displaying read counts for TATs (Y axis) and Tdrd1-associated piRNAs (X axis) for individual loci. A locus is defined as a region of the genome covered by at least 10 piRNAs and/or long RNAs, allowing for 100 base-pair gaps. Correlation coefficient R=0.72. (C) Heatmap showing strand biases of piRNAs from individual transposons in the indicated libraries. Map is relative from left to right and is sorted on strand ratios of Tdrd1-associated piRNAs. Low Z-score (red) indicates relative enrichment for antisense reads. (D) Number of transposon-derived Tdrd1-associated piRNAs (Y axis) that have a 5′-end overlap (X axis) with TATs from the opposite strand. (E) Bar diagram displaying the distance between the 5′-ends of TATs and Tdrd1-associated piRNAs from the same genomic strands. X axis reflects the most frequent distance of 5′-ends between any TAT species compared with all overlapping piRNAs. Y axis displays how often each distance is observed. (F) A schematic model displaying how TATs (solid black lines) may interact with Piwi proteins within a Tdrd1 environment. TATs may be base paired to piRNAs, may be cleavage products on their way to further destruction or may be bound by Piwi proteins to be processed further into mature piRNAs or any combination thereof.
To follow-up on this observation, we asked whether TATs display a ping-pong signal when they are compared with piRNAs derived from the opposite strand. An over-representation of 10 bases overlap between TATs and piRNAs was indeed observed (Figure 6D). Many overlaps also covered the complete piRNA length, possibly reflecting uncleaved targets or the fact that our libraries are not saturated (Supplementary Table S3). We also probed the distances between the 5′-ends of TATs and piRNAs derived from the same strand. This revealed that most TATs contain a 5′-end that is identical to that of a mature piRNA (Figure 6E). Together, these data strongly suggest that TATs represent cleaved piRNA target fragments that may be converted into mature piRNAs through 3′-end trimming activities.
Discussion
Collectively, our data provide a model for the molecular function of the vertebrate Piwi-pathway component Tdrd1. We show that Tdrd1 associates with RNA and protein components of the Piwi pathway and that in absence of Tdrd1 the Piwi pathway operates with a much lower efficiency. Loss of Tdrd1 results in partial piRNA target activation and defects in germ-cell development. In addition, we identify RNA transcripts that display hallmarks of piRNA targets and/or cleavage products. Below we further discuss both protein and RNA interactions of Tdrd1.
Tdrd1 and nuage formation
We describe strong defects in nuage formation in tdrd1 mutants. Electron-dense structures are largely absent from tdrd1 mutant germ cells and those that are present do not associate with mitochondria. Yet, like has been observed in mice (Vagin et al, 2009), perinuclear foci of Piwi proteins can still be observed in tdrd1 mutants. How can this conundrum be resolved? One potential explanation for these findings may be found in the fact that in Drosophila the Piwi protein Aubergine, but also Vasa and the arginine-protein-methyltransferase-5 cofactor Valois genetically act upstream of the tudor domain protein Tud (Ephrussi and Lehmann, 1992; Cavey et al, 2005; Thomson and Lasko, 2005). This could imply that Piwi-pathway activity triggers the onset of nuage formation and that Tdrd proteins are subsequently required to boost this process into formation of ‘mature’ nuage as we can detect it in wild-type germ cells. The Piwi-positive granules detected in the absence of Tdrd1 may thus represent immature nuage with insufficient RNA and protein content to allow detection in TEM experiments.
Specificity in the interactions between Tdrd1 and Zili
Tdrd1 has four tudor domains. We find that the most C-terminal domain can retrieve Zili from testis extracts the best. This differs from a previous study on mouse Tdrd1 in which the N-terminal part of Tdrd1 was implicated in Mili binding (Wang et al, 2009). This may be due to differences in the mouse and zebrafish Piwi pathways or to the fact that Wang et al investigated the Mili–Tdrd1 interaction in 293T cells, which could lead to sDMA formation on Mili residues that mediate binding to the N-terminal Tdrd1 tudor domains. Related to this hypothesis, we find that the Tdrd1–Zili interaction indeed displays sequence specificity. We identify two closely positioned sDMA sites in Zili that associate strongly with Tdrd1, while other sDMA containing peptides display a much weaker interaction. Furthermore, we have not been able to detect another tudor domain-containing protein, Tdrd6, coming down in our peptide pull-down assays (Huang and Ketting, unpublished data). With the caveat that our experiments can report indirect interactions, these results suggest that sequence specificity in the interactions between tudor domains and sDMA residues in Piwi proteins may be a general phenomenon. In case of Tdrd1–Zili interactions, the most C-terminal tudor domain of Tdrd1 may interact with sDMAs at positions 221 and 228 in Zili. Due to the poor sequence conservation in the N-terminal regions of Piwi proteins, it is difficult to directly relate these residues to homologous arginines in Zili homologues. The available structures of tudor domains in complex with sDMA containing peptides (Liu et al, 2010a, 2010b) have revealed rather broad groves in which peptides are bound, suggesting that the specificity here observed may not result from sequence-specific interactions between the residues surrounding the sDMA and the tudor domain. Rather it may be regional electrostatic and hydrophobic interactions that may determine how well a specific peptide will interact with a specific tudor domain.
How do TATs interact with Tdrd1?
We have identified long RNA molecules associated with Tdrd1 (TATs) and their characteristics suggest they may be piRNA targets. Such a target RNA association function for tudor domain proteins may be conserved, as for example Drosophila tudor has been found associated with long RNA transcripts, although these have not been analysed in detail (Nishida et al, 2009). How would Tdrd1 interact with these transcripts? Since the domain structure of Tdrd1 suggests it is not itself an RNA-binding protein, TATs are most likely associated with Tdrd1 indirectly. This may imply that TATs are associated with Tdrd1 through base-pairing interactions with mature, Piwi-bound piRNAs. This is consistent with our observation that overall the strand biases of TATs are opposite to those of mature piRNAs. Alternatively, TATs may be bound by Piwi proteins directly (Figure 6F). This would most likely involve binding of the 5′-end of TATs by the phosphate-binding pocket in the Piwi protein MID domain. In this context, TATs would be piRNA precursors, requiring 3′-end trimming and 2′-O methylation to become a mature piRNA. Further biochemical characterization of TATs in diverse mutant backgrounds will be required to address these issues. Unfortunately, the extremely small size of zebrafish gonads at the time when ziwi, zili and tdrd1 mutants still contain relatively healthy germ cells prevents detailed biochemical analysis of TATs in these mutant backgrounds. Perhaps such experiments will be feasible in other model systems, especially the recently developed cell culture-based systems (Kawaoka et al, 2009; Saito et al, 2009) that recapitulate specific aspects of the Piwi pathway.
TATs: piRNA biogenesis intermediates?
As already eluded to above, TATs may be considered as piRNA precursors for the following reasons. First, by definition, TATs are found in association with Tdrd1, suggesting they may be localized in nuage, a site where ping-pong amplification of piRNAs is believed to occur (Harris and Macdonald, 2001; Lim and Kai, 2007; Pane et al, 2007; Aravin et al, 2009; Li et al, 2009; Lim et al, 2009; Malone et al, 2009; Shoji et al, 2009; Patil and Kai, 2010). Direct testing of this hypothesis will require high-resolution RNA in situ hybridization techniques. Second, TAT 5′-ends match those of mature piRNAs and they display a ping-pong signal with mature piRNAs, suggesting that TATs are generated through piRNA-mediated cleavage. It will be of interest to see how TATs relate to other potential piRNA intermediates (named piR-ILs) described by Saito et al (2010). There are some differences between the two potential piRNA precursor populations: TATs are quite heterogeneous in size and likely have 5′-phosphate ends that match to 5′-ends of mature piRNAs, while piR-ILs lack 5′-phosphate groups and their 5′-ends do not align well with mature piRNAs. Since piR-ILs have been detected in the context of the primary piRNA biogenesis pathway an explanation for the differences between piR-ILs and TATs could be that piR-ILs are specific to primary piRNA biogenesis and that the TATs described here relate specifically to the ping-pong mechanism.
Since we find a clear anticorrelation between the strand ratios of TATs and Tdrd1-associated piRNAs, it is clear that not all TATs can be converted into piRNAs, at least not into Tdrd1-associated piRNAs. We currently entertain two mutually non-exclusive explanations for this observation. First, as suggested above, the majority of TATs may be quickly degraded, preventing further participation in the ping-pong cycle. Second, piRNAs generated in the context of Tdrd1 may not all remain associated with Tdrd1 (Figure 6F). The latter option is consistent with significant, non-Tdrd1-associated Piwi pools that have been observed in many studies. For example, in the adult ovary and testis of zebrafish most Ziwi is not found in perinuclear nuage but rather in a diffuse distribution in the cytoplasm (Houwing et al, 2008). In Xenopus oocytes, Xiwi has been found associated with the Balbiani body (Lau et al, 2009), a structure that we do not find to contain Tdrd1 in the zebrafish. Finally, in mice Miwi2 associates with piP bodies that lack Tdrd1 (Aravin et al, 2009). These observations strongly suggest that Tdrd1 is not the only scaffold for the Piwi pathway and that additional sites of piRNA accumulation or activity await identification and characterization. Obviously, other tudor domain-containing proteins represent good candidates for making such platforms.
Materials and methods
Detailed procedures can be found in the Supplementary Material.
Zebrafish strains and genetics
Zebrafish were kept under standard conditions. The tdrd1fh244+/− mutant allele zebrafish was generated by TILLING and obtained from Zebrafish International Resource Center (ZIRC). Animals carrying tdrd1fh244+/− were out crossed against wild-type fish (TL) or vas::EGFP transgenic fish (Krovel and Olsen, 2004) and subsequently in-crossed to obtain tdrd1fh244−/− offspring. Experiments involving zebrafish were done with permission from the Animal Experiments Committee of the KNAW.
Tdrd1 antibody
Tdrd1 antibodies were raised in rabbits against the synthetic peptide H2N-RRPATGPSSLSPRGPC-CONH2. Antisera were affinity purified (Eurogentec).
Peptide pull down
Peptide pull downs were done with biotinylated synthetic peptides at a concentration of 2 mM. Peptides were incubated with 500 μl lysate from three testes for 2 h at 4 °C while rotating. After washing, beads were eluted using SDS–PAGE loading buffer.
Sequencing
RNA was poly(A)-tailed using poly(A) polymerase followed by ligation of a RNA adaptor to the 5′-phosphate of the small RNAs. First-strand cDNA synthesis was performed using an oligo(dT)-linker primer and M-MLV-RNase H-reverse transcriptase. After amplification, the cDNA was sent for sequencing on an Illumina/Solexa platform. The four-base adaptor sequences were trimmed from generated data and reads were mapped to the Zv8 zebrafish genome assembly. Sequencing data has been submitted to GEO, accession number GSE29418.
Supplementary Material
Acknowledgments
We thank Willi Salvenmoser, Jeroen Korving and Marc van de Wetering for technical assistance; Richard van Schaik and Marc Timmers for assistance in peptide synthesis; Erma van Asselt and Mark Reijnen for fish care, members of the Ketting lab for discussions and suggestions; and the Hubrecht Imaging Center (HIC) for supporting microscopy experiments. TILLING was done with the support of NIH grant HG002995 to CBM. This work was further supported by grants to RFK: a VIDI (864.05.010) and ECHO (700.57.006) grant from the Netherlands Organization for Scientific Research, an ERC Starting Grant from the Ideas Program of the European Union Seventh Framework Program (202819) and the European Union Sixth Framework Program Integrated Project SIROCCO (LSHG-CT-2006-037900).
Author contributions: HH performed experiments, analysed data and wrote the manuscript. LJTK performed experiments and analysed data. SH performed experiments and analysed data. AM performed experiments. EB analysed data. SR performed experiments and analysed data. PL analysed data. HV contributed and analysed reagents. BMB contributed reagents. JK analysed data. BWD performed experiments. CBM contributed reagents and funding. SG performed experiments. RFK designed experiments, provided funding, analysed data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Cavey M, Hijal S, Zhang X, Suter B (2005) Drosophila valois encodes a divergent WD protein that is required for Vasa localization and Oskar protein accumulation. Development 132: 459–468 [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, Pawson T (2009) Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N (2006) Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA 103: 15894–15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Richard S (2005) Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280: 28476–28483 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by Piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H (2000) Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H (2002) Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R (1992) Induction of germ cell formation by oskar. Nature 358: 387–392 [DOI] [PubMed] [Google Scholar]
- Friberg A, Corsini L, Mourao A, Sattler M (2009) Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J Mol Biol 387: 921–934 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM (2001) Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N (2007) Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol 301: 38–52 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF (2008) Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S (2009) The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW (2002) RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev 16: 1884–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z (2009a) Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z (2007) Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z (2010) Arginine methylation of vasa protein is conserved across phyla. J Biol Chem 285: 8148–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z (2009b) Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovel AV, Olsen LC (2004) Sexual dimorphic expression pattern of a splice variant of zebrafish vasa during gonadal development. Dev Biol 271: 190–197 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD (2009) Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J 28: 2945–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T (2009) piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM (2010a) Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, Min J (2010b) Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA 107: 18398–18403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H (2011) PAPI, a novel Tudor-domain protein, complexes with AGO3, Me31b and Tral in the nuage to silence transposition. Development 138: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Bhati M, Lehtomaki E, Mansfield RE, Cubeddu L, Mackay JP (2009) It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr Pharm Des 15: 3681–3696 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC (2009) Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T (2010) Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20: 724–730 [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR (2000) P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219: 315–333 [DOI] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H (2011) The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem 286: 3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS (2009) Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC (2010) Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S (2009) The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H (2010) How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 24: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H (2009) The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lasko P (2005) Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res 15: 281–291 [DOI] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S (2011) P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol 192: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA (2009) Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R (2009) Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H (2009) Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M (2011) TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol 192: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.