Abstract
TorsinA is a membrane-associated enzyme in the endoplasmic reticulum (ER) lumen that is mutated in DYT1 dystonia. How it remains in the ER has been unclear. We report that a hydrophobic N-terminal domain (NTD) directs static retention of torsinA within the ER by excluding it from ER exit sites, as has been previously reported for short transmembrane domains (TMDs). We show that despite the NTD's physicochemical similarity to TMDs, it does not traverse the membrane, defining torsinA as a lumenal monotopic membrane protein and requiring a new paradigm to explain retention. ER retention and membrane association are perturbed by a subset of nonconservative mutations to the NTD, suggesting that a helical structure with defined orientation in the membrane is required. TorsinA preferentially enriches in ER sheets, as might be expected for a lumenal monotopic membrane protein. We propose that the principle of membrane-based protein sorting extends to monotopic membrane proteins, and identify other proteins including the monotopic lumenal enzyme cyclooxygenase 1 (prostaglandin H synthase 1) that share this mechanism of retention with torsinA.
Keywords: endoplasmic reticulum, membranes, protein sorting
Introduction
Early-onset (DYT1) torsion dystonia is a neurological movement disorder (Fahn, 1988) caused by a glutamic acid deletion (ΔE) in the catalytic domain of torsinA (Ozelius et al, 1997). TorsinA is an AAA+ ATPase of the endoplasmic reticulum (ER) and contiguous nuclear envelope (NE). The specific cellular functions ascribed to torsinA vary widely despite the fact that it has been a decade since the protein was first described and linked to dystonia (Breakefield et al, 2008). Based on its membership in the AAA+ family of ATPases (Ozelius et al, 1997; Hanson and Whiteheart, 2005), it is likely that torsinA disassembles or changes the conformation of a protein or protein complex in the ER or NE. The ΔE mutation is thought to compromise this function (Dang et al, 2005; Goodchild et al, 2005).
TorsinA is targeted to the ER lumen by an N-terminal signal peptide. Analyses of torsinA's subcellular localization, processing, and glycosylation show that the signal peptide is cleaved and the mature protein resides in the lumen of the ER (Kustedjo et al, 2000; Hewett et al, 2003; Liu et al, 2003), where it is a stable protein (Gordon and Gonzalez-Alegre, 2008; Giles et al, 2009). TorsinA's binding partners include the transmembrane proteins LULL1 in the ER and LAP1 in the NE (Goodchild and Dauer, 2005; Naismith et al, 2009). Abnormalities in NE structure (Naismith et al, 2004; Goodchild et al, 2005) and effects on NE-localized LINC complex proteins (Nery et al, 2008; Vander Heyden et al, 2009) when torsinA levels are perturbed suggest an important function for this enzyme specifically at the NE. Other studies point to additional functions elsewhere in the ER (Hewett et al, 2008; Bragg et al, 2011).
The steady-state localization of torsinA in the ER and NE demands that it escapes forward flux out of the ER into the secretory pathway. This is a significant issue for all ER resident proteins, and the underlying mechanisms are what define the composition of the ER. Proteins intended for efficient secretion are concentrated in nascent COPII vesicles at ER exit sites (ERES) by specific interactions with subunits of the COPII coat or, in the case of lumenal proteins, with transmembrane receptors that in turn interact with COPII subunits (Dancourt and Barlowe, 2010). However, proteins without specific export signals also leave the ER in COPII vesicles at a rate referred to as bulk flow. For soluble proteins in the ER lumen, quantitative measurements indicate that bulk flow empties the equivalent of half of the lumenal volume every 40 min (Thor et al, 2009). Proteins that leave the ER by bulk flow may return to the ER in COPI vesicles if they are recognized by a recycling receptor. The prototype for this is the KDEL receptor that recycles lumenal proteins with the tetrapeptide KDEL at their C-terminus (Lewis et al, 1990). For membrane proteins, there is less quantitative information about the rate of bulk flow. There are some membrane proteins that never leave the ER and are statically retained by their transmembrane domain (TMD) (Rayner and Pelham, 1997; Yang et al, 1997; Duvet et al, 1998; Ronchi et al, 2008; Daubner et al, 2010; Hsieh et al, 2010). This retention is attributed to preferential partitioning of short TMDs into the thinner and less ordered membrane of the ER, and the best characterized of these TMDs has been shown to partition differentially among subdomains of the ER (Ronchi et al, 2008). However, that there is a bulk flow of membrane proteins is clear from the need for the TMD-specific recycling receptor, Rer1 (Sato et al, 2003), and from studies showing that small increases in the hydrophobicity of ER-retained TMDs allow their escape from the ER (Rayner and Pelham, 1997; Yang et al, 1997; Ronchi et al, 2008; Hsieh et al, 2010).
How does torsinA achieve its localization to the ER? An early study indicated that a hydrophobic domain at the N terminus of the protein was required for ER localization of human torsinA in heterologous cells (Liu et al, 2003). However, the mechanism underlying localization was unclear as neither the N-terminal domain (NTD) nor other regions of the torsinA sequence contain any canonical targeting motifs. A subsequent proposal was that torsinA remains in the ER lumen because of protein–protein interactions with other resident proteins (Callan et al, 2007). The facts that the N terminus is not involved in interactions with known abundant binding partners including LULL1 in the ER and LAP1 in the NE (Vander Heyden et al, 2009), and that even highly overexpressed torsinA remains in the ER (Kustedjo et al, 2000), suggest that this is unlikely. These discrepancies raise the question of whether a previously unknown mechanism might be responsible for keeping torsinA and similar proteins in the ER. In this study, we provide evidence that torsinA's NTD is a monotopic membrane-associating domain that is directly responsible for static retention in the ER. Further, we identify other membrane proteins that appear to behave similarly, providing new insight into protein sorting in the early secretory pathway.
Results
TorsinA is a life-long ER resident
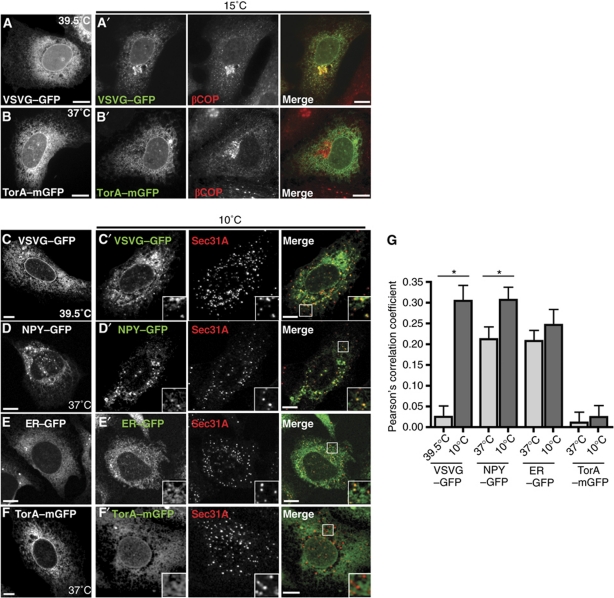
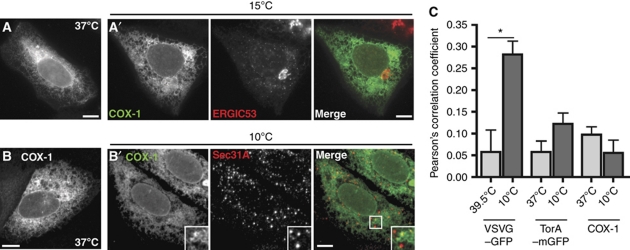
While all transmembrane and secreted proteins are transiently found in the ER, proteins that reside in the ER either never leave and are said to be statically retained or are retrieved from the ER-to-Golgi intermediate compartment (ERGIC) by COPI-mediated retrograde transport. Both are commonly referred to as ER retention mechanisms. To determine how torsinA remains in the ER, we incubated cells expressing mGFP-tagged torsinA or cargo proteins at temperatures that selectively block different trafficking steps. At 15°C, COPII-mediated transport from the ER proceeds but recycling and forward transport from the ERGIC do not, causing proteins that leave the ER to accumulate in the ERGIC (Kuismanen and Saraste, 1989) while statically retained ER proteins are unchanged. At 10°C, COPII components assemble together with cargo at ERES but do not generate free vesicles (Tartakoff, 1986; Lotti et al, 1996; Mezzacasa and Helenius, 2002). VSVG-(ts045)–mGFP, a plasma membrane-targeted transmembrane protein that directly interacts with COPII, accumulated dramatically with the COPI component βCOP in the ERGIC (Figure 1A). TorsinA–mGFP, in contrast, remained distributed throughout the ER and was absent from the ERGIC after incubation at 15°C (Figure 1B). After incubation at 10°C, GFP fusions of VSVG-(ts045), the secreted neuropeptide Y (NPY), and ER–GFP appeared at ERES marked by the COPII component Sec31A, but torsinA–mGFP did not (Figure 1C′–F′). Quantitating colocalization confirmed that VSVG-(ts045), NPY, and ER–GFP are present in ERES at 10°C, while torsinA is not (Figure 1G). Notably, the large increase in colocalization of VSVG-(ts045) with Sec31A at 10 versus 39.5°C is because VSVG-(ts045) is a temperature-sensitive mutant protein that misfolds at 39.5°C and interacts with folding chaperones that keep it away from ERES; once folded at a permissive temperature, its diacidic COPII interacting motif promotes rapid accumulation at ERES (Mezzacasa and Helenius, 2002). The fact that torsinA avoids ERES to an extent comparable to misfolded VSVG, while ER–GFP is readily detectable in ERES despite the lack of a forward transport signal, suggests specific exclusion of torsinA from ERES. This exclusion is similar to that previously described for short TMD-containing proteins (Ronchi et al, 2008). Finally, overexpressed torsinA–mGFP is efficiently retained in the ER at physiological temperature (Figure 1B and F) and is completely EndoH sensitive (Kustedjo et al, 2000), implying that torsinA rarely leaves the ER. Altogether, these data indicate that torsinA is statically retained in the ER.
Figure 1.
TorsinA is a static ER resident and is excluded from ERES. (A, B) Epifluorescence microscopy of U2OS cells expressing VSVG-(ts045)–GFP (at 39.5°C) or torsinA–mGFP (at 37°C). (A′, B′) Costaining with βCOP after 2 h incubation at 15°C. (C–F) Confocal microscopy of cells expressing VSVG-(ts045)–GFP (at 39.5°C), NPY–GFP, ER–GFP, or torsinA–mGFP (at 37°C). (C′–F′) Costaining with Sec31A after 2 h incubation at 10°C. Scale bars, 10 μm. (G) Quantification of colocalization of the indicated GFP-tagged proteins with Sec31A. N>20 cells for each condition. Bars indicate standard error of the mean. *Significant difference between conditions (P<0.05).
An NTD directs ER retention
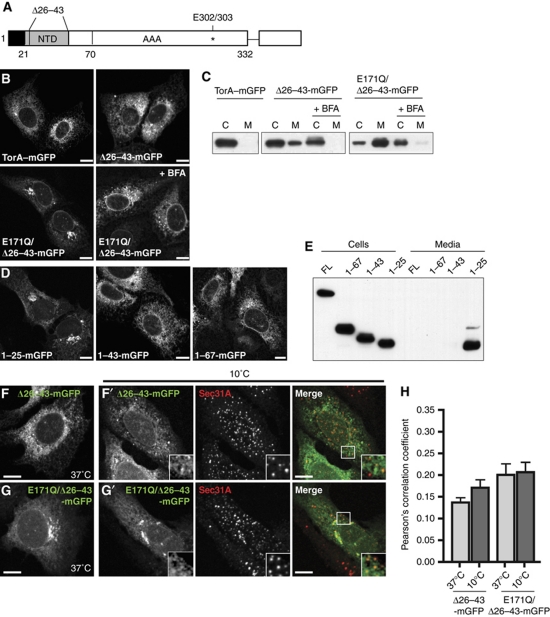
Mature torsinA consists of a hydrophobic NTD, a short linker region, and the AAA+ domain (Figure 2A). Building on an earlier report showing that deleting the NTD led to secretion of human torsinA from Drosophila S2 cells (Liu et al, 2003), we deleted residues 26–43 from human torsinA and found that the mutant protein appeared in the Golgi (Figure 2B) and in the cell medium (Figure 2C) when expressed in human U2OS cells. Both changes were blocked by brefeldin A (BFA), indicating that without its NTD, torsinA traffics through the classical secretory pathway. Deletion of the N terminus allows torsinA to access ERES to an extent comparable to the lumenal marker ER–GFP (Figure 2F–H, compare to Figure 1E and G), consistent with truncated torsinA exiting the ER by bulk flow transport.
Figure 2.
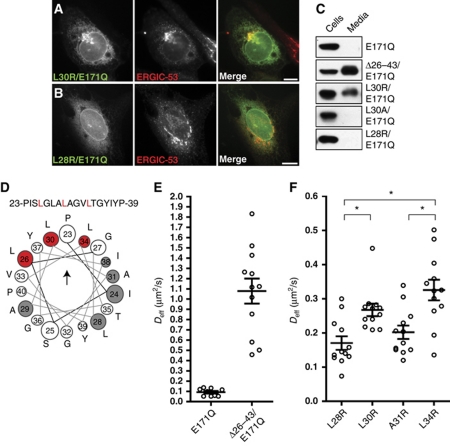
Residues 26–43 of torsinA's NTD are necessary and sufficient for ER retention. (A) Schematic view of torsinA sequence: signal peptide (1–20, black), NTD (21–43, grey), linker region (44–70), AAA domain (71–332), ΔE mutation (*, 302/303), and C-terminal mGFP tag. (B) Confocal microscopy of torsinA–mGFP, Δ26–43-torsinA–mGFP, E171Q/Δ26–43-torsinA–mGFP, and E171Q/Δ26–43-torsinA–mGFP in the presence of BFA. (C) Immunoblot of the indicated GFP fusion proteins in cell lysates or media immunoprecipitates. (D) Confocal microscopy of torsinA's signal sequence (1–25), NTD (1–43), or NTD plus linker region (1–67) fused to mGFP. Scale bars, 10 μm. (E) Immunoblot of the indicated GFP fusion proteins in cell lysates or media immunoprecipitates. (F,G) Confocal microscopy of cells expressing Δ26–43-torsinA–mGFP or E171Q/Δ26–43-torsinA–mGFP at 37°C. (F′,G′) Costaining with Sec31A after 2 h incubation at 10°C. Scale bars, 10 μm. (H) Quantification of colocalization of the indicated GFP-tagged proteins with Sec31A. N>20 cells for each condition. Bars indicate standard error of the mean.
While the deletion led to secretion of all torsinA variants analysed (Figure 2C), changes in intracellular distribution were most apparent in cells expressing the Δ26–43 deletion in combination with an E171Q mutation in the AAA+ domain (Figure 2B). A likely explanation for this difference is that the E171Q mutant is trapped in its ATP-bound state (Naismith et al, 2004) and may therefore be more conformationally stable than the wild-type enzyme, which can cycle through different nucleotide states. In support of this, OOC-5, the Caenorhabditis elegans orthologue of torsinA, is more thermostable in the presence of nucleotide (Zhu et al, 2008) and protein stability often correlates with the efficiency of secretion (Kowalski et al, 1998).
To determine whether the NTD is sufficient for ER retention or whether the AAA+ domain is also required, we attached torsinA's signal sequence (1–25), the signal sequence plus the NTD (1–43), or the entire pre-AAA sequence (1–67) to mGFP. (1–43)-mGFP and (1–67)-mGFP localized to the ER while (1–25)-mGFP was present in the Golgi (Figure 2D). Furthermore, only (1–25)-mGFP was detectable in the cell medium (Figure 2E). The NTD is thus both necessary and sufficient for ER retention, and determining why it stays in the ER should delineate the mechanism controlling torsinA localization.
Physical properties of the NTD
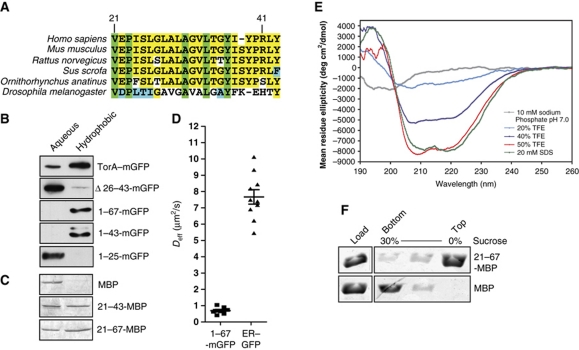
The NTD contains a pattern of hydrophobic and nonpolar residues that is well conserved among torsinA orthologues (Figure 3A), suggestive of a membrane-associating domain. As previously shown, torsinA partitions almost completely into the hydrophobic phase in Triton-X-114 phase separation experiments; this is reversed by deleting the NTD (Figure 3B) (Liu et al, 2003; Vander Heyden et al, 2009). The NTD alone controls this partitioning (Figure 3B) providing hydrophobic behaviour typical of a TMD (Bordier, 1981). To determine whether cellular factors or modifications contribute to this hydrophobicity, we purified the NTD fused to maltose binding protein from Escherichia coli. Residues 21–43 or 21–67 of torsinA shifted a fraction of this 45 kDa soluble protein into the hydrophobic phase (Figure 3C), confirming that hydrophobicity is intrinsic to the NTD and is not the result of post-translational modification or binding to another protein.
Figure 3.
TorsinA's NTD is intrinsically hydrophobic, helical, and stably associates with membranes. (A) Alignment of torsinA's NTD (residues 21–43 of human torsinA). Completely conserved residues are indicated in green, conserved residues in yellow, conservative substitutions in cyan, and nonconservative substitutions in white. (B) Anti-GFP Immunoblot of equal proportions of aqueous and Triton-X-114 detergent phases of the indicated constructs. (C) Coomassie stain of equal proportions of aqueous and Triton-X-114 detergent phases of recombinant MBP chimeras. (D) Diffusion coefficients determined by FRAP (ER–GFP, 7.67±0.44 μm2/s, n=10; 1–67-mGFP, 0.69±0.07 μm2/s, n=8). (E) Far UV circular dichroism of a synthetic peptide of residues 21–43 of human torsinA in aqueous buffer, in increasing concentrations of TFE, and in 20 mM SDS. The 50% TFE and 20 mM SDS conditions induced 28.5 and 29.3% helicity, respectively (see Materials and methods for calculation). (F) Coomassie stain showing the distribution of MBP chimeras following sucrose flotation of DOPC liposomes generated by detergent dialysis in the presence of the indicated proteins.
Since α-helices are the predominant secondary structure found in membrane-associating domains (Fiedler et al, 2010), we analysed the secondary structure of torsinA's NTD. Far UV circular dichroism shows that a synthetic peptide corresponding to residues 21–43 of torsinA is unstructured in aqueous buffer, but becomes partially helical as trifluoroethanol (TFE) is added (Figure 3E). To test whether helicity also increases in a membranous environment, we added detergent. In the presence of 20 mM SDS, the NTD peptide became as helical as in 50% TFE (Figure 3E). This stabilization of NTD structure in detergent micelles implies that the peptide may also be partially helical in the lipid bilayer (Tulumello and Deber, 2009).
The NTD associates directly and stably with membranes
To study the behaviour of the NTD in cellular membranes, we analysed the diffusion of an NTD–mGFP fusion protein by fluorescence recovery after photobleaching (FRAP) (Snapp et al, 2003). While free GFP in the ER lumen has a diffusion coefficient (Deff) of 7.67±0.44 μm2/s (Figure 3D), (1–67)-mGFP diffuses at least an order of magnitude slower (0.69±0.07 μm2/s) (Figure 3D). Single-pass TMD proteins of the ER have a similar Deff (Snapp et al, 2003). The NTD's behaviour in cellular membranes is thus most comparable to that of a membrane protein.
To determine whether the NTD associates directly with membranes, we asked whether residues 21–67 of torsinA fused to MBP could be coreconstituted into proteoliposomes with phosphatidylcholine, which is a major component of the ER membrane (van Meer et al, 2008). We combined protein with 1,2-dioleoyl-sn-glycerol-3-phosphocholine (DOPC) lipid in the presence of n-octyl glucoside, then generated liposomes by dialysing away the detergent. Liposomes were separated from soluble protein by flotation through a sucrose step gradient. The MBP–NTD fusion protein floated to the top of a sucrose gradient with the lipids, while the MBP protein remained at the bottom (Figure 3F). This experiment shows that the NTD associates directly with membranes in the absence of other proteins or modifications, and thus has the ability to mediate stable and direct association with the ER membrane.
The NTD is a monotopic membrane interacting domain
Proteins known to be statically retained in the ER appear to rely on preferential partitioning of short and/or polar TMDs into the thinner and less ordered ER bilayer (Rayner and Pelham, 1997; Yang et al, 1997; Ronchi et al, 2008). We have established that torsinA's NTD is necessary and sufficient for ER retention and associates directly with membranes. We do not, however, know how the NTD is positioned in the membrane. While originally thought to be a transmembrane protein (Breakefield et al, 2001; Liu et al, 2003), torsinA has more recently been termed a peripheral membrane protein based on partial extraction from membranes by alkaline wash and modification of a glycosylation site inserted into the NTD (Callan et al, 2007). However, bona fide TMD proteins can be sensitive to alkaline extraction (Karsten et al, 2004) and inserting an Asn residue could itself perturb handling of a TMD by the translocon (Hessa et al, 2007). Indeed, based on a biologically based TMD prediction algorithm (Snider et al, 2009), torsinA's NTD contains a 23-residue sequence (Ile24–Phe46) that might traverse the bilayer (see Figure 5A below). Because both torsinA's ER retention (Figure 2) and LULL1-dependent NE targeting (Vander Heyden et al, 2009) rely on the NTD, we set out to definitively assess its membrane topology.
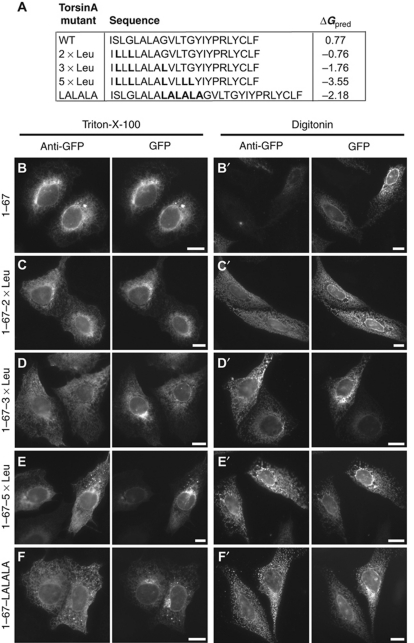
Figure 5.
TorsinA's NTD can be converted into a TMD. (A) MPEx predicted TM segment in torsinA and ΔGpred for membrane insertion of wild-type and mutated NTD sequences. (B–F) Epifluorescence microscopy of cells expressing the indicated GFP fusion proteins, stained with anti-GFP antibody after either selective permeabilization in digitonin (B–F) or complete permeabilization in Triton-X-100 (B′–F′). Scale bars, 10 μm.
The bulk of torsinA's sequence is in the lumen of the ER, as demonstrated by its protection from protease digestion (Kustedjo et al, 2000; Hewett et al, 2003). Importantly, if the NTD were to function as a stop-transfer sequence, the protein's C terminus would be in the cytosol, the reverse of what is seen in cells. However, the unusually close apposition of torsinA's signal peptide and NTD could influence topogenesis in more complex ways (Goder and Spiess, 2001). In addition, there are examples of post-translational insertion of hydrophobic domains across the membrane, particularly when that domain falls at a protein terminus (Renthal, 2010). We, therefore, wanted to directly test whether torsinA's N terminus is exposed to the cytosol. To do this, we asked whether a polyhistidine tag inserted between the signal peptide and the NTD (Figure 4A) is accessible from the cytosol. We verified that the epitope tag did not disrupt lumenal targeting (Figure 4B) and confirmed that selective permeabilization with digitonin allows detection of the cytosolic Sec31A epitope (Figure 4C) but not of the lumenal PDI epitope (Figure 4D). We found that the His tag was accessible to antibody when membranes were fully permeabilized with Triton-X-100 (Figure 4E), but was inaccessible when only the plasma membrane was permeabilized with digitonin (Figure 4F). To test whether the NTD is read as a TMD when not adjacent to torsinA's signal peptide, we inserted residues 21–43 of torsinA into a TMD reporter construct (Saaf et al, 1998) and assessed whether the NTD integrates into the membrane or translocates into the lumen by monitoring glycosylation after in vitro translation (Supplementary Figure S2). Comparing the behaviour of the NTD to test segments that do (Lep 13A/6L) or do not (Lep 18A/1L) insert into the ER membrane (Hessa et al, 2005, 2007) shows that the NTD is most similar to the latter segment (Supplementary Figure S1). These results suggest that in any context, torsinA's NTD does not traverse the bilayer, but rather associates with the lumenal side of the ER membrane. Together with its direct membrane association demonstrated above, these experiments define the NTD as a monotopic membrane interacting domain.
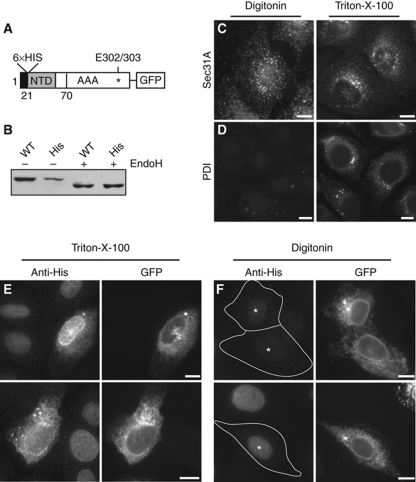
Figure 4.
TorsinA's N terminus is not accessible from the cytosol and is therefore monotopically associated with the ER bilayer. (A) Schematic diagram of His–torsinA–mGFP construct. (B) His–torsinA–mGFP migrates at the same apparent molecular weight as wild-type torsinA–mGFP and is similarly sensitive to EndoH. (C, D) Selective permeabilization with digitonin allows detection of the cytosolic Sec31A epitope (C) but not the lumenal PDI epitope (D). (E, F) Cells expressing His–torsinA–mGFP were treated with Triton-X-100 (E) or with digitonin under conditions that permeabilize only the plasma membrane (F). The N-terminal His tag was detected using anti-His antibody and gave specific signal in Triton-X-100 but not in digitonin-treated samples. Transfected cells are indicated with asterisks (*); note that background nuclear staining by the anti-His antibody is unrelated to His–TorA–GFP expression. Scale bars, 10 μm.
Mutations to the NTD have distinct effects on topology and ER retention
Given that the NTD is not behaving as a TMD, we wondered what distinguishes it from a TMD and as a TMD, how it would be sorted within the secretory pathway. Energetic predictions suggest that small changes to the sequence will decrease the energy barrier to membrane insertion (Figure 5A); we increased hydrophobicity either by inserting the sequence LALALA between Ala 31 and Gly 32, or by replacing polar residues in the NTD itself with Leu. Both inserting the LALALA sequence and replacing two or more polar residues with Leu caused the domain to be read as a stop-transfer sequence, monitored by cytosolic exposure of the C-terminal GFP tag to antibody (Figure 5B–F). This shows that the NTD is close to the edge of recognition as a type I TMD. Significantly, there were noticeable differences in the steady-state subcellular distribution of these hydrophobic mutant proteins. The LALALA and 5 × Leu mutants clearly accumulated in a perinuclear region suggestive of the Golgi in many cells (Figure 5E and F). Upon closer inspection, (1–67)-LALALA–GFP, (1–67)-5 × Leu–GFP, and (1–67)-3 × Leu–GFP all partially colocalize with the intermediate compartment marker ERGIC-53 (Supplementary Figure S2). In contrast, the 2 × Leu construct did not colocalize with ERGIC-53 (Supplementary Figure S2) despite its transmembrane orientation (Figure 5C). Thus, the small increase in hydrophobicity created by two leucine substitutions promotes integration as a TMD optimized for retention in the ER, while further hydrophobic substitution or extension of the sequence allows the same TMD to escape to post-ER compartments. These data indicate that both topology and sorting of torsinA is affected by changes in NTD amino-acid composition. The changes in sorting once the NTD becomes a TMD parallel those previously reported when the hydrophobicity of other ER TMDs was increased (Rayner and Pelham, 1997; Yang et al, 1997; Ronchi et al, 2008; Hsieh et al, 2010).
A map of the orientation of the NTD in the ER bilayer
We next performed scanning mutagenesis along the NTD to explore its orientation with respect to the membrane. Ala or Gly substitutions had no effect on targeting (Supplementary Figure S4A) or topology (data not shown) of torsinA. We therefore introduced Arg residues, which are typically tolerated on the surface and in interfacial regions of the bilayer but not in the hydrophobic core (Hessa et al, 2007). We analysed the effects of these Arg substitutions on E171Q–torsinA's steady-state localization, secretion, and diffusion in cellular membranes. Arg substitutions at positions 24, 28, 29, 31, and 38 had little to no effect on localization (Figure 6B; Supplementary Figure S3C), ER retention (Figure 6C; Supplementary Figure S3D), or diffusive behaviour in cells (Figure 6F and data not shown). In contrast, Arg substitutions at positions 26, 30, and 34 caused partial relocalization to an ERGIC-53-labelled compartment (Figure 6A; Supplementary Figure S3B) and some secretion into the cell medium (Figure 6C; Supplementary Figure S3D). These mutations also caused an increase in diffusion as measured by FRAP (Figure 6F and data not shown), although not to the extent observed when the NTD is deleted (Figure 6E). Altogether, these data indicate that selected mutations perturb torsinA's association with the membrane and retention in the ER. Importantly, the residues most affected by single Arg substitutions would fall along one face of a helix formed by the NTD (Figure 6D). This less tolerant helical face where Leu 26, 30, and 34 lie is therefore likely to be buried in the hydrophobic core of the lipid bilayer, while the more tolerant helical surface where Ile 24, Leu 28, Ala 29 and 31, and Ile 38 lie may be positioned in the interfacial region or on the membrane surface (Hessa et al, 2007). While the NTD is not a strikingly amphipathic sequence, the asymmetry in distribution of its small nonpolar residues versus leucine residues results in a low hydrophobic moment (μH=0.22) that may define the orientation of the domain in the membrane. Hydrophobic residues along one surface of this domain are necessary for retention of torsinA in the ER.
Figure 6.
ER retention correlates with membrane association. (A, B) Epifluorescence microscopy of E171Q–torsinA–mGFP with the indicated mutations, costained with ERGIC-53. Scale bars, 10 μm. (C) Immunoblot of cell lysates or media immunoprecipitates from cells expressing the indicated torsinA–mGFP constructs. (D) Helical wheel plot summarizing the results of nonconservative mutations placed throughout the NTD. Sensitive residues marked in red, insensitive residues marked in grey, and untested residues marked in white. See Supplementary Figure S3 for supporting data. Arrowhead indicates direction of hydrophobic moment of wild-type sequence (μH=0.22). (E, F) Diffusion coefficients of the indicated mutations in E171Q–torsinA–mGFP determined by FRAP. (E) E171Q, 0.09±0.01 μm2/s, n=9; E171Q/Δ26–43, 1.08±0.12 μm2/s, n=12. (F) L28R, 0.17±0.02 μm2/s, n=12; L30R, 0.27±0.02 μm2/s, n=12; A31R, 0.20±0.02 μm2/s, n=12; L34R, 0.33±0.03 μm2/s, n=12. Mean and s.e.m. values are indicated by lines and brackets. *P<0.05; L28R versus A30R, A31R versus L34R, and L28R versus L34R are significantly different, as determined by ANOVA followed by Tukey's post test.
Other lumenal membrane proteins are also excluded from ERES
To identify proteins that might share torsinA's topology and sorting mechanism, we took advantage of the proteomic analysis of secretory pathway components published a few years ago by Bergeron and colleagues (Gilchrist et al, 2006) (see Materials and methods). From a group of hydrophobic ER-enriched proteins, we selected those for which the bulk of the protein is known to be in the ER lumen and which were either known to be monotopic lumenal membrane proteins or had less well-characterized hydrophobic domains that could be either monotopic membrane-associating domains or TMDs (Table I). The mechanism(s) underlying the ER localization of these proteins are unknown, although data in each case pointed to a role for their association with the membrane in ER retention.
Table 1. Identification of proteins with similar characteristics to torsinA from Gilchrist et al (2006).
| GI accession | Protein name | Topology | References | PDB ID |
|---|---|---|---|---|
| Group I | ||||
| 603052 | Cyclooxygenase 1 (COX-1) (prostaglandin G/H synthase 1) | Monotopic lumenal | Picot et al (1994); Li et al (1998); Guo and Kulmacz (2000); Balali-Mood et al (2009) | 1PRH |
| 78214365 | 11-β-Hydroxysteroid dehydrogenase | Monotopic lumenal | Odermatt et al (1999); Hosfield et al (2005) | 1XU7 |
| Group II | ||||
| 8347733 | Arylacetamide deacetylase | Lumenal, N-terminal HD | Mziaut et al (1999); Frick et al (2004) | |
| 695162 89276780 27545358 | UDP-GT 1A1UDP-GT 1A6UDP-GT 2B1 | Lumenal, internal HD, C-terminal TMD | Meech and Mackenzie (1998) Ouizzine et al (1999) Radominska-Pandya et al (2005) | |
| Group III | ||||
| 34872654 | Malectin | Lumenal, C-terminal HD | Schallus et al (2008); Galli et al (2011) | |
| 27664728 | Peptidyl-prolyl cis–trans isomerase FKBP19 | Lumenal, C-terminal HD | Rulten et al (2006) | |
| Abbreviations: HD, hydrophobic domain; FKBP19, FK506-binding protein of 19 kDa; TMD, transmembrane domain; UDP-GT, UDP-glucuronosyltransferase. | ||||
| Hydrophobic ER-enriched proteins with experimentally confirmed ER localization and lumenal orientation are listed. | ||||
| Group I: structurally characterized as monotopic lumenal membrane proteins with defined membrane domains. Group II: deletion mutagenesis analyses indicate hydrophobic domains that are necessary and/or sufficient for ER localization. Group III: confirmed ER localization and proposed membrane associating domains based on hydrophobicity. | ||||
The best studied of these proteins is COX-1, a key enzyme in prostaglandin synthesis (Picot et al, 1994) (Table I, group I). Structures and simulations show that it uses segments of four amphipathic helices to associate monotopically with the lumenal bilayer leaflet (Picot et al, 1994; Balali-Mood et al, 2009). COX-1 was originally proposed to reside in the ER because of a KDEL-like S/P-TEL sequence at its C terminus (Song and Smith, 1996). However, mutation or deletion of this sequence had no effect on ER localization (Regier et al, 1995; Ren et al, 1995; Guo and Kulmacz, 2000). Rather, deletion studies suggest that the membrane-associating region of COX-1 is independently retained in the ER (Li et al, 1998), although a mechanistic explanation for this result has been lacking. A similar situation applies to the unrelated ER protein 11-β-hydroxysteroid dehydrogenase, which produces cortisol and uses an amphipathic helix to associate monotopically with the lumenal bilayer leaflet (Hosfield et al, 2005; Balali-Mood et al, 2009). To test whether one of these known lumenal monotopic membrane proteins exhibits torsinA-like ER retention, we analysed the effects of low temperature blocks on the localization of transfected COX-1 (Figure 7). Immunostained COX-1 is distributed throughout the ER at physiological temperature. Like torsinA, COX-1 is absent from the ERGIC after incubation at 15°C and is efficiently excluded from ERES after incubation at 10°C (Figure 7). These data indicate that COX-1 is statically retained in the ER by exclusion from ERES.
Figure 7.
COX-1 is a lumenal membrane protein that is excluded from ERES. (A, B) Confocal microscopy of cells expressing untagged COX-1 at 37°C. (A′) Costaining with ERGIC-53 after 2 h at 15°C. (B′) Costaining with Sec31A after 2 h at 10°C. Scale bars, 10 μm. (C) Quantification of colocalization of the indicated proteins with Sec31A. N>50 cells for each condition. Bars indicate standard error of the mean. *Significant difference between conditions (P<0.05).
We also identified a number of less well-characterized ER membrane proteins that may share a similar retention mechanism, although detailed studies of their hydrophobic membrane interacting domains are lacking (Table I, groups II and III). Mutagenesis analyses on group II proteins point to a role for their hydrophobic domains in ER localization. This group includes arylacetamide deacetylase, whose N-terminal hydrophobic domain is responsible for localization in the ER and bears similarity to the monotopic hydrophobic domain of 11-β-hydroxysteroid dehydrogenase (Mziaut et al, 1999). The UDP-glucuronosyltransferases (UDP-GTs) contain a lumenal hydrophobic domain that is responsible for ER localization independently of a C-terminal TMD and dilysine motif (Meech and Mackenzie, 1998). Proteins in group III are minimally studied, having confirmed ER localization and proposed membrane-associating domains based on hydrophobicity. These include malectin, a recently identified lectin involved in protein N-glycosylation in the ER, and the peptidyl-prolyl cis–trans isomerase FKBP19; each has a proposed N-terminal signal peptide and C-terminal hydrophobic domain (Rulten et al, 2006; Schallus et al, 2008; Galli et al, 2011). Our finding that both torsinA and COX-1 are excluded from ERES and thereby retained in the ER suggest a general mechanism for ER retention that can be tested in future studies of these other proteins.
Preferential partitioning of lumenal monotopic membrane proteins into ER sheets
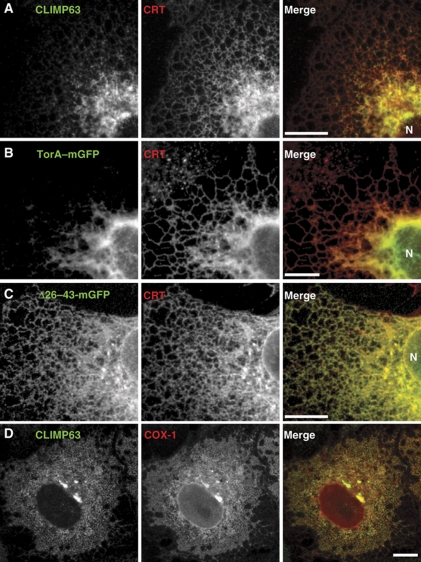
The fact that torsinA and COX-1 are monotopically associated with the ER membrane via short helical domains predicts preferential positioning of their membrane interacting domains in the lumenal leaflet of the ER membrane. Partial insertion of a protein into a bilayer is known to influence and/or respond to membrane curvature (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006). The ER consists of an anastamosing network of flat sheets and curved tubules (Shibata et al, 2009). Reticulons are monotopic membrane proteins that face the cytoplasm and distribute preferentially into ER tubules (Voeltz et al, 2006; Shibata et al, 2010). Because of the opposing curvature on the inside of ER tubules, one might expect monotopic proteins that face the lumen to avoid tubules and distribute preferentially into the flatter sheets. To test this prediction, we analysed the distribution of torsinA–mGFP as well as the known sheet-preferring protein CLIMP63 in COS-7 cells (Figure 8) (Shibata et al, 2010). The localization of both proteins was compared with that of calreticulin, which is a soluble lumenal protein present throughout the ER. We found that both torsinA and CLIMP63 colocalized with calreticulin in the perinuclear ER with its predominant population of sheets but were largely absent from the tubules of the peripheral ER (Figure 8A and B; Supplementary Figure S4). At high levels of expression, torsinA could in some cases be observed in tubules while in other cases, highly expressed torsinA promoted formation of additional sheet structures, similar to what has been described for CLIMP63 (Shibata et al, 2010). Importantly, sheet preference of torsinA was abolished when its NTD was deleted (Figure 8C; Supplementary Figure S4); the Δ26–43-torsinA–mGFP signal was superimposable with that of calreticulin. We also examined cells expressing COX-1 and saw robust colocalization of it with endogenous CLIMP63 in ER sheets (Figure 8D). In many cases, COX-1 overexpression also appeared to cause sheet proliferation. Overall, this analysis indicates that the monotopic lumenal proteins torsinA and COX-1 segregate into ER sheets, and at least in the case of torsinA, that this segregation correlates with the presence of its ER retention determinants. Notably, in a previous study of TMD-based ER retention, a plasma membrane-targeted TMD partitioned into ER tubules before it exited the ER, while a shorter ER-retained TMD preferred ER sheets (Ronchi et al, 2008). Thus, it appears that preference for less curved membranes correlates with decreased likelihood of entering ERES.
Figure 8.
Preference of torsinA and COX-1 for ER sheets. (A–C) Confocal microscopy of COS-7 cells stained for or expressing the indicated GFP-tagged proteins, zoomed in to show a section of the perinuclear and peripheral ER. ‘N’ indicates the nucleus. (D) Confocal microscopy of COS-7 cells expressing untagged COX-1, costained for CLIMP-63. Scale bars, 10 μm.
Discussion
Protein flux into and out of the ER is governed by the cell's need to create, fold, and secrete proteins. Direct or indirect interaction of secreted proteins with the COPII coat results in concentration into transport vesicles and efficient secretion; alternatively, proteins may passively enter COPII vesicles and exit the ER by bulk flow. Retention and retrieval mechanisms counter the forward flux of the secretory pathway to keep resident proteins in the ER. Although many sorting signals have been defined, how the unique protein and lipid composition of the ER is established and maintained remains a topic of study and debate (Thor et al, 2009). Here, we identify a monotopic membrane interacting domain in the lumenal enzyme torsinA that allows it to escape bulk flow out of the ER, and is both necessary and sufficient for static retention in the ER. Importantly, this sorting behaviour is attributable to the domain's direct association with the membrane, and results in exclusion of the protein from ERES. We find evidence for similar behaviour in other lumenal monotopic proteins.
TorsinA has over the years been called either a transmembrane (Breakefield et al, 2001; Liu et al, 2003) or a peripheral membrane-associated protein (Callan et al, 2007). Our data show that its NTD is hydrophobic (Figure 3B and C) and interacts directly with membranes in the absence of any other proteins (Figure 3F) but in a monotopic and not transmembrane configuration (Figure 4). Several observations indicate that the NTD controls ER retention independently of other proteins. First, retention of torsinA or the NTD alone in the ER is not readily saturated by overexpression. Second, torsinA achieves ER localization by static retention rather than by retrieval from post-ER compartments (Figure 1). As has been previously noted (Ronchi et al, 2008), a receptor-mediated mechanism for excluding a protein from ERES is currently considered unlikely and would both require enough statically retained receptor protein to interact with torsinA over its entire lifetime and move the problem of defining a retention mechanism from torsinA to the receptor. In addition, although the NTD controls ER retention, it is not involved in interaction with the predominant torsinA binding partners LAP1 and LULL1 (Naismith et al, 2009; Vander Heyden et al, 2009) (data not shown), ruling these out as potential retention factors. Finally, conservative substitutions along the length of the NTD (Supplementary Figure S3A and B) have no effect on ER retention, which would be unexpected if the NTD were to harbour a protein–protein interacting motif. We, therefore, conclude that ER retention is mediated by physical features of the NTD.
While proteins that associate with the lumenal leaflet of the ER membrane have not been extensively studied, we identified a few that share torsinA's topology and its exclusion from ERES (Table I; Figure 7). Two of these—COX-1 and 11-β-hydroxysteroid dehydrogenase—have been structurally characterized as lumenal monotopic proteins. Molecular modelling of one of these proteins, COX-1, in membranes showed that its monotopic membrane association disorders the lumenal leaflet (Nina et al, 2000) and affects membrane curvature (Wan and Coveney, 2009). Both of these effects could influence partitioning of the domain within the lipid bilayer (as schematized in Figure 9) and ultimately protein sorting. Although a structure of torsinA's NTD in a membrane will be important to define its relationship to the ER membrane, our finding that it tolerates nonconservative mutations on only one face of the helix involved in membrane association suggests that it adopts a defined position in the membrane, perhaps as an in-plane helix.
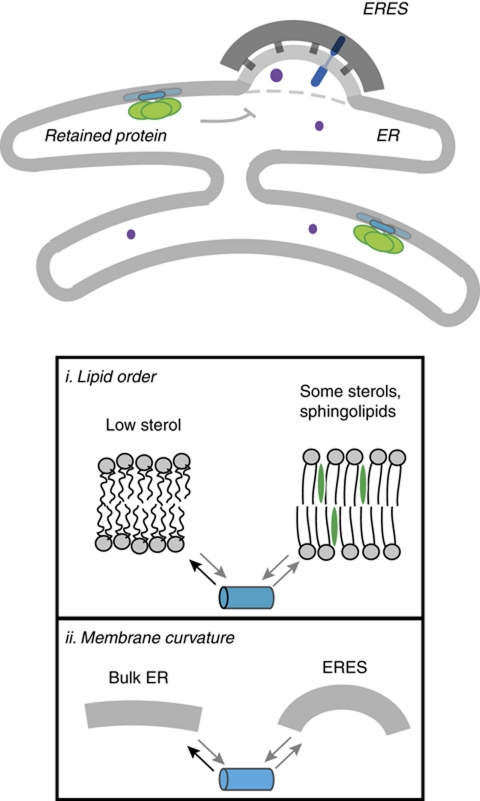
Figure 9.
Model of retention of monotopic lumenal proteins within the ER. Monotopic lumenal proteins associate with the lumenal leaflet of the ER. This membrane association favours partitioning away from sites of ER-to-Golgi transport. This effect on protein sorting may be achieved by monotopic domains selectively partitioning into less ordered bilayers (box i) or less curved bilayers (box ii).
Membrane-based sorting of proteins along the secretory pathway is a well-characterized phenomenon for transmembrane proteins, generally explained by a matching of TMD length and composition to bilayer thickness, which in turn depends on lipid composition (Bretscher and Munro, 1993; Sharpe et al, 2010). The ER bilayer is generally thinner, less ordered, and less charged than post-ER membranes (van Meer et al, 2008). Based on our analysis of torsinA and COX-1, we suggest that the characteristics of membranes that result in sorting of TMDs (Sharpe et al, 2010) also influence the sorting of monotopic membrane interacting domains (Figure 9). We propose that there are two potentially complementary effects arising from association of monotopic domains with bilayers that could contribute to ER retention. First, the energetic cost of inserting a monotopic domain into the lumenal bilayer leaflet is likely to be lower in the loosely packed membrane of the ER relative to the more ordered membranes of the Golgi and later secretory pathway (Figure 9, box i). Second, monotopic domains may preferentially partition into less curved bilayers because the inner leaflet of curved ER domains, such as tubules and budding vesicles, will be relatively contracted (Figure 9, box ii). Crowding of coat-associated cargo in the membrane may amplify these effects in COPII vesicles in vivo.
As is typical for AAA+ family members (Hanson and Whiteheart, 2005), torsinA is an oligomeric enzyme (Vander Heyden et al, 2009) held together by interactions among its AAA+ domains. It is possible that the NTD helices on such an oligomer will function together to perturb lipid structure and thereby amplify the effects of the NTD on protein distribution among different subdomains of the ER. The rigidity of a flat AAA+ ring structure (Hanson and Whiteheart, 2005), which torsinA is expected to adopt, may further restrict the oligomer to regions of low curvature. Notably, the dimeric COX-1 enzyme contributes four helices per subunit to anchor itself in the membrane (Picot et al, 1994). The cooperative effects of membrane interacting domains on oligomers may thus amplify the partitioning of monotopic membrane interacting domains between subdomains of the ER.
The exclusion of torsinA (this study) and other statically retained ER membrane proteins (Ronchi et al, 2008) from the ERES microenvironment and their preferential concentration in ER sheets raises questions about the composition and structure of ERES and the transitional ER. This membrane domain lacks ribosomes and is enriched in COPII components and cargo, consistent with its role in secretory pathway trafficking (Bannykh et al, 1996; Hammond and Glick, 2000). Phosphatidylinositol 4-phosphate is found on the cytosolic surface of ERES (Blumental-Perry et al, 2006) and long-chain phosphatidylserine is concentrated in COPII vesicles relative to the bulk ER (Sturbois-Balcerzak et al, 1999), suggesting that the transitional ER may also be a distinct lipid environment. Notably, the transitional ER is a stable ER subdomain that is marked throughout the cell cycle by Sec16 (Hughes and Stephens, 2010). Further studies will be needed to delineate the relationships between ER subdomains with differing composition, membrane curvature, and membrane protein sorting.
Materials and methods
Plasmids
TorsinA–mGFP and Δ26–43-torsinA–mGFP are as described previously (Vander Heyden et al, 2009). (1–67), (1–43), and (1–25) torsinA–mGFP were made by PCR amplification of the indicated sequences with primers containing XhoI and EcoRI sites, followed by ligation into the mGFP-containing pEGFP-N1 vector. All NTD substitution and insertion mutations were introduced by QuikChange mutagenesis. MBP-(21–43) and MBP-(21–67) were made by PCR amplification of the corresponding sequence with primers containing EcoRI and BamHI restriction sites, followed by ligation into pMAL-c (NEB). The Lep reporter vector, Lep 13A/6L, and 18A/1L constructs were generously provided by Gunnar von Heijne (Stockholm University). The NTD was inserted into the Lep reporter vector as described (Hessa et al, 2005): oligonucleotides encoding residues 21–43 of human torsinA in forward (Nlum–Ccyto orientation) or reverse (Ncyto–Clum orientation) sequence were annealed together and ligated into Lep vector that had been digested with SpeI and KpnI. The COX-1 expression vector was provided by Robert J Kulmacz (University of Texas, Houston). VSVG-(ts045)–GFP (Presley et al, 1997) and NPY–GFP (Perrais et al, 2004) were as described.
Reagents
A peptide consisting of residues 21–43 of human torsinA, an aminohexanoic acid linker and a biotin tag was synthesized and verified by mass spectrometry by the WM Keck Foundation Biotechnology Resource Laboratory (Yale University). 7-Methyl diguanosine triphosphate cap structure analogue and amylose resin were from NEB (Ipswich, MA). Rabbit reticulocyte lysate, canine rough microsomes (RMs), and NTPs were from Promega (Madison, WI). Complete protease inhibitor cocktail was from Boehringer (Ridgefield, CT). Protein-G-Sepharose was from GE/Amersham (Piscataway, NJ). Antibodies used include mouse monoclonal anti-βCOP (clone maD, Sigma), mouse monoclonal anti-Sec31A (BD Biosciences, San Jose, CA), mouse monoclonal anti-GFP (clone B-2, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-GFP (Dalal et al, 2004), rabbit polyclonal anti-His (Cell Signaling, Beverley, MA), mouse monoclonal ERGIC-53 (Alexis Biochemicals, San Diego, CA), and rabbit polyclonal anti-COX-1 (Cayman Chemical, Ann Arbor, MI). All other chemicals were from Sigma (St Louis, MO).
Cell culture
U2OS cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and L-glutamine. Transient transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Immunofluorescence
For temperature block experiments, coverslips were transferred to HEPES-buffered media supplemented with 10% FBS and L-glutamine, and incubated for 2 h in a water bath set to 10 or 15°C. For BFA treatment, cells were transfected ∼16 h before adding 1 mM BFA to the medium for 4 h. Cells were washed in PBS and then fixed with 4% paraformaldehyde in PBS for 10 min, followed by permeabilization in 0.2% Triton-X-100 for 10 min. Coverslips were blocked in 2% goat serum in PBS for 1 h before incubation with primary and Alexa Fluor-conjugated secondary antibodies. For selective permeabilization with digitonin, cells were washed once in PBS before transfer to 0.0025% digitonin in PBS for 5 min at 4°C. Cells were then fixed, blocked, and stained as described above. Coverslips were mounted in Mowiol (Calbiochem, San Diego, CA). Epifluorescence imaging was performed with a Diaplan microscope (Leica Microsystems, Bannockburn, IL) using a × 63 1.4 NA objective and a Zeiss MRm camera (Thornwood, NY). Confocal imaging was performed on an Olympus FV500 microscope using a × 60 1.4 NA objective. Brightness and contrast were adjusted with Adobe Photoshop (Adobe Systems, San Jose, CA) and composite figures were prepared in Adobe Illustrator.
Quantifying colocalization
To monitor localization of candidate cargo proteins to ERES, cells expressing the indicated candidate proteins were costained for Sec31A and viewed by confocal microscopy. Colocalization analysis was restricted to ERES as described (French et al, 2008). Briefly, a binary mask of the Sec31A signal was created, manually thresholded, and added as an additional channel in the RGB image. Colocalization was then quantified in the regions defined by the mask, and expressed as Pearson's correlation coefficient. Colocalization was analysed in >20 cells for each condition reported. Statistical comparison of data sets was performed by t-test.
Immunoprecipitation from culture medium
In all, 60 mm plates of U2OS cells were transiently transfected with the indicated plasmids and incubated for ∼16 h. For experiments involving BFA treatment, fresh media was added with or without 1 mM BFA and cells were incubated for an additional 6 h. The media was then collected, placed on ice, and supplemented with 1 mM PMSF, 1 × complete protease inhibitors, and 5 mM EDTA. The media was centrifuged at 4000 r.p.m. for 10 min at 4°C to remove cell debris. Protein-G-sepharose beads covalently conjugated to affinity-purified rabbit anti-GFP antibody were then added to the supernatant and incubated 2 h at 4°C. The beads were then pelleted and washed 3 × in PBS/0.5% Triton-X-100 with PMSF and protease inhibitors, then boiled in 50 μl SDS–PAGE sample buffer for analysis. The cells, meanwhile, were scraped and pelleted in PBS and solubilized in ∼300 μl PBS/0.5% Triton-X-100 with PMSF and protease inhibitors for 30 min at 4°C. The insoluble material was pelleted at 15 000 g for 10 min at 4°C. An aliquot of the supernatant was boiled in SDS–PAGE sample buffer. A total of 15 μl each of the cell lysate and immunoprecipitate were separated on SDS–PAGE and analysed by immunoblotting.
Far UV circular dichroism
Far UV CD spectra were recorded using a 0.2-mm pathlength cuvette in a Jasco J715 spectropolarimeter at ambient temperature, scanning from 260 to 190 nm in 0.2 nm steps at 100 nm/min, averaging five spectra per condition. Peptide was diluted to 1 mg/ml in 10 mM sodium phosphate buffer, pH 7.0 supplemented with TFE (Sigma) or SDS as indicated. Solvent spectra gave negligible signal and were subtracted from sample spectra. Data shown are expressed as mean residue ellipticity (deg cm2/dmol). The percentage of α-helix was estimated from the molar ellipticity at 222 nm (θ222) using the equation fh=(θ222/θ222a)+(iκ/N), where fh is the fraction in the α-helical form, θ222 is the mean residue ellipticity at 222 nm, θ222a is the molar ellipticity at 222 nm for an infinitely long α-helix (−39 500 deg cm2/dmol), i is the number of helices (assumed to be one), κ is a wavelength-specific constant (2.6 at 222 nm), and N is the number of peptide bonds in the peptide (defined as 26 for the 23 residues of torsinA plus aminohexanoic acid linker and biotin) (Bernstein et al, 2000).
Purification of MBP fusion proteins
BL21(DE3) E. coli transformed with MBP, MBP-21–43, or MBP-21–67 were grown in 0.5 l of Terrific Broth and induced to express protein by addition of isopropyl 1-thio-β-D-galactopyranoside and shaking for 3 h at room temperature. After pelleting, bacteria were lysed in 20 mM Tris pH 7.4, 200 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM PMSF by sonication. n-Octyl glucoside was added to 1% w/v and samples were incubated for 30 min at 4°C, followed by centrifugation at 9000 g for 20 min at 4°C. Amylose resin was added to the supernatant and incubated overnight at 4°C. Unbound material was removed by washing in lysis buffer, and proteins were eluted in lysis buffer supplemented with 10 mM maltose. MBP-21–67 was incubated sequentially with 10 mM DTT (30 min at room temperature) and 50 mM NEM (30 min at room temperature) to reduce and alkylate cysteines. For use in proteoliposome preparations, proteins were dialysed into 10 mM sodium phosphate pH 7.0, 150 mM NaCl, 30 mM n-octyl glucoside overnight at 4°C. Proteins were clarified by centrifugation at 200 000 g for 1 h at 4°C before use in proteoliposome preparations. Protein concentrations were quantitated by Bradford assay with BSA as standard and snap frozen.
Proteoliposome preparation and characterization
DOPC was purchased from Avanti Polar Lipids (Alabaster, AL). In all, 60 μg of DOPC in chloroform was dried under a stream of nitrogen and then under vacuum for several hours. The lipid film was resuspended in 1 ml of 10 mM sodium phosphate, pH 7.0, 150 mM NaCl, and 30 mM n-octyl glucoside, followed by addition of MBP fusion proteins for a final protein:lipid ratio of 2:1 by weight. Liposomes were generated by removing detergent by dialysis at 37°C in 2000 Da molecular weight cutoff dialysis cassettes (Pierce) against 3 × 1 l changes of buffer lacking detergent for 48 h. To determine how much protein was associated with liposomes, samples were loaded on the bottom of a sucrose cushion and centrifuged to separate liposomes from unincorporated protein by flotation. A 150-μl aliquot of the proteoliposome preparation was mixed with 100 μl 2.2 M sucrose/10 mM sodium phosphate pH 7.0/150 mM NaCl in a polycarbonate centrifuge tube. This mixture was overlaid with a 200-μl cushion of 0.75 M sucrose/10 mM sodium phosphate pH 7.0/150 mM NaCl, then with a 50-μl layer of buffer lacking sucrose. The step gradients were centrifuged for 80 min at 240 000 g (55 000 r.p.m.) in a Beckman TLS 55 rotor. Three fractions were collected from the bottom using a syringe (250, 150, and 100 μl). Aliquots of each fraction were analysed by SDS–PAGE and Coomassie stain.
Sorting proteomics data
Gilchrist et al (2006) identified 1430 proteins of the secretory pathway proteome by mass spectrometry analysis of rat liver RM, smooth microsome, Golgi, and COPI vesicle fractions. To identify ER resident proteins similar to torsinA, these 1430 proteins were sorted using the following criteria: (1) presence of a predicted cleavable signal peptide (Supplementary Table S1C); (2) similar biochemical behaviour to torsinA in salt extraction and Triton-X-114 partitioning experiments (Supplementary Table S3B); and (3) enrichment in rough microsomal and smooth microsomal fractions over Golgi and COPI fractions (Supplementary Table S3D). Approximately 150 proteins including torsinA matched these criteria. Literature and database searches were then used to eliminate proteins known to be localized to different compartments and identify proteins for which the cell biological data are consistent with a lumenal orientation in the ER, and for which other established ER retention or retrieval mechanisms either do not exist or do not fully explain sorting behaviour. Selected proteins of interest from this set are highlighted in Table I.
Transmembrane segment prediction
To explore the potential for hydrophobic sequences in torsinA to adopt a TMD configuration, the full sequence of human torsinA was analysed in the MPEx program (http://blanco.biomol.uci.edu/mpex/) (Snider et al, 2009) in biological hydrophobicity scale mode (‘translocon TM analysis’). The predicted ΔG for membrane insertion (ΔGpred) was calculated for the MPEx-identified protein sequence and mutants to that region using the dGpred prediction algorithm (http://dgpred.cbr.su.se/) (Hessa et al, 2007); these values are reported in Figure 5A.
Triton-X-114 phase separation and FRAP were performed as described in Vander Heyden et al (2009). SDS–PAGE and immunoblotting were performed as described (Dalal et al, 2004).
Supplementary Material
Acknowledgments
We thank the members of the Hanson laboratory for helpful discussions. Daved Fremont and members of his laboratory at Washington University are gratefully acknowledged for assistance with circular dichroism spectroscopy. We also thank Dennis Oakley at the Bakewell Neuroimaging Laboratory at Washington University for helpful discussions and assistance with quantitation of colocalization on subcellular structures. This work was supported by grants from the National Institute of Neurological Disorders and Stroke (R01 NS050717, PIH; F31 NS065608, ABV).
Author contributions: AV, TN, ES, and PH participated in designing the experiments; AV and TN performed most of the experiments; ES performed FRAP analysis; AV, TN, ES, and PH analysed the data; and AV and PH wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balali-Mood K, Bond PJ, Sansom MS (2009) Interaction of monotopic membrane enzymes with a lipid bilayer: a coarse-grained MD simulation study. Biochemistry 48: 2135–2145 [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE (1996) The organization of endoplasmic reticulum export complexes. J Cell Biol 135: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein LS, Grillo AA, Loranger SS, Linder ME (2000) RGS4 binds to membranes through an amphipathic alpha-helix. J Biol Chem 275: 18520–18526 [DOI] [PubMed] [Google Scholar]
- Blumental-Perry A, Haney CJ, Weixel KM, Watkins SC, Weisz OA, Aridor M (2006) Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export. Dev Cell 11: 671–682 [DOI] [PubMed] [Google Scholar]
- Bordier C (1981) Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256: 1604–1607 [PubMed] [Google Scholar]
- Bragg DC, Armata IA, Nery FC, Breakefield XO, Sharma N (2011) Molecular pathways in dystonia. Neurobiol Dis 42: 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakefield XO, Blood AJ, Li Y, Hallett M, Hanson PI, Standaert DG (2008) The pathophysiological basis of dystonias. Nat Rev Neurosci 9: 222–234 [DOI] [PubMed] [Google Scholar]
- Breakefield XO, Kamm C, Hanson PI (2001) TorsinA: movement at many levels. Neuron 31: 9–12 [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S (1993) Cholesterol and the Golgi apparatus. Science 261: 1280–1281 [DOI] [PubMed] [Google Scholar]
- Callan AC, Bunning S, Jones OT, High S, Swanton E (2007) Biosynthesis of the dystonia-associated AAA+ ATPase torsinA at the endoplasmic reticulum. Biochem J 401: 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal S, Rosser MF, Cyr DM, Hanson PI (2004) Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol Biol Cell 15: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C (2010) Protein sorting receptors in the early secretory pathway. Annu Rev Biochem 79: 777–802 [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, McNaught KS, Jengelley TA, Jackson T, Li J, Li Y (2005) Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp Neurol 196: 452–463 [DOI] [PubMed] [Google Scholar]
- Daubner T, Fink A, Seitz A, Tenzer S, Muller J, Strand D, Seckert CK, Janssen C, Renzaho A, Grzimek NK, Simon CO, Ebert S, Reddehase MJ, Oehrlein-Karpi SA, Lemmermann NA (2010) A novel transmembrane domain mediating retention of a highly motile herpesvirus glycoprotein in the endoplasmic reticulum. J Gen Virol 91: 1524–1534 [DOI] [PubMed] [Google Scholar]
- Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J (1998) Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem 273: 32088–32095 [DOI] [PubMed] [Google Scholar]
- Fahn S (1988) Concept and classification of dystonia. Adv Neurol 50: 1–8 [PubMed] [Google Scholar]
- Fiedler S, Broecker J, Keller S (2010) Protein folding in membranes. Cell Mol Life Sci 67: 1779–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Frick C, Atanasoz AB, Arnold P, Ozols J, Odermatt A (2004) Appropriate function of 11beta-hydroxysteroid dehydrogenase type 1 in the endoplasmic reticulum lumen is dependent on its N-terminal region sharing similar topological determinants with 50-kDa esterase. J Biol Chem 279: 31131–31138 [DOI] [PubMed] [Google Scholar]
- Galli C, Bernasconi R, Solda T, Calanca V, Molinari M (2011) Malectin participates in a backup glycoprotein quality control pathway in the mammalian ER. PLoS One 6: e16304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127: 1265–1281 [DOI] [PubMed] [Google Scholar]
- Giles LM, Li L, Chin LS (2009) TorsinA protein degradation and autophagy in DYT1 dystonia. Autophagy 5: 82–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Spiess M (2001) Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett 504: 87–93 [DOI] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT (2005) The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol 168: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT (2005) Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48: 923–932 [DOI] [PubMed] [Google Scholar]
- Gordon KL, Gonzalez-Alegre P (2008) Consequences of the DYT1 mutation on torsinA oligomerization and degradation. Neuroscience 157: 588–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Kulmacz RJ (2000) Distinct influences of carboxyl terminal segment structure on function in the two isoforms of prostaglandin H synthase. Arch Biochem Biophys 384: 269–279 [DOI] [PubMed] [Google Scholar]
- Hammond AT, Glick BS (2000) Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell 11: 3013–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433: 377–381 [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hewett J, Ziefer P, Bergeron D, Naismith T, Boston H, Slater D, Wilbur J, Schuback D, Kamm C, Smith N, Camp S, Ozelius LJ, Ramesh V, Hanson PI, Breakefield XO (2003) TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J Neurosci Res 72: 158–168 [DOI] [PubMed] [Google Scholar]
- Hewett JW, Nery FC, Niland B, Ge P, Tan P, Hadwiger P, Tannous BA, Sah DW, Breakefield XO (2008) siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cells. Hum Mol Genet 17: 1436–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Wu Y, Skene RJ, Hilgers M, Jennings A, Snell GP, Aertgeerts K (2005) Conformational flexibility in crystal structures of human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation. J Biol Chem 280: 4639–4648 [DOI] [PubMed] [Google Scholar]
- Hsieh SC, Tsai WY, Wang WK (2010) The length of and nonhydrophobic residues in the transmembrane domain of dengue virus envelope protein are critical for its retention and assembly in the endoplasmic reticulum. J Virol 84: 4782–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Stephens DJ (2010) Sec16A defines the site for vesicle budding from the endoplasmic reticulum on exit from mitosis. J Cell Sci 123: 4032–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten V, Hegde RS, Sinai AP, Yang M, Joiner KA (2004) Transmembrane domain modulates sorting of membrane proteins in Toxoplasma gondii. J Biol Chem 279: 26052–26057 [DOI] [PubMed] [Google Scholar]
- Kowalski JM, Parekh RN, Mao J, Wittrup KD (1998) Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J Biol Chem 273: 19453–19458 [DOI] [PubMed] [Google Scholar]
- Kuismanen E, Saraste J (1989) Low temperature-induced transport blocks as tools to manipulate membrane traffic. Methods Cell Biol 32: 257–274 [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF (2000) Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem 275: 27933–27939 [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Sweet DJ, Pelham HR (1990) The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell 61: 1359–1363 [DOI] [PubMed] [Google Scholar]
- Li Y, Smith T, Grabski S, DeWitt DL (1998) The membrane association sequences of the prostaglandin endoperoxide synthases-1 and -2 isozymes. J Biol Chem 273: 29830–29837 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zolkiewska A, Zolkiewski M (2003) Characterization of human torsinA and its dystonia-associated mutant form. Biochem J 374: 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti LV, Torrisi MR, Erra MC, Bonatti S (1996) Morphological analysis of the transfer of VSV ts-045 G glycoprotein from the endoplasmic reticulum to the intermediate compartment in vero cells. Exp Cell Res 227: 323–331 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596 [DOI] [PubMed] [Google Scholar]
- Meech R, Mackenzie PI (1998) Determinants of UDP glucuronosyltransferase membrane association and residency in the endoplasmic reticulum. Arch Biochem Biophys 356: 77–85 [DOI] [PubMed] [Google Scholar]
- Mezzacasa A, Helenius A (2002) The transitional ER defines a boundary for quality control in the secretion of tsO45 VSV glycoprotein. Traffic 3: 833–849 [DOI] [PubMed] [Google Scholar]
- Mziaut H, Korza G, Hand AR, Gerard C, Ozols J (1999) Targeting proteins to the lumen of endoplasmic reticulum using N-terminal domains of 11beta-hydroxysteroid dehydrogenase and the 50-kDa esterase. J Biol Chem 274: 14122–14129 [DOI] [PubMed] [Google Scholar]
- Naismith TV, Dalal S, Hanson PI (2009) Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem 284: 27866–27874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI (2004) TorsinA in the nuclear envelope. Proc Natl Acad Sci USA 101: 7612–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO (2008) TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci 121: 3476–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nina M, Berneche S, Roux B (2000) Anchoring of a monotopic membrane protein: the binding of prostaglandin H2 synthase-1 to the surface of a phospholipid bilayer. Eur Biophys J 29: 439–454 [DOI] [PubMed] [Google Scholar]
- Odermatt A, Arnold P, Stauffer A, Frey BM, Frey FJ (1999) The N-terminal anchor sequences of 11beta-hydroxysteroid dehydrogenases determine their orientation in the endoplasmic reticulum membrane. J Biol Chem 274: 28762–28770 [DOI] [PubMed] [Google Scholar]
- Ouizzine M, Magdalou J, Burchell B, Fournel-Gigleux S (1999) An internal signal sequence mediates the targeting and retention of the human UDP-glucuronosyltransferase 1A6 to the endoplasmic reticulum. J Biol Chem 274: 31401–31409 [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO (1997) The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet 17: 40–48 [DOI] [PubMed] [Google Scholar]
- Perrais D, Kleppe IC, Taraska JW, Almers W (2004) Recapture after exocytosis causes differential retention of protein in granules of bovine chromaffin cells. J Physiol 560: 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot D, Loll PJ, Garavito RM (1994) The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature 367: 243–249 [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J (1997) ER-to-Golgi transport visualized in living cells. Nature 389: 81–85 [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Ouzzine M, Fournel-Gigleux S, Magdalou J (2005) Structure of UDP-glucuronosyltransferases in membranes. Meth Enzymol 400: 116–147 [DOI] [PubMed] [Google Scholar]
- Rayner JC, Pelham HR (1997) Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J 16: 1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier MK, Otto JC, DeWitt DL, Smith WL (1995) Localization of prostaglandin endoperoxide synthase-1 to the endoplasmic reticulum and nuclear envelope is independent of its C-terminal tetrapeptide-PTEL. Arch Biochem Biophys 317: 457–463 [DOI] [PubMed] [Google Scholar]
- Ren Y, Loose-Mitchell DS, Kulmacz RJ (1995) Prostaglandin H synthase-1: evaluation of C-terminus function. Arch Biochem Biophys 316: 751–757 [DOI] [PubMed] [Google Scholar]
- Renthal R (2010) Helix insertion into bilayers and the evolution of membrane proteins. Cell Mol Life Sci 67: 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi P, Colombo S, Francolini M, Borgese N (2008) Transmembrane domain-dependent partitioning of membrane proteins within the endoplasmic reticulum. J Cell Biol 181: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Kinloch RA, Tateossian H, Robinson C, Gettins L, Kay JE (2006) The human FK506-binding proteins: characterization of human FKBP19. Mamm Genome 17: 322–331 [DOI] [PubMed] [Google Scholar]
- Saaf A, Wallin E, von Heijne G (1998) Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur J Biochem 251: 821–829 [DOI] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (2003) Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol Biol Cell 14: 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallus T, Jaeckh C, Feher K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, Pieler T, Muhle-Goll C (2008) Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell 19: 3404–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S (2010) A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA (2009) Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010) Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Altan N, Lippincott-Schwartz J (2003) Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr Protoc Cell Biol Chapter 21, Unit 21.1 [DOI] [PubMed] [Google Scholar]
- Snider C, Jayasinghe S, Hristova K, White SH (2009) MPEx: a tool for exploring membrane proteins. Protein Sci 18: 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Smith WL (1996) C-terminal Ser/Pro-Thr-Glu-Leu tetrapeptides of prostaglandin endoperoxide H synthases-1 and -2 target the enzymes to the endoplasmic reticulum. Arch Biochem Biophys 334: 67–72 [DOI] [PubMed] [Google Scholar]
- Sturbois-Balcerzak B, Vincent P, Maneta-Peyret L, Duvert M, Satiat-Jeunemaitre B, Cassagne C, Moreau P (1999) ATP-dependent formation of phosphatidylserine-rich vesicles from the endoplasmic reticulum of leek cells. Plant Physiol 120: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff AM (1986) Temperature and energy dependence of secretory protein transport in the exocrine pancreas. EMBO J 5: 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor F, Gautschi M, Geiger R, Helenius A (2009) Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic 10: 1819–1830 [DOI] [PubMed] [Google Scholar]
- Tulumello DV, Deber CM (2009) SDS micelles as a membrane-mimetic environment for transmembrane segments. Biochemistry 48: 12096–12103 [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI (2009) LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell 20: 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wan S, Coveney PV (2009) A comparative study of the COX-1 and COX-2 isozymes bound to lipid membranes. J Comput Chem 30: 1038–1050 [DOI] [PubMed] [Google Scholar]
- Yang M, Ellenberg J, Bonifacino JS, Weissman AM (1997) The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem 272: 1970–1975 [DOI] [PubMed] [Google Scholar]
- Zhu L, Wrabl JO, Hayashi AP, Rose LS, Thomas PJ (2008) The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol Biol Cell 19: 3599–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM (2006) How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7: 9–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.