Abstract
RAD52 protein has an important role in homology-directed DNA repair by mediating RAD51 nucleoprotein filament formation on single-stranded DNA (ssDNA) protected by replication protein-A (RPA) and annealing of RPA-coated ssDNA. In human, cellular response to DNA damage includes phosphorylation of RAD52 by c-ABL kinase at tyrosine 104. To address how this phosphorylation modulates RAD52 function, we used an amber suppressor technology to substitute tyrosine 104 with chemically stable phosphotyrosine analogue (p-Carboxymethyl-L-phenylalanine, pCMF). The RAD52Y104pCMF retained ssDNA-binding activity characteristic of unmodified RAD52 but showed lower affinity for double-stranded DNA (dsDNA) binding. Single-molecule analyses revealed that RAD52Y104pCMF specifically targets and wraps ssDNA. While RAD52Y104pCMF is confined to ssDNA region, unmodified RAD52 readily diffuses into dsDNA region. The Y104pCMF substitution also increased the ssDNA annealing rate and allowed overcoming the inhibitory effect of dsDNA. We propose that phosphorylation at Y104 enhances ssDNA annealing activity of RAD52 by attenuating dsDNA binding. Implications of phosphorylation-mediated activation of RAD52 annealing activity are discussed.
Keywords: c-ABL, DNA repair, RAD52, ssDNA annealing, tyrosine phosphorylation
Introduction
If left unchecked, DNA double-strand breaks (DSBs) can cause chromosome fragmentation, loss and rearrangement of the genetic information (Kanaar et al, 1998; Rich et al, 2000), which are frequently associated with tumour formation and cancer progression (Lengauer et al, 1998; Kolodner et al, 2002; Hanahan and Weinberg, 2011). DSBs can be repaired by non-homologous DNA end joining (Lieber, 2010), which mends the damage but has a high risk of inducing oncogenic translocations. In contrast, the process of homologous genetic recombination (HR) repairs DSBs most accurately (Couedel et al, 2004; Moynahan and Jasin, 2010). The third pathway of DSB repair, single-strand annealing (SSA), uses sequence repeats and is, therefore, potentially mutagenic (Weinstock et al, 2006). RAD52 protein has an important role in the two homology-directed DNA repair pathways, HR and SSA. Yeast Rad52 has two distinct roles as recombination mediator and by facilitating annealing of the complementary DNA strands and importantly, of the ssDNA–replication protein-A (RPA) complexes (Sung, 1997; New et al, 1998; Shinohara and Ogawa, 1998). In contrast, mediator activity of human RAD52 is dispensable for recombination under most cellular conditions (Rijkers et al, 1998; Yamaguchi-Iwai et al, 1998). This is because a number of mammalian genes such as BRCA2 (Esashi et al, 2007) and RAD51 paralogues (Thompson and Schild, 2001) encode functions that may be complementary with yeast Rad52 protein. For example, human BRCA2 protein functions as a recombination mediator by facilitating RAD51 nucleoprotein filament formation (San Filippo et al, 2006; Jensen et al, 2010; Liu et al, 2010; Thorslund et al, 2010). In contrast to its well-studied counterpart from Ustilago maydis, Brh2 (Mazloum and Holloman, 2009), human BRCA2 is unable to facilitate annealing of RPA-coated ssDNA (Jensen et al, 2010). RAD52 acts independently of BRCA2 and its inactivation is synthetically lethal with BRCA2 deficiency (Feng et al, 2011). Additionally, unlike BRCA2 which acts early, RAD52 is involved in late response to the DSBs at stalled or collapsed replication forks (Wray et al, 2008). These observations leave human RAD52 to have a unique role by catalysing ssDNA annealing in homology-directed DNA repair (Paques and Haber, 1999; Symington, 2002).
Ubiquitously expressed, c-ABL tyrosine kinase is activated in response to various types of DNA damage including DSBs and crosslinks (Kharbanda et al, 1995) in an ATM- and DNA-PKCS-dependent manner (Baskaran et al, 1997; Kharbanda et al, 1997; Shafman et al, 1997). Activated c-ABL promotes cell-cycle arrest and apoptosis that depend on p53 (Yuan et al, 1996), p73 (Agami et al, 1999) and Rad9 (Yoshida et al, 2002). Additionally, in response to DNA damage c-ABL mediates phosphorylation of several DNA repair proteins such as RAD51 (Yuan et al, 1998; Chen et al, 1999), RAD52 (Kitao and Yuan, 2002), WRN (Cheng et al, 2003), DDB1 (Cong et al, 2002) and BRCA1 (Foray et al, 2002). Interaction with c-ABL promotes RAD51 and RAD52 localization at the sites of DNA damage and modulates homology-directed repair (Chen et al, 1999; Kitao and Yuan, 2002; Shimizu et al, 2009). However, the mechanistic details underlying the role of c-ABL in DNA repair remain unclear.
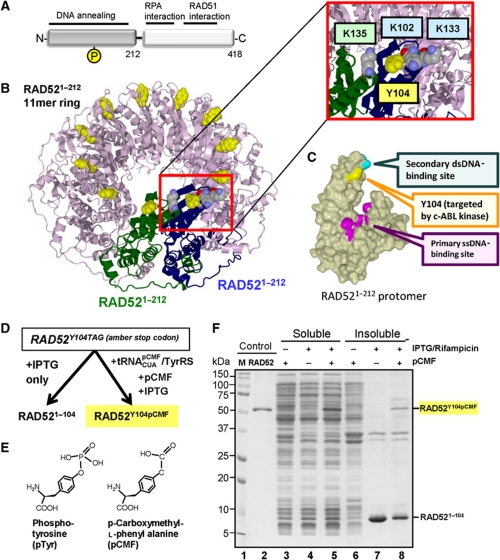
Tyrosine 104 targeted by c-ABL kinase is located in a highly conserved N-terminal domain of RAD52 (amino acids 1–212) that is responsible for DNA annealing and oligomerization (Figure 1A; Kagawa et al, 2001; Lloyd et al, 2002). In the structure of the N-terminal domain of RAD52, Y104 is positioned at the subunit interface of the undecameric (11mer) ring between positively charged K102 and K133 of the same subunit and K135 from the adjacent subunit (Figure 1B; Kagawa et al, 2002; Singleton et al, 2002). Although the full-length RAD52 predominantly forms heptameric (7mer) rings (Stasiak et al, 2000; West, 2003), the arrangement of the subunit–subunit interface is believed to be similar to that observed in the structure of the N-terminal domain. K102 and K133 residues of RAD52 are directly involved in dsDNA binding (Kagawa et al, 2008). Substitution of RAD52 K135 counterpart in Saccharomyces cerevisiae Rad52 (K150) for alanine results in very low inter-chromosomal recombination rates and in a mild γ-ray sensitivity, suggesting its importance for Rad52 function (Mortensen et al, 2002). Phosphorylation of Y104 will place a negatively charged phosphate group within the Debye length of positively charged lysines, and therefore may attenuate DNA binding in addition to modulating subunit–subunit interface within the RAD52 ring. The Y104 modification should not affect binding of ssDNA into the positively charged groove of the primary ssDNA-binding site (Mortensen et al, 2002; Lloyd et al, 2005) located below and away from Y104 (Figure 1C).
Figure 1.
Incorporation of genetically encoded phosphotyrosine mimetic in place of Y104 of RAD52. (A) RAD52 domain organization. Regions responsible for DNA annealing and RPA/Rad51 interactions are indicated above schematic representation of the two domains constituting RAD52 monomer. Position of Y104 phosphorylation site in the N-terminal domain is marked by yellow circle labelled with P. (B) Ribbon diagram of the undecameric ring of truncated RAD521–212 (PDB ID: 1KN0). Y104 is shown in yellow space-filling model. Two adjacent subunits, chains A and B are coloured green and blue. Y104 of chain B and three surrounding lysines are shown in the inset. (C) Locations of primary and secondary DNA binding sites on the surface of RAD521–212 protomer. The primary ssDNA and secondary dsDNA binding sites are coloured in magenta and cyan, respectively. Previously identified ssDNA binding residues (R55, Y65, K152, R153 and R156, magenta) are gathered at the bottom of the narrow ssDNA-binding groove, while K102/K103 secondary DNA-binding region is located above the groove (cyan). The Y104 is coloured in yellow. (D) Experimental strategy for pCMF incorporation using amber suppressor technology. (E) Chemical structure of phosphotyrosine and pCMF. (F) SDS–PAGE confirming successful RAD52Y104pCMF expression. Lane 1, M (standards); lane2, wild-type RAD52; lanes 3–5, soluble fractions of E. coli lysate and lanes 6–8, insoluble fractions.
Although regulation by c-ABL kinase points to RAD52 phosphorylation as a means to regulate its engagement in DNA repair, how Y104 phosphorylation affects RAD52 activities remains unclear. Moreover, acquiring homogenously phosphorylated protein is technically challenging, which impedes in vitro study of phosphotyrosine effect. To address how Y104 phosphorylation modulates RAD52 function, we used an orthogonal pair of amber suppressor tRNA and aminoacyl tRNA synthetase (Xie et al, 2007) to successfully incorporate a stable phosphotyrosine analogue p-Carboxymethyl-L-phenylalanine (pCMF) into RAD52. The RAD52Y104pCMF displayed diminished dsDNA-binding capacity that contributed to targeting of RAD52Y104pCMF specifically to ssDNA and facilitated annealing of ssDNA and ssDNA–RPA complexes. These findings support the mechanism whereby c-ABL kinase transiently activates RAD52 in response to DNA damage by targeting it to the DNA repair intermediates requiring annealing.
Results
Incorporation of phosphotyrosine mimetic at the Y104 site of RAD52
To mimic both aromatic residue and negative charge of phosphotyrosine, we substituted Y104 of RAD52 with chemically stable phosphotyrosine analogue, pCMF. Uniform incorporation was achieved by using an artificially evolved orthogonal pair of amber suppressor transfer RNA (MjtRNACUApCMF) and aminoacyl tRNA synthetase MjTyrRS (schematically depicted in Figure 1D; Xie et al, 2007). pCMF has been established as a good structural and functional mimetic of phosphotyrosine because its side chain (CH2COO−) spatially overlaps with the two of phosphate oxygen atoms and carries a negative charge similar to that of phosphate group (Tong et al, 1998; Burke et al, 1999; Figure 1E). RAD52Y104pCMF was expressed using E. coli Rosetta strain transformed with both pET15b-RAD52Y104TAG(amber stop codon) and pSUPT-UaaRS (encodes MjtRNACUApCMF/MjTyrRS expressed from proK and araBAD promoters, respectively) plasmids. The production of soluble full-length RAD52Y104pCMF required supplementation of E. coli culture with IPTG, Rifampicin and pCMF (Figure 1F, lanes 3–5). There was no detectable incorporation of natural amino acids at position 104, as the full-length RAD52 (49.4 kDa) was not observed in the absence of pCMF (Figure 1F, lane 4). Instead, IPTG induction in the absence of pCMF yielded insoluble RAD521–104 truncation product (Figure 1F, lane 7), which was easily separated from soluble RAD52Y104pCMF (Figure 1F, lanes 5 and 8). Uniform incorporation of pCMF in the purified protein was confirmed by MALDI-TOFMS analysis. Notably, we found that Rifampicin, an inhibitor of bacterial RNA polymerase, significantly enhances expression of the full-length RAD52Y104pCMF (Supplementary Figure S1). The reduced RAD52Y104pCMF expression in the absence of Rifampicin was likely due to improper incorporation of pCMF into native E. coli proteins. Roughly 10% of E. coli genes contain TAG (amber) stop codon (Nakamura et al, 2000).
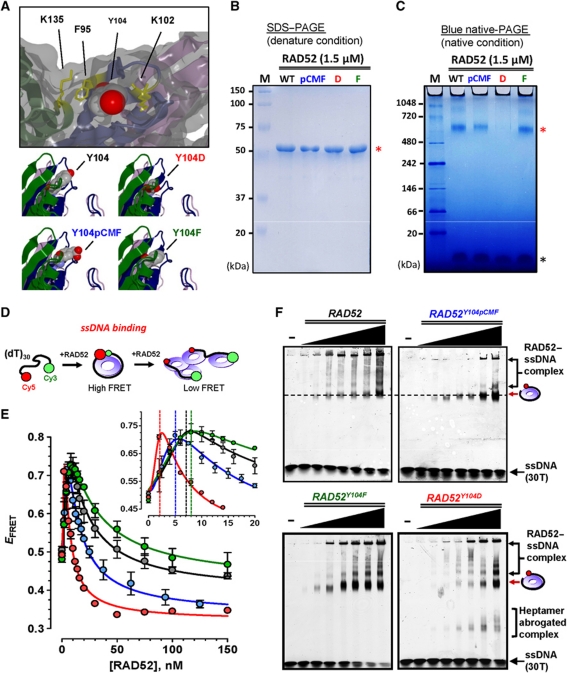
Tyr104 replacement with pCMF, Asp and Phe differentially modulates RAD52 oligomeric state
In general, negatively charged Asp and Glu serve as convenient mimetics of phosphorylated Ser and Thr. These residues, however, are not ideal substitutions for phosphotyrosine, which contains an aromatic ring that can participate in stacking and hydrophobic interactions (Muratore and Cole, 2007). Upper panel of Figure 2A shows local environment of Tyr104 in N-terminal domain of RAD52 oligomer (chain colour corresponds to Figure 1B). Although the hydroxyl group of Tyr104 targeted by c-ABL is exposed on the protein surface and is close to the basic residues forming the dsDNA-binding site of RAD52, most of the aromatic ring is buried in the hydrophobic core. Replacement of aromatic ring with negatively charged short side chain might affect interactions at the subunit–subunit interface (Figure 2A, bottom panels). To confirm this notion, we purified RAD52Y104D and RAD52Y104F along with wild-type RAD52 and RAD52Y104pCMF. Equal amounts (1.5 μM) of four proteins were separated on denaturing SDS–PAGE to confirm protein purity (Figure 2B). Non-denaturing blue native-PAGE (BN-PAGE) was performed to compare distributions of oligomeric states for the native and mutant RAD52 proteins (Figure 2C). Wild-type RAD52 migrated primarily as a 677-kDa species, as determined by the molecular mass standards (Figure 2C, red asterisk). At this protein concentrations, RAD52 exists in predominantly heptameric form (7mer) revealed by analytical ultracentrifugation (Kagawa et al, 2002) and scanning transmission electron microscopy (Stasiak et al, 2000). However, size exclusion chromatography, which, similar to BN-PAGE, is sensitive to the shape of the molecule, estimated molecular weights for RAD52 between 540 kDa (Deng et al, 2009) and 960 kDa (Lloyd et al, 2002). Slower than expected migration of 346 kDa RAD52 heptamer is likely due to its doughnut-like ring structure and/or ‘floppy’ C-terminal region (Ranatunga et al, 2001). Therefore, we concluded that the species migrating as 677 kDa band represent heptamers of RAD52. Minor slower migrating species (30.3% of total RAD52) are likely double and triple heptamers produced by stacking of RAD52 rings. Majority of RAD52Y104pCMF migrated similar to the RAD52 heptamer, with 12.2% displays a slower migrating species. The band corresponding to the heptamer was barely noticeable for RAD52Y104D and was significantly broadened. The diffused migration profile of RAD52Y104D likely reflects heterogeneity of its oligomeric states. The difference in oligomeric state distributions between RAD52 and RAD52Y104D was additionally supported by analysis of bleaching transitions within Cy3-labelled surface-tethered proteins (Supplementary Figure S2). Migration profile of RAD52Y104F was identical to that of the wild-type RAD52.
Figure 2.
Both charge and shape of pCMF are important for mimicking Y104 phosphotyrosine of RAD52. (A) A close-up view of Y104 site in ribbon and surface representation of the RAD52 N-terminal domain. Y104 is shown as space-filling model while surrounding residues are depicted as yellow stick models (upper panel). A side view of Y104 with Y104pCMF, Y104D and Y104F substitutions (lower panels). (B) Analysis of RAD52, RAD52Y104pCMF, RAD52Y104D and RAD52Y104F by SDS–PAGE stained with Coomassie Brilliant Blue. Molecular mass markers are indicated on the left. Red asterisk marks position of 6 × His–RAD52. (C) Analysis by non-denaturing BN-PAGE. Molecular weight markers are indicated on the left. Red asterisk marks position of RAD52 heptamer. Black asterisk marks position of Digitonin, which was added to BN-PAGE to improve solubility of the sample. (D) Cartoon depiction of the ssDNA-binding assay. RAD52 heptamer is shown as a purple ring. (E) Titration experiments were performed by additions of indicated concentrations of RAD52 (black circles), RAD52Y104pCMF (blue circles), RAD52Y104D (red circles) or RAD52Y104F (green circles) to a reaction mixture containing 1 nM Cy5-(dT)30-Cy3. The data points and error bars represent averages and standard deviations for three independent titrations. EFRET denotes FRET efficiency. (F) Gel mobility shift assays using 50 nM Cy5-(dT)30-Cy3 for wild-type RAD52 and RAD52 mutants. Protein concentrations are 50, 100, 200, 350, 500, 650 and 800 nM from lanes 2 to 8. The respective positions of the protein–DNA complexes are indicated by arrows on the right of the gel.
RAD52Y104pCMF maintains a biphasic ssDNA-binding mode characteristic of wild-type RAD52 protein
The ssDNA-binding site spans the perimeter of predominantly heptameric RAD52 rings (Singleton et al, 2002; Lloyd et al, 2005). We previously determined that search for complementary sequences and ssDNA annealing proceed via sequential rearrangements of the RAD52–ssDNA complexes (Rothenberg et al, 2008) and that RAD52–wrapped ssDNA complexes are the active species in ssDNA annealing (Grimme et al, 2010). These studies also allowed us to establish the FRET-based assays that reflect on the wrapped form of RAD52 oligomers. To examine the effect of Y104pCMF substitution on ssDNA binding and wrapping, we carried out FRET-based analyses of RAD52 binding to poly (dT)30 ssDNA labelled with a donor Cy3 fluorophore at one terminus and with an acceptor Cy5 fluorophore at the other (shown as cartoon illustration in Figure 2D and described in detail in Grimme et al, 2010 and Grimme and Spies, 2011).
Protein titration to 1 nM Cy5-(dT)30-Cy3 showed that at subsaturating concentrations RAD52 and all RAD52 mutants readily bound to and distorted the ssDNA substrate, resulting in a linear increase in FRET with increasing protein concentration (high FRET phase; Figure 2E). The highest EFRET was achieved for the substrate at approximately stoichiometric amounts of DNA (1 nM) to RAD52 (8 nM), RAD52Y104pCMF (5 nM) and RAD52Y104F (8 nM), respectively (Figure 2E, inset). A distinct protein concentration dependence of EFRET signal produced by RAD52Y104D (Figure 2E, red circles) suggests that its binding mode or a fraction of intact protein rings is substantially different from that of other RAD52 variants.
Further increase in RAD52 concentration resulted in EFRET decrease, which represents the unwrapping and stretching of the ssDNA and was attributed to the binding of multiple RAD52 oligomers to the same DNA molecule (low FRET phase). Difference in protein concentration corresponding to maximum EFRET between RAD52 (8 nM) and RAD52Y104pCMF (5 nM) suggests that Y104pCMF substitution could have induced conformational rearrangement of the oligomeric ring. Similar to the wild-type RAD52, RAD52Y104pCMF was capable of binding to and wrapping of ssDNA–RPA complex (Supplementary Figure S3).
To evaluate the distributions of RAD52–ssDNA bound species, we carried out the electrophoretic DNA mobility shift assays using substoichiometric concentrations of RAD52 (Figure 2F). In all cases, the dominant mobility shifted species corresponded to a single RAD52 heptamer bound to ssDNA (position indicated by red arrow on the right of the gels). In addition, complexes where multiple RAD52 heptamers were associated with the same ssDNA molecule formed a ladder of slower migrating RAD52–ssDNA complexes. The amount of these slower migrating high-order nucleoprotein complexes was significantly reduced in the case of RAD52Y104pCMF (upper-right panel of Figure 2F). This could be an indication that either pCMF modification affects RAD52 heptamer–heptamer interaction and thereby attenuates high-order complex formation, or that the phosphomimetic containing protein binds tighter to ssDNA in the wrapped conformation reducing the fraction of mobility shifted species where the same ssDNA molecule is partially wrapped around the two protein rings. The gel-based ssDNA-binding assays revealed the presence of RAD52Y104D–ssDNA complexes migrating faster than ssDNA bound to an intact heptamer, which we attributed to ssDNA binding by the abrogated heptamers.
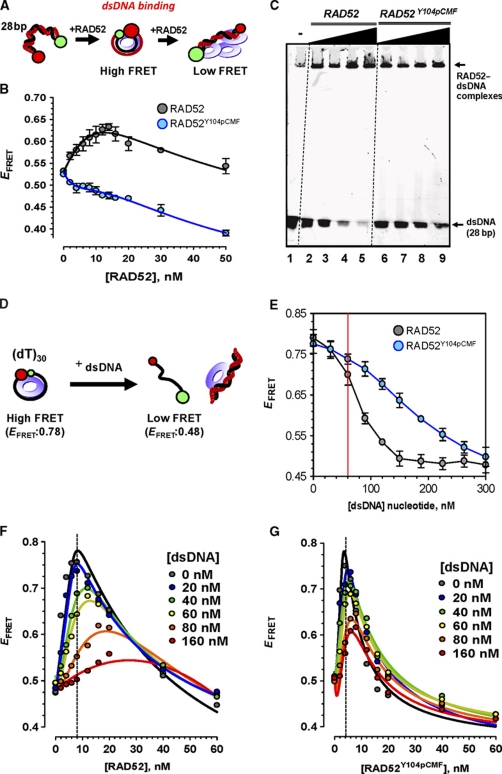
Y104pCMF substitution negatively affects dsDNA binding by RAD52
RAD52 binds to dsDNA and distorts it resulting in increase in FRET between two dyes incorporated at the ends of oligonucleotide size duplex (Grimme et al, 2010). While RAD52 bound Cy5-28bp-Cy3 in a bent conformation (as depicted in Figure 3A) and increased EFRET (Figure 3B, black circles), RAD52Y104pCMF exhibited no increase in EFRET (Figure 3B, blue circles), indicating lower capacity to bind or to distort dsDNA. At higher concentration of either RAD52 or RAD52Y104pCMF, we observed decrease in EFRET, which is likely due to the stretching of dsDNA by multiple RAD52 rings bound to the duplex or the ends that are not completely paired. Gel mobility shift assay confirmed that RAD52 bound and shifted dsDNA in a concentration-dependent manner with most RAD52–dsDNA complexes trapped in the well (Figure 3C, lanes 2–5). Capacity of RAD52Y104pCMF to bind and shift dsDNA was lower compared with the wild-type RAD52 (Figure 3C, lanes 6–9). Only 5.2% of 50 nM dsDNA remained free in the presence of 600 nM wild-type RAD52, while 41.5% of dsDNA remained free in the same concentration of RAD52Y104pCMF (Figure 3C, lanes 5 and 9).
Figure 3.
RAD52Y104pCMF is deficient in dsDNA binding compared with unmodified RAD52. (A) Cartoon representation of dsDNA–RAD52 complexes and corresponding FRET states. (B) Titrations were performed by additions of RAD52 (black circles) or RAD52Y104pCMF (blue circles) at indicated concentrations to a solution containing 1 nM Cy5-28bp-Cy3. (C) Gel mobility shift assays using 50 nM Cy5-28bp-Cy3. Protein concentrations are 100, 200, 400 and 600 nM from lanes 2 to 5 and lanes 6–9 for RAD52 and RAD52Y104pCMF, respectively. (D) Cartoon representation of the experimental strategy used in competition titration experiments. (E) Stoichiometric RAD52–Cy5-(dT)30-Cy3 complex was challenged with linearized ΦX174 dsDNA. Release of ssDNA from RAD52 (black circles) or RAD52Y104pCMF (blue circles) was observed as decrease in FRET between Cy3 and Cy5 dyes. EFRET value of protein-free ssDNA substrate (0.48) is indicated by dotted line. (F, G) EFRET dependence on RAD52 (F) and RAD52Y104pCMF (G) concentrations in the presence of various dsDNA concentrations. Stoichiometric amounts of RAD52 (8 nM) and RAD52Y104pCMF (5 nM) are marked by dotted line.
RAD52Y104pCMF overcomes the inhibitory effect of dsDNA on ssDNA wrapping
Kagawa et al (2001, 2008) proposed that dsDNA binding to the secondary DNA-binding site on RAD521–212 prevents ssDNA binding into the primary ssDNA-binding site of the RAD52 ring. In agreement with this model, competition titration experiments (Figure 3D) in which linearized dsDNA (5386 bp, ΦX174 RFI digested with XhoI) was titrated to stoichiometric complex of RAD52 (8 nM) bound to dually labelled ssDNA (1 nM molecules; 30 nM nucleotides) showed ssDNA release (unwrapping) detected as decrease in EFRET (Figure 3E, black circles). When concentration of competitor dsDNA reached 60 nM nucleotides (same molar concentration as ssDNA), 29.5% of ssDNA was unwrapped (Figure 3E, red line). Almost complete (95.1%) release of ssDNA was achieved when dsDNA concentration reached to 150 nM (nucleotides). In contrast, much smaller inhibitory effect was observed when a stoichiometric RAD52Y104pCMF (5 nM) complex with ssDNA (1 nM molecules; 30 nM nucleotides) was challenged with the dsDNA (Figure 3E, blue circles). When concentration of dsDNA reached 60 nM (nucleotides), 17.2% of ssDNA was unwrapped. Moreover, only half (49.2%) of the ssDNA was released in the presence of 150 nM dsDNA. The difference between inhibition of the wild-type RAD52 and RAD52Y104pCMF for ssDNA wrapping by dsDNA confirmed that Y104pCMF substitution affected dsDNA-binding activity of RAD52.
If dsDNA does indeed compete with binding and wrapping of ssDNA by RAD52 by blocking access to the ssDNA-binding groove as proposed (Kagawa et al, 2008), we expected that increase in RAD52 concentration can compensate for EFRET decrease by recovering population of RAD52–ssDNA complexes. In agreement with this prediction, maximum EFRET was observed at higher concentrations of RAD52 when dsDNA was present in the reaction (Figure 3F). In contrast, all EFRET binding curves for RAD52Y104pCMF showed EFRET maximum at the same protein concentration (Figure 3G), suggesting that dsDNA binding is impaired in RAD52Y104pCMF.
Competition experiment was performed by adding unlabelled non-complementary and complementary ssDNA to stoichiometric (7 nM protein per 1 nM of DNA substrate) wrapped RAD52 or RAD52Y104pCMF and Cy5-(dT)30-Cy3 complex (Supplementary Figure S4). In both cases, labelled ssDNA and annealed dsDNA product were more efficiently released from RAD52Y104pCMF than from unmodified RAD52. This observation provides additional confirmation that pCMF incorporation in RAD52Y104pCMF affects secondary DNA-binding site, which serve as dsDNA-binding and secondary ssDNA-binding region.
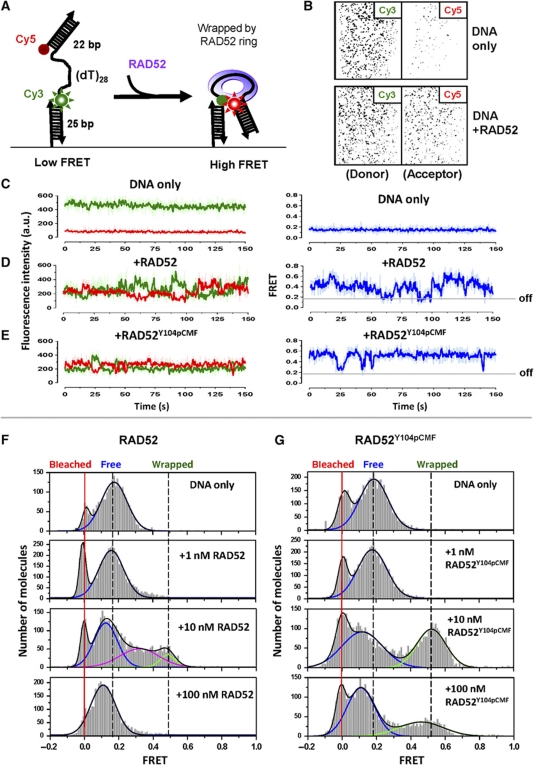
Y104pCMF modification targets RAD52 to single-stranded region of DNA molecules
Y104 phosphorylation facilitates localization of RAD52 protein at the sites of damaged DNA in vivo (Kitao and Yuan, 2002). To examine whether this localization is related to selective binding of phosphotyrosine mimetic RAD52 to ssDNA and not to dsDNA, we employed single-molecule FRET (Ha et al, 1996) based on total internal reflection fluorescence microscopy (TIRFM). Single-molecule FRET analysis can follow binding of RAD52 and RAD52Y104pCMF to and their dynamics on the gapped DNA. The DNA substrate was dually labelled with Cy3 and Cy5 fluorophores at the opposite ends of 28-nt-long ssDNA region flanked by dsDNA regions (Figure 4A). Gapped DNA substrate was immobilized on a PEG-coated surface of the TIRFM flow chamber via biotin–neutravidin interaction. Similar to the bulk experiments as Figure 2E, binding of RAD52 to and wrapping of ssDNA yielded high FRET signal due to proximity of Cy3 (donor) to Cy5 (acceptor), allowing us to distinguish between free and wrapped conformations of (dT)28 ssDNA region of individual surface-tethered DNA substrates. Figure 4B shows donor (Cy3) and acceptor (Cy5) channel images of the gapped DNA substrate, before and after 5 min incubation with 10 nM of RAD52. Each spot represents single dually labelled DNA molecule. Addition of RAD52 results in wrapping of ssDNA region, which brings the Cy3 and Cy5 dyes located on the flanking dsDNA closer together manifesting an increase in FRET between them, decrease in emission in the Cy3 donor channel and corresponding increase in the emission in the acceptor Cy5 channel. To monitor changes in FRET efficiency in real time (time resolution of 100 ms), we extracted and analysed Cy3, Cy5 and FRET trajectories for individual molecules (Figure 4C–E). Protein-free gapped DNA showed stable low FRET trajectories (Figure 4C). Upon binding of RAD52, FRET increased and continuously fluctuated between intermediate and high FRET states, while the RAD52 oligomer remained bound to DNA (Figure 4D). Binding of RAD52Y104pCMF yielded higher FRET values and less fluctuation compared with RAD52 (Figure 4E).
Figure 4.
Single-molecule analysis of RAD52 and RAD52Y104pCMF binding to gapped DNA reveals different DNA-binding dynamics. (A) A gapped DNA was immobilized on a PEG-coated surface of the slide via biotin–neutravidin interaction. Binding of RAD52 or RAD52Y104pCMF to ssDNA region was expected to bring Cy3 (donor) to Cy5 (acceptor) to proximity resulting in a high FRET signal. (B) Immobilization of gapped DNA to the surface gives rise to hundreds of fluorescent spots in donor channel (upper panels). Addition of RAD52-induced high FRET state manifested in appearance of fluorescent signals in the acceptor channel (lower panels). (C–E) Representative Cy3 and Cy5 trajectories (left; fluorescent intensity is plotted in arbitrary units) and FRET efficiency trajectory (right) from a single DNA molecule. (C) 50 pM DNA only, (D) in the presence of 10 nM RAD52 and (E) in the presence of 10 nM RAD52Y104pCMF. Raw time trajectories are shown in light colour, while five-point averaged traces are shown in dark colour. (F, G) FRET histograms of gapped DNA substrates at different RAD52 (F) and RAD52Y104pCMF (G) concentrations. Note that FRET values around and below zero (marked by red line) represent incomplete Cy5-22 annealed or Cy5 photo-bleached DNA molecules and were excluded from the analysis.
The distributions of distinct FRET species can also be analysed by examining a large number (2000–6000) of molecules collectively (Figure 4F and G). The low-to-high FRET transition was observed when protein concentration increased up to 10 nM of either RAD52 or RAD52Y104pCMF proteins (Figure 4F and G, panel 10 nM). FRET histogram recorded in the presence of 10 nM RAD52 was fitted to a three-state Gaussian distribution, resulting in low FRET peak with a median value of 0.18 (free DNA, individual peak shown in blue), high FRET peak with a median value of 0.49 (corresponds to the wrapped DNA, green), which included 9.47% of the molecules) and the third intermediate FRET peak with a median value of 0.32 (magenta), which included 36% of molecules. In contrast, FRET histograms recorded for RAD52Y104pCMF fitted well to a two-state Gaussian distribution with distinct high FRET peak with mean FRET value around 0.52. At 10 nM RAD52Y104pCMF, ∼43.5% molecules were found on average in this high FRET state (Figure 4G, panel 10 nM). This difference in FRET distributions suggested that RAD52Y104pCMF wraps ssDNA region of gapped DNA forming a more stable complex than unmodified RAD52. When the protein concentration was increased to 100 nM, the population of molecules in high FRET states decreased and majority of molecules assumed a new FRET state, which was lower than the initial FRET. Similar to the bulk binding experiments (Figure 2E), high concentration of RAD52 or RAD52Y104pCMF caused extension of DNA due to binding of multiple RAD52 oligomers.
High frequency of FRET fluctuations (Figure 4D) and less stable wrapping (Figure 4F) of unmodified RAD52–gapped DNA complex can be explained by two alternative scenarios: it can be generated by the internal dynamics of RAD52–gapped DNA complex, where equilibrium between multiple conformational states and slow transitions between the states lead to the FRET fluctuation; it can also be explained as association with and dissociation of RAD52 protomers and/or oligomers from RAD52–gapped DNA complex. To clarify which of the two explanations is correct, we carried out flow experiments. After 5 min incubation of RAD52 with surface-tethered gapped DNA to form RAD52–DNA complex, free RAD52 remaining in the reaction mixture was removed by extensive washing with protein-free buffer (Supplementary Figure S5A and B). As evident from the FRET histograms, most of RAD52 or RAD52Y104pCMF remained bound to DNA after washing. The FRET trajectories displayed fluctuations, which could now be attributed only to partial DNA wrapping and unwrapping of the bound RAD52 or RAD52Y104pCMF oligomers. Moreover, binding experiments carried out in the presence of low (5 nM) RAD52 or RAD52Y104pCMF concentrations displayed similar fluctuations with less frequent binding events (Supplementary Figure S5C and D). Based on these observations, we concluded that these FRET fluctuations reflect the internal dynamics of complexes formed by the oligomers of RAD52 or RAD52Y104pCMF on the gapped DNA substrate.
Y104pCMF modification confines RAD52 diffusion to the ssDNA region of gapped DNA, resulting in a stable wrapped complex
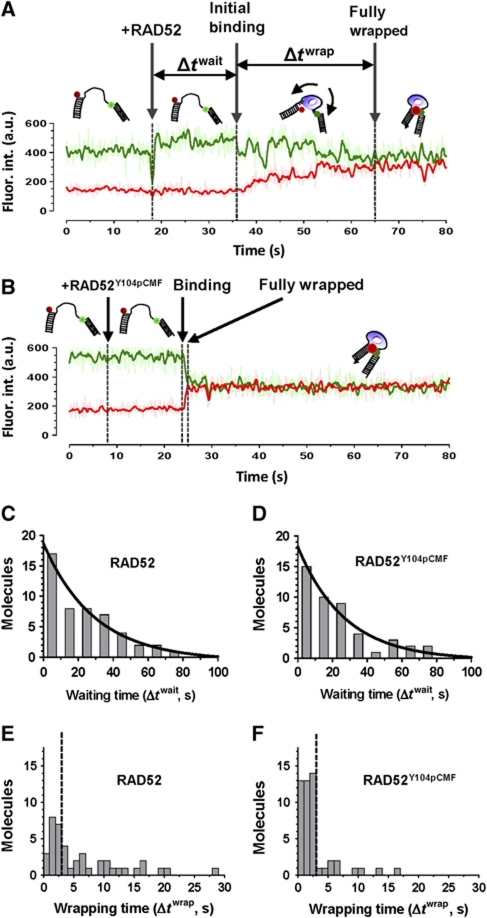
To capture the initiation of DNA binding and wrapping events by RAD52 or RAD52Y104pCMF, we carried out flow experiments where RAD52 or RAD52Y104pCMF was injected into the reaction chamber after recording the signal produced by the protein-free DNA (Figure 5A and B). Distributions of waiting times between protein injection and initiation of protein binding to DNA (Δtwait) followed single exponential decay with the apparent rate (inverse time constant twait multiplied by 10 nM protein concentration) of 0.49 (μs−1) and 0.53 (μs−1) for RAD52 and RAD52Y104pCMF, respectively (Figure 5C and D). Initiation and propagation of DNA binding events was observed by monitoring transition from the constant low FRET signal of protein-free DNA (high Cy3 intensity and low Cy5 intensity) to the fully wrapped high FRET state. The initial wrapping time (Δtwrap) was different for each molecule, and the distributions were different for unmodified RAD52 and RAD52Y104pCMF (Figure 5E and F). In wild-type RAD52, 57% (24/42) of recorded binding events displayed slow wrapping (defined as Δtwrap >3 s, shown as dotted line in Figure 5E and F), while only 12% (8/50) of the events recorded for RAD52Y104pCMF represented slow wrapping.
Figure 5.
Initiation of DNA wrapping by RAD52 and RAD52Y104pCMF. (A, B) Representative fluorescent trajectories depicting flow experiments probing initiation of RAD52-gapped DNA binding. After recording was started, 10 nM of RAD52 (A) or Rad52Y104pCMF (B) in standard buffer was injected in the reaction chamber. The time point of RAD52 injections, initiation of binding and complete wrapping are indicated by arrows. (C, D) Distribution of waiting times (Δtwait, waiting time for protein binding) displays a single exponential behaviour, with time constant is half lifetime. (E, F) Distributions of wrapping times.
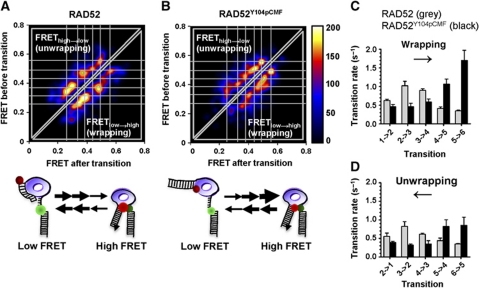
To examine dynamics of DNA wrapping and unwrapping by RAD52, steps and transition rates between number of FRET states were analysed using a hidden Markov model (HMM)-based analysis (McKinney et al, 2006; example of fitting shown as Supplementary Figure S6). HMM method analyses and fits the data statistically without imposing a preconceived model (described in Materials and methods). In all, 2385 and 1837 FRET transitions were extracted from FRET trajectories of 50 gapped DNA molecules for RAD52 and RAD52Y104pCMF, respectively, and visualized as a two-dimensional transition density plots (TDPs; Joo et al, 2006; McKinney et al, 2006) shown in Figure 6A and B. Constant low FRET trajectories representing protein-free DNA were omitted from the analysis. Five FRET transition peaks in both FRETlow → high (wrapping) and FREThigh → low (unwrapping) region of TDP were identified with six FRET states (0.24, 0.3, 0.38, 0.45, 0.51 and 0.57, indicated by white lines). Symmetrical pattern of the observed transitions indicates that the reaction reached equilibrium. The overall TDP patterns were similar between RAD52 and RAD52Y104pCMF. The sequential and periodic FRET increase or decrease correlation pattern in TDP suggests that DNA binding and release by both RAD52 and RAD52Y104pCMF involves step-by-step wrapping or unwrapping. The six FRET states identified for 28 nt ssDNA region of gapped DNA substrate suggest a step size comparable to previously reported four nucleotide DNA-binding site size of RAD52 protomer determined in DNA foot printing assays (Singleton et al, 2002). Figure 6C and D depicts wrapping and unwrapping modes explaining the FRET transitions for the wild-type RAD52 (grey) and RAD52Y104pCMF (black). Six FRET states identified from TDP analysis were labelled as 1–6 from low to high FRET and the transition rate between the individual FRET states was calculated from the number of FRET transition (in corresponding FRET transition peaks in Figure 6A and B). Overrepresentation of low and intermediate FRET states in the TDP for the unmodified RAD52 arise from decreased rates of 2 → 3, 3 → 4, 4 → 5 and 5 → 6 transitions and slightly increased rates of 1 → 2 to 2 → 3 transitions for both wrapping and unwrapping (Figure 6C and D, grey bars). As results, the higher FRET states were the least stable for RAD52–gapped DNA complex. In contrast, there was a clear bias towards highest FRET state indicative of fully wrapped ssDNA portion of the substrate defined by faster wrapping towards high FRET states for RAD52Y104pCMF–gapped DNA complex (Figure 6C and D, black bars). We assumed that inhibition of dsDNA binding by Y104pCMF substitution is likely responsible for efficient RAD52Y104pCMF localization on ssDNA region while partial unwrapping of wild-type RAD52 from ssDNA region was stabilized by RAD52–dsDNA interaction as illustrated in lower panel of Figure 6A and B. This distinct type of nucleoprotein dynamics was observed only on the gapped DNA, which at least in part, restricts the proteins to ssDNA region: When 58 nt ssDNA with Cy3 and Cy5 separated by (dT)28 spacer was used as the substrate (Supplementary Figure S7), both RAD52 and RAD52Y104pCMF induced similar FRET fluctuations which we attributed to diffusion of RAD52 and RAD52Y104pCMF on ssDNA as was observed for E. coli SSB using a similar approach (Roy et al, 2009).
Figure 6.
RAD52Y104pCMF localizes to and stably wraps ssDNA region of gapped DNA. (A, B) Transition density plot (TDP) analyses. Occurrences of distinct FRET transitions are plotted against two-dimensional FRET (before) and FRET (after) plane. Six FRET states were observed for both RAD52 (A) and RAD52Y104pCMF (B) and are indicated by white lines. Cartoons under each TDP represent models explaining gapped DNA-binding mode of RAD52 and RAD52Y104pCMF, respectively. (C, D) Comparison of both wrapping (C) and unwrapping (D) transition rates between different FRET states for RAD52 (grey) and RAD52Y104pCMF (black).
RAD52Y104pCMF displayed faster ssDNA annealing rate
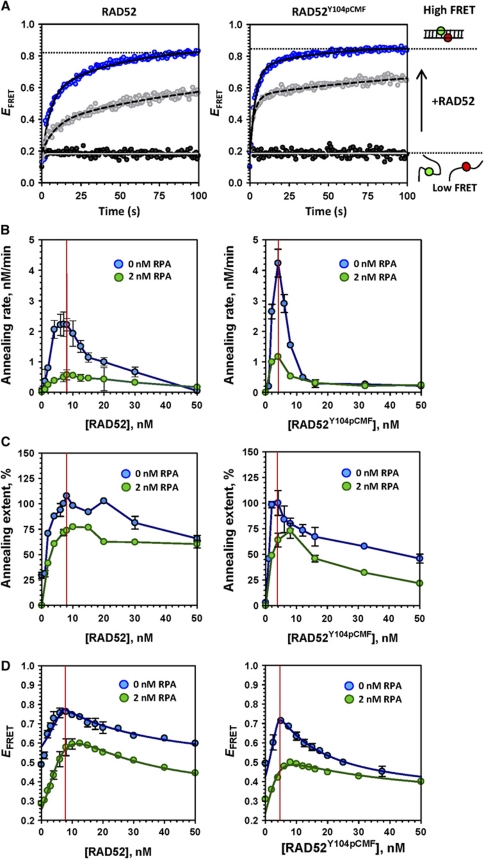
To determine whether difference in substrate selectivity between RAD52 and RAD52Y104pCMF affects DNA annealing activity, we compared the ability of two proteins with anneal complementary oligonucleotides and ssDNA–RPA complexes using bulk annealing experiments (Grimme et al, 2010). Two complementary oligonucleotides (0.5 nM each) were internally labelled with Cy3 and Cy5 dyes and were separately pre-incubated with RAD52 or RAD52Y104pCMF. Annealing reaction was initiated by mixing the two half-reactions. Increase in EFRET over time indicated formation of DNA duplex which brings the two dyes in close proximity (Figure 7A). The assays yielded the initial rates of annealing and the annealing extents (fraction of dsDNA%, as the reaction reaches the equilibrium). Initial annealing rate of the RAD52Y104pCMF (4.23 nM/min) was ∼2-fold higher than that of unmodified RAD52 (2.22 nM/min; Figure 7A, blue circles). Addition of 2 nM non-complementary linearized ΦX174 dsDNA significantly reduced annealing rate (2.22–1.18 nM/min) of unmodified RAD52, while more moderate decrease in the annealing rate was observed for RAD52Y104pCMF (4.23–3.54 nM/min; Figure 7A, grey circles). The faster annealing by phosphomimetic containing protein is likely due to alleviation of the inhibition by the dsDNA, which is also a product of annealing reaction.
Figure 7.
Y104pCMF substitution promotes RAD52-mediated annealing of ssDNA and ssDNA–RPA complexes. (A) FRET-based annealing assays were performed using 0.5 nM of T-28(Cy3) and complementary P-28(Cy5). Increase in EFRET over time indicated ssDNA annealing (formation of DNA duplex which brings the two dyes in close proximity) and the data were fitted to a double exponential. Representative annealing curves depict reactions with 8 nM of RAD52 and 4 nM of RAD52Y104pCMF in the absence (blue circles) or presence (grey circles) of non-complementary dsDNA (2 nM molecule). Black circles represent complementary DNA substrates in the absence of RAD52 as negative control. (B) Initial rates of annealing reactions using RPA-free ssDNA (blue circles) and 2 nM RPA-coated ssDNA (green circles). (C) Extents of the annealing reactions were calculated from the amplitude of EFRET change for each condition after reaction got plateau. (D) The EFRET trends for RAD52 and RAD52Y104pCMF binding to Cy5-(dT)30-Cy3 (1 nM) in the absence (blue) or presence (green) of 2 nM RPA are shown for comparison of optimal binding and annealing conditions.
The dependencies of initial rates of annealing (Figure 7B, blue circles) and the annealing extents (Figure 7C, blue circles) on RAD52 or RAD52Y014pCMF concentration were examined. The fastest annealing was achieved with 8 nM of RAD52 and 4 nM of RAD52Y014pCMF (Figure 7B). Y104pCMF modification of RAD52 may facilitate dsDNA product dissociation and increase the turnover rate of ssDNA annealing or progression of the homology search, since the positive effect of phosphomimetic for ssDNA annealing rate was only observed at substoichiometric protein–DNA ratios (Figure 7B). At oversaturating protein concentrations (>8 nM), the annealing rate as well as extent was reduced especially in the presence of RAD52Y104pCMF (Figure 7B and C). Notably, the fastest annealing was observed at stoichiometric protein–DNA ratios for both RAD52 and RAD52Y104pCMF (Figure 7B–D, red line), indicating that RAD52–wrapped ssDNA complexes are the active species in ssDNA annealing by RAD52Y104pCMF similar to that previously reported for the unmodified RAD52 (Grimme et al, 2010). Similar experiments were carried out using ssDNA saturated with RPA (Figure 7B–D, green circles). Although annealing still occurred, the presence of RPA (2 nM) negatively affected both the rate and extent of annealing by RAD52 and RAD52Y104pCMF. ssDNA annealing rate of RAD52Y104pCMF at stoichiometric binding conditions (1.17 nM/min) was higher than that of unmodified RAD52 (0.57 nM/min), suggesting that Y104pCMF substitution enhances RAD52-mediated ssDNA annealing even in the presence of RPA.
In contrast to RAD52Y104pCMF, ssDNA annealing activity of RAD52Y104D was significantly reduced compared with the wild-type RAD52 (Supplementary Figure S8), confirming our prediction that the shape and negative charge of pCMF make it a better mimetic of phosphotyrosine than aspartic acid.
Previously, we proposed (Rothenberg et al, 2008) that the reason why efficient RAD52-mediated annealing requires both DNA strands to be wrapped around two RAD52 oligomeric rings is to initiate reaction by testing and extending homology starting from a short stretch (likely four nucleotides) to prevent annealing between heterologous sequences. We compared ssDNA annealing activity of RAD52 and RAD52Y104pCMF using complementary oligos containing 0, 3 and 6 mismatches (Supplementary Figure S9). Both rate and extent of annealing by either RAD52 or RAD52Y104pCMF were affected by the presence of mismatches. Because homology search occurs at the peripheral outer curved surface of RAD52 ring, limited length over which the homology is probed would be expected. After the initial homology is found, the newly formed short duplex would be excluded from the narrow DNA-binding groove and the two protein rings will shift gear to probe whether the homology is extended over the next few nucleotides. The presence of heterology will shift the thermodynamic balance towards ssDNA bound by the RAD52 ring versus an imperfect duplex. Similar effect of DNA mismatches on annealing by unmodified and phosphotyrosine mimetic RAD52 suggested that the homology search mechanism and the shape of the oligomer are likely to be very similar for RAD52 and RAD52Y104pCMF.
Discussion
We demonstrated here that amber suppressor technology for site-specific incorporation of non-hydrolysable phosphotyrosine analogue pCMF (Xie et al, 2007) can be applied to investigate post-translational regulation of DNA repair protein by tyrosine kinase. Although not identical, both pCMF and phosphotyrosine contain an aromatic ring and similarly positioned negative charges making pCMF a more suitable phosphotyrosine mimetic than traditionally used Asp and Glu. Our observation that Y104D substitution interfered with the RAD52 ring integrity (Figure 2C and F) and ssDNA annealing (Supplementary Figure S8) suggests that the aromatic ring of tyrosine (preserved in phosphotyrosine and pCMF) has a role in stabilizing the ring structure of RAD52 competent for annealing. Genetic incorporation of pCMF allows uniform modification of the protein in comparison with in vitro phosphotyrosine modification by purified kinase, which rarely achieves high modification efficiency. To confirm that the effect elicited by pCMF incorporation is similar to actual phosphorylation, we carried out in vitro phosphorylation of RAD52 by a constitutively active v-ABL kinase fragment (Supplementary Figure S10A). The phosphorylated RAD52 displayed bound gapped DNA. Its binding behaviour was somewhat in between that of the wild-type and pCMF containing proteins and was different from RAD52Y104D mutant (Supplementary Figure S10B and C). This intermediate behaviour is likely due to incomplete phosphorylation—that is, not all monomers within each ring are phosphorylated and different individual oligomers may have different fraction of phosphorylated and unmodified monomers. In contrast to non-uniform and somewhat promiscuous in vitro phosphorylation, pCMF provides more controlled way to address the effect of tyrosine phosphorylation in vitro.
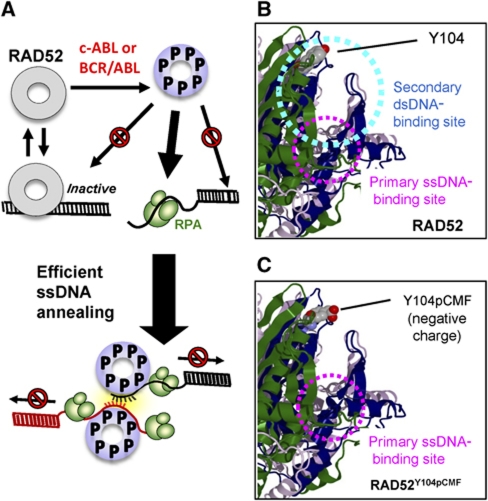
Figure 8A shows a cartoon model of the mechanism by which tyrosine phosphorylation upregulates ssDNA annealing by RAD52. Annealing activity of unmodified RAD52 is reduced due to sequestration of the RAD52 to dsDNA and to the double-stranded products of the annealing reaction. When the cell experiences DSBs, ATM kinase is activated and in turn activates c-ABL kinase by Ser/Thr phosphorylation. Activated c-ABL kinase phosphorylates RAD52, which was mimicked in our studies by RAD52 protein containing phosphotyrosine mimetic residue (pCMF) at Y104 site. The most critical distinction between unmodified RAD52 and RAD52Y104pCMF was a marked reduction in dsDNA-binding activity of RAD52Y104pCMF and ensuing ability to overcome the inhibitory effect of dsDNA on ssDNA binding and annealing (Figures 3 and 6). Figure 8B and C shows a close view of RAD52 N-terminal domain of unmodified RAD52 and RAD52Y104pCMF. Y104 residue targeted by c-ABL kinase and Y104pCMF substitution are shown in space-filling model; ssDNA (magenta) and dsDNA (cyan) binding regions are indicated by dotted circles. Relative spatial arrangement of the two binding sites suggests that the dsDNA molecule bound to the secondary DNA-binding site can sterically hinder the access of ssDNA into the primary ssDNA-binding site (Kagawa et al, 2008). Appearance of a negative charge among the residues comprising the secondary DNA-binding site is likely to inhibit dsDNA binding by the ionic repulsion between negatively charged phosphotyrosine (or pCMF) and DNA phosphodiester backbone, resulting in an unhindered path for ssDNA to bind in the primary ssDNA-binding site. This allows phosphorylated RAD52 to preferentially localize to the resected ssDNA region of damaged DNA, possibly promoting ssDNA annealing during SSA and the second-end capture step of recombinational DSB repair.
Figure 8.
Model of c-ABL mediated RAD52 activation through Y104 phosphorylation. (A) Y104 phosphorylation increases the active and free RAD52 heptamer population and decreases the population of ssDNA annealing-inefficient high-order complex as well as RAD52 trapped by dsDNA. Consequently, RAD52Y104pCMF localizes to RPA-coated ssDNA substrates and promote efficient ssDNA annealing. (B) A close view of ribbon representation of the RAD52 N-terminal domain. Putative ssDNA (magenta) and dsDNA (cyan) binding regions are indicated by dotted circles. Y104 residue targeted by c-ABL kinase is shown in space-filling model. (C) Negative charge at the gating path for ssDNA activates RAD52 by preventing dsDNA from blocking ssDNA access to primary binding site.
We previously reported that the search for regions of extended complementarity and ssDNA annealing proceed via sequential rearrangements of the RAD52–ssDNA complexes (Rothenberg et al, 2008). Single-molecule analyses in this study revealed that spontaneous repositioning of RAD52 on gapped DNA occurs with discrete step size (Figure 6) and that RAD52 or RAD52Y014pCMF can diffuse on ssDNA (Supplementary Figure S7) similar to SSB diffusion on ssDNA (Roy et al, 2009). This dynamic nature of RAD52 bound to DNA may help RAD52 to distribute along resected DNA while searching for the region of adequate complementarity as well as expanding ssDNA annealing region after initial pairing. Y104pCMF modification stimulates RAD52 localization to and wrapping of ssDNA region of DNA by inhibiting RAD52 diffusion into dsDNA region. Furthermore, the modification accelerated ssDNA annealing rate. The ring shape of RAD52 and its ability to mediate annealing of two complementary strands (or complementary ssDNA–RPA complexes) wrapped around the two RAD52 rings would ensure that the initial pairing of ssDNA annealing occurs by probing homology a few nucleotides at a time, and therefore would only involve highly homologous repeats (Supplementary Figure S9). Interestingly, K102A and K133A mutant of RAD521–212 are defective in ssDNA annealing (Kagawa et al, 2008). Therefore, placing a negative charge between these positively charged residues of the secondary dsDNA-binding site produces the effect distinct from removal of these residues.
RAD52 is unique among the many proteins that promote annealing of complementary DNA strands in that it can mediate annealing of ssDNA molecules protected by ssDNA-binding protein, RPA (New et al, 1998; Shinohara and Ogawa, 1998; McIlwraith and West, 2008; Grimme et al, 2010). In contrast to yeast Rad52 protein which functions both as a strand annealing protein and as a recombination mediator, the presence of distinct HR mediators such as BRCA2 likely dedicate human RAD52 primarily to ssDNA annealing (McIlwraith et al, 2000; Jensen et al, 2010; Liu et al, 2010). Tyrosine phosphorylation events are transient with <4 min lifetime after the tyrosine kinase is turned off due to cellular tyrosine phosphatase activity (Qiao et al, 2006). The mechanism whereby rapid signalling occurs via transient tyrosine phosphorylation and dephosphorylation is a characteristic of multicellular organisms that need to control cell proliferation (Lim and Pawson, 2010). Tyrosine 104 of RAD52 is not conserved in yeast (Supplementary Figure S11). Neither is c-ABL kinase. Brief lifetime of the phosphorylated RAD52 may restrict its activity to ssDNA annealing steps of homology-directed repair. Saccharomyces cerevisiae Rad52 is also phosphorylated. This phosphorylation, however, occurs in the species-specific C-terminal domain, is cell-cycle dependent and DNA damage independent (Antunez de Mayolo et al, 2006; Barlow and Rothstein, 2009), and therefore is likely to have role completely different from tyrosine phosphorylation in human RAD52.
Constitutive activation of the c-ABL due to BCR–ABL fusion is a key event in the pathogenesis of chronic myeloid leukaemia and other myeloproliferative disease (Shtivelman et al, 1986; Clark et al, 1988). BCR/ABL enhances RAD51 expression and phosphorylates Y315, resulting in drug resistance such as cisplatin and mitomycin c (Slupianek et al, 2001). Among other consequences of constitutive c-ABL activation, BCR/ABL expressing leukaemia cells display upregulated SSA (Fernandes et al, 2009) accompanied by enhanced nuclear localization of RAD52 (Cramer et al, 2008). The two pathways of homology-directed DNA repair, HR and SSA, compete with each other and the ratio between the two pathways is balanced by assembly of stable RAD51 filaments (Kass and Jasin, 2010). RAD52-meadiated annealing of relatively short homologous sequences located in the vicinity of a DNA break serves important function in the RAD51-independent SSA pathway of homology-directed DNA repair (Stark et al, 2004). Therefore, BCR/ABL kinase may break the balance towards HR to mutagenic SSA through RAD52 phosphorylation. Further study will be required to clarify how c-ABL influence homology-directed repair and the consequence of BCR/ABL expression.
Materials and methods
FRET-based DNA binding and annealing assays
FRET-based DNA binding and annealing analyses were carried out essentially as previously described (Grimme et al, 2010). A detailed description of the substrates and methods is provided in Supplementary data. Standard deviation <4% of main value is overlapped with the data point, and therefore blinded from the graph.
Gel-based DNA-binding assay
The Cy5-(dT)30-Cy3 ssDNA (50 nM) was mixed with RAD52 (50, 100, 200, 350, 500, 650 and 800 nM) in 10 μl of standard reaction buffer, containing 30 mM HEPES-Acetate (pH 7.5) and 1 mM DTT. The reaction mixtures were incubated at 37°C for 10 min, fixed with 0.1% glutaraldehyde and were analysed using non-denaturing 3.5% polyacrylamide gel electrophoresis in TBE buffer (90 mM Tris (pH 8.0), 64.6 mM boric acid and 2 mM EDTA). The resolved species were visualized using a Typhoon 9400 fluorescence imager (GE Healthcare) by exciting and monitoring Cy5 fluorescence, and analysed using ImageQuant software. The Cy5-28bp-Cy3 duplex DNA (50 nM) was mixed with RAD52 (100, 200, 4000 and 600 nM) and subjected to DNA-binding assay as described above.
Reaction conditions for the single-molecule assay
Gapped DNA was produced by annealing of Bio-25, 22-(dT)28-Cy3-25 and Cy5-22 substrates. Gapped DNA was immobilized on a quartz surface (Finkenbeiner), which was coated with polyethyleneglycol in order to eliminate non-specific surface adsorption of proteins (Ha et al, 2002). The immobilization was mediated by biotin–neutravidin interaction between biotinylated DNA, neutravidin (Pierce), and biotinylated polymer (PEG-MW 5000, Nectar Therapeutics). The standard buffer contained 50 mM Tris-Acetate (pH 7.5), 1 mM DTT and the oxygen scavenging system consisting of 1 mg/ml glucose oxidase (Sigma), 0.4% (w/v) D-glucose (Sigma), 0.04 mg/ml catalase (Roche) and 1 mM Trolox (6-hydroxy-2,5,7,8-tetramethyl-chromane-2-carboxylic acid, Sigma) (Rasnik et al, 2006). Cy3 dye was excited using 532 nm laser and the density of tethered DNA was confirmed by counting the surface-associated Cy3 moieties. RAD52 was then added and incubated for 5 min at 25°C in the standard buffer before start recording unless otherwise indicated.
Single-molecule data acquisition and data analysis
TIRFM was used to excite fluorophores present on the immobilized molecules. Cy3 fluorescence was excited by a frequency-doubled Nd:YAG laser (532 nm, 75 mW, Crysta-Laser). The fluorescence signals originated from the Cy3 and Cy5 dyes were collected by a water immersion objective × 60 (Olympus), passed through a 550-nm long-pass filter to block out laser scattering, separated by a 630 nm dichroic mirror and detected by EMCCD camera (Andor) with a time resolution of 100 ms. Single-molecule trajectories were extracted from the recorded video file by IDL software. FRET transitions were globally analysed using HMM-based statistically approach with an initial assumption of 10 distinct FRET states (McKinney et al, 2006).
BN-PAGE analysis
Recombinant RAD52 and RAD52 mutants were suspended in standard buffer (50 mM Bis-Tris (pH 7.2), 50 mM NaCl, 10% glycerol and 0.001% Ponceau S) containing 0.5% Digitonin. The mixtures were separated by BN-PAGE using 4–16% gradient Native PAGE gels at 25°C as described previously (Swamy et al, 2006). A molecular weight calibration kit (NativeMark Protein Standards, Invitrogen) was used as a standard for molecular weight estimation.
Supplementary Material
Acknowledgments
We thank JM Grimme and S Lin for experimental help; P Yau (Proteomics facility, UIUC) for mass spectroscopic analysis; Dr P Shultz (Scripps Research Institute) for providing pSUPT-UaaRS plasmid and AsisChem Inc. for pCMF synthesis. MS is an Early Career Scientist with Howard Hughes Medical Institute (HHMI). TH is an investigator with HHMI. This work was also supported by American Cancer Society grant RSG-09-182-01-DMC to MS and by the NIH grant GM 065367 and NSF grant 0646550 to TH.
Author contributions: MH and MS designed research; MH and YO performed research; JY contributed new reagents/analytic tools; MH analysed data; and MH, TH and MS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agami R, Blandino G, Oren M, Shaul Y (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399: 809–813 [DOI] [PubMed] [Google Scholar]
- Antunez de Mayolo A, Lisby M, Erdeniz N, Thybo T, Mortensen UH, Rothstein R (2006) Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res 34: 2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R (2009) Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J 28: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB, Wang JY (1997) Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature 387: 516–519 [DOI] [PubMed] [Google Scholar]
- Burke TR Jr, Luo J, Yao ZJ, Gao Y, Zhao H, Milne GW, Guo R, Voigt JH, King CR, Yang D (1999) Monocarboxylic-based phosphotyrosyl mimetics in the design of GRB2 SH2 domain inhibitors. Bioorg Med Chem Lett 9: 347–352 [DOI] [PubMed] [Google Scholar]
- Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff SP, Wu Y, Arlinghaus R, Baltimore D, Gasser PJ, Park MS, Sung P, Lee EY (1999) Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem 274: 12748–12752 [DOI] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA (2003) Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cell Biol 23: 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SS, McLaughlin J, Timmons M, Pendergast AM, Ben-Neriah Y, Dow LW, Crist W, Rovera G, Smith SD, Witte ON (1988) Expression of a distinctive BCR-ABL oncogene in Ph1-positive acute lymphocytic leukemia (ALL). Science 239 (4841 Part 1): 775–777 [DOI] [PubMed] [Google Scholar]
- Cong F, Tang J, Hwang BJ, Vuong BQ, Chu G, Goff SP (2002) Interaction between UV-damaged DNA binding activity proteins and the c-Abl tyrosine kinase. J Biol Chem 277: 34870–34878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, Alt FW, Jasin M (2004) Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev 18: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer K, Nieborowska-Skorska M, Koptyra M, Slupianek A, Penserga ET, Eaves CJ, Aulitzky W, Skorski T (2008) BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res 68: 6884–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Prakash A, Dhar K, Baia GS, Kolar C, Oakley GG, Borgstahl GE (2009) Human replication protein A-Rad52-single-stranded DNA complex: stoichiometry and evidence for strand transfer regulation by phosphorylation. Biochemistry 48: 6633–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC (2007) Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14: 468–474 [DOI] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN (2011) Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci USA 108: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes MS, Reddy MM, Gonneville JR, DeRoo SC, Podar K, Griffin JD, Weinstock DM, Sattler M (2009) BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood 114: 1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foray N, Marot D, Randrianarison V, Venezia ND, Picard D, Perricaudet M, Favaudon V, Jeggo P (2002) Constitutive association of BRCA1 and c-Abl and its ATM-dependent disruption after irradiation. Mol Cell Biol 22: 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res 38: 2917–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme JM, Spies M (2011) FRET-based assay to monitor DNA binding and annealing by RAD52 recombination mediator protein. Methods Mol Biol 745: 463–483 [DOI] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S (1996) Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci USA 93: 6264–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419: 638–641 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T (2006) Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126: 515–527 [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kagawa A, Saito K, Ikawa S, Shibata T, Kurumizaka H, Yokoyama S (2008) Identification of a second DNA binding site in the human Rad52 protein. J Biol Chem 283: 24264–24273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T (2001) Homologous pairing promoted by the human Rad52 protein. J Biol Chem 276: 35201–35208 [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S (2002) Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell 10: 359–371 [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8: 483–489 [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M (2010) Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett 584: 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S, Pandey P, Jin S, Inoue S, Bharti A, Yuan ZM, Weichselbaum R, Weaver D, Kufe D (1997) Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature 386: 732–735 [DOI] [PubMed] [Google Scholar]
- Kharbanda S, Ren R, Pandey P, Shafman TD, Feller SM, Weichselbaum RR, Kufe DW (1995) Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature 376: 785–788 [DOI] [PubMed] [Google Scholar]
- Kitao H, Yuan ZM (2002) Regulation of ionizing radiation-induced Rad52 nuclear foci formation by c-Abl-mediated phosphorylation. J Biol Chem 277: 48944–48948 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Pawson T (2010) Phosphotyrosine signaling: evolving a new cellular communication system. Cell 142: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JA, Forget AL, Knight KL (2002) Correlation of biochemical properties with the oligomeric state of human rad52 protein. J Biol Chem 277: 46172–46178 [DOI] [PubMed] [Google Scholar]
- Lloyd JA, McGrew DA, Knight KL (2005) Identification of residues important for DNA binding in the full-length human Rad52 protein. J Mol Biol 345: 239–249 [DOI] [PubMed] [Google Scholar]
- Mazloum N, Holloman WK (2009) Second-end capture in DNA double-strand break repair promoted by Brh2 protein of Ustilago maydis. Mol Cell 33: 160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Van Dyck E, Masson JY, Stasiak AZ, Stasiak A, West SC (2000) Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J Mol Biol 304: 151–164 [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, West SC (2008) DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell 29: 510–516 [DOI] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91: 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Erdeniz N, Feng Q, Rothstein R (2002) A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161: 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore KE, Cole PA (2007) A lock on phosphotyrosine signaling. ACS Chem Biol 2: 454–456 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T (2000) Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Molina H, Pandey A, Zhang J, Cole PA (2006) Chemical rescue of a mutant enzyme in living cells. Science 311: 1293–1297 [DOI] [PubMed] [Google Scholar]
- Ranatunga W, Jackson D, Flowers IR II, Borgstahl GE (2001) Human RAD52 protein has extreme thermal stability. Biochemistry 40: 8557–8562 [DOI] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T (2006) Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods 3: 891–893 [DOI] [PubMed] [Google Scholar]
- Rich T, Allen RL, Wyllie AH (2000) Defying death after DNA damage. Nature 407: 777–783 [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol 18: 6423–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E, Grimme JM, Spies M, Ha T (2008) Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc Natl Acad Sci USA 105: 20274–20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Kozlov AG, Lohman TM, Ha T (2009) SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P (2006) Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem 281: 11649–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, Egerton M, Shiloh Y, Kharbanda S, Kufe D, Lavin MF (1997) Interaction between ATM protein and c-Abl in response to DNA damage. Nature 387: 520–523 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Popova M, Fleury F, Kobayashi M, Hayashi N, Sakane I, Kurumizaka H, Venkitaraman AR, Takahashi M, Yamamoto K (2009) c-ABL tyrosine kinase stabilizes RAD51 chromatin association. Biochem Biophys Res Commun 382: 286–291 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407 [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E (1986) Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell 47: 277–284 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA 99: 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T (2001) BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell 8: 795–806 [DOI] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M (2004) Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH (2000) The human Rad52 protein exists as a heptameric ring. Curr Biol 10: 337–340 [DOI] [PubMed] [Google Scholar]
- Sung P (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197 [DOI] [PubMed] [Google Scholar]
- Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW (2006) Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci STKE 2006: pl4. [DOI] [PubMed] [Google Scholar]
- Symington LS (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–670, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Schild D (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res 477: 131–153 [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Warren TC, Lukas S, Schembri-King J, Betageri R, Proudfoot JR, Jakes S (1998) Carboxymethyl-phenylalanine as a replacement for phosphotyrosine in SH2 domain binding. J Biol Chem 273: 20238–20242 [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M (2006) Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 5: 1065–1074 [DOI] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Wray J, Liu J, Nickoloff JA, Shen Z (2008) Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res 68: 2699–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Supekova L, Schultz PG (2007) A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol 2: 474–478 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol 18: 6430–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Komatsu K, Wang HG, Kufe D (2002) c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol Cell Biol 22: 3292–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Fan MM, Sawyers C, Kharbanda S, Kufe D (1996) Genotoxic drugs induce interaction of the c-Abl tyrosine kinase and the tumor suppressor protein p53. J Biol Chem 271: 26457–26460 [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, Kufe D (1998) Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem 273: 3799–3802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.