Abstract
In multicellular organisms, morphogenesis relies on a strict coordination in time and space of cell proliferation and differentiation. In contrast to animals, plant development displays continuous organ formation and adaptive growth responses during their lifespan relying on a tight coordination of cell proliferation. How developmental signals interact with the plant cell-cycle machinery is largely unknown. Here, we characterize plant A2-type cyclins, a small gene family of mitotic cyclins, and show how they contribute to the fine-tuning of local proliferation during plant development. Moreover, the timely repression of CYCA2;3 expression in newly formed guard cells is shown to require the stomatal transcription factors FOUR LIPS/MYB124 and MYB88, providing a direct link between developmental programming and cell-cycle exit in plants. Thus, transcriptional downregulation of CYCA2s represents a critical mechanism to coordinate proliferation during plant development.
Keywords: A2-type cyclins, differentiation, G2-to-M, proliferation, transcriptional repression
Introduction
After germination, the minimal body plan of the seedling is elaborated by iterative organ development that will shape the adult plant. Each new organ is formed according to a predictable pattern, which reflects a complex interplay between plant hormones and developmental programs (De Veylder et al, 2007). One of the targets of morphogenetic cues is the modulation of local cell proliferation and differentiation. Because plant cells cannot move within the plant body due to their rigid cell walls, cell proliferation must be highly controlled in time and space. While recent studies provide insights into the coordination of plant development and cell-cycle regulation, only a few connections between these processes have been identified at the molecular level (Brownfield et al, 2009; Sozzani et al, 2010; Xie et al, 2010).
Cell proliferation is characterized by consecutive cycles of DNA replication (Synthesis; S-phase) and cell division (Mitosis; M-phase). S-phase is preceded by G1-phase, when cells prepare for DNA synthesis, and M-phase by G2-phase, when cells prepare to divide. The orderly transition between phases depends largely on oscillations of Cyclin-Dependent Kinase (CDK) activity. Recently, it was shown that thresholds of CDK activity delineate independent cell-cycle phases (Coudreuse and Nurse, 2010), providing support for a quantitative model of cell-cycle progression. Importantly, CDK activity is modulated at multiple levels. As monomers, CDKs are usually inactive due to a steric blockage of their catalytic cleft. Binding to a cyclin partner removes this block, and thus represents a major regulatory switch of CDK activity (Jeffrey et al, 1995). Further fine-tuning of CDK activity is achieved by phosphorylation, dephosphorylation and binding to several cofactors and/or inhibitors (Morgan, 1995, 1997; Inzé and De Veylder, 2006).
Compared with the relatively simple cell-cycle regulatory module in yeast, which includes just one major CDK and a few cyclins (CYC), higher eukaryotes harbour an elaborate repertoire of CDKs and cyclins. Here, the specialized phase- and tissue-specific expression of multiple CDKs and cyclins provides a wide combinatorial range that enables to deal with the increased complexity associated with multicellularity (De Veylder et al, 2007; Satyanarayana and Kaldis, 2009).
Animals utilize well-characterized D- and E-type cyclins which are expressed at the onset of cell division (G1-to-S) and which connect extracellular signals with the cell cycle (Matsushime et al, 1991; Koff et al, 1992; Motokura and Arnold, 1993; Payton and Coats, 2002). Moreover, A- and B-type cyclins are primarily restricted to G2-to-M phase, with A-type cyclins being more broadly expressed, starting as early as S-phase (Pines and Hunter, 1990; Fung and Poon, 2005). Such expression patterns suggest that they function specifically in respective phases of the cell cycle. However, in some cases the loss of one cyclin type can be compensated for by the expression of another cyclin type (Fisher and Nurse, 1996).
Based on sequence homology and conserved motifs, many core cell-cycle regulators have been annotated in plant genomes (Vandepoele et al, 2002). Interestingly, plants have many more cyclins compared with animals. As an example, the Arabidopsis genome encodes 10 A-type, 11 B-type and 10 D-type cyclins, but no E-type cyclins, whereas animal genomes usually code for 1 or 2 of each type. In plants, D-type and A3-type cyclins have been implicated in G1-to-S regulation (Dewitte et al, 2003, 2007; Takahashi et al, 2010), while subgroups of A- and B-type cyclins likely act in G2-to-M regulation (Schnittger et al, 2002; Imai et al, 2006; Boudolf et al, 2009; Ishida et al, 2010). The expanded number of cyclins in plants, compared with animals, might represent a mechanism that integrates a broader range of signals to control of proliferation. However, much of what is known about cyclins and plant cell-cycle regulation derives from gain-of-function analyses (Schnittger et al, 2002; Dewitte et al, 2003; Yu et al, 2003; Boudolf et al, 2009; Takahashi et al, 2010). Quantitative models suggest that the timing of cyclin expression controls differences in cell-cycle regulation (Fisher and Nurse, 1996; Coudreuse and Nurse, 2010), including in plants (Schnittger et al, 2002). Therefore, it is essential to define the phenotypic effects of loss of cyclin gene functions to understand their role in plant development.
Although there have been many advances in understanding the regulation of the plant cell cycle, it is still unclear how cell cycling is coordinated with differentiation during development. Components of the G1-to-S transition have been shown to control cell proliferation and differentiation events in shoots (Dewitte et al, 2003, 2007) and roots (Wildwater et al, 2005; Caro et al, 2007; Sozzani et al, 2010), which emphasizes the key role of this transition in the cell's decision to exit the cell cycle and activate differentiation. In addition, some differentiated plant cell types are known to undergo multiple rounds of DNA duplication without mitosis (endoreduplication; Melaragno et al, 1993), suggesting that cyclin downregulation at the G2-to-M transition could be part of a developmental mechanism that coordinates the switch between proliferation and endoreduplication.
Among putative G2-to-M regulatory cyclins, A2-type cyclins are poorly characterized in plants. In synchronized cell suspensions, their expression starts in S-phase and peaks during the G2-to-M transition (Reichheld et al, 1996; Shaul et al, 1996; Menges et al, 2005). Plant A2 cyclins have been shown to rescue the growth of yeast cyclin-deficient mutants (Setiady et al, 1995), and also induced Xenopus oocyte maturation (Renaudin et al, 1994), suggesting they act during entry into mitosis. Developmentally, CYCA2 expression is not obligately associated with cell proliferation, as it is also expressed in seemingly differentiated cells, such as the vascular tissues (Burssens et al, 2000) and developing trichomes (Imai et al, 2006). In the vascular tissues, it was proposed that CYCA2;1 expression reflects a competence to divide, while in trichomes CYCA2;3 acts to terminate endoreduplication. Indeed, cyca2;1, cyca2;3 and ilp1-1D mutants displaying reduced CYCA2 expression, exhibit increased ploidy levels (Imai et al, 2006; Yoshizumi et al, 2006), whereas overexpression of CYCA2;3 shows lower ploidy levels, combined with increased proliferation (Imai et al, 2006; Boudolf et al, 2009). Recently, auxin signalling has been implicated in the switch from proliferation to endoreduplication as it stimulates CYCA2;3 expression (Ishida et al, 2010). However, it is not clear if this is a direct or indirect effect.
Biochemical interaction studies revealed that plant CYCA2s can interact with a diverse set of CDKs as well as other cell-cycle regulatory proteins (Imai et al, 2006; Boudolf et al, 2009; Boruc et al, 2010b), suggesting that CYCA2s contribute to multiple CDK complexes that might reflect a broad array of biochemical events. Importantly, different CYCA2s have distinct and overlapping expression patterns (Burssens et al, 2000; Imai et al, 2006) corroborating the idea that tissue-specific co-expression with interaction partners is key to their function. Besides transcriptional regulation, CYCA2s degradation is an equally regulatory mechanism. The Anaphase Promoting Complex (APC) regulates CYCA and CYCB turnover via their destruction boxes (Marrocco et al, 2009). Moreover, CCS52A1-dependent activation of the APC mediates proteolysis of CYCA2;3 during the switch to endoreduplication (Boudolf et al, 2009). These complex regulatory mechanisms highlight the importance of tight control over the cell cycle.
Here, we address the functional requirement of the subfamily of plant A2-type cyclins in plant cell-cycle regulation in different developmental contexts and report a novel transcriptional repression mechanism that acts during terminal differentiation of guard cells.
Results
Sequence similarity (Vandepoele et al, 2002), co-regulation during the cell cycle (Menges et al, 2005), subcellular colocalization (Boruc et al, 2010a), common interaction partners (Boruc et al, 2010b) and mild phenotypes in single mutants (Imai et al, 2006; Yoshizumi et al, 2006) collectively suggest redundancy among the four CYCA2s in the Arabidopsis genome obscuring their functional analysis. To circumvent this obstacle, phenotypic effects of various combinations of multiple cyca2 loss-of-function mutants were analysed (Supplementary Figure S1).
CYCA2s regulate the G2/M transition in roots
Since CYCA2 expression is strongly associated with proliferative tissues, such as primary and lateral root meristems (Supplementary Figure S2), we probed the impact of their loss-of-function on root growth. Growth defects were apparent when three out of the four CYCA2s were mutated. Because the post-embryonic growth of the quadruple mutant was extremely slow, we preferentially analysed triple mutant combinations (Supplementary Figure S3). Triple mutants cyca2;134 and cyca2;234 had shorter roots and were impaired in lateral root formation compared with WT (Supplementary Figure S4). To determine whether these growth defects arose from an abnormal cell proliferation, root meristem phenotypes were analysed. Primary root meristems of cyca2;134 and cyca2;234 were smaller (Figure 1A; Supplementary Figure S5A and B) and contained fewer dividing cells than WT, as detected by antibodies to the cytokinesis-specific syntaxin KNOLLE (Lauber et al, 1997; Figure 1B; Supplementary Figure S5C). Similarly, developing lateral root primordia in cyca2;234 contained fewer cells than WT (Figure 1C), suggesting that cell proliferation defects underlie both reduced root length and lateral root formation.
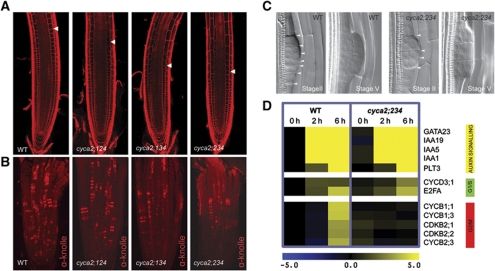
Figure 1.
cyca2 triple mutants have defects in cell-cycle progression. (A) Propidium iodide stained root meristems of WT, cyca2;124 (cyca2;1-1 cyca2;2-1 cyca2;4-1), cyca2;134 (cyca2;1-1 cyca2;3-1 cyca2;4-1) and cyca2;234 (cyca2;2-1 cyca2;3-1 cyca2;4-1) 10 days after germination. Arrowheads indicate the ends of meristems, defined as the position where cells start elongating. (B) Immunolocalization of the cytokinesis-specific syntaxin KNOLLE, labelling cells undergoing cytokinesis in roots of 7-day-old WT, cyca2;124, cyca2;134 and cyca2;234. (C) Stage II and stage V lateral root primordia of WT and cyca2;234 cleared with chloral hydrate. Lateral root primordia of cyca2;234 are composed of fewer cells than WT. Arrowheads indicate periclinal cell walls. Stages as defined previously (Malamy and Benfey, 1997). (D) Transcriptional responses of auxin signalling genes, G1/S and G2/M regulators in WT and cyca2;234 root segments during auxin-induced lateral root initiation. In all, 0, 2 and 6 h correspond to time of auxin treatment (10 μM) after being germinated in presence of the auxin transport inhibitor NPA (10 μM). Range indicator from blue to yellow represents expression levels on a log2 scale relative to NPA germinated WT (0 h).
To determine at which cell-cycle stage CYCA2s are prominently involved, cell-cycle progression was compared during synchronized lateral root initiation in WT versus cyca2;234 triple mutants (Figure 1D). In WT, expression of both auxin signalling and G1-to-S regulatory genes preceded the expression of G2-to-M regulatory genes (Figure 1D), as previously reported (Himanen et al, 2002, 2004; Vanneste et al, 2005). By contrast, expression of mitotic regulators, such as B-type cyclins, was no longer induced within the same time course in cyca2;234 mutants, whereas the expression of auxin signalling and G1-to-S regulatory genes was unaffected (Figure 1D). This delay in activation of mitotic regulators indicates that plant CYCA2s function early in the G2/M transition, as was predicted based on sequence homology (Vandepoele et al, 2002) and on expression patterns in synchronized cell suspensions (Menges et al, 2005). Moreover, it is likely that CYCA2s also function in S-phase, given that CYCA2;2/CDKA;1 can phosphorylate the S-phase regulator E2Fc in vitro (del Pozo et al, 2002). However, the lack of appropriate markers hampers such determination.
CYCA2s drive proliferation in leaves, while repressing endoreduplication
To obtain its characteristic final size and shape, leaf morphogenesis depends upon a tight coordination between cell proliferation, cell-cycle exit and differentiation. Early leaf development displays high cell division activity that is followed by a gradual tip-to-base deceleration of proliferation and the start of differentiation-associated endoreduplication and cell expansion (Donnelly et al, 1999; Beemster et al, 2006). The expression pattern of several CYCA2s also showed a comparable and dynamic gradient of expression (Supplementary Figure S6; Imai et al, 2006). Dramatic increases in ploidy levels and cell sizes were observed in the mature first true leaves of cyca2 triple mutants (Figure 2A and B). To address the mechanism driving enhanced ploidy levels and cell sizes, the development of cyca2;234 leaves was analysed in greater detail. Kinematic analysis of leaf growth showed lower cell division rates in cyca2;234 leaves compared with the WT (Figure 2C; Supplementary Figure S7). In addition, as soon as the first leaf pair became macroscopically visible (after 8 days of growth, Stage 1.02; Boyes et al, 2001), DNA content was already dramatically higher than the WT (Figure 2D; %2C, %4C and %8C). Moreover, ploidy levels continued to rise in cyca2;234 (after 14 days of growth), a period when endoreduplication had already stopped in the WT (Figure 2D; %16C and %32C). Thus, enhanced ploidy levels in cyca2;234 are the combined result of an early onset and extended duration of endoreduplication. Collectively, these phenotypic and molecular analyses in roots and shoots of cyca2 triple mutants demonstrate that plant CYCA2s are fundamental elements of the plant cell cycle, and, like their animal counterparts, function in early G2-to-M transition. Furthermore, the enhanced endoreduplication in these mutants is consistent with the observation that low CDK activity allows yeast cells in G2 to (re)enter the G1-to-S program without undergoing mitosis (Coudreuse and Nurse, 2010), suggesting that plant CYCA2s contribute to CDK activities that are required for mitosis.
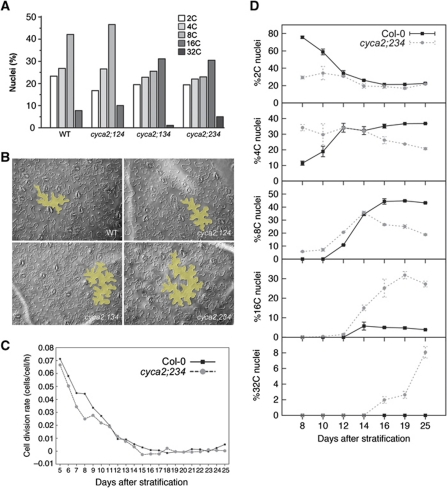
Figure 2.
Leaf development shows enhanced endoreduplication and slowed down cell-cycle progression in cyca2 triple mutants. (A) Distribution of nuclear ploidy in mature primary leaves of WT, cyca2;124, cyca2;134 and cyca2;234. Triple mutants cyca2;134 and cyca2;234 show highest ploidy levels. (B) Pavement cell size in mature primary leaves of WT, cyca2;124, cyca2;134 and cyca2;234. Yellow overlays highlight representative cells. (C) Kinematic analysis reveals a slowdown in cell division rates in developing primary leaves cyca2;234 compared with WT. (D) Evolution of ploidy levels during the development of WT and cyca2;234 primary leaves. In early stages, WT has predominantly 2C nuclei and a low 4C fraction. Later, the 2C fraction drops rapidly, while higher ploidy fractions increase until ∼16 days after stratification. In cyca2;234, the 2C fraction is already low at the earliest stage analysed, while the 4C fraction is already high and even a small fraction 8C nuclei can be detected. At later stages, higher ploidy fractions continue to increase, and do not saturate within the time frame of our analysis. Data are represented as mean±s.e.
Tissue-specific CYCA2 expression contributes to vascular proliferation near hydathodes
In addition to their expression in meristems, CYCA2s were expressed in the leaf; however, while CYCA2;2 and CYCA2;3 were expressed throughout the organ (Supplementary Figures S6, S8 and S9), the expression pattern of CYCA2;1 and CYCA2;4 remarkably mimicked the reticulate vein pattern of the leaf (Figure 3A; Supplementary Figure S9). Moreover, the promoter activities of these two genes in the leaf overlapped with one of the earliest hallmarks of the vascular precursor (‘preprocambial’) cell state (Figure 3B), the promoter activity of the homeodomain-leucine zipper (HD-ZIP) III gene, ATHB8 (Scarpella et al, 2004; Donner et al, 2009). The tissue-specific expression of CYCA2;1 and CYCA2;4 suggests that these CYCA2 genes function in leaf vascular development. Indeed, cyca2;234 leaves showed fewer vascular hypertrophy zones than the WT (Figure 3C and D); however, vascular defects in cyca2;234 were seemingly associated with changes in leaf shape resulting in leaves with fewer serration tips (Figure 3C and E). Additional mutation of the vascular-specific CYCA2;1 in the cyca2;234 background further reduced the number of vascular hypertrophy zones without additional effects on the number of serration tips (Figure 3D and E), data which are consistent with the tissue-specific expression pattern of CYCA2;1. Thus, vascular cell proliferation defects in cyca2 mutants likely derive from tissue-specific modulation of CYCA2 levels, rather than being secondary consequences of disrupted leaf growth.
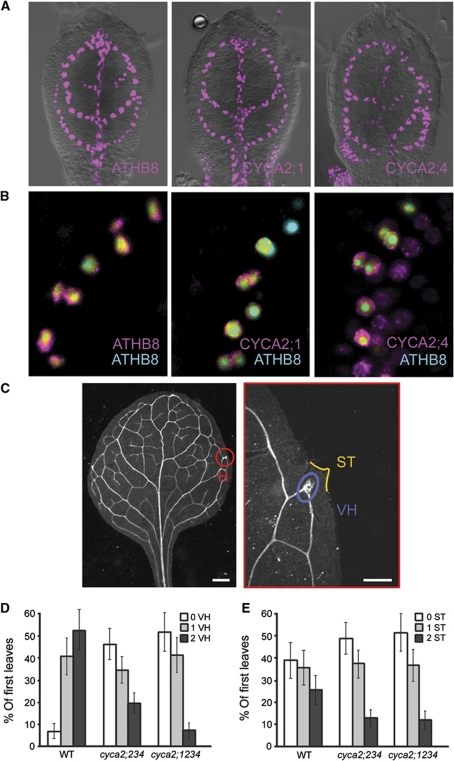
Figure 3.
Tissue-specific expression of CYCA2s is required for vascular cell proliferation. (A) Expression patterns of CYCA2;1pro:HTA6:EYFP and CYCA2;4pro:HTA6:EYFP in 4-DAG first leaves resemble that of ATHB8pro:HTA6:EYFP, which is an early hallmark of vascular development. (B) Co-expression of ATHB8pro:HTA6:EYFP, CYCA2;1pro:HTA6:EYFP and CYCA2;4pro:HTA6:EYFP with ATHB8pro:ECFP-Nuc in 4-DAG first leaves. Note how CYCA2;4 expression is initiated slightly earlier than ATHB8, and in wider expression domains that over time narrow to single cell files. In contrast, CYCA2;1 expression is initiated slightly later than ATHB8, but its expression is always confined to single cell files. Images colour-coded with a dual-channel LUT from cyan to magenta through green, yellow and red (Demandolx and Davoust, 1997). Preponderance of cyan signal over colocalized magenta signal is encoded in green, opposite in red and colocalized cyan and magenta signals of equal intensity in yellow. (C) Overview of cleared, mature first WT leaf and detail of hydathode (H) that shows vascular hypertrophy (VH) and serration tip (ST). (D, E) Percentage of mature first leaves showing zero, one or two zones of vascular hypertrophy (VH) (D) and serration tips (STs) (E). Plots represent mean values±s.e. Experiments were done in triplicate, and VH and ST were counted on the primary leaves (19⩽n⩽37) of each of the three genotypes. A generalized linear model with the multinomial distribution was fitted to the data, as implemented in Genstat (Payne, 2010). VH is significantly affected by genotype (P=3.79E−11), while ST is not (P=0.24).
Stomatal formation requires CYCA2 activity
Stomata consist of two guard cells around a pore whose regulation controls gas exchange between the shoot and the atmosphere. Their development requires at least one asymmetric division as well as a single symmetric division. After the latter division, which occurs in a guard mother cell (GMC) precursor, stomatal differentiation and morphogenesis take place (Bergmann and Sack, 2007). The leaf epidermis of cyca2;134 and cyca2;234, but not those of WT and cyca2;124, showed frequent occurrence of unpaired oval cells, displaying cell wall thickenings and plastid accumulations, trait characteristics of wild-type guard cells (Figure 4A). As in normal stomatal guard cells, these single cells were positioned above large intercellular spaces in the subjacent mesophyll (Figure 4B). Moreover, they expressed mature guard cell identity markers, KAT1pro:GUS (Nakamura et al, 1995) and ET1728 (Gardner et al, 2009; Figure 4C). Thus, these cells correspond to aberrant, single guard cells (SGCs) that are located where stomata would normally be found. These SGCs had twice the nuclear-DNA content (4C) of normal guard cells (2C) (Supplementary Figure S10), suggesting they are arrested in G2-phase. Yet, the aberrant cells attained a guard cell identity and formed SGCs instead of a pair.
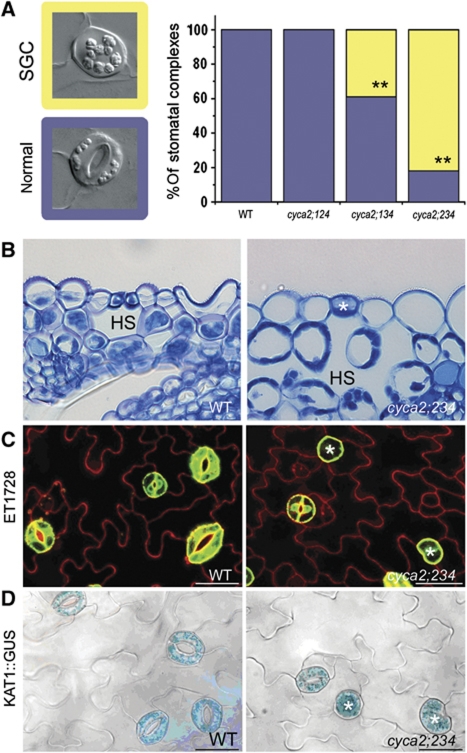
Figure 4.
Stomatal expression of CYCA2s is required for guard mother cell division. (A) Stomatal phenotypes (left) of WT and representative triple mutant. Bar chart: quantification of stomatal phenotypes. Asterisks indicate P<0.001; Fisher’s exact test (comparison with WT). The cyca2;234 triple mutant displays the highest frequency of single guard cells (SGCs). Blue=normal stoma; yellow=SGC. (B) Anatomical section through a WT stomatal complex and a cyca2;234 SGC showing correct placement of abnormal SGC (asterisk) over a hypostomatal space (HS). (C, D) Expression of mature guard cell identity markers; (C) ET1728 (GFP) and (D) KAT1pro∷GUS in WT and cyca2;234 (asterisks indicate SGCs).
Strikingly, SGCs could only be found in cyca2;3 mutant alleles and derived higher order cyca2 mutant combinations (Supplementary Table SI), suggesting that CYCA2;3 is a major contributing factor to this phenotype. However, while in single mutants the frequency of SGC formation is very low, additional mutations of other CYCA2 members resulted in dramatic increases in SGC frequencies (Supplementary Table SI). Collectively, these data demonstrate that CYCA2s are synergistically required for the symmetric division that is a prerequisite for stomatal formation, and that acquisition of guard cell identity occurs independently from GMC division.
CYCA2s and CDKB1s synergistically promote GMC division
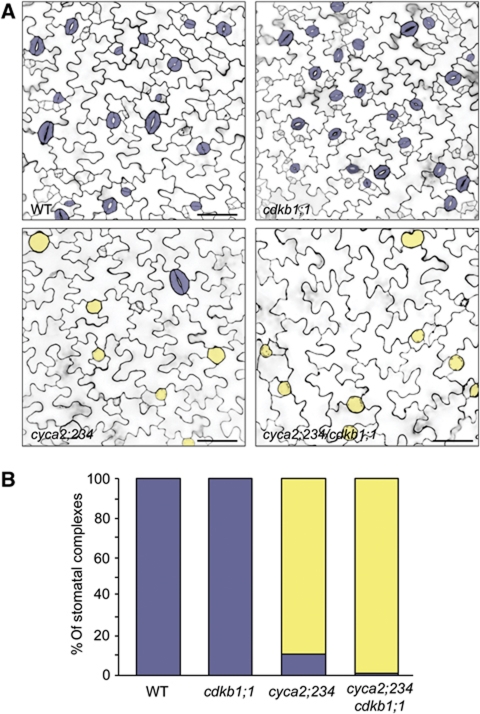
SGCs were previously reported in transgenic plants harbouring a CDKB1;1-N161 dominant-negative construct (Boudolf et al, 2004), as well as cdkb1;1 cdkb1;2 double mutants (Xie et al, 2010). Moreover, CDKB1;1 can form a functional complex with CYCA2;3 (Boudolf et al, 2009) and CDKB1;1 is expressed around the time of GMC symmetric division (Boudolf et al, 2004), suggesting that CYCA2s and CDKB1s directly interact in promoting the formation of a two-celled stoma. Indeed, while cdkb1;1 single mutants only had normal stomata, cyca2;234 cdkb1;1 quadruple mutants displayed even less SGCs than cyca2;234 triple mutants (Figure 5; Supplementary Figure S11). Thus, all four genes act synergistically in promoting GMC symmetric division, and thus stomatal morphogenesis.
Figure 5.
CYCA2;2, CYCA2;3, CYCA2;4 and CDKB1;1 genes synergistically promote guard mother cell symmetric division. (A) Micrographs of WT, cdkb1;1, cyca2;234 and quadruple cyca2;234/cdkb1;1 cotyledons of 4-day-old seedlings. Cell walls in 4-day-old developing cotyledons were visualized using propidium iodide and laser scanning confocal microscopy. False colouring highlights stomatal complexes: blue=normal stoma; yellow=SGC. (B) Quantification of stomatal phenotypes. Quadruple cyca2;234 cdkb1;1 mutant displays more SGCs than cyca2;234. (Fisher’s exact test, P=0.0004).
FLP and MYB88 regulate the timely repression of CYCA2;3 during terminal guard cell differentiation
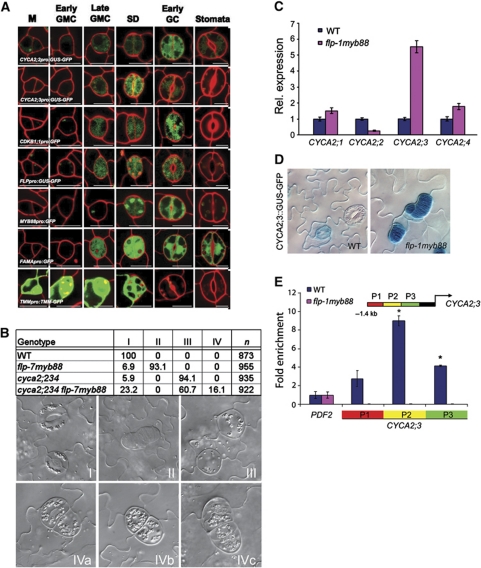
While for CYCA2;1 and CYCA2;4 no stomatal expression could be observed (Supplementary Figure S12), CYCA2;3 expression along with CDKB1;1 and CYCA2;2, was induced in late GMCs, remained high in young guard cells, but was strongly reduced or did even disappear in mature stomata (Figure 6A). Together with previously identified mutants that have supernumerary guard cells in stomatal complexes (Lai et al, 2005; Ohashi-Ito and Bergmann, 2006), the observed decline in cell-cycle gene expression at the end of stomatal development hints at the existence of an active repression mechanism. Loss-of-function mutations in two MYB transcription factors, FOUR LIPS (FLP/MYB124) and its paralogue MYB88 induce clusters of four or more guard cells (Lai et al, 2005). Loss-of-function in the basic helix-loop-helix protein FAMA (Ohashi-Ito and Bergmann, 2006) also results in cell clusters, but unlike those of flp myb88, without guard cell identity. The apparent independency from the stomata differentiation process renders FLP/MYB124 and MYB88 as potential candidate CYCA2 repressors, the more because they are expressed at roughly the same stages of stomatal development as CYCA2;2, and CYCA2;3 (Figure 6A).
Figure 6.
FLP/MYB124 and MYB88 are direct repressors of CYCA2;3 expression during guard mother cell (GMC) division. (A) Expression analysis of transcriptional promoter:reporter fusions (except for TMMpro:TMM:GFP translational fusion). FLP, MYB88 and FAMA, which encode transcription factors, are expressed in late GMCs, during symmetric division, and in young guard cells. TMM expression marks an earlier phase of stomatal development. CYCA2;2, CYCA2;3 and CDKB1;1 are expressed at similarly stages to FLP, MYB88 and FAMA. Each meristemoid (M) develops into a GMC. Late GMCs have thickened end walls that are usually bisected by the symmetric division (SD) that produces two young guard cells (GCs). The latter undergo further morphogenesis including stomatal pore formation. (B) Chart showing frequencies of different stomatal phenotypes in WT, flp-7 myb88, cyca2;234, and in cyca2;234 flp-7 myb88. Stomata in the wild type are normal by definition (type I stomata). In flp-7 myb88, many stomata are arranged in clusters (type II), while in cyca2;234 single guard cells (type III) predominate. In a cyca2;234 flp-7 myb88 quintuple mutant, most stomata are single-celled (type III), but some small clusters of stomata next to apparent single guard cells are present (type IV), suggesting a ‘fusion’ phenotype (IVa–c). (C) Mean relative expression levels of CYCA2;1, CYCA2;2, CYCA2;3, CYCA2;4 determined using Q-RT–PCR from cotyledons of WT and flp-7 myb88 seedlings 10 days after germination. CYCA2;3 expression in flp-7 myb88 was markedly higher than in WT (Col-0) (used for reference levels). (D) CYCA2;3pro:GUS:GFP in WT and in flp-1 myb88. CYCA2;3pro:GUS:GFP GUS levels are low or absent from mature guard cells in WT plants, but is strongly expressed in flp-1 myb88 stomatal clusters. (E) ChIP-Q–PCR on three fragments upstream (−1.4 kb) of the translational start of CYCA2;3 (P1–P3). PCR conducted on ChIPed DNA samples from 10-day-old wild-type and flp-1 myb88 shoots using FLP/MYB88 antibody. PDF2 is a negative control. The positions for PCR products in CYCA2;3 promoter are indicated. Strongest, specific binding was observed for P2. The error bars indicate the standard error from two biological replicates. Asterisk denotes a statistically significant difference (P<0.05).
To determine whether CYCA2 expression is required for the extra divisions found in flpmyb88, and/or fama backgrounds, we generated cyca2;234 fama-1 quadruple and cyca2;234 flp-7myb88 quintuple mutants. The cyca2;234 fama-1 plants did not show any SGCs, instead they formed clusters of cells that lacked guard cell identity; however, these clusters had fewer cells than fama-1 suggesting that fama-1 is only partly epistatic to cyca2;234 (Supplementary Figure S13). By contrast, the formation of stomatal clusters in a flp-7myb88 cyca2;234 background was completely suppressed (Figure 6B), demonstrating that CYCA2 gene products are required for the flp-7myb88 stomatal phenotype and that FLP and MYB88 might represent transcriptional regulators of CYCA2 expression. Therefore, we analysed CYCA2 expression in a flpmyb88 background. Ten days after sowing, cotyledons from flp-7myb88 seedlings showed about five-fold higher CYCA2;3 expression than the WT (Figure 6C). Moreover, in flp-1myb88 stomata, CYCA2;3 promoter activity remained high after the GMC division (Figure 6D), suggesting that FLP and MYB88 repress CYCA2;3 promoter activity. To test if this was a direct effect, we performed ChIP-Q-PCR using polyclonal antibodies raised against FLP and MYB88 (Xie et al, 2010). In the WT, CYCA2;3 promoter chromatin fragments were enriched after ChIP, while these were lost in flp-1myb88 mutants (Figure 6E), demonstrating a specific, direct interaction of FLP and MYB88 with CYCA2;3 chromatin. Thus, FLP and MYB88 appear to restrict CYCA2;3 transcription after GMC division via direct interaction with its promoter.
Discussion
CYCA2s modulate the G2-to-M transition
Several findings led to the initial assumption that plant A-type cyclins function in S-phase and in the G2-to-M transition, in analogy to the animal and yeast cell-cycle model. These findings include CYCA2 expression patterns in synchronized suspension cells (Reichheld et al, 1996; Shaul et al, 1996; Menges et al, 2005), their ability to rescue the growth of yeast cyclin mutants (Setiady et al, 1995) and their ability to induce Xenopus oocytes maturation (Renaudin et al, 1994). In addition, the ectopic expression of plant cyclins is sufficient to drive cells into mitosis (Imai et al, 2006; Boudolf et al, 2009).
Recently, it was shown that engineered yeast cells arrested in G2 are able to skip mitosis and re-acquire a G1 status when CDK activity is low (Coudreuse and Nurse, 2010). Therefore, if CYCA2s affect mitotic CDK activity, one could expect ectopic endoreduplication and reduced proliferation in the absence of CYCA2 function. Previously, single mutants in cyca2;1 and cyca2;3 were shown to have increased levels of endoreduplication (Imai et al, 2006; Yoshizumi et al, 2006). Consistent with these data, we found that cyca2 triple mutants displayed greatly increased endoreduplication levels, reduced cell proliferation in developing leaves and G2-arrest of GMCs resulting in SGCs with 4C DNA levels. Together, these data demonstrate that CYCA2s contribute to the CDK activity that is required for mitosis.
In animal systems, it is well established that B-type cyclins in complex with a CDK act as mitosis-promoting factor (MPF). MPF activity is further regulated by A-type cyclins through effects on transcription, activation, localization and stability (Lindqvist et al, 2009). In plants, ectopic expression of CYCB1;2 in differentiated cell types such as trichomes was sufficient to trigger ectopic cell divisions, suggesting a MPF-like function of CYCBs in plants (Schnittger et al, 2002). Using an in planta synchronized cell cycle-inducible system, we found that the onset of B-type cyclin expression was delayed in cyca2 triple mutants. Thus, mitotic entry involves the sequential activity of CYCA2-CDK and CYCB-CDK complexes.
Tissue specificity and redundancy among CYCA2s
Each cell type and tissue, within complex organs such as developing leaves, needs a custom-tailored cell-cycle regulation for the organ to reach its typical size and shape. This complexity is reflected in the large number of cell-cycle regulatory genes in plants. In Arabidopsis, four CYCA2 genes are encoded in its genome. Each individual CYCA2 shows its own peculiar expression patterns across developing organs, displaying tissue- and cell type-specific expression, such as in vascular tissues and the stomatal lineage. Their expression patterns also show variable degrees of overlap in certain tissues, suggesting local redundancies. Striking examples are the vascular expression of CYCA2;1 and CYCA2;4 and the stomatal expression of CYCA2;2 and CYCA2;3. In both tissues, the individual genes contribute locally to proliferation in a specific tissue or cell type.
Besides the expression pattern-dependent redundancy, the mutant analyses revealed differential contributions of individual CYCA2s to proliferation. The analysis of the phenotypes of different triple mutants allowed the estimation of their relative importance for specific processes. In the case of root meristem size, lateral root formation, endoreduplication and stomatal development, CYCA2;3 seemed to be most relevant; during stomatal formation, only single cyca2;3 mutants resulted in SGC formation. Moreover, in combination with cyca2;3, other cyca2 mutations synergistically enhanced the frequency of SGC formation.
Observed differences in penetrance can be explained in part by tissue-specific expression and relative expression levels. However, our study does not allow to exclude effects on protein stability and differences in biochemical properties, as additional regulatory mechanisms.
Developmental control over cell cycle through repression of CYCA2
Proliferation and differentiation are largely mutually exclusive processes. While some cells exit the cell cycle after mitosis and remain in G1-phase, other differentiating cells undergo several rounds of a modified cell cycle, in which the G2-to-M transition is omitted and only DNA synthesis occurs (endoreduplication). In animals, some developmental programs coordinate cell-cycle exit during differentiation through transcription factor activity (Myster and Duronio, 2000; Buttitta and Edgar, 2007). One strategy is to induce CDK inhibitory proteins, while another is to repress cell-cycle activating proteins. Interestingly, the transcription of A-type cyclins is often actively repressed during differentiation processes (Li and Vaessin, 2000; James et al, 2006; Martinez et al, 2006; Sebastian et al, 2009; Pan et al, 2010). In plants, it is not known how developmental signals can modulate the switch between a full cell cycle and the endocycle or cell-cycle exit during differentiation. Previously, increased level of polyploidy1 (ILP1) was found to act as repressor of CYCA2 expression (Yoshizumi et al, 2006). Here, we show that FLP and MYB88 repress CYCA2;3 expression during cell-cycle exit in differentiating guard cells. This mechanism resembles the PROSPERO-dependent mechanism in Drosophila that links neuronal lineage development with the transcriptional regulation of cell-cycle regulatory genes (Li and Vaessin, 2000).
Mutations that affect CYCA2 function display higher than normal ploidy levels (Imai et al, 2006; Yoshizumi et al, 2006), whereas CYCA2;3 overexpression strongly suppresses endoreduplication (Imai et al, 2006; Boudolf et al, 2009), indicating that CYCA2 levels are major negative determinants of endoreduplication in leaves. Early stages of leaf development involve high proliferation rates, while later stages gradually switch to differentiation-associated endoreduplication and cell expansion (Donnelly et al, 1999; Beemster et al, 2006). Interestingly, CYCA2;3 expression is rapidly repressed during the switch from proliferation to endoreduplication in differentiating leaves (Imai et al, 2006). Similarly, antagonizing auxin signalling also enhances endoreduplication via reduced CYCA2;3 expression (Ishida et al, 2010). However, it remains to be seen whether this effect is directly mediated by differentiation-induced transcription factors, and how auxin is involved in this.
Stomatal development ends after a single symmetric division of a GMC, each of whose daughter cells terminally differentiate into individual guard cells (Bergmann and Sack, 2007). Mutants in the stomatal transcription factors FLP and MYB88 do not stop dividing after the GMC has divided, even though guard cell identity markers are expressed (Lai et al, 2005). We found that downregulation of CYCA2;3 after the first GMC division, normally seen in wild-type plant, was absent in flpmyb88 double mutants. Direct interaction with CYCA2;3 promoter chromatin corroborate that FLP and MYB88 act as direct repressors of CYCA2;3 expression in guard cells. Similarly, the expression of an interacting CDK (Boudolf et al, 2009; Boruc et al, 2010b), CDKB1;1 was also shown to be directly repressed by FLP and MYB88 (Xie et al, 2010). These data are consistent with a model in which FLP and MYB88 enforce cell-cycle exit during terminal guard cell differentiation by direct repression of CYCA2/CDKB1;1 kinase complexes. This mechanism ensures that stomata consist of only two guard cells, a condition required for their proper functioning as adjustable air valves.
Materials and methods
Plant material and growth conditions
We used Arabidopsis seedlings of the accession Col-0 and Ler and mutants for the various A2-type cyclins from publicly available collections (SALK (Alonso et al, 2003), GABI-KAT (Rosso et al, 2003) and EXOn Trapping Insert Consortium (EXOTIC; http://www.jic.bbsrc.ac.uk/science/cdb/exotic/index.htm)), and stomatal lineage mutant alleles flp-1myb88, flp-7myb88 (Lai et al, 2005) and fama-1 (Ohashi-Ito and Bergmann, 2006). Cyclin mutant alleles used are cyca2;1-1 (SALK_121077; Yoshizumi et al, 2006), cyca2;1-2 (SALK_123348), cyca2;2-1 (GABI_120D03), cyca2;3-1 (SALK_092515; Imai et al, 2006), cyca2;3-2 (SALK_086463; Imai et al, 2006), cyca2;3-3 (SALK_043246), cyca2;4-1 (SALK_070301) and cyca2;4-2 (GAT_5.10009; Supplementary Figure S1). Promoter∷reporter lines for FLP (Lai et al, 2005), CDKB1;1 (Xie et al, 2010) and CYCA2;1 (Burssens et al, 2000) have been reported previously. For detection of T-DNA inserts, we used primers specific to the left border of the T-DNAs used for mutagenesis (LBC1, LB_GABI and LB_EXOTIC) in combination with gene-specific primers (Supplementary Table SII). The alleles cyca2;1-1, cyca2;2-1, cyca2;3-1 and cyca2;4-1 are representative knockout alleles and have been used for generating triple mutants. After surface sterilization, seeds were sown on half-strength MS medium supplemented with 1% sucrose and 0.8% agar. After stratification, plates were moved to cooled benches in a growth chamber (temperature, 22°C; irradiation, 65 μE/m2/s photosynthetically active radiation; photoperiod, 16 h light/8 h dark or continuous light).
Immunofluorescence localization
One-week-old seedlings, grown on 0.5 × MS medium under continuous illumination, were fixed in paraformaldehyde. Immunolocalization was performed as described (Sauer et al, 2006). The rabbit anti-knolle antibody (1:2000; Lauber et al, 1997) and the fluorochrome-conjugated secondary antibody anti-rabbit-Cy3 (1:600; Dianova) were used. Fluorescence detection was done with a confocal laser-scanning microscope Zeiss 710.
Cloning
Promoter∷GUS-GFP fusions of MYB88, CYCA2;2, CYCA2;3 and CYCA2;4 were generated through Gateway cloning of promoter fragments into pKGWFS7. PCR fragments of CYCA2;2, CYCA2;3 and CYCA2;4 promoters were described previously (Benhamed et al, 2008). To generate the CYCA2;1 and CYCA2;4 transcriptional fusions (CYCA2;1pro:HTA6:EYFP and CYCA2;4pro:HTA6:EYFP, respectively), 1808 bp upstream of the CYCA2;1 start codon and 1963 bp upstream of the CYCA2;4 start codon were amplified from Arabidopsis (Arabidopsis thaliana) ecotype Col-0 genomic DNA and cloned into the Gateway-adapted pFYTAG binary vector, which contains a translational fusion between the coding region of histone 2A (HTA6; At5g59870) and that of the enhanced YFP (EYFP) (Zhang et al, 2005).
Vascular expression analysis
The origin of the ATHB8pro:HTA6:EYFP and the ATHB8pro:ECFP-Nuc has been described (Sawchuk et al, 2007, 2008). Seeds were sterilized and germinated, and seedlings and plants were grown, transformed and selected as described (Sawchuk et al, 2007, 2008). For CYCA2;1pro:HTA6:EYFP and CYCA2;4pro:HTA6:EYFP, the progeny of eight independent, single insertion transgenic lines were inspected to identify the most representative expression pattern. We define ‘days after germination’ (DAG) as days following exposure of imbibed seeds to light. Dissected seedling organs were mounted and imaged as described (Sawchuk et al, 2007, 2008; Donner et al, 2009). Brightness and contrast were adjusted through linear stretching of the histogram in ImageJ (National Institutes of Health; http://rsb.info.nih.gov/ij). Signal levels and colocalization were visualized as described (Sawchuk et al, 2008).
Q-RT–PCR
Total RNA was extracted with the RNeasy Mini kit (Qiagen, Venlo, The Netherlands). Poly(dT) cDNA was prepared from 1 μg total RNA with the Superscript III First Strand Synthesis System for RT–PCR (Invitrogen, Carlsbad, CA) and quantified on an iCycler apparatus (Bio-Rad, Hercules, CA) with the Platinum SYBR Green qPCR Supermix-UDG kit (Invitrogen, Merelbeke, Belgium). PCR was carried out in 96-well optical reaction plates heated for 10 min to 50°C to allow UNG activity, followed by 10 min of 95°C to activate hot start Taq DNA polymerase, and 40 cycles of denaturation for 20 s at 95°C and annealing–extension for 20 s at 58°C. Target quantifications were performed with specific primer pairs designed using Beacon Designer 4.0 (Premier Biosoft International, Palo Alto, CA). Expression levels were normalized to At5g25760 (Q_PEX4) and At4g16100 (Q_UNKN1), which showed constitutive expression across samples. All Q-RT–PCR experiments were performed in triplicates and the data were processed using qBase v1.3.4 (Hellemans et al, 2007).
Histochemical staining and anatomical analysis
The β-glucuronidase (GUS) assays were performed as described (Beeckman and Engler, 1994). For microscopic analysis, chlorophyll was removed by EtOH treatment and further cleared by mounting in 90% lactic acid (Acros Organics, Brussels, Belgium). All samples were analysed by differential interference contrast microscopy.
For anatomical sections, samples were fixed overnight in 1% glutaraldehyde and 4% paraformaldehyde in 50 mM phosphate buffer (pH 7). Samples were dehydrated and embedded in Technovit 7100 resin (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer's protocol. Sections of 5 μm were cut with a microtome (Minot 1212; Leitz, Wetzlar, Germany), dried on Vectabond-coated object glasses, stained with Toluidine Blue for 8 min (Fluka Chemica, Buchs, Switzerland), and rinsed in tap water for 30 s. After drying, the sections were mounted in DePex medium (British Drug House, Poole, UK).
Flow cytometry
Primary leaves of 3-week-old seedlings were chopped with a razor blade in 300 μl of buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-[N-morpholino]propanesulphonic acid, pH 7, and 1% Triton X-100). To the supernatants, 1 μl of 4′,6-diamidino-2-phenylindole from a stock of 1 mg/ml was added, which was filtered over a 30-μm mesh. The nuclei were analysed with a CyFlow® ML (Partec) flow cytometer.
Guard cell nuclear content measurement
Nuclei were stained fluorescently by fixing 3-week-old cotyledons in a mixture of 9:1 ethanol:acetic acid (v/v). After the samples had been rinsed, they were stained for 24 h with 0.1 μg/ml of 4′,6-diamidino-2-phenylindole, mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA), and observed under a × 63 oil immersion objective on a Zeiss Axioskop equipped with an Axiocam CCD camera (Zeiss). Images were obtained using the Axiovision software and were analysed in grey scale with the public domain image analysis program ImageJ (version 1.28; http://rsb.info.nih.gov/ij/). Relative fluorescence units were reported as integrated density, which are the product of the area and the average fluorescence of the selected nucleus.
Kinematic analysis of leaf development
Plants of the wild-type and the cyca2 triple mutants were sown in quarter sections of round 12 cm Petri dishes filled with 100 ml of half-strength Murashige and Skoog medium (Duchefa, Haarlem, The Netherlands) and 0.6% plant tissue culture agar (Lab M, Bury, UK). At relevant time points after sowing, plants or primary leaves (depending on the size) of the respective genotypes were harvested. All healthy plants were placed in methanol overnight to remove chlorophyll, and subsequently they were cleared and stored in lactic acid for microscopy.
The following parameters were determined: total area of all cells in the drawing, total number of cells and number of guard cells. From these data, we calculated the average cell area. We estimated the total number of cells per leaf by dividing the leaf area by the average cell area (averaged between the apical and basal positions). Finally, average cell division rates for the whole leaf were determined as the slope of the log2-transformed number of cells per leaf, which was done using five-point differentiation formulas (Erickson, 1976).
FLP/MYB88 ChIP experiment
Polyclonal antibodies against the FLP/MYB88 proteins were generated by inoculating rabbits with Ni-NTA affinity purified NHis6-MYB88. ChIP experiments were performed essentially as before (Xie et al, 2010). In brief, 10-day-old shoots of wild-type, flp-1 myb88 (200 mg fresh weight for each) were crosslinked in 1% formaldehyde for 20 min by vacuum filtration, and the crosslinking reaction was stopped by the addition of 0.1 M glycine (final concentration) for additional 5 min. Tissues were ground to a fine powder using mortar and pestle in liquid nitrogen and then suspended in 300 μl of lysis buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 1 mM EDTA, pH 8.0; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; 1 mM phenylmethanesulphonylfluoride; 10 mM sodium butyrate; 1 × protein protease inhibitor from Sigma), and sonicated to achieve an average DNA size of 0.3–1 kb. The sonication conditions using the Bioruptor (Diagenode) were as follows: at high power; 30 s of sonication followed by 30 s of break; change ice every 10 min; 30 min in total. After cleared using 30 μl salmon sperm DNA/protein-A agarose (Upstate) at 4°C for at least 1 h, the supernatant fractions were incubated, respectively, with 1 μl FLP/MYB88 rabbit polyclonal antibody or 1 μg rabbit IgG (Abcam) at 4°C overnight. At the same time, 10% of the supernatant was saved as the input fraction. The chromatin–antibody complex was incubated with salmon sperm DNA/protein-A agarose (Upstate) at 4°C for at least 3 h, washed with lysis buffer, LNDET buffer (0.25 M lithium chloride; 1% NP40; 1% sodium deoxycholate and 1 mM EDTA, pH 8.0) and TE buffer (10 mM Tris–Cl, pH 7.5; 1 mM EDTA, pH 8.0) twice, respectively, and the complex was reverse crosslinked in elution buffer (1% SDS; 0.1 M NaHCO3; 1 mg/ml proteinase K) overnight at 65°C. DNA was extracted using the PCR Cleaning Kit (Qiagen). The presence of the promoter of CYCA2;3 gene was examined by real-time PCR using SYBR-Green chemistry. The housekeeping gene PDF2/PP2A (At1g13320) was used as an internal control for normalization. The fold enrichment was normalized to the internal control PDF2/PP2A using the 2−ΔΔCt method. Two biological replicates were conducted for each real-time PCR experiment. The ChIP-PCR primers used are listed in Supplementary Table II.
Supplementary Material
Acknowledgments
We thank Dominique Bergmann, David Galbraith and Gerd Jürgens for kindly providing mutant seeds, plasmids and antibodies; Karel Spruyt for assistance with photography; Martine De Cock for help in preparing the manuscript; and NASC for providing T-DNA insertion mutants. The T-DNA mutant GABI_120D03 was generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (MPI for Plant Breeding Research; Cologne, Germany). This work was supported by EMBO and Research Foundation of Flanders grants to SV, by an Excellence Graduate Fellowship from the Plant Molecular Biology/Biotechnology Program at the Ohio State University to ZX, by a National Science Foundation grant to EG and by a Discovery Grant of the Natural Sciences and Engineering Research Council of Canada (NSERC) to ES and FS; ES was supported, in part, by the Canada Research Chairs Program; TJD was supported by an NSERC CGS-M Scholarship, an NSERC CGS-D Scholarship and an Alberta Ingenuity Student Scholarship. SD is indebted to the Agency for Innovation through Science and Technology for a predoctoral fellowship. This work was supported by grants from Ghent University (‘Bijzonder Onderzoeksfonds Methusalem project’ No. BOF08/01M00408) and the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office.
Author contributions: SV, FC, GVI, FD, BDR, JF, GTS and TB conceived the general idea, isolated higher order mutants and performed cell division-related experiments. SD, LDV and DI generated and provided promoter:GUS:GFP lines. TJD and ES conceived and performed vascular-related experiments. SV, EL, FS and TB conceived and performed the stomatal-related experiments. ZX and EG conceived and performed ChIP-PCR. MV performed statistical analyses on the data. SV, FC and TB wrote the paper with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Engler G (1994) An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep 12: 37–42 [Google Scholar]
- Beemster GT, Vercruysse S, De Veylder L, Kuiper M, Inzé D (2006) The Arabidopsis leaf as a model system for investigating the role of cell cycle regulation in organ growth. J Plant Res 119: 43–50 [DOI] [PubMed] [Google Scholar]
- Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, De Meyer B, Buysschaert C, Rombauts S, Villarroel R, Aubourg S, Beynon J, Bhalerao RP, Coupland G, Gruissem W, Menke FL, Weisshaar B, Renou JP, Zhou DX, Hilson P (2008) Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J 56: 493–504 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD (2007) Stomatal development. Annu Rev Plant Biol 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Boruc J, Mylle E, Duda M, De Clercq R, Rombauts S, Geelen D, Hilson P, Inzé D, Van Damme D, Russinova E (2010a) Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol 152: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010b) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler Jde A, Verkest A, Beeckman T, Naudts M, Inzé D, De Veylder L (2004) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, De Jaeger G, Inzé D, De Veylder L (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L, Hafidh S, Borg M, Sidorova A, Mori T, Twell D (2009) A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S, de Almeida Engler J, Beeckman T, Richard C, Shaul O, Ferreira P, Van Montagu M, Inzé D (2000) Developmental expression of the Arabidopsis thaliana CYCA2;1 gene. Planta 211: 623–631 [DOI] [PubMed] [Google Scholar]
- Buttitta LA, Edgar BA (2007) Mechanisms controlling cell cycle exit upon terminal differentiation. Curr Opin Cell Biol 19: 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro E, Castellano MM, Gutierrez C (2007) A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217 [DOI] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P (2010) Driving the cell cycle with a minimal CDK control network. Nature 468: 1074–1079 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demandolx D, Davoust J (1997) Multicolor analysis and local image correlation in confocal microscopy. J Microsc 185: 21–36 [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E (2009) Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246 [DOI] [PubMed] [Google Scholar]
- Erickson RO (1976) Modeling of plant growth. Annu Rev Plant Physiol 27: 407–434 [Google Scholar]
- Fisher DL, Nurse P (1996) A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J 15: 850–860 [PMC free article] [PubMed] [Google Scholar]
- Fung TK, Poon RY (2005) A roller coaster ride with the mitotic cyclins. Semin Cell Dev Biol 16: 335–342 [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AA (2009) GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot 60: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier GR, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K (2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- James CG, Woods A, Underhill TM, Beier F (2006) The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol Biol 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313–320 [DOI] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM (1992) Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 257: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, Lee EK, Nakagawa T, Zhao L, Geisler M, Sack FD (2005) The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Vaessin H (2000) Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev 14: 147–151 [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH (2009) The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol 185: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC (2009) The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development 136: 1475–1485 [DOI] [PubMed] [Google Scholar]
- Martinez AM, Colomb S, Dejardin J, Bantignies F, Cavalli G (2006) Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev 20: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime H, Roussel MF, Ashmun RA, Sherr CJ (1991) Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65: 701–713 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1995) Principles of CDK regulation. Nature 374: 131–134 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Motokura T, Arnold A (1993) Cyclin D and oncogenesis. Curr Opin Genet Dev 3: 5–10 [DOI] [PubMed] [Google Scholar]
- Myster DL, Duronio RJ (2000) To differentiate or not to differentiate? Curr Biol 10: R302–R304 [DOI] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL Jr, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR (1995) Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol 109: 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Nakade K, Huang YC, Zhu ZW, Masuzaki S, Hasegawa H, Murata T, Yoshiki A, Yamaguchi N, Lee CH, Yang WC, Tsai EM, Obata Y, Yokoyama KK (2010) Suppression of cell-cycle progression by Jun dimerization protein-2 (JDP2) involves downregulation of cyclin-A2. Oncogene 29: 6245–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RW (2010) Genstat Release 13 Reference Manual, Part3: Procedure Library PL21. VSN International: Oxford, UK [Google Scholar]
- Payton M, Coats S (2002) Cyclin E2, the cycle continues. Int J Biochem Cell Biol 34: 315–320 [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T (1990) Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature 346: 760–763 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Chaubet N, Shen WH, Renaudin JP, Gigot C (1996) Multiple A-type cyclins express sequentially during the cell cycle in Nicotiana tabacum BY2 cells. Proc Natl Acad Sci USA 93: 13819–13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin JP, Colasanti J, Rime H, Yuan Z, Sundaresan V (1994) Cloning of four cyclins from maize indicates that higher plants have three structurally distinct groups of mitotic cyclins. Proc Natl Acad Sci USA 91: 7375–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P (2009) Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939 [DOI] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Sawchuk MG, Donner TJ, Head P, Scarpella E (2008) Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol 148: 1908–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchuk MG, Head P, Donner TJ, Scarpella E (2007) Time-lapse imaging of Arabidopsis leaf development shows dynamic patterns of procambium formation. New Phytol 176: 560–571 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Francis P, Berleth T (2004) Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131: 3445–3455 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Stierhof YD, Hulskamp M (2002) Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol 12: 415–420 [DOI] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J (2009) MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA 106: 4719–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiady YY, Sekine M, Hariguchi N, Yamamoto T, Kouchi H, Shinmyo A (1995) Tobacco mitotic cyclins: cloning, characterization, gene expression and functional assay. Plant J 8: 949–957 [DOI] [PubMed] [Google Scholar]
- Shaul O, Mironov V, Burssens S, Van Montagu M, Inze D (1996) Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc Natl Acad Sci USA 93: 4868–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I, Kojima S, Sakaguchi N, Umeda-Hara C, Umeda M (2010) Two Arabidopsis cyclin A3s possess G1 cyclin-like features. Plant Cell Rep 29: 307–315 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, Inzé D, Fukaki H, Beeckman T (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E (2010) Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi T, Tsumoto Y, Takiguchi T, Nagata N, Yamamoto YY, Kawashima M, Ichikawa T, Nakazawa M, Yamamoto N, Matsui M (2006) Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 18: 2452–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gong FC, Lambert GM, Galbraith DW (2005) Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.