Abstract
The Polycomb repressive complex 1 (PRC1) mediates gene silencing, in part by monoubiquitination of histone H2A on lysine 119 (uH2A). Bmi1 and Ring1b are critical components of PRC1 that heterodimerize via their N-terminal RING domains to form an active E3 ubiquitin ligase. We have determined the crystal structure of a complex between the Bmi1/Ring1b RING–RING heterodimer and the E2 enzyme UbcH5c and find that UbcH5c interacts with Ring1b only, in a manner fairly typical of E2–E3 interactions. However, we further show that the Bmi1/Ring1b RING domains bind directly to duplex DNA through a basic surface patch unique to the Bmi1/Ring1b RING–RING dimer. Mutation of residues on this interaction surface leads to a loss of H2A ubiquitination activity. Computational modelling of the interface between Bmi1/Ring1b–UbcH5c and the nucleosome suggests that Bmi1/Ring1b interacts with both nucleosomal DNA and an acidic patch on histone H4 to achieve specific monoubiquitination of H2A. Our results point to a novel mechanism of substrate recognition, and control of product formation, by Bmi1/Ring1b.
Keywords: Bmi1, nucleosome, Polycomb repression, Ring1b, UbcH5c
Introduction
Bmi1 and Ring1b are critical members of the Polycomb group (PcG) proteins, a set of transcriptional repressors that maintain chromatin in a silenced state, thus repressing target genes (Schwartz and Pirrotta, 2008). During the process of development, PcG proteins and the opposing Trithorax group proteins work together to establish transcriptional patterns of key developmental regulators, such as the Hox genes (Ringrose and Paro, 2004). In addition, the activity of PcG complexes is implicated in epigenetic inheritance, X-chromosome inactivation, stem cell pluripotency, senescence, and tumourigenesis (Sparmann and van Lohuizen, 2006; Pietersen and van Lohuizen, 2008; Bracken and Helin, 2009). In particular, the loss of Bmi1 leads to defects in stem cell self-renewal, highlighting the importance of PcGs in determining proper cell fate (Park et al, 2003; Bruggeman et al, 2005; Liu et al, 2006).
PcG proteins assemble into several distinct multiprotein complexes. The Polycomb repressive complex 2 (PRC2) is a histone methyltransferase that catalyses the tri-methylation of lysine 27 on histone H3 (H3K27me3) (Cao et al, 2002; Czermin et al, 2002). A second complex, PRC1, binds specifically to the H3K27me3 mark and catalyses the monoubiquitination of histone H2A on lysine 119 (uH2A) (de Napoles et al, 2004; Wang et al, 2004). This modification leads to the stalling of RNA polymerase at the promoter of uH2A-modified genes, shutting down transcription (Stock et al, 2007). In Drosophila, PRC1 contains stoichiometric Polycomb (Pc), Sex Combs Extra (Sce), Posterior Sex Combs (Psc), and Polyhomeotic (Ph) (Shao et al, 1999; Francis et al, 2001). By contrast, duplications of the PcG genes have complicated the picture in humans, where there are five Pc proteins (CBX2, CBX4, CBX6, CBX7, and CBX8), two Sce proteins (Ring1a and Ring1b), six Psc proteins (Bmi1/PCGF4, Mel-18/PCGF2, PCGF1, PCGF3, PCGF5, and PCGF6), and three Ph proteins (PHC1, PHC2, and PHC3) (Gil and Peters, 2006). Each PRC1 complex member has a distinct role. For example, the Pc/CBX proteins contain a chromodomain, which is responsible for recruitment of the complex to H3K27me3 (Bernstein et al, 2006).
The uH2A activity of PRC1 is due to the combined activity of the Bmi1 and Ring1b proteins, which together form a heterodimeric E3 ubiquitin ligase (de Napoles et al, 2004; Wang et al, 2004; Cao et al, 2005; Ben-Saadon et al, 2006; Buchwald et al, 2006; Li et al, 2006). Classical ubiquitin conjugation occurs via a sequential pathway, wherein a cascade of reactions by E1, E2, and E3 enzymes leads to the formation of an isopeptide bond between the C-terminus of ubiquitin and a lysine sidechain of the target (Pickart, 2001; Kerscher et al, 2006). The C-terminus of ubiquitin is first linked via a thioester bond with a cysteine residue of the E1 activation enzyme. Next, the ubiquitin is transferred to the catalytic cysteine of an E2 conjugation enzyme. In the final step, the E2-ubiquitin conjugate interacts with an E3 ligase. There are two general classes of E3s: (1) HECT domain E3s, in which ubiquitin is transferred from the E2 onto a cysteine residue of the E3 before linkage to the substrate and (2) RING-domain E3s, which promote transfer of ubiquitin directly from the active site of the E2 to an acceptor lysine residue in the substrate (Deshaies and Joazeiro, 2009). Bmi1/Ring1b is a member of this latter class.
RING domains are stabilized structurally through the binding of two zinc atoms, and they often form dimeric pairs as in the case of BRCA1/BARD (Jackson et al, 2000; Brzovic et al, 2001). Bmi1 and Ring1b each contain an N-terminal RING domain through which they heterodimerize, both in vitro and in vivo, to form a functional E3 ligase. In addition, this heterodimer interacts directly with E2-ubiquitin conjugate (Buchwald et al, 2006). Two structures of the Bmi1/Ring1b RING–RING heterodimer (Buchwald et al, 2006; Li et al, 2006) revealed an unusual mode of dimerization, with the N-terminal arm of Ring1b wrapped around a groove on the surface of Bmi1. This dimerization is critical for the function of Bmi1/Ring1b: although Ring1b alone has weak intrinsic ubiquitin ligase activity, the binding of Bmi1 to Ring1b both stabilizes the structure of Ring1b and greatly stimulates ligase activity (Wang et al, 2004; Cao et al, 2005; Buchwald et al, 2006; Li et al, 2006).
While much is known about the Bmi1/Ring1b dimer interface, the nature of the E2 and substrate binding sites are less well understood. The Bmi1/Ring1b complex is active only when paired with E2 enzymes of either the UbcH5 subtype (a, b, or c), or UbcH6 (Buchwald et al, 2006). Mutation of a putative E2-binding site on Ring1b has been shown to impair the catalytic activity of Bmi1/Ring1b, while mutation of the corresponding site on Bmi1 had no effect (Buchwald et al, 2006). Molecular modelling studies further suggested that the binding site for the E2 enzyme lies on Ring1b (Buchwald et al, 2006); however, this has not yet been confirmed through direct binding or structural studies. In addition, the manner in which Bmi1/Ring1b specifies monoubiquitination of H2A is not understood. The Drosophila homologue of Bmi1 (Psc) has been shown to bind to DNA (Francis et al, 2001), as has the Bmi1 paralogue Mel-18 (Kanno et al, 1995), suggesting DNA binding as a possible mechanism for nucleosomal targeting. However, it was not established which regions of Psc or Mel-18 were required for DNA binding.
Intriguingly, the minimal RING–RING heterodimer of Bmi1/Ring1b has the same level of ubiquitin ligase activity, the same specificity for monoubiquitination of H2A, and the same requirement for a nucleosomal substrate as the intact complex (Buchwald et al, 2006). This implies that the minimum specificity determinants for the ligase reaction must lie within the RING domains of Bmi1 and Ring1b. To identify these specificity determinants, we have used a combination of X-ray crystallography, site-directed mutagenesis, and molecular modelling to map the residues involved in both the E2 and nucleosomal interfaces. Notably, we find that the Bmi1/Ring1b RING–RING heterodimer binds directly to short DNA duplexes, and on this basis, we propose a new model for the site-selective monoubiquitination of histone H2A by the Bmi1/Ring1b ubiquitin ligase complex.
Results
Crystal structure of the Bmi1/Ring1b–UbcH5c complex
To better understand the recognition of Bmi1/Ring1b by its cognate E2 enzyme, we determined the crystal structure of Bmi1(1−109)/Ring1b(1−116) in complex with UbcH5c. Crystals of a 1:1:1 complex between Bmi1, Ring1b, and UbcH5c grew rapidly. The X-ray structure was determined to 2.65 Å by molecular replacement using ensembles of previous structures of the Bmi1/Ring1b heterodimer and of UbcH5c as search models. The final structure is well refined, with an Rwork of 21.7% and an Rfree of 24.3% (see Table I). Electron density was absent for the N-terminal 15 amino acids of Ring1b, as was the case in previous structures of the RING–RING heterodimer (Buchwald et al, 2006; Li et al, 2006).
Table 1. Summary of crystallographic analysis.
| Data collection a | |
|---|---|
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 107.9, 107.9, 77.6 |
| α, β, γ (deg) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50–2.65 (2.74–2.65) |
| Unique reflections | 15 512 (1512) |
| Redundancy | 10.8 (10.5) |
| I/σI | 25.5 (5.75) |
| Completeness (%) | 100.0 (100.0) |
| Rsym | 10.0 (45.9) |
| Refinement | |
| Resolution (Å) | 50–2.65 |
| Rwork/Rfree (%) | 21.7/24.3 |
| Protein atoms | 2785 |
| Zn2+ atoms | 4 |
| Water atoms | 116 |
| R.m.s.d. bonds (Å) | 0.004 |
| R.m.s.d. angles (deg) | 0.900 |
| aNumbers in brackets give values for highest resolution bin. | |
The structure of Bmi1/Ring1b within the ternary complex is similar to previous structures of the Bmi1/Ring1b heterodimer, with an average r.m.s.d. of 0.44 Å (PDB: 2H0D) and 0.40 Å (PDB: 2CKL) over all backbone atoms. The main difference between the models lies in the α2 helix of Ring1b (Ser44–Met50), where the average B-factors are lower in our structure (35.7 Å2) compared with the apo-structures (66.5 and 49.2 Å2 in 2H0D and 2CKL, respectively), consistent with a more well-ordered helix in the E2–E3 complex (Supplementary Figure S1). The Bmi1/Ring1b interface is identical to that observed in previous structures, with prominent salt bridges being formed by the Bmi1/Ring1b pairs of Asp72/Arg70, Glu11/Lys112, Lys81/Glu48, and Thr41/Arg26.
UbcH5c interacts only with Ring1b (Figure 1A), explaining the strong effects of Ring1b mutations on activity (Buchwald et al, 2006; Li et al, 2006). The Bmi1/Ring1b heterodimer interface buries a total of ≈2500 Å2 surface area from the two proteins while the UbcH5c/Ring1b interface buries only a total of 507 Å2. This small interface is reflected in the low-affinity interaction between Bmi1/Ring1b and UbcH5c (Buchwald et al, 2006). The Ring1b-binding surface on UbcH5c consists of two loops (L4 and L7), as well as residues from the N-terminal α helix (α1) (Figure 1A). Salt bridges are formed between Lys4 and Lys8 from the α1 helix of UbcH5c (ULys4 and ULys8) with Asp56 on Ring1b (RAsp56) (Figure 1B, right panel). An equivalent interaction was previously observed between Asp562 of the cIAP2 RING domain and Lys4 of UbcH5b (Mace et al, 2008). UAsp12 interacts via a water molecule with RHis46, analogous to the Asp12/Glu553 interaction in the UbcH5b/cIAP2 structure.
Figure 1.
Structure of the Bmi1/Ring1b–UbcH5c complex. (A) (Left) Ribbon representation of the overall complex architecture, with UbcH5c in grey, Ring1b(1−116) in light blue, and Bmi1(1−109) in orange. Zn2+ ions are shown in pink. The three Ring1b-binding regions on the surface of UbcH5c are coloured in teal (N-terminal α-helix), yellow (L4 loop), and purple (L7 loop), respectively. The sidechain of the catalytic Cys85 residue (site of ubiquitin attachment) is shown in green in stick format. (Right) Bmi1/Ring1b are shown in surface representation, highlighting the UbcH5c-binding groove on Ring1b. (B) Interactions between Ring1b and UbcH5c along the L4 and L7 loops (left), and N-terminal α-helix (right). The sidechains involved are shown in stick format. Hydrogen-bond distances are given in angstroms. Loops are coloured as in part (A). Key: nitrogen=blue, oxygen=red, sulphur=yellow, Zn2+=pink spheres, H2O=tan spheres.
The SPA motif (L7 residues USer94, UPro95, and UAla96) has been previously implicated in E2–E3 recognition (Christensen et al, 2007; Huang et al, 2009). In our structure, the sidechain hydroxyl of USer94 makes a hydrogen bond with the backbone carbonyl of RPro88 (Figure 1B). Hydrophobic interactions between UPro95 and UAla96 with RIle53 and RPro88 further stabilize the interface. Likewise, contacts between the PFK motif (L4 residues UPro61, UPhe62, and ULys63) and RING domains have been observed in several previous structures (Zheng et al, 2000; Dominguez et al, 2004; Zhang et al, 2005). In the case of UbcH5c, Phe62 from the L4 loop is buried in the hydrophobic pocket formed between the Zn2+-coordination loops of Ring1b, where it packs against RIle53.
One key difference between the Ring1b/UbcH5c interface and other E2/RING complexes lies in the interaction between RArg91 and UGln92. RArg91 makes hydrogen bonds with both the backbone and the sidechain of UGln92, helping to position the interface. By contrast, the equivalent residues do not interact directly in the c-Cbl/UbcH7 complex (Zheng et al, 2000), and they make only a single interaction in the cIAP2/UbcH5b complex (Mace et al, 2008). Comparison of these complex structures reveals subtle differences in the orientation of E2 relative to E3, and an unusual interaction like that observed between RArg91 and UGln92 may be a means by which different E3 enzymes position the E2 ubiquitin thioester for efficient transfer.
The Bmi1/Ring1b complex requires a nucleosomal substrate for E3 ligase activity
The Bmi1/Ring1b complex is an E3 ligase for histone H2A monoubiquitination both in vivo and in vitro (Wang et al, 2004; Cao et al, 2005). Recently, it has been shown that the minimal RING–RING heterodimer of Bmi1/Ring1b is as active as the full complex (Buchwald et al, 2006). This suggests that chromodomain binding to H3K27me3 by Pc (CBX proteins) in PRC1 is not the only mechanism for achieving substrate targeting; in particular, it implies that the RING-domain heterodimer is itself capable of recognizing and binding to the nucleosomal substrate.
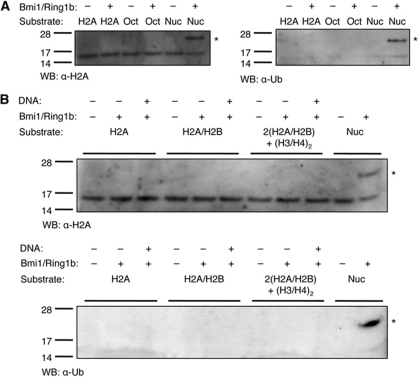
First, we confirmed that the RING–RING heterodimer (Bmi1(1−109)/Ring1b(1−116)) is an active histone H2A monoubiquitinase. In vitro reactions using a nucleosomal substrate (Supplementary Figure S2) produced a product of ≈22 kDa (14 kDa H2A+8 kDa ubiquitin) that is detected using antibodies against both H2A and ubiquitin (Figure 2A, sixth lane of each gel). Mass spectrometric analysis of this sample confirmed that is H2A containing a single ubiquitin moiety (uH2A) (Supplementary Figure S3). We further confirmed that, as previously reported, the ubiquitin-conjugating activity of Bmi1/Ring1b is specific to nucleosome substrates (Buchwald et al, 2006): in vitro reactions using either recombinant histone H2A or core histone octamers (Wang et al, 2004; Buchwald et al, 2006) failed to produce uH2A (Figure 2A).
Figure 2.
Substrate preference of the Bmi1/Ring1b E3 ligase. (A) Reactions were incubated in ligase buffer containing 30 nM human E1, 1.5 μM UbcH5c, 25 μM ubiquitin, 3 mM ATP, 500 nM Bmi1(1−109)/Ring1b(1−116), and 5 μg of recombinant histone H2A (H2A), core histone octamers (Oct), or nucleosomes (Nuc). Reactions were incubated for 1 h at 30 °C, then separated by SDS–PAGE and analysed by western blot using either an anti-histone H2A (left) or anti-ubiquitin (right) antibody. Symbol: * represents ubiquitin H2A (22 kDa). (B) Added DNA is not sufficient to rescue uH2A activity towards non-nucleosomal substrates. Recombinant histone H2A (H2A), recombinant H2A/H2B dimer (H2A/H2B), 2:1 H2A/H2B dimer:H3/H4 tetramer (2(H2A/H2B)+(H3/H4)2), or nucleosomes were treated in the presence or absence of equimolar concentrations of a 146-bp DNA duplex. All reactions contained 2.5 μM histone H2A, and all other conditions were the same as in (A). Symbol: * represents ubiquitin H2A (22 kDa).
Given the lack of activity of Bmi1/Ring1b against both histone H2A and core octamers, we reasoned that the presence of DNA might be a requirement for H2A monoubiquitination. To test this, we prepared a 146-bp palindromic DNA duplex (the sequence from the crystal structure of the human nucleosome core particle (PDB: 2CV5) (Tsunaka et al, 2005)). This duplex was then added in stoichiometric amounts to Bmi1/Ring1b assays containing recombinant H2A, H2A/H2B dimer, or two equivalents of the H2A/H2B dimer with one equivalent of the H3/H4 tetramer: 2(H2A/H2B)+(H3/H4)2. The presence of DNA was not sufficient to render any of the histone samples a substrate for Bmi1/Ring1b (Figure 2B). Only nucleosomes, containing well-ordered histone octamers wrapped with DNA, were substrates for the reaction. Given this, and the previously reported ability of PRC1 to bind to DNA (Francis et al, 2001), we hypothesized that the Bmi1/Ring1b heterodimer might bind directly to nucleosomal DNA, and thus promote ubiquination of H2A.
Bmi1/Ring1b binds to short duplex DNA in a sequence-independent manner
To test the hypothesis that Bmi1/Ring1b binds to DNA, we synthesized a fluorescently labelled duplex DNA probe and assessed binding of Bmi1/Ring1b by fluorescence polarization (FP). A 12-bp probe was selected, with the sequence based on an H2AK119-proximal sequence from the 146-bp duplex used above. Bmi1/Ring1b was able to bind to the 12-bp duplex probe, with an apparent KD of 3.0 μM (Figure 3A and B). Fluorescence-electrophoretic mobility shift assays (F-EMSAs) were used to confirm formation of a DNA–Bmi1/Ring1b complex (Supplementary Figure S4).
Figure 3.
Bmi1/Ring1b binds to short duplex DNA. (A, B) In all, 70 nM 5′-FAM-labelled DNA probe was incubated with indicated concentrations of Bmi1/Ring1b at room temperature for 10 min. Polarization data are plotted as average values±s.d. at each concentration. (A) Direct binding assay, showing increase in FP as a function of [Bmi1/Ring1b] KD,app=3.2±0.3 μM. (B) Competition binding assay. Bmi1/Ring1b (2.5 μM) was mixed with 70 nM probe, and unlabelled DNA (same sequence as probe) was used to compete for binding. IC50=3.0±1.0 μM. (C) DNA binding is non-sequence specific. Four scrambled sequences (see Table II) were tested in the competition assay as in (B). (D) Length dependence of DNA binding. 3′ truncations of the original 12 bp sequence were assayed as in (B). In (C, D), data were normalized and plotted as average values±s.d. IC50 values were determined from fitting the average values to a four-parameter fit.
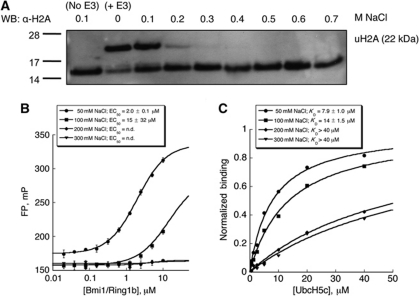
We next asked whether DNA binding by Bmi1/Ring1b was sequence specific. To test this, we made four additional 12 bp duplexes in which the original sequence was scrambled (see Table II). Each of these sequences was able to compete with the original probe for binding to Bmi1/Ring1b, with a KD,app similar to that of the original sequence (Figure 3C). These results indicate that binding of Bmi1/Ring1b to DNA is not sequence specific. Next, we investigated the length dependence of DNA binding. We designed 3′ truncations of the original sequence, generating duplexes of 10, 8, and 6 bp each. The 10-bp sequence was able to efficiently compete for binding with the original (12 bp) probe, the 8-bp sequence was somewhat impaired in binding, and the 6-bp sequence was strongly impaired (Figure 3D). Taken together, these data indicate that ≈10 bp are required for effective binding to Bmi1/Ring1b. Interestingly, this length corresponds to a single turn of double helical B-form DNA, suggesting that Bmi1/Ring1b might require at least one turn of DNA for recognition.
Table 2. Oligonucleotide probes used in this study.
| Probe | Sequence |
|---|---|
| 5′-FAM (12 bp) | 5′-FAM-TCAGCTGAACAT-3′ 3′-AGTCGACTTGTA-5′ |
| Comp1 (12 bp) | 5′-TCAGCTGAACAT-3′ 3′-AGTCGACTTGTA-5′ |
| Comp2 | 5′-GGACCTGATGAC-3′ 3′-CCTGGACTACTG-5′ |
| Comp3 | 5′-CTAGCCTAGCGA-3′ 3′-GATCGGATCGCT-5′ |
| Comp4 | 5′-AAGACATACGAG-3′ 3′-TTCTGTATGCTC-5′ |
| Comp5 | 5′-GTGTTACTAGCT-3′ 3′-CACAATGATCGA-5′ |
| 10 bp | 5′-TCAGCTGAAC-3′ 3′-AGTCGACTTG-5′ |
| 8 bp | 5′-TCAGCTGA-3′ 3′-AGTCGACT-5′ |
| 6 bp | 5′-TCAGCT-3′ 3′-AGTCGA-5′ |
The E3 ligase activity of Bmi1/Ring1b is highly salt dependent
Because the observed binding of DNA to Bmi1/Ring1b is not sequence specific, we surmised that Bmi1/Ring1b might recognize DNA through electrostatic interactions between basic residues on Bmi1/Ring1b and the phosphodiester backbone of DNA. Such a mechanism for DNA recognition has previously been observed in nucleosome-binding proteins, for example in the RanGEF protein RCC1 (Makde et al, 2010). We might expect that a predominantly electrostatic interaction of Bmi1/Ring1b with the nucleosome would be highly sensitive to added salt. The enzymatic activity of Bmi1/Ring1b is indeed greatly decreased by concentrations of NaCl in excess of 200 mM (Figure 4A). Similarly, the binding of Bmi1/Ring1b to short duplex DNA is highly salt sensitive (Figure 4B). An abrupt decrease in DNA binding occurs between 100 and 200 mM NaCl, the same concentration range over which enzymatic activity is lost.
Figure 4.
Salt dependence of Bmi1/Ring1b ubiquitin ligase activity, UbcH5c binding, and DNA binding. (A) Histone H2A ubiquitin ligase assay in the presence of added salt. Bmi1/Ring1b was incubated in ligase buffer with nucleosomes under the conditions described in Figure 2, with [NaCl] allowed to vary from 0.1 to 0.7 M. H2A ubiquitin ligase activity is highly salt dependent, with complete loss of product formation at 0.3 M NaCl. (B) DNA binding as a function of NaCl concentration. The FP assay was conducted as described in Figure 3A at each indicated [NaCl]. (C) UbcH5c binding as a function of added NaCl. Immobilized Bmi1/Ring1b was exposed to increasing concentrations of UbcH5c, and binding was detected by BLI. Normalized steady-state responses at equilibrium are plotted as a function of UbcH5c concentration, and KD values were determined as described in Materials and methods.
It is possible that the loss of enzymatic activity of Bmi1/Ring1b in response to increasing concentrations of NaCl is due not only to loss of affinity for DNA but also to a disruption of the E2–E3 interaction between Bmi1/Ring1b and UbcH5c. In fact, several key interactions between Ring1b and UbcH5c are electrostatic. To test this possibility, we used biolayer interferometry (BLI) to measure the steady-state affinity between the Bmi1/Ring1b heterodimer and UbcH5c as a function of salt concentration (Figure 4C). KD is affected by NaCl, rising from 8 μM at 50 mM NaCl to 14 μM at 100 mM NaCl, and to >40 μM at concentrations of 200 mM NaCl and above. Thus, both the E2–E3 and the E3–DNA interactions are sensitive to salt concentration.
Mutation of basic surface residues on Bmi1/Ring1b disrupts uH2A activity
Having established that the Bmi1/Ring1b RING–RING heterodimer can bind directly to DNA, we next sought to identify residues involved in the DNA-binding interface. Analysis of the structure of the Bmi1/Ring1b heterodimer identified several basic patches on the surface that might plausibly be involved in DNA binding (Supplementary Figure S5A). In addition, alignment of the sequences of RING-domain E3 ligases identified basic sites that are conserved in Ring1b and Bmi1, but not in other E3s (Supplementary Figure S5B). Based on these analyses, we generated a set of mutants with basic surface residues substituted by alanines (see Supplementary Figure S5C; Table III). We reasoned that a single mutation might not be sufficient to disrupt DNA-binding activity; thus, most mutations were made two or three residues at a time. For comparison, we mutated RAsp56, as this residue salt bridges to ULys4 and ULys8 (see above). Mutation of RAsp56 to lysine would be expected to destabilize the E2–E3 interface, rather than the E3 interaction with DNA. All mutants were co-expressed in Escherichia coli with the wild-type (wt) partner, and, like the wt RING domains, formed stable 1:1 Bmi1/Ring1b heterodimers as assessed by gel filtration (Supplementary Figure S6).
Table 3. Bmi1/Ring1b mutants generated in this study.
| Name | Bmi1(1−109) | Ring1b(1−116) | uH2A activity | DNA KD (μM) | UbcH5c KD (μM) |
|---|---|---|---|---|---|
| Bwt/Rwt | wt | wt | +++ | 3.1±0.4 | 5±1 |
| BwtRD56K | wt | D56K | − | 1.8±0.6 | >40 |
| Bwt/RK15A | wt | K15A | +++ | 6.5±0.6 | |
| Bwt/RK92A.RK93A | wt | K92A.K93A | + | 56±6 | |
| Bwt/RK97A.RR98A | wt | K97A.R98A | − | 32±2 | 9.6±0.4 |
| BK62A.BR64A/Rwt | K62A.R64A | wt | + | 25±3 | |
| BK88A.BK92A.BR95A/Rwt | K88A.K92A.R95A | wt | +++ | 3.7±0.3 |
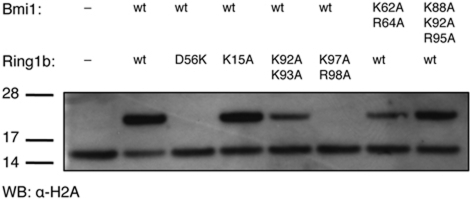
Several of the mutations had large effects on the ubiquitin ligase activity of the Bmi1/Ring1b heterodimer (Figure 5). Mutation of RAsp56 to lysine completely abolished the H2A ubiquitin ligase activity of the complex, as expected for disruption of the E2–E3 interface. Among the basic surface residues, mutation of RLys97/RArg98 to alanine also abolished ubiquitin ligase activity, while mutation of the RLys92/RLys93 and Bmi Lys64/Arg64 (BLys62/BArg64) pairs to alanine each had a smaller, but noticeable, effect. However, mutation of three basic surface residues along the C-terminal α helix of Bmi1 (BLys88/BLys92/BArg95) to alanine had no discernable effect on catalysis, nor did mutation of RLys15 on the N-terminal arm of Ring1b.
Figure 5.
Ubiquitin ligase activity of Bmi1/Ring1b mutants. The indicated mutants (Bmi1/Ring1b heterodimers) were incubated in ligase buffer (1 h; 30°C) with E1, UbcH5c, ubiquitin, ATP, and nucleosomes. Products were analysed as described above.
Correlation between DNA-binding and ligase activity
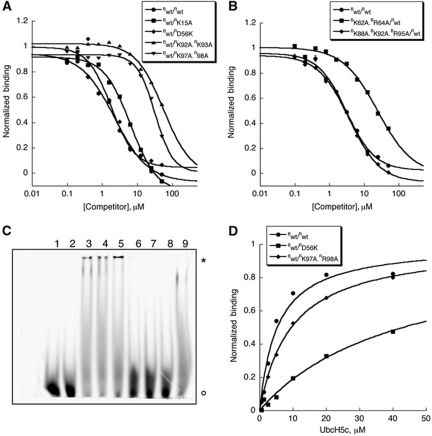
To determine the effect of Bmi1/Ring1b mutations on DNA binding, we used the FP DNA-binding assay described above (Figure 6A and B). As we might expect, mutations that do not affect the ligase reaction (RK15A and BK88A.BK92A.BR95A) do not significantly perturb the interaction of the Bmi1/Ring1b complex with DNA. The mutation designed to disrupt the E2–E3 interface (RD56K) also failed to disrupt binding to DNA. By contrast, the mutations RK92A.RK93A, RK97A.RR98A and BK62A.BR64A, each of which shows a defect in the ligase reaction, significantly impair DNA binding (at least eight-fold increase in KD,app). These results were confirmed using the F-EMSA assay: Bwt/Rwt, Bwt/RD56K, Bwt/RK15A, and BK88A.BK92A.BR95A/Rwt all bind strongly to DNA, whereas Bwt/RK92A.RK93A, Bwt/RK97A.RR98A, and BK62A.BR64A/Rwt do not (Figure 6C).
Figure 6.
Mutation of basic surface residues on Bmi1/Ring1b disrupts DNA binding without affecting UbcH5c binding. (A, B) FP DNA-binding assay of Bmi1/Ring1b mutants. Binding of heterodimers bearing mutations in Ring1b is shown in (A), and in (B) for those bearing mutations in Bmi1. Complex (at a concentration that gave rise to a 50-mP change in signal from a free probe baseline) was incubated with 70 nM 5′-FAM-labelled probe, and unlabelled probe was used to compete for binding. Raw mP values were converted to normalized binding curves. Data are plotted as average values±s.d. (C) F-EMSA assay of Bmi1/Ring1b mutants. In all, 20 μM wt or mutant protein was incubated with 70 nM 5′-FAM probe for 10 min at room temperature before electrophoretic separation. Symbols: * represents Bmi1/Ring1b–probe complex and ° represents free probe. Lane key: 1=blank (H2O), 2=blank (buffer), 3=Bwt/Rwt, 4=Bwt/RD56K, 5=Bwt/RK15A, 6=Bwt/RK92A.RK93A, 7=Bwt/RK97A.RR98A, 8=BK62A.BR64A/Rwt, 9=BK88A.BK92A.BR95A/Rwt. (D) BLI (Octet)-binding assay of UbcH5c to Bwt/Rwt, Bwt/RD56K, or Bwt/RK97A.R98A. Normalized responses (binding) are plotted as a function of UbcH5c concentration, and KD values were determined as described in Materials and methods.
The E2 and DNA-recognition surfaces of Bmi1/Ring1b are distinct
To assess the effect of Ring1b mutations on the E2–E3 interaction, we next used BLI to evaluate the binding of biotinylated Bmi1/Ring1b to UbcH5c under low salt conditions (50 mM NaCl). The wt complex bound to UbcH5c with a steady-state KD of 5±1 μM (Figure 6D), and a complex containing a Ring1b mutant defective in DNA binding (RK97A.RR98A) bound to UbcH5c with nearly full affinity (KD=9.6±0.4 μM). As expected, the RD56K mutant showed a clear defect in binding to UbcH5c, as evidenced by its steady-state KD of >40 μM. Taken together with the DNA-binding experiments, these data indicate that the RD56K mutation disrupts the ability of Bmi1/Ring1b to bind to UbcH5c, but not to DNA. On the other hand, the RK97A.RR98A double mutation has little to no effect on the binding of UbcH5c, but it disrupts DNA binding. Overall, this indicates that the mutations do not globally perturb the structure of the heterodimer and that the UbcH5c and DNA-binding faces of Bmi1/Ring1b are distinct.
Computational modelling of the Bmi1/Ring1b–UbcH5c interaction with the nucleosome
To provide insight into the geometry of the complex between the Bmi1/Ring1b–UbcH5c ligase and its nucleosome substrate, we performed experimentally guided computational docking, using HADDOCK2.0 (Dominguez et al, 2003; de Vries et al, 2007). Briefly, we docked the Bmi1/Ring1b–UbcH5c heterotrimer against the nucleosome core particle, allowing full flexibility of part of the H2A C-terminal tail (residues 118–120 only; see Materials and methods). To limit the conformational search to catalytically competent geometries, we enforced an unambiguous constraint such that UCys85 must be located within 2 Å of the H2A acceptor lysine. Lys119 has been reported to be the in vivo site of modification (Goldknopf and Busch, 1977), although we could also detect ubiquitination of Lys118 by mass spectrometry (Supplementary Figure S3). Nevertheless, we obtained similar docking results when using either Lys118 or Lys119 as a restraint, and so we describe below only models obtained using the Lys119 restraint. Mutagenesis data were incorporated as ambiguous interaction restraints to bring biochemically important residues of Bmi1 (BLys62 or BArg64) or Ring1b (RArg97 or RArg98) near the surface of the nucleosome.
The docked models partition into two distinct groups, rotated 180° relative to one another about the UbcH5c catalytic cysteine (Supplementary Figure S7A and B). The most-populated group (197/200 models), contains the top scoring decoys by intermolecular interaction energy (−3327 kcal/mol), constraint violation penalty (0.76 kcal/mol), and buried surface area (1400 Å2). Additionally, the C-terminal extensions present in both full-length Bmi1 and Ring1b would be accommodated by the orientation of the most-populated cluster, but members of the 180°-rotated cluster appear likely to clash with the nucleosome in their full-length forms. For the reasons above, we focused on placements of Bmi1/Ring1b–UbcH5c found in the most-populated cluster (Supplementary Figure S7A).
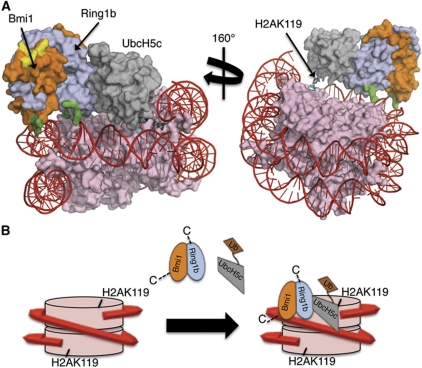
Requiring that UCys85 be near H2A Lys119 restricts the ligase complex to one region of the nucleosome, with UbcH5c located adjacent to where nucleic acid exits the nucleosomal wrap (Figure 7A). In the docked model, there are no direct interactions of UbcH5c with H2A except in the immediate vicinity of the modification site (residues 118–120). Ring1b appears to reach both nucleic acid and histone H4. The surface of histone H4 in this region contains several acidic residues, most notably Asp24 and Glu2, as well as the polar residue Gln27, which interact with the basic surface of Ring1b. Bmi1 makes almost no contacts with the histone, and instead interacts with nucleic acid via BLys62 and BArg64. Interestingly, the tertiary structure of this basic face on Bmi1/Ring1b forms a saddle, which is well suited to sit on top of the helical DNA duplex and to make contacts with the backbone phosphate groups. When compared with the BRCA1/BARD heterodimer or the cIAP2 homodimer, this saddle feature is a notable difference (see Supplementary Figure S5A). Taken together, the docking results suggest that Bmi1 and Ring1b are nucleosome recognition elements, with UbcH5c functioning as a modular catalytic component.
Figure 7.
Model for the interaction of the Bmi1/Ring1b–UbcH5c complex with the nucleosome. (A) Computational docking model generated using HADDOCK2.0. Colour coding is as follows: histones=pink, DNA=red, Ring1b=light blue, Bmi1=orange, UbcH5c=grey. Basic surface residues of Bmi1/Ring1b that were mutated in this study are shaded according to their effect on DNA binding: green=mutation that affected DNA binding, yellow=mutation that had no effect on DNA binding. The C-terminus of histone H2A (cyan stick representation) is the site of ubiquitin modification (K119). UCys85 is coloured by element: nitrogen=blue, oxygen=red, sulphur=yellow. (B) Cartoon model for recognition of the nucleosome by Bmi1/Ring1b–UbcH5c∼Ub complex. Colour scheme is the same as above (ubiquitin is shown in brown). Bmi1/Ring1b uses the basic saddle region to recognize and dock onto the nucleosome through contacts to both DNA and histone H4, positioning UbcH5c∼Ub for ubiquitin transfer to H2AK119. Following transfer of a single ubiquitin, UbcH5c∼Ub is no longer able to access both the E2-binding site on Ring1b and the nucleosome at the same time, leading to termination of the cycle after a single ubiquitination event.
Discussion
Bmi1 was originally identified as an oncogene cooperating with a myc transgene in lymphomagenesis (van Lohuizen et al, 1991). It eventually became clear that the Bmi1 protein is part of an E3 ubiquitin ligase complex (PRC1) (Wang et al, 2004) and that this complex has an important role in gene regulation during development (van der Lugt et al, 1994). Later in life, misregulation of PRC1 can lead to repression of important tumour suppressors, such as p16 or INK4a/ARF (Jacobs et al, 1999a, 1999b). The ubiquitin C-terminal hydrolase protein BAP1 has been identified as a deubiquitinase for uH2A (Scheuermann et al, 2010), thus reversing the function of PRC1. BAP1 itself had been identified previously as a tumour suppressor (Jensen et al, 1998; Nijman et al, 2005; Ventii et al, 2008), and in one recent study, inactivating mutations of BAP1 were found in 84% of metastatic uveal melanoma samples (Harbour et al, 2010). Significantly, single missense point mutations were found in the BAP1 catalytic residues (C91G and H169Q), suggesting that it is the loss of H2A deubiquitinase activity that is associated with the metastatic phenotype in these tumours, rather than loss of a protein–protein interaction. Given the links between excess Bmi1 activity and cancer (Sparmann and van Lohuizen, 2006; Bracken and Helin, 2009), the finding that H2A deubiquitination activity is required for tumour suppression by BAP1 provides a strong rationale for exploring the molecular interactions of the H2A ubiquitin ligase complex and its potential tractability as a therapeutic target.
To better understand the mechanism of Bmi1/Ring1b ubiquitin ligase activity, we have structurally characterized the interaction between the E2 UbcH5c and the Bmi1/Ring1b RING–RING heterodimer E3. Our results confirm that Ring1b alone interacts with UbcH5c, and that the binding site is localized to the two Zn2+-binding loops and the central α-helix of Ring1b, similar to the interface between UbcH5c and BRCA1 (Brzovic et al, 2003). Previous work had established that UbcH5c was an active E2 for Bmi1/Ring1b, but a stable complex between E2 and E3 was not detected by gel filtration (Buchwald et al, 2006). We find that the interaction between UbcH5c and the Bmi1/Ring1b heterodimer is both low affinity (≈7 μM) and highly salt sensitive, either of which might affect its isolation. The highly polar nature of the E2–E3 interface provides an explanation for the salt dependence of binding. Interactions between the RING-domain E3 ligase SCF and the E2 enzyme Cdc34 are also largely polar, and binding has been shown to be highly salt dependent for this complex, as well (Kleiger et al, 2009).
The ubiquitin ligase activity of the Bmi1/Ring1b heterodimer depends upon the presence of DNA and a well-ordered nucleosome. No combination of recombinant histone proteins (H2A, H2A/H2B dimer, or histone octamers) is a substrate for Bmi1/Ring1b, and addition of DNA is insufficient to rescue ubiquitination of any of these substrates (Figure 2). Further, we have demonstrated that the Bmi1/Ring1b heterodimer binds to short duplex DNA in a non-sequence-specific manner, and that mutation of DNA-binding residues disrupts E3 ligase activity without affecting either RING–RING heterodimer formation or E2 binding. Taken together, our data show that the direct interaction of the RING domains with nucleosomal DNA is crucial for the ubiquitin ligase activity of Bmi1/Ring1b. Interestingly, RING domains were first discovered in proteins with functions involving DNA, and, before it was shown that they function as E3 ubiquitin ligases, it was thought that they might mediate DNA binding (Deshaies and Joazeiro, 2009). In the case of Bmi1/Ring1b, it appears that the RING-domain heterodimer functions as both a DNA-binding platform and an E3 ligase. To our knowledge, this is the first example of a RING-domain E3 ligase binding directly to its substrate via the RING domain. It seems likely that other RING domains will be found to assist in steering E2s to the appropriate substrate lysines. However, in those RING-domain E3 ligases presently characterized, the substrate is recruited either through an additional domain of the E3, as in the case of the IAPs, or through a separate adaptor protein in a multiprotein complex, as in the APC/C and SCF complexes (Takahashi et al, 1998; Zheng et al, 2002; Varfolomeev et al, 2007; Schreiber et al, 2011).
Through site-directed mutagenesis we have been able to uncouple the E2 binding and the DNA-binding activities of Bmi1/Ring1b, indicating that Bmi1/Ring1b uses distinct binding surfaces to recognize E2 and the nucleosomal substrate. A mutation that impairs DNA binding has no effect on the binding of UbcH5c, whereas a mutation that disrupts UbcH5c binding does not affect the ability of Bmi1/Ring1b to bind to duplex DNA (Figure 6). Basic residues involved in DNA binding are located both on Ring1b and on Bmi1, and they map to the same general region of the complex surface. Importantly, mutations of basic residues on different faces of the complex do not affect either DNA binding or uH2A activity. This argues that association with DNA is not simply a matter of overall positive charge on Bmi1/Ring1b, but instead depends on a specific interaction site on the surface of the heterodimer. This putative DNA-recognition element is close to the E2-binding surface and would allow Bmi1/Ring1b to bind to both the nucleosome and ubiquitin-charged UbcH5c at the same time, bringing them into close proximity for ubiquitin transfer.
There has been some debate regarding the autoubiquitination of Bmi1 and Ring1b. One study suggested that full-length Bmi1 and Ring1b are capable of extensive autoubiquitination, and that this autoubiquitination activity is required for uH2A ligase activity (Ben-Saadon et al, 2006). Subsequently, it was found that the RING–RING heterodimer of Bmi1/Ring1b is only autoubiquitinated on a single site (RK112), and that this modification is dispensable for H2A ligase activity (Buchwald et al, 2006). We failed to detect autoubiquitination of the RING domains of either Bmi1 or Ring1b under our normal assay conditions. Based on our mutagenesis results, it seems instead that the primary role of many of the conserved lysines in the RING domains of Bmi1 and Ring1b is to interact with the nucleosome.
Given that Bmi1/Ring1b uses distinct faces to interact with E2 and the nucleosome, we wondered what the structure of the transfer complex might be. To this end, we performed biochemically driven docking using HADDOCK2.0 to generate a model of the Bmi1/Ringb–UbcH5c complex bound to the nucleosome core particle. It is important to note that the model was generated using only residues implicated in DNA recognition as restraints; no negative restraints of any kind were included. To cross-validate the final model, we assessed its compatibility with additional data that were not included in the docking. As can be seen in Figure 7A, several basic residues on Bmi1 (BLys88, BLys92, BArg95) that were not included as restraints point out towards solvent in the final HADDOCK model. When a triple mutation of these residues (BK88A.BK92A.BR95A/Rwt) was tested, it was found to be fully competent to bind DNA and an active ligase. In addition, mutation of RLys15, which points away from the nucleosome in our model, had no effects on catalysis or DNA binding. Thus, our docking model is consistent with our biochemical data. Furthermore, our model appears largely compatible with the low-resolution structure of a tetranucleosome core particle and derived chromatin fibre models (Schalch et al, 2005), although some remodelling might be required for PRC1 to fully access sites in the fibre.
Taken together, our data are consistent with a model for nucleosomal recognition as presented in Figure 7, in which the Bmi1/Ring1b heterodimer (as part of the larger PRC1 complex, or as part of dRAF) uses a basic saddle to interact both with an acidic region on histone H4 and with nucleosomal DNA in a non-sequence-specific manner. Upon binding of the Bmi1/Ring1b complex at this location, the E2-binding site on Ring1b would be positioned such that interaction with the E2∼Ub conjugate would place the C-terminal thioester of activated ubiquitin within a few angstroms of the Lys119 sidechain on histone H2A. Alternatively, the E2–E3 complex might slide along nucleosomal DNA until encountering the suitably positioned lysine of H2A. Following transfer of a single ubiquitin to Lys119, the steric bulk of the ubiquitin would prevent another E2∼Ub thioester from simultaneously accessing both the Ring1b-binding site and an acceptor lysine on either H2A or the H2AK119-Ub conjugate. Therefore, this arrangement would be expected to lead to terminal monoubiquitination of H2AK119, as is observed both in vitro and in vivo (Wang et al, 2004).
Our model suggests that the RING domain of Bmi1 is critical for the activity of the complex in two ways: (1) it provides conformational stability to the catalytic Ring1b subunit, stabilizing its structure and preventing aggregation and (2) it helps to provide some of the DNA-binding interface, through the BLys62 and BArg64 sidechains. Interestingly, these residues are not fully conserved among Bmi1 homologues, as they are Asn and Lys in the Drosophila protein Psc. However, mutation of BLys62 and BArg64 has less of an effect on DNA binding than does mutation of DNA-binding residues in Ring1b; thus, the Bmi1/Psc residues may be playing more of a supporting role in the interaction with the nucleosome, with Ring1b providing the majority of the binding affinity.
Bmi1 has long been appreciated as a critical player in the maintenance of repressed chromatin, and its overexpression has been linked to several different types of cancers (Mills, 2010; Sauvageau and Sauvageau, 2010). However, the mechanism by which Bmi1 and the PRC1 complex are targeted to chromatin has remained unclear. Recently, Wang et al (2010) showed that the C-terminal domain of Ring1b (C-RING1B) uses the same binding site to recognize either the Pc cbox domain or RYBP, an adaptor protein that links Ring1b to YY1, a sequence-specific DNA-binding protein. In this study, we have focused on the N-terminal RING domains of Bmi1 and Ring1b. However, we note that our model predicts a binding mode for the Bmi1/Ring1b heterodimer that leaves room for higher-order structure to form through their non-RING domains. It is tempting to speculate that PRC1 will utilize several recognition modes for specific domains of chromatin: Pc or YY1 first target the complex to a region of chromatin, and this is followed by a short-range search along the nucleosome by the Bmi1/Ring1b RING–RING heterodimer that directs the ubiquitin ligase activity to H2AK119. Future studies with the PRC1 complex will no doubt provide new insights into the mechanisms of its recruitment and assembly, and will shed light on the function of this critical family of epigenetic regulators.
Materials and methods
Protein expression and purification
UbcH5c, Bmi1, and Ring1b were cloned, expressed, and purified as described in the Supplementary data, that accompany this paper.
Mutagenesis
Mutants of Ring1b and Bmi1 were generated using the QuikChangeII Site-Directed Mutagenesis kit (Stratagene), using the manufacturer's protocol. All mutations were confirmed by DNA sequencing. All mutant protein complexes were expressed and purified in the same manner as the corresponding wt protein complexes.
Crystallization of Bmi1/Ring1b–UbcH5c complex
For co-crystallization, the Bmi1/Ring1b complex and UbcH5c were combined at a 1:1 ratio (150 μM each), then the mixture was concentrated to 12.5 mg/ml. total protein. Initial crystals of the UbcH5c–Bmi1/Ring1b complex were grown at 4 °C by vapour diffusion from sitting drops, formed by equal volumes (100 nl) of protein and crystallization buffer (40.0% MPD, 0.1 M MES pH 6.0), suspended over a reservoir of 100 μl. Small crystals appeared within 24 h. Crystals were optimized using hanging drops (1+1 μl) with a 1- ml well volume, and a final condition of 30.0% MPD, 0.1 M MES, pH 6.0 yielded crystals that grew to a typical size of 100 × 50 × 20 μm3. Crystals belonged to space group P3221, with cell dimensions of a=b=107.9 Å, c=77.6 Å, α=β= 90.0°, γ=120.0°, and a solvent content of 63%.
Data collection/structure determination and refinement
Diffraction data were collected at 100 K on a Rigaku MicroMax 007HF (1.541 Å) equipped with a Saturn 994+CCD detector and processed using HKL2000 (Otwinowski and Minor, 1997). The structure was solved by molecular replacement with PHASER (McCoy et al, 2007) within the CCP4i program suite (Collaborative Computational Project, 1994) using an ensemble of two structures of the Bmi1/Ring1b complex (PDB: 2CKL and 2HOD) and an ensemble of the UbcH5c E2-conjugating enzyme (PDB: 2FUH and 1X23). One copy of the Bmi1/Ring1b complex and one UbcH5c molecule were found in the asymmetric unit. The structure was rebuilt using Coot (Emsley et al, 2010) and refined using PHENIX (Adams et al, 2010) to a resolution of 2.65 Å, with final Rwork of 21.7% and Rfree of 24.3%. No residues fall in disallowed regions of Ramachandran space (96.5% in favoured, 3.5% in additionally allowed), as evaluated by ProCheck (Laskowski et al, 1996) and Molprobity (Chen et al, 2010).
Histone and nucleosome preparation
Recombinant histones H2A, H2B, H3.1, and H4 were purchased from New England Biolabs (catalogue # M2502S, M2505S, M2503S, M2504S, respectively). Recombinant human histone H2A/H2B dimer and (H3.1/H4)2 tetramer were purchased from New England Biolabs (catalogue # M2508S, M2509S, respectively). Nucleosomes were prepared from HeLa cell nuclei, digested with micrococcal nuclease, and purified by gel filtration according to the procedure of Schnitzler (2001). The quality of the purified nucleosomes was analysed by both SDS–PAGE and agarose gel electrophoresis (Supplementary Figure S2). Digestion of the nucleosomes with Proteinase K released a band of ≈400 bp, corresponding to tri-nucleosomes. Nucleosomes were frozen at −80 °C, and thawed immediately before use.
In vitro ubiquitin ligase assays
Human E1 and ubiquitin were obtained from in-house sources (Dong et al, 2011), and purified human nucleosomes were a generous gift from Sebastien Guelman (Genentech). Reactions were carried out by incubating human E1 (30 nM), UbcH5c (E2; 1.5 μM), Bmi1/Ring1b complex (E3; 500 nM), ubiquitin (25 μM), ATP (3 mM), and nucleosomes (7.5 μg, corresponds to 2 μM H2A) in 50 mM HEPES pH 7.2, 100 mM NaCl, 10 mM MgCl2, 1 μM ZnCl2, 1 mM DTT at 30°C for 1 h. Reactions were quenched by the addition of NuPAGE® LDS sample buffer (Invitrogen). Reaction products were separated by SDS–PAGE using 4–12% NuPAGE® Bis–Tris precast gels (Invitrogen), transferred to a nitrocellulose membrane using an iBlot® device (Invitrogen), probed using either rabbit polyclonal anti-H2A (Millipore; catalogue #07-146; 1:1000 dilution) or mouse monoclonal anti-ubiquitin (Clone PD41, Cell Signaling; catalogue #3936; 1:1000 dilution) antibodies, and detected by ECL+ chemiluminescence reagents (GE Healthcare).
FP DNA-binding assays
Oligonucleotides were synthesized by the Genentech DNA synthesis facility. Probes were combined with their (unlabelled) reverse complement and then annealed to form duplex DNA containing a FAM label on a single strand. The FAM probe was used at a concentration of 70 nM in all binding assays.
For direct binding measurements, FAM probe was combined with increasing concentrations of target protein in black 384-well ProxiPlate-F Plus plates (Perkin-Elmer, Inc.) in 25 mM HEPES, pH 7.2, 50 mM NaCl, 0.5% (w/v) BSA, 0.05% (v/v) Triton X-100, and incubated for 10 min at room temperature. Following incubation, fluorescence was measured at excitation and emission wavelengths of 482 and 535 nm, respectively, using a Wallac Victor3V 1420 Multilabel Counter (Perkin-Elmer, Inc.). FP was determined according to the equation:
 |
where Ipara and Iperp are the fluorescence intensities in the parallel and perpendicular planes, respectively.
Competition assays were performed using a fixed concentration of target protein that corresponded to the KD,app in a direct binding FP assay. Unlabelled probe containing the same sequence as the FAM-labelled probe was used for competition. Raw mP values were normalized against the range of signal, according to the relation:
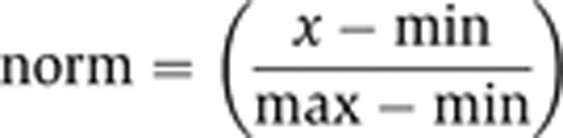 |
where norm is the normalized signal, x is the observed signal in mP, max is the maximum signal with no competition, and min is the minimum signal at full competition. 10, 8, and 6 bp probes were truncations of the 12-bp probe from the 3′ end. All plots were generated using KaleidaGraph version 4.03 (Synergy Software). Each reported value is the average of at least three data points. IC50 values were determined by fitting the average data to the four-parameter equation:
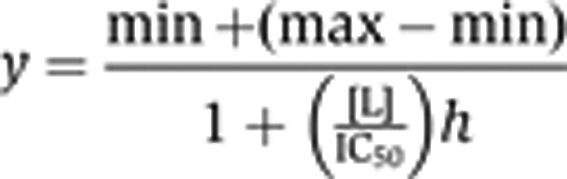 |
where max is the maximum normalized signal, min is the minimum normalized signal, [L] is the concentration of ligand, and h is the Hill slope.
Fluorescence-electrophoretic mobility shift assays
Following the FP assay, an equal volume of 50% glycerol was added to each well of the assay plates described above, and the samples were loaded onto 6% DNA Retardation Gels (Invitrogen, Inc.) that had been pre-run for 20 min in 0.5 × TBE buffer (44.5 mM Tris base, 44.5 mM boric acid, 1 mM EDTA pH 8.0) at 150 V on ice. Gels were run at 150 V for 38 min on ice. FAM-labelled DNA was visualized using excitation and emission wavelengths of 488 and 526 nm, respectively, and a PMT intensity of 600V using a Typhoon Trio Imager (GE Healthcare).
BLI-binding assays
BLI-binding assays were performed on an Octet Red384 system (ForteBio, Inc.). All binding studies were carried out at 30°C. Streptavidin high binding (SA) biosensors were loaded with biotinylated Bmi1/Ring1b complex (either Bwt/Rwt, Bwt/RD56K, or Bwt/RK97A.RR98A), in a buffer containing 50 mM HEPES pH 7.2, 50 mM NaCl, 0.5% (w/v) BSA, and 0.05% (v/v) Triton X-100. After loading, biosensors were washed in the same buffer, and then association and disassociation kinetic measurements were carried out for 900 s each. For measuring the salt dependence of UbcH5c binding, assays were run in the same manner as above, but the concentration of NaCl in the buffer was varied from 50 to 300 mM. Steady-state binding responses were determined by the overall response (nm) on each sensor at the end of the association phase of the binding experiment. For normalized measurements, the overall steady-state response at a given concentration of ligand was divided by the maximum response at saturation (Rmax) for that experiment, and the resulting percentages (fraction bound) were plotted as a function of ligand concentration. Plots were fit to a modified form of the Langmuir isotherm:
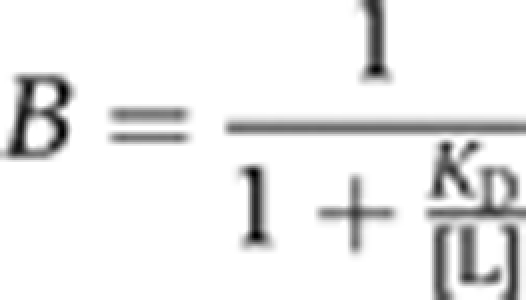 |
where B is the fraction bound and [L] is the concentration of UbcH5c. For all plotted experimental curves, the correlation coefficient (R2) of the fit is >0.97. All plots were generated using KaleidaGraph version 4.03 (Synergy Software).
Molecular docking studies using HADDOCK2.0
The. structure of the Bmi1/Ring1b–UbcH5c complex (reported here) was docked against a core nucleosome using HADDOCK2.0 (Dominguez et al, 2003; de Vries et al, 2007) and CNS (Brunger et al, 1998; Brunger, 2007), so as to yield a complex with potential to transfer ubiquitin to Lys119 of histone H2A. Since Lys119 is disordered in all reported crystal structures of human nucleosomes, the structure of the highly related Xenopus laevis nucleosome core particle was used (PDB ID: 3B6F). This structure is composed of a human α-satellite DNA sequence wrapped around Xenopus histones, which are ∼97% identical to human histones. The strained Lys119 rotamer present in the crystal structure was replaced with the most commonly populated lysine rotamer, which yielded no clashes with the rest of the nucleosome. An unambiguous interaction restraint with an effective upper limit of 2.0 Å was defined between UCys85 and H2A Lys119, while ambiguous interaction restraints were created between all solvent-exposed residues of the nucleosome and residues of Bmi1 or Ring1b whose mutation produced strong effects in an H2A ubiquitin ligase reaction (BLys62 or BArg64, RArg97, or RArg98). Half of these ambiguous constraints were randomly discarded in each docking trial. In total, 2000 rigid-body trials were performed, followed by semi-flexible refinement of the top 200 scoring trajectories. This refinement included automatic interface detection within the nucleosome, but kept Bmi1/Ring1b–UbcH5c fixed to avoid rigid-body movements within the ligase complex. The C-terminal tail of histone H2A (residues 118–120) was allowed to be fully flexible during the refinement. Resulting structures were sorted by intermolecular interaction and restraint energies and aligned on the nucleosome. Almost all structures belong to a single cluster (197/200), with the top 168 models by interaction energy falling in this category.
Supplementary Material
Acknowledgments
We thank Sarah Hymowitz for critical review of the final structure, Sebastian Guelman for his gift of purified human nucleosomes, and the Genentech DNA synthesis facility for providing labelled DNA probes. Atomic coordinates and structure factors for the Bmi1/Ring1b–UbcH5c complex have been deposited with the PDB, accession number 3RPG.
Author contributions: MLB designed and performed most of the experiments; MLB and AGC analysed the data; JEC performed HADDOCK computational modelling; KCD performed X-ray structure determination; QP and TKC performed mass spectrometry experiments; MLB, JEC and AGC wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr 66(Part 2): 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell 24: 701–711 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD (2006) Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9: 773–784 [DOI] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr 54(Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D 3rd, Fukuda M, Ohta T, Klevit R (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (2001) Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 8: 833–837 [DOI] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25: 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr 66(Part 1): 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol 14: 941–948 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr 50(Part 5): 760–763 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69: 726–733 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Ann Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R (2004) Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12: 633–644 [DOI] [PubMed] [Google Scholar]
- Dong KC, Helgason E, Yu C, Phu L, Arnott DP, Bosanac I, Compaan D, Huang OW, Fedorova AV, Kirkpatrick DS, Hymowitz SG, Dueber EC (2011) Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure (in press). [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr 66 (Part 4): 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE (2001) Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H (1977) Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci USA 74: 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM (2010) Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330: 1410–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, de Jong RN, Wienk H, Winkler GS, Timmers HT, Boelens R (2009) E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity. J Mol Biol 385: 507–519 [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10: 429–439 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999a) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M (1999b) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13: 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ 3rd (1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16: 1097–1112 [DOI] [PubMed] [Google Scholar]
- Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M (1995) mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J 14: 5672–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139: 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM (2006) Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem 281: 20643–20649 [DOI] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS (2006) Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 66: 6063–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL (2008) Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem 283: 31633–31640 [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467: 562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40(Part 4): 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA (2010) Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 10: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276 (Macromolecular Crystallography, Part A): 307–326 [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Ann Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M (2008) Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 20: 201–207 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J (2010) Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler GR (2001) Isolation of histones and nucleosome cores from mammalian cells. In Current Protocols in Molecular Biology, Frederick MA et al. (ed). Chapter 21: Unit 21 25 [DOI] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D (2011) Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470: 227–232 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2008) Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC (1998) A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem 273: 7787–7790 [DOI] [PubMed] [Google Scholar]
- Tsunaka Y, Kajimura N, Tate S, Morikawa K (2005) Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res 33: 3424–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, Berns A (1994) Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8: 757–769 [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A (1991) Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65: 737–752 [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131: 669–681 [DOI] [PubMed] [Google Scholar]
- Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD (2008) BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 68: 6953–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, Schirf V, Hart PJ, Lafer EM, Demeler B, Hinck AP, McEwen DG, Kim CA (2010) Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18: 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH (2005) Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.