Abstract
Production of non-canonical D-amino acids (NCDAAs) in stationary phase promotes remodelling of peptidoglycan (PG), the polymer that comprises the bacterial cell wall. Impairment of NCDAAs production leads to excessive accumulation of PG and hypersensitivity to osmotic shock; however, the mechanistic bases for these phenotypes were not previously determined. Here, we show that incorporation of NCDAAs into PG is a critical means by which NCDAAs control PG abundance and strength. We identified and reconstituted in vitro two (of at least three) distinct processes that mediate NCDAA incorporation. Diverse bacterial phyla incorporate NCDAAs into their cell walls, either through periplasmic editing of the mature PG or via incorporation into PG precursor subunits in the cytosol. Production of NCDAAs in Vibrio cholerae requires the stress response sigma factor RpoS, suggesting that NCDAAs may aid bacteria in responding to varied environmental challenges. The widespread capacity of diverse bacteria, including non-producers, to incorporate NCDAAs suggests that these amino acids may serve as both autocrine- and paracrine-like regulators of chemical and physical properties of the cell wall in microbial communities.
Keywords: D-amino acids; L,D-transpeptidase; peptidoglycan; stationary phase; Vibrio cholerae
Introduction
Nearly all bacteria synthesize a cell wall that is found outside of the cell membrane. This strong yet elastic network counteracts osmotic pressure, maintains cell shape and serves as a protective barrier against physical, chemical and biological threats (Holtje, 1998; Vollmer et al, 2008a). In both gram-positive and gram-negative bacteria, the cell wall is composed of a peptidoglycan (PG) polymer (also known as murein) that consists of glycan chains (alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc)) crosslinked by short peptides. The peptide chains vary somewhat among bacterial species, but normally include the D-amino acids D-Ala and D-Glu (or its amidated form, D-Gln). Unlike L-amino acids, D-amino acids are not used for ribosomal synthesis of proteins; in bacteria, their principal role is as cell wall constituents (Holtje, 1998; Vollmer et al, 2008a).
Synthesis of PG requires many enzymes and takes place in two cellular compartments—the cytosol and the periplasm/extracytoplasm. D-Ala and D-Glu are made by racemases from the corresponding L-amino acids, and the D-alanyl-D-alanine dipeptide (D-Ala-D-Ala) is subsequently produced by D-alanyl-D-alanine ligase (Ddl) (Walsh, 1989; Duncan et al, 1990; Choi et al, 1992). UDP-muramyl-L-Ala-D-γ-Glu-meso-diaminopimelate (UDP-M3) is formed in sequential steps requiring MurA-MurE; MurF then adds the D-Ala-D-Ala to generate UDP-MurNAc-pentapeptide (UDP-M5) (Barreteau et al, 2008). UDP-M5 is transferred to a C55 lipid phosphate (bactoprenyl phosphate) on the cytoplasmic face of the cell membrane generating lipid I; finally, UDP-GlcNAc is added to lipid I, yielding lipid II, the lipid-disaccharyl pentapeptide. All steps up to this point occur within the cytosol, and many require ATP. Lipid II is ‘flipped’ from the cytoplasmic to the periplasmic face of cell membrane for PG assembly and further modification (Bouhss et al, 2008; Mohammadi et al, 2011).
In the periplasmic space of gram-negative bacteria (or extracytoplasmic space of gram-positive organisms), membrane-bound transglycosylases and transpeptidases, such as high molecular weight penicillin-binding proteins (PBPs), assemble disaccharyl pentapeptide subunits into the PG polymer (Young, 2001; Scheffers and Pinho, 2005). Peptide chains are linked and modified, typically by D,D-transpeptidation reactions that link the D-Ala at position 4 in one peptide to the D-center of the meso-diaminopimelate residue [DAP] of a second peptide, either directly or through a short peptide bridge (Holtje, 1998; Vollmer et al, 2008a). As such transpeptidation reactions transfer energy from one peptide bond to another, they proceed in the absence of ATP, which is not present in the periplasm. The D,D-transpeptidases that generate these 4 → 3 crosslinks contain active site serines. Recently, a new class of cysteine transpeptidases, which are able to transfer L,D-peptide bonds to appropriate acceptors, have been identified and characterized. These L,D-transpeptidases (Ldts) can be subdivided in two types: those that mediate formation of DAP-DAP (3 → 3) crosslinks (Mainardi et al, 2005; Biarrotte-Sorin et al, 2006; Magnet et al, 2008) and those that attach lipoproteins to DAP (only reported in some gram-negatives species) (Magnet et al, 2007b).
PG is a dynamic polymer. Glycan chains and peptide crosslinks are cleaved by glycosylases and peptidases in order to permit expansion during cell growth (Vollmer et al, 2008b). Conversely, PG is often modified as cells enter stationary phase. Quiescent cells are often smaller than their rapidly growing counterparts (Nystrom, 2004). However, there is relatively scant knowledge of the factors and mechanisms that regulate the composition, architecture and amount of PG. Recently, we found that stationary phase bacteria from diverse phyla release distinct sets of D-amino acids other than those classically thought to be part of PG (‘non-canonical’ D-amino acids, in short NCDAAs). Furthermore, such D-amino acids, which were not previously known to be synthesized by bacteria, regulate the chemistry, amount and strength of PG. In Vibrio cholerae, NCDAAs (principally D-Met and D-Leu) are produced by BsrV, a periplasmic broad spectrum racemase (Lam et al, 2009). Chromosome sequence analyses suggest that other bacterial species may also encode amino acid racemases that enable synthesis of NCDAAs (Lam et al, 2009). D-Met was previously shown to be incorporated into V. cholerae PG during stationary phase; however, the mechanism and physiologic consequences of such incorporation were not determined. It has been hypothesized that NCDAAs alter cell wall properties, particularly its strength, via their incorporation into the PG polymer and/or via altering the activity of PBPs, key enzymes for PG synthesis (Lam et al, 2009).
Here, we explored the sites, mechanisms and consequences of incorporation of NCDAAs into bacterial cell walls. All bacterial species tested can incorporate NCDAAs into their PG; however, the sites and mechanisms utilized vary among species. Incorporation can be mediated both by periplasmic enzymes, predominantly Ldts, that ‘edit’ polymerized PG and by cytoplasmic enzymes that incorporate NCDAAs into PG precursors. In V. cholerae, interference with NCDAA incorporation leads to hypersensitivity to osmotic shock and excess accumulation of PG in the cell wall. NCDAA production is dependent upon the alternative sigma factor RpoS, a stress-associated regulator, suggesting that D-amino acid synthesis and incorporation into the bacterial exoskeleton may promote bacterial resistance to diverse environmental threats.
Results
Bacteria from diverse phyla incorporate NCDAAs into their cell walls
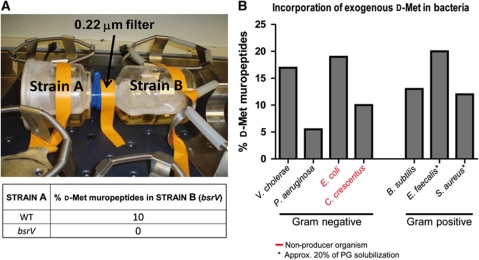
Diverse gram-negative and gram-positive bacteria produce NCDAAs and release them into the extracellular media (Lam et al, 2009). For example, supernatants from stationary phase V. cholerae cultures contain ∼1 mM of D-amino acids, primarily D-Met and D-Leu. HPLC analyses revealed that these unusual D-amino acids can become incorporated into PG (Lam et al, 2009). Such incorporation is not directly coupled to D-amino acid production; strains that fail to produce D-amino acids can nonetheless incorporate them into PG. For example, when bsrV V. cholerae (a non-producer; Lam et al, 2009) was grown for 16 h in a culture chamber linked to another chamber containing wild-type (WT) V. cholerae (a D-amino acid producer), using an apparatus that allows for the passage of small molecules but not cells, 10% of muropeptides isolated from the bsrV strain contained D-Met (Figure 1A). In contrast, no D-Met incorporation was detected when both chambers of the co-culture apparatus contained the bsrV mutant. Addition of supra-physiological (5 mM) D-Met to cultures of the bsrV mutant also resulted in its incorporation to the PG, at a level (19%) comparable to that observed with WT V. cholerae (17%) under equivalent conditions (Figure 1B). Exogenously supplied D-Met (5 mM) was likewise incorporated into the cell wall muropeptides of all bacterial species assayed, both those that synthesize NCDAAs, although not necessarily D-Met (e.g., Pseudomonas aeruginosa, Bacillus subtilis, Enterococcus faecalis and Staphylococcus aureus) and those do not (e.g., Escherichia coli and Caulobacter crescentus) (Figure 1B; Supplementary Figure S1). The extent of incorporation did not correlate with the capacity to synthesize these molecules. These observations suggest that the ability to incorporate NCDAAs into PG is widespread throughout the bacterial kingdom, and is mechanistically independent of the synthesis of such molecules.
Figure 1.
Incorporation of NCDAAs into PG muropeptides of diverse bacteria is not linked to their synthesis. The percentage of D-Met-containing peptides was calculated relative to total muropeptide isolated. (A) Co-culture system that permits chemical communication between two strains while they are kept physically apart (top). Percentage of D-Met-containing muropeptides in PG from strain B (bsrV V. cholerae) following co-culture with either WT or bsrV V. cholerae (bottom); note that the strain A culture was inoculated 2 h before the strain B culture to permit higher NCDAAs accumulation and hence better yields of incorporation into the PG. (B) Percentage of D-Met-containing muropeptides in PG from diverse bacteria following growth in the presence of exogenous 2 mM (C. crescentus) or 5 mM (remaining species) D-Met. The data shown are representative of two independent experiments.
D-Met is incorporated in the fourth and fifth positions of the peptide moiety of V. cholerae cell wall muropeptides
HPLC analyses revealed that D-Met is incorporated at two locations within PG subunits from stationary phase V. cholerae (Figure 2). Most of the D-Met replaces D-Ala in the fourth position of muropeptides ([muro4M]; Figure 2A and B; Supplementary Figure S2A), either within monomers [mono4M] or within 4 → 3 crosslinked dimers [di4,4M], as previously reported (Lam et al, 2009). However, further analyses of minor PG components from stationary phase samples revealed that D-Met can also replace D-Ala in the fifth position of muropeptides [muro5M], either within monomers [mono5M] or 4 → 3 crosslinked dimers [di4,5M] (Figure 2A and B; Supplementary Figure S2A). As expected, when exogenous D-Met is not supplied, all incorporation of D-Met into the cell wall is dependent upon its production by the BsrV racemase (Figure 2B). Analyses of exponential phase V. cholerae grown with non-deleterious supra-physiological concentrations (up to 10 mM) of D-Met did not lead to identification of additional D-Met-containing muropeptides, suggesting that incorporation is restricted to these sites within PG (muro4M and muro5M; Figure 2C; Supplementary Figure S2A).
Figure 2.
L,D-transpeptidases incorporate non-canonical D-amino acids into tetrapeptides of V. cholerae PG. Muropeptides within purified PG were identified and quantified using HPLC. (A) Schematic representation of muropeptide structures, illustrating amino acid content and peptide chain length. The canonical pentapeptide structure is shown on the left. (B) Percentage of muro4M and muro5M peptides, relative to total muropeptides, in PG purified from the indicated V. cholerae strains at stationary phase following growth in LB. (C) Percentage of muro4M and muro5M peptides, relative to total muropeptides, in PG purified from WT V. cholerae following the exponential phase growth in LB supplemented with the indicated concentration of D-Met. (D) Abundance of DAP-DAP muropeptides (muro-DAP-DAP), Lpp muropeptides (muro-Lpp) and D-Met muropeptides (muro-D-Met), relative to total amount of muropeptides from the indicated strains. Muro-Lpp includes the lipoprotein-attached muropeptides mono3-Lpp (GlcNAc-MurNAc-L-Ala-D-Glu-γ-meso-DAP-ε-L-Lys-L-Thr) and the crosslinked dimer GlcNAc-MurNAc-L-Ala-D-Glu-γ-meso-DAP-D-Ala-meso-DAP-(ε-L-Lys-L-Thr)-γ-D-Glu-L-Ala-MurNAc-GlcNAc. The Thr-Lys dipeptide corresponds to the C-terminal dipeptide of the V. cholerae homologue for E. coli Braun's lipoprotein. Digestion of sacculi with pronase E, one step of HPLC sample processing, degrades the covalently bound lipoprotein molecules leaving the C-terminal dipeptide bound to the connecting muropeptide which can therefore be easily differentiated. (E) Immunodetection of D-cysteine-labelled murein in sacculi from the indicated strains of V. cholerae. For each strain, the inset box was magnified to generate the subpanel on the right. The data shown are representative of three independent experiments.
Multifunctional Ldts incorporate D-amino acids into the cell wall
The most likely pathway for the generation of muro4M is an amino acid exchange reaction mediated by an Ldt in which the L,D-peptide bond from the L-center of DAP is transferred to the amino group of the acceptor D-amino acid, as previously hypothesized (Caparros et al, 1992). Recent studies have revealed that Ldts catalyse the formation of DAP-DAP crosslinks between muropeptides as well as the linkage between DAP and Braun's lipoprotein (Lpp) in some Gram-negative bacteria (Magnet et al, 2007a, 2007b, 2008; Supplementary Figure S2B). However, to date they have not been shown to catalyse amino-acid exchanges in vivo. We found two homologues of the five recently characterized E. coli Ldts (Magnet et al, 2007b, 2008) in the genome of V. cholerae N16961. One locus, vc1268, encodes a protein homologous to the Ldts YcbB and YnhG, whereas the other, vca0058, encodes a protein homologous to the Ldts ErfK, YcfS and YbiS (Supplementary Figure S2C). Cell fractionation and immunoblotting experiments revealed that, like other characterized Ldts, the two putative V. cholerae Ldts are membrane proteins (Supplementary Figure S2D). Furthermore, topologic predictions suggest that both VC1268 and VCA0058 are primarily periplasmic proteins anchored to the inner membrane by a single transmembrane helix (Supplementary Figure S2E).
Analysis of mutants lacking these putative transpeptidases revealed that V. cholerae lacking vc1268 (here renamed ldtA) is devoid of DAP-DAP crosslinks, while it maintains WT levels of Braun's Lpp-linked muropeptides (muro-Lpp: mono3-Lpp and di4,3-Lpp; Figure 2D). In contrast, V. cholerae lacking vca0058 (here renamed ldtB) has normal levels of DAP-DAP-linked muropeptides, but lacks Lpp covalently bound to the PG (Figure 2D). Thus, in these assays, LdtA and LdtB appear to have mutually exclusive roles. Neither class of crosslink was detected in PG from V. cholerae lacking both ldtA and ldtB. However, each could be restored in this background by expression of the relevant gene, thereby confirming the roles inferred from analysis of the individual mutants (Figure 2D).
Further analyses revealed that both LdtA and LdtB contribute to incorporation of NCDAAs, such as D-Met, at the fourth position within muropeptides. Deletion of either ldtA or ldtB significantly reduced the percentage of muro4M peptides in PG from stationary phase V. cholerae, and deletion of both genes rendered muro4M peptides undetectable, suggesting that they account for most, if not all, of the DAP-D-Met linkages in V. cholerae PG (Figure 2B; Supplementary Figure S3A). Complementation analyses confirmed the roles of LdtA and LdtB in production of muro4M (Figure 2B). In contrast, neither LdtA nor LdtB appears important for generation of muro5M peptides, as the level of such muropeptides in PG from the ldtA and ldtB mutants was equivalent to that of the WT strain (Figure 2B; Supplementary Figure S3A). This result is consistent with the fact that muro5M peptides contain a D,D-peptide bond between D-Met and the adjacent D-Ala, rather than the L,D-bond typically synthesized by Ldts. LdtA and LdtB were also shown to be required for incorporation of D-Met into muro4M peptides by exponential phase cultures of V. cholerae grown in the presence of exogenous D-Met (Supplementary Figure S3B).
D-Cysteine labelling of sacculi is mediated by Ldts
Almost two decades ago, de Pedro et al (1997) demonstrated that D-Cys could be incorporated into the murein of several bacterial species, but the mechanism by which this occurs has not been elucidated. Nevertheless, D-Cys-mediated labelling of PG has facilitated important studies of the dynamics of PG synthesis in diverse organisms (de Pedro et al, 2003; Aaron et al, 2007). Based on our observation that Ldts mediate incorporation of D-Met into V. cholerae PG, we explored whether these enzymes are also responsible for incorporation of D-Cys (another NCDAA). We found that D-Cys is incorporated into the V. cholerae sacculus via a process that is largely dependent on ldtA and ldtB, since virtually no D-Cys incorporation was detected in the ldtA ldtB double mutant (Figure 2E). D-Cys labelling of the ldtA ldtB mutant was at least partially restored by expression of plasmid-encoded ldtA or ldtB, providing further evidence that these genes contribute to the labelling process in V. cholerae (Figure 2E). Similarly, deletion of Ldts that crosslink DAP residues in E. coli (YnhG and YcbB) greatly reduced immunodetection of D-Cys in the sacculi (Supplementary Figure S3C). However, it is likely that processes other than L,D-transpeptidation contribute to D-Cys-mediated labelling in other bacterial species (discussed later).
LdtA substrate specificity
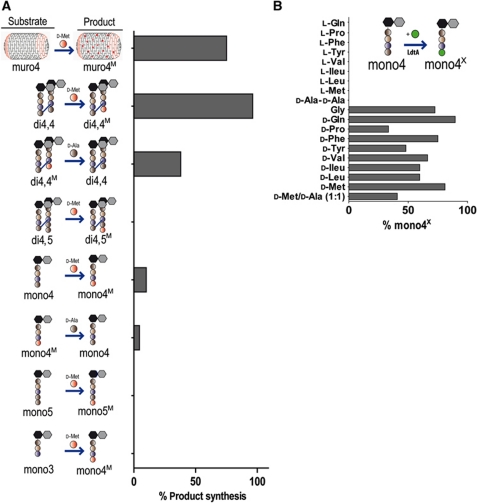
His-tagged LdtA and LdtB were purified to enable in vitro characterization of these proteins' biochemical properties. Purified LdtB showed no activity in D-amino-acid incorporation assays; consequently, we focused on elucidating LdtA's activity. Initially, we tested whether sacculi from exponential phase V. cholerae, which contain no D-Met (Lam et al, 2009), could serve as a substrate for LdtA. After purified sacculi were incubated with LdtA and D-Met for 2 h, 75% of the dimer and monomer tetrapeptides (muro4) contained D-Met rather than D-Ala in position 4 (muro4M; Figure 3A; Supplementary Figure S4A). No incorporation of D-Met was found in position 5, consistent with the in vivo observations presented above that the Ldts only incorporate D-Met at position 4. Thus, LdtA can recognize and modify native V. cholerae PG in the absence of additional cofactors.
Figure 3.
In vitro characterization of LdtA. Muropeptides were identified and quantified using HPLC; no reaction products other than those shown in the figures were detected. (A) The percentage of substrate converted into product via in vitro L,D-transpeptidation by LdtA is shown for various muropeptides. Reactions were incubated for 30 min (purified muropeptides) or 2 h (sacculi). (B) The percentage of mono4 altered by in vitro LdtA transpeptidation to have a different terminal amino acid when incubated (2 h) in the presence of the indicated L- or D-amino acid is shown. The data shown in (A) and (B) are representative of three and two independent experiments, respectively.
We then used crosslinked dimer (di) and monomer (mono) muropeptides purified from PG to further assess the substrate specificity of LdtA. Assay conditions (see Materials and methods) were optimized with mono4 and D-Met as substrates (Supplementary Figure S4B). Under our standard conditions, about 10% of mono4 was converted into mono4M in 30 min at 37°C, a value corresponding to 10 μg of muropeptide converted per minute and per microgram of enzyme. Assays with di4,4 as the donor substrate revealed that LdtA acted far more efficiently on this muropeptide than on mono4, with a conversion into di4,4M of 96% under otherwise identical conditions. Neither mono5 nor di4,5 was accepted as substrates for the exchange reaction even after an extended (2 h) incubation period (Figure 3A; Supplementary Figure S4C).
Since D-Ala is present in the periplasm as a consequence of PG remodelling and turnover (Park and Uehara, 2008), we tested whether LdtA could catalyse the reverse reaction, with D-Met muropeptides (mono4M and di4,4M) as donor substrates and D-Ala as acceptor. LdtA was indeed able to do so, although at significantly lower rates than the forward (D-Ala → D-Met) reaction and, as before, the crosslinked muropeptide di4,4M was a better substrate than the monomer, showing 38 and 4.5% conversion, respectively (Figure 3A; Supplementary Figure S4C). The difference between efficiencies for the forward (D-Ala → D-Met) and reverse (D-Met → D-Ala) reactions suggests that accumulation of D-Met muropeptides would be favoured in vivo, even if both D-Met and D-Ala were simultaneously present at similar concentrations. Therefore, in stationary phase, when D-Met is present, LdtA is likely to promote accumulation of muro4M peptides, rather than a continual exchange of the C-terminal amino acid within tetrapeptides.
In natural environments, bacterial communities typically contain multiple species (Straight and Kolter, 2009), which could produce a variety of D-amino acids (Lam et al, 2009). Therefore, we tested whether LdtA could incorporate other amino acids besides D-Met or D-Cys into PG. Indeed, LdtA accepted, at varying efficiencies, all the D-amino acids tested as acceptors for the exchange reaction using mono4 as the donor substrate (Figure 3B; Supplementary Figure S4D). However, none of the corresponding L-amino acids was accepted, as previously reported for the E. faecium Ldt (Ldtfm) (Mainardi et al, 2005). Interestingly, both D-Pro, which has a secondary amino group, and Gly, which lacks a chiral centre, were accepted as substrates for the in vitro reaction (Figure 3B; Supplementary Figure S4D). Simultaneous addition of equal concentrations of D-Met and D-Ala to an LdtA in vitro reaction with mono4 as donor substrate resulted in about a 50% reduction in the incorporation of D-Met (Figure 3B), suggesting that using this substrate LdtA does not preferentially utilize D-Met rather than D-Ala as an acceptor. Finally, in contrast to findings reported in Magnet et al (2007a), LdtA did not use a D-Ala-D-Ala for the exchange reaction with mono4 as the donor substrate, suggesting that there are structural constraints upon its substrates beyond the requirement for a D-amino acid acceptor molecule.
Analysis of the LdtA protein sequence revealed a YkuD domain that has been observed in other Ldts (Supplementary Figure S5A; Bielnicki et al, 2006). This domain contains three invariant residues (H, G and C), including the presumed catalytic cysteine residue (Biarrotte-Sorin et al, 2006; Supplementary Figure S5). When all three residues were mutated to Alanine, LdtA activity was abolished (Supplementary Figure S5B), consistent with this domain containing the enzyme's active site and suggesting that the catalytic mechanism of LdtA is similar to previously characterized homologues (Mainardi et al, 2005; Biarrotte-Sorin et al, 2006).
Synthesis of muro5M in V. cholerae is dependent on cytoplasmic processes
Generation of muro5M could be explained as the result of a D,D-transpeptidase mediated D-amino acid exchange reaction on normal muro5. However, the scarcity of muro5 in V. cholerae PG argues against it. Therefore, we hypothesized that the D-Ala-D-Met-peptide bond in muro5M could be formed via the biosynthetic pathway for generation of cytoplasmic PG precursors. Consistent with this hypothesis, previous studies reported that cytoplasmic Ddl ligases are not highly specific for incorporation of D-Ala at the C-terminal position of dipeptides, at least in unbalanced media (Barreteau et al, 2008). Therefore, we assessed D-Met incorporation into V. cholerae PG in early stationary phase cells treated with sublethal concentrations of D-cycloserine, a D-amino-acid analogue that inhibits both Ddl and the alanine racemase (Barreteau et al, 2008; Figure 4A). D-Cycloserine rendered muro5M peptides undetectable but had a minimal influence on muro4M formation (Figure 4A). The latter observation suggests that cycloserine's effect on muro5M formation is not simply a consequence of reducing D-Met availability, for example, due to inhibition of racemase activity, and is instead likely to ensue from Ddl inhibition.
Figure 4.
Incorporation of non-canonical D-amino acids into muropeptides occurs via several Ldt-independent pathways. (A) The relative percentage of muro4M and muro5M peptides in PG isolated from stationary phase cultures of V. cholerae grown in the absence (control) or presence of D-cycloserine (100 μg/ml). (B) Schematic representation of the in vitro conversion of D-amino acids (D-Ala and/or D-Met) and UDP-M3 into UDP-M5 and UDP-M5M by Ddl and MurF. (C) Yield, based on HPLC quantification of UDP muropeptides following A262 (UDP detection), of the reaction depicted in (B), when the indicated amino acid substrates are supplied. (D) The percentage of muropeptides that contain D-Met (muro4M or muro5M) in various bacterial species and their ldt mutants following growth in 5 mM D-Met, or 2 mM in the case of C. crescentus. (E, F) The effects of D-cycloserine (100 μg/ml) and/or penicillin G (50 μg/ml) on the relative level of muro5M peptides in PG isolated from C. crescentus (E) or B. subtilis (F) following growth in the presence of 2 mM or 5 mM D-Met, respectively. The data shown are representative of three independent experiments.
Cytoplasmic ligases Ddl and MurF catalyse formation of muro5M precursors
In vitro analyses provided additional support for the hypothesis that muro5M formation is dependent upon utilization of D-Met by Ddl and MurF, as depicted in Figure 4B. We purified Ddl and MurF from V. cholerae (see Materials and methods) and reconstituted their coupled reaction, which generates UDP-M5, in vitro in the presence of UDP-M3 as substrate. HPLC analysis of the final contents of these reactions showed that, in the presence of D-Ala, ∼9% of UDP-M3 was transformed into UDP-M5 (Figure 4C; Supplementary Figure S5C), and this process was inhibited by D-cycloserine. A similar yield was obtained using MurF alone if D-Ala-D-Ala was supplied in place of the D-Ala monomer, suggesting that MurF may catalyse the rate-limiting step in this synthetic process. When D-Met was used to replace an increasing proportion of the D-Ala within the reaction, the yield of UDP-M5 was steadily reduced. However, when a high proportion (80%) of D-Ala was replaced by D-Met, a new peak was evident in HPLC analyses (Figure 4C; Supplementary Figure S5C), which was shown to be UDP-M5M, with the D-Met residue at position 5 according to electrospray-ion trap MS/MS. Thus, although in these in vitro conditions D-Met is not the preferred substrate, it can be incorporated into PG precursors by Ddl and MurF, and such incorporation suggests formation of muro5M peptides via the cytosolic biosynthetic pathway in V. cholerae.
Different processes control formation of D-Met muropeptides in bacteria
To begin to explore whether the pathways for incorporation of NCDAAs are conserved, we assessed whether D-Met was targeted to equivalent muropeptides within diverse species, and whether mutations and antibiotics have uniform effects upon D-Met incorporation. Unexpectedly, these assays revealed that only V. cholerae and C. crescentus incorporate NCDAAs into both tetrapeptides and pentapeptides, although C. crescentus, unlike V. cholerae, contains predominantly muro5M (Figure 4D; Supplementary Figure S1). PG from all other organisms tested contained just one of the two classes of peptides. Following growth in media supplemented with D-Met, muro4M peptides were detected in PG from E. coli and P. aeruginosa, while muro5M peptides were detected in PG from B. subtilis, E. faecalis and S. aureus (Figure 4D; Supplementary Figure S1).
Since Ldts are exclusively involved in muro4M synthesis in V. cholerae, we tested whether LdtA orthologues in other species account for the same function. Consistently, deletion of Ldts reduced the abundance of muro4M in bacteria that produce it (Figure 4D). All of E. coli's five Ldts appear to contribute to D-Met incorporation into its PG, although two of them (YcbB and YnhG) seem to account for >75% of this process (Figure 4D). Similarly, in P. aeruginosa, the ldt PA_27180 appears to mediate most of the D-Met incorporation into its PG. On the contrary, ldt deletion did not impair incorporation of D-Met into B. subtilis PG, which occurs exclusively at position 5 in this bacterium (Lam et al, 2009; Supplementary Figure S1).
To assess whether the process for muro5M production in V. cholerae is similarly conserved among bacterial species, we studied the impact of antibiotics upon muropeptide abundance in C. crescentus and B. subtilis. As in V. cholerae, muro5M levels in C. crescentus PG were markedly inhibited by D-cycloserine, suggesting their production is also dependent upon a cytoplasmic process mediated by Ddl (Figure 4E). However, muro5M accumulation in B. subtilis was not affected by D-cycloserine; instead, it was efficiently inhibited by penicillin G (Figure 4F). Given the observed antibiotic sensitivity, it is possible that muro5M in B. subtilis is the product of an exchange reaction catalysed by penicillin-binding D,D-peptidyl transferases. Thus, unlike the case for muro4M peptides, it appears that muro5M peptides can arise via multiple distinct processes.
Incorporation of NCDAAs into V. cholerae PG controls stationary phase PG strength and amount
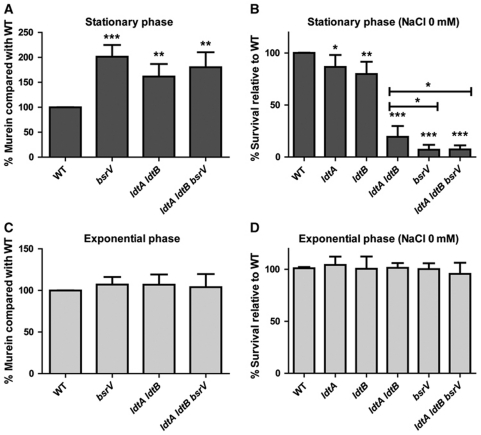
In V. cholerae, production of D-Met and other NCDAAs in stationary phase negatively regulates the amount of PG per cell and is required to maintain resistance against low osmolarity (Lam et al, 2009), but it is not known whether these phenotypes are a function of D-amino acid incorporation or instead reflect the activity of the free amino acids. To test if Ldt-mediated incorporation of NCDAAs into position 4 of the peptide moiety is actually required to manifest these phenotypes, we measured the amount of PG accumulated by stationary phase cells of both the ldtA ldtB mutant and the wt and found 1.7 times more PG per cell in the mutant than in the wt, an increment similar to that described for the bsrV mutant (Figure 5A; Lam et al, 2009). Additionally, at stationary phase, the ldtA ldtB mutant was significantly more sensitive to low osmolarity challenge than the WT strain, although it was still somewhat less sensitive than the bsrV mutant (Figure 5B). A triple ldtA ldtB bsrV mutant was not more sensitive to low osmolarity than the single bsrV mutant, suggesting that the mutations might have overlapping effects, rather than independent (additive) consequences. Notably, none of the mutants showed altered PG abundance or resistance to a low osmolarity challenge during exponential growth phase (Figure 5C and D), although both ldtA and ldtB are expressed and active during exponential growth (Figure 6A and B). Instead, the effects of ldtA and ldtB deletion were only evident at times when bsrV is expressed (i.e., stationary phase; Figure 6B). Furthermore, none of the mutants showed altered viability upon incubation in isotonic (150 mM) NaCl solutions (Supplementary Figure S6), in accordance with our previous results for the bsrV mutant (Lam et al, 2009). Collectively, these data are consistent with the hypothesis that the alterations in PG content, composition and resistance to osmotic challenge in the ldtA ldtB mutants results from their incapacity to incorporate the NCDAAs produced by BsrV into the macromolecular PG, rather than the absence of DAP-DAP or DAP-Lpp crosslinks.
Figure 5.
Influence of D-amino acids and Ldts on PG abundance and osmotic tolerance of stationary phase V. cholerae. (A, C) Relative abundance of PG in stationary (A) and exponential (C) V. cholerae cells. Abundance of murein from the indicated mutants was normalized to that from wild-type (WT) cells in each growth phase. Mean and standard deviation values are derived from six independent experiments. Asterisks above the error bars in the mutants represent significant P-values from comparisons of the WT strain and the corresponding mutant (***=0.0001, **<0.003). (B, D) Relative survival of stationary (B) or exponential (D) V. cholerae cells following a hypo-osmotic challenge. Survival of the indicated mutants following a 15-min incubation in distilled water is shown relative to the survival of the WT strain. Asterisks represent significant P-values from comparisons of mutants with the WT strain or from comparisons of the indicated pairs; *<0.03, **<0.002, ***<0.0001.
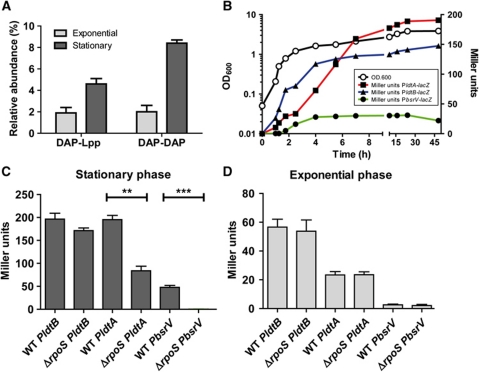
Figure 6.
The alternative sigma factor RpoS contributes to growth phase-dependent expression of bsrV, ldtA and ldtB. (A) The abundance of Lpp muropeptides (DAP-Lpp) and DAP-DAP muropeptides (DAP-DAP) in PG from exponential and stationary phase V. cholerae. (B) The culture density (OD600; open circles) and β-galactosidase activities (Miller units; closed symbols) of strains containing chromosomal transcription reporter fusions for bsrV, ldtA and ldtB are shown as a function of time. Note that in (B) the culture densities of all three strains overlap, and therefore it has been represented with a single growth curve. (C, D) Comparison of the β-galactosidase activities of the reporter strains shown in (B) with their ΔrpoS derivative strains in exponential (OD600=0.4) (C) and stationary (OD600=3.5) (D) phase. Mean and standard deviation values reflect three (A, C and D) or two (B) independent experiments; **=0.0012, ***=0.0003.
Stationary phase expression of BsrV and LdtAB is governed by RpoS
Since the NCDAAs synthesized by V. cholerae BsrV are only generated upon entry into stationary phase, and the DAP-Lpp and DAP-DAP linkages formed by Ldts are more abundant in stationary phase (Figure 6A), we explored whether RpoS, an alternative sigma factor active predominantly during stationary phase, contributes to expression of these enzymes. Transcription of bsrV, ldtA and ldtB was assessed using transcription reporter fusions comprising lacZ fused to the chromosomal loci, both in the presence and absence of rpoS. These assays revealed that expression of BsrV is exclusively triggered when bacterial growth slows (Figure 6B), consistent with the time when NCDAAs begin to accumulate (Lam et al, 2009), and that bsrV transcription is fully dependent upon rpoS (Figure 6D). In contrast, expression of ldtA and ldtB occurs in the exponential as well as stationary phase of growth, as noted above, although it is higher in stationary phase (Figure 6B). Expression of ldtA and ldtB is not exclusively dependent on rpoS in stationary phase, but is diminished by 57 and 15%, respectively, in the rpoS background (Figure 6C). As expected, deletion of rpoS had no effect on the activity of any of the reporter fusions during exponential phase of growth (Figure 6D). Collectively, these results indicate that RpoS augments, but is not critical for, production of Ldts and L,D-peptide bonds during stationary phase; however, RpoS is essential for generation of non-canonical muropeptides by Ldts, as it is required for expression of BsrV, which generates their D-amino acid substrates.
Discussion
PG, the primary constituent of the bacterial cell wall, is a strong and dynamic polymer that is crucial for maintaining cell integrity and for protecting against a variety of environmental challenges. Despite fairly extensive knowledge of the cytoplasmic pathways for generating the disaccharide-pentapeptide PG precursor subunits and the periplasmic enzymes that generate the PG polymer (Figure 7), little is known about the factors and mechanisms that modulate cell wall properties. Recently, we demonstrated that a variety of bacteria release NCDAAs upon entry into stationary phase, and that the presence of these amino acids was linked to changes in PG amount, strength and chemistry (Lam et al, 2009). Here, we characterized the sites, mechanisms and consequences of the incorporation of NCDAAs into PG. The capacity to incorporate such amino acids apparently is widespread among bacteria. However, multiple incorporation mechanisms seem to exist. Incorporation was detected at two sites in PG subunits, and seems to be mediated by at least three processes, including both periplasmic PG editing and cytoplasmic incorporation into PG precursors (Figure 7). Incorporation of D-amino acids into the cell wall during stationary phase is instrumental for V. cholerae to resist osmotic stress and to control the amount of PG per cell. Furthermore, at least in V. cholerae, production and incorporation of NCDAAs are largely governed by the sigma factor RpoS, which is active during stationary phase but also in response to numerous stresses (Hengge, 2008), raising the possibility that D-amino acids may aid bacteria in adapting to, and protecting themselves against, a wide variety of environmental challenges.
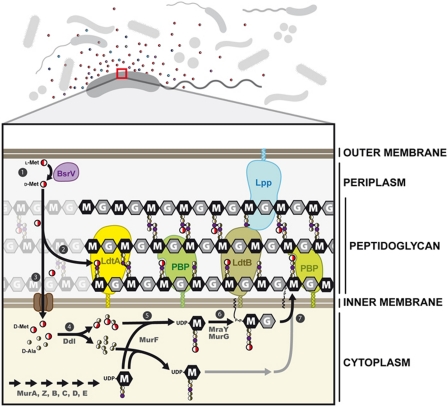
Figure 7.
Schematic representation of the pathways of incorporation of non-canonical D-amino acids into the V. cholerae cell wall. (1) Periplasmic BsrV racemase converts L-Met into D-Met. (2) Periplasmic L,D-transpeptidases LdtA and LdtB incorporate D-Met into the fourth position of the peptide moiety of the muropeptides. (3) Transport of D-Met into the cytoplasm through an ABC transporter, for example, a MetNIQ homologue (Kadaba et al, 2008). (4) Ddl forms D-Ala-D-Met dipeptides that (5) serve as substrates for MurF (Duncan et al, 1990) to generate a UDP-muramyl-(D-Met)-pentapeptide (UDP-M5M). Finally, (6) cytosolic activities generate the final precursor unit (UDP-disaccharide pentapeptide) (Barreteau et al, 2008) that (7) is flipped to the outer face of the cytoplasmic membrane to be incorporated into the macromolecular murein by PBPs with transglycosylase and transpeptidase activities.
NCDAAs, such as D-Met, are incorporated at two distinct sites within V. cholerae PG subunits. The majority of D-Met (80%) is present as the C-terminal amino acid within monomeric or dimeric tetramers (muro4M), while the remainder comprises the C-terminal amino acid within pentapeptides (muro5M). Formation of muro4M peptides is dependent upon Ldts, inner membrane-anchored periplasmic proteins that catalyse L,D crosslinking of muropeptides and linkage of lipoprotein Lpp to PG (Magnet et al, 2007b, 2008). Both of V. cholerae's two Ldts (LdtA and LdtB) contribute to incorporation of D-Met, although the two proteins have exclusive roles with respect to muropeptide crosslinking (Figure 7). Ldts have not previously been shown to modulate PG architecture in vivo by processes distinct from muropeptide crosslinking. Ldts also mediate incorporation of D-Met into PG tetrapeptides in E. coli and P. aeruginosa, even though E. coli, unlike the majority of bacterial species tested, does not produce NCDAAs. It remains to be seen why Ldt homologues in a subset of organisms (e.g., B. subtilis) apparently do not catalyse incorporation of NCDAAs.
In vitro characterization of a purified Ldt from V. cholerae (LdtA) revealed that it can exchange the C-terminal amino acid of a variety of tetrapeptide substrates. However, LdtA is more efficient at catalysing exchange in dimeric than in monomeric substrates, and it more readily catalyses replacement of D-Ala by D-Met than the reverse reaction. The latter finding indicates that LdtA should promote muro4M accumulation in stationary phase, when NCDAAs are produced. LdtA can accept many D-amino acids as substrates, a flexibility that may enable reaction to the range of D-amino acids produced during stationary phase by diverse organisms in the natural environment.
Deletion of V. cholerae ldts increased the quantity of PG but impaired the resistance of this organism to an osmotic challenge at stationary phase. Notably, these phenotypes mirror those of a mutant lacking bsrV, the periplasmic racemase that generates NCDAAs. Furthermore, deletion of ldts in the bsrV mutant did not augment its osmotic sensitivity, consistent with all mutations conferring sensitivity via the same pathway. Neither the bsrV mutant nor the Ldt-deficient mutant was more fragile than the WT strain during exponential growth, when D-Met is not available. Collectively, the similarities among these mutants suggest that incorporation of NCDAAs during stationary phase by Ldts is necessary to maintain the proper amount of PG and strength of the cell wall during transition into a resting state, and that such incorporation accounts for most of the effects of D-amino acids upon V. cholerae's resistance to osmotic challenge. It is not yet known why PG containing D-Met is stronger, as well as why strains that lack this incorporation lose control of the PG amount per cell. Such effects may be a consequence of the different physical properties of muro4 and muro4M peptides. For example, DAP within these peptides may differ in its accessibility for subsequent formation of crosslinks (Supplementary Figure S7), and muro4M may modulate the catalytic properties of PG synthetic and/or modifying enzymes.
Unlike the Ldt-mediated periplasmic incorporation of D-Met into muro4M, incorporation of D-Met into muro5M likely occurs within V. cholerae cytoplasm, via PG precursors. Presumably, D-Met reaches this compartment via a specific transporter, such as the MetNIQ D-Met transporter identified in E. coli (Kadaba et al, 2008; Figure 7). Both in vivo and in vitro analyses indicate that D-Met is an alternate substrate for Ddl and that MurF can utilize dipeptides that contain D-Met, although the yield of UDP-muramyl pentapeptides from these enzymes is partially inhibited by D-Met. Such inhibition might contribute to reduced PG accumulation in response to growth arrest. Muro5M peptides also appear to alter periplasmic PG editing. As previously observed in E. coli and other species, canonical pentapeptides are scarce in V. cholerae PG, as they are presumably transformed into tetrapeptides by D,D-carboxypeptidases. In contrast, muro5M is readily detected in stationary phase, suggesting that it is a poor substrate for these enzymes. It is possible that the alteration in the balance of pentapeptides and tetrapeptides also influences the accumulation and/or properties (e.g., strength) of PG.
Muro5M was also detected in PG from other bacteria; however, such muropeptides were generated by at least two different pathways. Formation of muro5M in C. crescentus is inhibited by D-cycloserine as in V. cholerae, suggesting that it is likewise mediated by Ddl and MurF. However, muro5M formation in B. subtilis appears to be dependent on extracytoplasmic processes, as it was blocked by penicillin G but unaffected by D-cycloserine. A D-amino acid exchange reaction at position 5 mediated by a D,D-peptidyl transferase, as described in Streptomyces R61 (Pollock et al, 1972), may account for muro5M formation in B. subtilis. Given that both Ddl and MurF are highly conserved proteins, it is unclear why they apparently catalyse D-Met incorporation only in a subset of bacterial species. Some enzymes might have more permissive catalytic sites. Alternatively, different species may have unequal capacities to transport NCDAAs into their cytoplasms.
The finding that NCDAAs can be incorporated into PG via multiple pathways may enable alternate approaches to visualization of PG dynamics. As established by de Pedro in 1997, PG can be uniformly labelled via incorporation of D-cysteine. Detection of cysteine-free and cysteine-rich regions following a chase period without D-cysteine allows identification of newly synthesized PG as well as zones where PG is largely inert. However, for organisms in which D-Met (and presumably D-Cys) is incorporated only into pentapeptide precursors, it should be possible to visualize newly synthesized PG directly via D-Cys-mediated labelling.
Finally, the observation that BsrV-mediated production of NCDAAs is fully dependent upon RpoS suggests that modification of PG with these amino acids may enhance PG strength and bacterial survival under a variety of conditions, rather than just stationary phase. D-amino acids may also alter the relative fitness of bacteria within mixed microbial communities, including bacteria that do not produce NCDAAs, as the capacity for incorporation seems to be widely spread. Such alteration could lead to a variety of phenotypes. For example, incorporation of D-amino acids has been linked to dissolution of biofilms (Kolodkin-Gal et al, 2010) and antibiotic resistance (Reynolds and Courvalin, 2005). Future studies are likely to reveal additional roles for D-amino acids in stress responses and bacterial communication.
Materials and methods
Bacterial strains and growth conditions
Strains, plasmids and primers are listed in Supplementary Tables S1–S3. C. crescentus was grown at 30°C in PYE broth or solid media (2 g Bacto peptone; 1 g yeast extract; 1 ml 1 M MgSO4; 0.5 ml 1 M CaCl2 per litre of distilled water). Other species were grown in Luria broth (LB) or on LB agar plates at 37°C. Antibiotics were used at the following concentrations (per millilitre): 200 μg streptomycin [Sm]; 50 μg ampicillin [Ap] or carbenicillin [Cb]; 50 μg kanamycin [Kn]; 20 μg/ml (E. coli) and 5 μg/ml (V. cholerae) chloramphenicol [Cam].
Co-culture assays
Strain A was inoculated in one of the two flasks connected by a 0.22-μm filter (Figure 1A). When strain A reached OD600=0.5, strain B was inoculated in the adjacent compartment and incubated overnight (16 h), then harvested for HPLC analyses of muropeptides (below). Inoculation of strain A before B permits a longer exposure of the later to the NCDAAs released by strain A (WT or bsrV), and therefore a higher level of incorporation within the B strain (bsrV) PG.
Quantification of total murein
Cells from 1 l cultures of exponential phase (OD600=0.4) and overnight stationary phase (OD600=4.0) cultures were resuspended in 5 ml of PBS, added to an equal volume of 10% SDS in a boiling water bath and vigorously stirred for 4 h, then stirred overnight at 37°C. The insoluble fraction (PG) was pelleted at 100 000 r.p.m. (400 000 g), 15 min, 30°C (TLA110 Beckman rotor; OptimaTM Max ultracentrifuge Beckman), boiled in 1% SDS for 2 h, then washed 4 × with H2O. Quantification of DAP (an indicator of total murein) was performed as previously reported (Work, 1957). Samples were hydrolysed overnight in 6 N HCl at 100°C in 1 ml capacity crystal ampoules (Wheaton). After hydrolysis, samples were dried and resuspended in water and mixed in a 1:1:1 ratio with pure acetic acid and ninhydrin reagent (250 mg ninhydrin, 4 ml phosphoric acid 0.6 M and 6 ml pure acetic acid). Samples were measured at OD434 nm and the amounts of DAP were calculated based on a standard curve. Total murein per cell was calculated based on colony forming units.
HPLC/MS analysis of PG
PG was isolated largely as above, then processed for HPLC analysis as previously described (Caparros et al, 1992) on a Hypersil ODS18 reverse-phase column. Samples were incubated ±1.5% (w/v) H2O2 for detection of D-Met-containing muropeptides, as oxidation of methionine causes a large shift in the HPLC elution time of D-Met-containing muropeptides while elution of normal muropeptides is unaffected. Individual D-Met muropeptides were identified using MALDI-TOF and electrospray-ion trap MS (Supplementary Table S4). Data shown in Figures 1, 2, 3, 4 correspond to representative experiments. Experimental variability in muropeptide quantification by HPLC is <5% for minor muropeptides and <1% for more abundant muropeptides. See supporting information for additional details.
Low osmolarity survival
V. cholerae cells were pelleted at 2000 g, washed with 150 mM NaCl, then resuspended in 150 mM NaCl or distilled water. Cells were incubated for 15 min at room temperature then plated on LB agar plates to assess viability.
D-Cysteine incorporation and immunolabelling
IPTG (1 mM) was added to V. cholerae cultures at OD600=0.3. After 1 h of protein induction, D-Cys (5 mM) was added and the culture was incubated for 1 h. E. coli cultures were grown to OD600=0.8, supplemented with D-Cys (1 mM), and incubated for 30 min before purification of sacculi. For electron microscopy, sacculi were processed as described (de Pedro et al, 1997).
In vitro L,D-transpeptidation assay
With pure muropeptide substrates, reactions contained 10 μg of muropeptide (purified from E. coli murein through collection of HPLC-separated muropeptide peaks as described in Caparros et al (1992)), 5 μg of purified Ldt and 20 mM of D-amino acid in 50 μl of 50 mM Phosphate buffer pH 7.5, 150 mM NaCl and 1% Triton X-100. Antibiotics (50 μg/ml final concentration), when used, were added 10 min before addition of the muropeptide. Reactions were performed at 37°C for 30 min and 2 h, then heat inactivated (95°C, 5 min) and centrifuged (15 000 r.p.m., 10 min). Peptides in the supernatant were identified by HPLC as above. Product synthesis (%) reflects the percentage of substrate transformed in the presence of the Ldt relative to parallel control reactions that lacked enzyme.
Sacculi substrates were prepared from exponential phase V. cholerae (1 L) as described (Caparros et al, 1992), and resuspended in 2 ml H2O. Reactions contained 250 μl sacculi, 10 μg Ldt, 20 mM of D-Met in 50 mM Phosphate buffer pH 7.5, 150 mM NaCl and 1% Triton X-100. The reaction was incubated overnight at 37°C, then stopped with SDS (1% w/v), boiled 20 min and ultracentrifuged (100 000 r.p.m., 15 min). In brief, pellets were washed, digested with muramidase, reduced with NaBH4, and subjected to HPLC analysis. Product synthesis (%) was calculated as above. See supporting information for additional details.
In vitro synthesis of UDP-muramyl-(D-Met)-pentapeptide (UDP-M5M)
Reactions (50 μl) contained 10 μg of pure UDP-M3 (UK-BaCWAN), 5 μg Ddl and 5 μg MurF in Tris–HCl 100 mM pH 8.6, 250 mM NaCl, 25 mM ATP, 12.5 mM MgCl2, 10 mM KCl with 25 mM of D-Ala/D-Met at various ratios. They were incubated overnight at 37°C, then heat inactivated (95°C, 5 min) and centrifuged (15 000 r.p.m., 10 min) and analysed via HPLC. When D-cycloserine was used, it was added to reaction mixes 10 min before addition of substrates. Product synthesis (%) reflects transformation of UDP-M3 relative to parallel control reactions that lacked enzymes.
Supplementary Material
Acknowledgments
We thank Chris Walsh for insightful comments on the manuscript. This work was supported by Howard Hughes Medical Institute (HHMI); NIH AI-R37-42347 (MKW) and Ministry of Education and Science, Spain (MEC) BFU2006-04574 and Fundación Ramón Areces (MAP); MEC Fellowship (FC) and Jane Coffin Childs Fellowship (HL).
Author contributions: FC designed experiments. FC, MAP and HL carried out experiments. FC, MAP and MKW analysed results. FC, BMD and MKW wrote the manuscript with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952 [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 32: 168–207 [DOI] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C (2006) Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol 359: 533–538 [DOI] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda U, Dauter Z, Joachimiak A, Derewenda ZS (2006) B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins 62: 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D (2008) The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev 32: 208–233 [DOI] [PubMed] [Google Scholar]
- Caparros M, Pisabarro AG, de Pedro MA (1992) Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol 174: 5549–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Esaki N, Yoshimura T, Soda K (1992) Reaction mechanism of glutamate racemase, a pyridoxal phosphate-independent amino acid racemase. J Biochem 112: 139–142 [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Holtje JV, Schwarz H (1997) Murein segregation in Escherichia coli. J Bacteriol 179: 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Young KD, Holtje JV, Schwarz H (2003) Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol 185: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K, van Heijenoort J, Walsh CT (1990) Purification and characterization of the D-alanyl-D-alanine-adding enzyme from Escherichia coli. Biochemistry 29: 2379–2386 [DOI] [PubMed] [Google Scholar]
- Hengge R (2008) The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv Exp Med Biol 631: 40–53 [DOI] [PubMed] [Google Scholar]
- Holtje JV (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62: 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC (2008) The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R (2010) D-amino acids trigger biofilm disassembly. Science 328: 627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK (2009) D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325: 1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Arbeloa A, Mainardi JL, Hugonnet JE, Fourgeaud M, Dubost L, Marie A, Delfosse V, Mayer C, Rice LB, Arthur M (2007a) Specificity of L,D-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J Biol Chem 282: 13151–13159 [DOI] [PubMed] [Google Scholar]
- Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, Marie A, Mengin-Lecreulx D, Arthur M, Gutmann L (2007b) Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol 189: 3927–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Dubost L, Marie A, Arthur M, Gutmann L (2008) Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol 190: 4782–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M (2005) A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem 280: 38146–38152 [DOI] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Disteche M, de Kruijff B, Breukink E (2011) Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J 30: 1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T (2004) Stationary-phase physiology. Annu Rev Microbiol 58: 161–181 [DOI] [PubMed] [Google Scholar]
- Park JT, Uehara T (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev 72: 211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock JJ, Ghuysen JM, Linder R, Salton MR, Perkins HR, Nieto M, Leyh-Bouille M, Frere JM, Johnson K (1972) Transpeptidase activity of Streptomyces D-alanyl-D-carboxypeptidases. Proc Natl Acad Sci USA 69: 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PE, Courvalin P (2005) Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. Antimicrob Agents Chemother 49: 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG (2005) Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69: 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight PD, Kolter R (2009) Interspecies chemical communication in bacterial development. Annu Rev Microbiol 63: 99–118 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA (2008a) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S (2008b) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32: 259–286 [DOI] [PubMed] [Google Scholar]
- Walsh CT (1989) Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem 264: 2393–2396 [PubMed] [Google Scholar]
- Work E (1957) Reaction of ninhydrin in acid solution with straight-chain amino acids containing two amino groups and its application to the estimation of alpha epsilon-diaminopimelic acid. Biochem J 67: 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD (2001) Approaching the physiological functions of penicillin-binding proteins in Escherichia coli. Biochimie 83: 99–102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.