Abstract
Mammalian target of rapamycin (mTOR) complex 1 (mTORC1) is an important, highly conserved, regulator of cell growth. Ancient among the signals that regulate mTORC1 are nutrients. Amino acids direct mTORC1 to the surface of the late endosome/lysosome, where mTORC1 becomes receptive to other inputs. However, the interplay between endosomes and mTORC1 is poorly understood. Here, we report the discovery of a network that links mTORC1 to a critical component of the late endosome/lysosome, the V-ATPase. In an unbiased screen, we found that mTORC1 regulated the expression of, among other lysosomal genes, the V-ATPases. mTORC1 regulates V-ATPase expression both in cells and in mice. V-ATPase regulation by mTORC1 involves a transcription factor translocated in renal cancer, TFEB. TFEB is required for the expression of a large subset of mTORC1 responsive genes. mTORC1 coordinately regulates TFEB phosphorylation and nuclear localization and in a manner dependent on both TFEB and V-ATPases, mTORC1 promotes endocytosis. These data uncover a regulatory network linking an oncogenic transcription factor that is a master regulator of lysosomal biogenesis, TFEB, to mTORC1 and endocytosis.
Keywords: autophagy, lysosome, microarray, RCC, Tcfeb
Introduction
Target of rapamycin (TOR) complex 1 (TORC1) is a pivotal regulator of cell growth conserved from yeast to humans and implicated in cancer (Wullschleger et al, 2006; Abraham and Eng, 2008). Mammalian TORC1 (mTORC1) includes the atypical serine/threonine kinase mTOR and an adaptor protein, regulatory-associated protein of mTOR (Raptor) (Hara et al, 2002; Kim et al, 2002; Loewith et al, 2002). mTORC1 regulates cell growth, at least in part, by promoting cap-dependent translation and its best-characterized substrates are the eukaryotic initiation factor 4E binding protein-1 (4E-BP1) (Sonenberg and Hinnebusch, 2009) and S6 kinase 1 (S6K1) (Fingar and Blenis, 2004). mTORC1 is specifically inhibited by rapamycin (also called sirolimus) (Jacinto et al, 2004; Sarbassov et al, 2004) and rapamycin analogues are used clinically for many applications including cancer treatment (Abraham and Eng, 2008). Indeed, two rapamycin analogues, temsirolimus (Hudes et al, 2007) and everolimus (Motzer et al, 2008), are approved by the FDA for the treatment of advanced RCC (renal cell carcinoma).
mTORC1 is regulated by a complex formed by the proteins tuberous sclerosis complex 1 (TSC1) and 2 (TSC2), which is essential for the relay of signals from oxygen (Brugarolas et al, 2004; Connolly et al, 2006; Kaper et al, 2006; Liu et al, 2006; DeYoung et al, 2008), energy stores (Inoki et al, 2003b; Corradetti et al, 2004; Shaw et al, 2004) and growth factors (Jaeschke et al, 2002; Kwiatkowski et al, 2002; Zhang et al, 2003a). mTORC1 regulation by hypoxia involves the protein regulated in development and DNA damage response 1 (REDD1), a conserved protein with a novel fold (Vega-Rubin-de-Celis et al, 2010), which, when overexpressed is sufficient to inhibit mTORC1 in a TSC1/TSC2-dependent manner (Brugarolas et al, 2004). Interestingly, in some settings, hypoxia signals are transduced via AMP-activated protein kinase (Liu et al, 2006; Wolff et al, 2011), which phosphorylates TSC2 and is normally involved in the relay of energy signals (Inoki et al, 2003b; Gwinn et al, 2008). mTORC1 regulation by growth factors involves TSC2 phosphorylation by Akt (Dan et al, 2002; Inoki et al, 2002; Manning et al, 2002; Potter et al, 2002), extracellular signal-regulated kinase (Erk) (Ma et al, 2005) and ribosomal S6 kinase (Rsk) (Roux et al, 2004). TSC2 functions as a GTPase-activating protein towards Ras homologue enriched in brain (Rheb) (Castro et al, 2003; Garami et al, 2003; Tee et al, 2003; Inoki et al, 2003a; Zhang et al, 2003b), a small GTPase that directly interacts with and activates mTORC1 (Sancak et al, 2007; Avruch et al, 2009).
mTORC1 regulation by nutrients is independent of TSC1/TSC2 (Zhang et al, 2003a; Smith et al, 2005; Roccio et al, 2006) and involves its localization to a cellular compartment where it becomes receptive to activation by Rheb (Sancak et al, 2008, 2010). Amino-acid stimulation drives mTORC1 to the surface of the late endosome/lysosome in a manner that depends on a Rag GTPase heterodimer (RagA [or B] bound to RagC [or D]) and a multimeric complex termed the Ragulator (Kim et al, 2008; Sancak et al, 2010). Importantly, constitutive mTORC1 targeting to this compartment renders mTORC1 insensitive to amino-acid withdrawal (Sancak et al, 2010). The significance of the late endosome/lysosome in mTORC1 activation has been recently established in experiments manipulating endosome maturation (Flinn et al, 2010; Li et al, 2010). Disrupting endosome maturation with a constitutively active Rab5 or through depletion of hVps39 inhibited the activation of mTORC1 by growth factors and amino acids (Flinn et al, 2010; Li et al, 2010).
A critical component of the endosome required for its acidification and maturation is the vacuolar H+-ATPase (V-ATPase) (Marshansky and Futai, 2008). V-ATPases are multisubunit complexes formed by a membrane-embedded V0 domain (a, d, e, c, c′ and c″ subunits) responsible for proton translocation, and a cytosolic V1 domain (A–H subunits), which provides energy through ATP hydrolysis (Forgac, 2007). V-ATPases are present in virtually every eukaryotic cell and V-ATPase function is essential for survival under a variety of conditions (Beyenbach and Wieczorek, 2006). However, little is known about the regulation of V-ATPases and in particular about how their expression is controlled.
Here, we have uncovered a regulatory network linking V-ATPases and endocytosis to mTORC1 that involves the transcription factor EB (TFEB). TFEB is a basic helix-loop-helix (bHLH) leucine zipper transcription factor of the Myc family, microphthalmia transcription factor subfamily (Steingrimsson et al, 2004), that is translocated in a subset of renal tumours (Davis et al, 2003; Kuiper et al, 2003; Srigley and Delahunt, 2009).
Results
V-ATPase enrichment among mTORC1-regulated genes
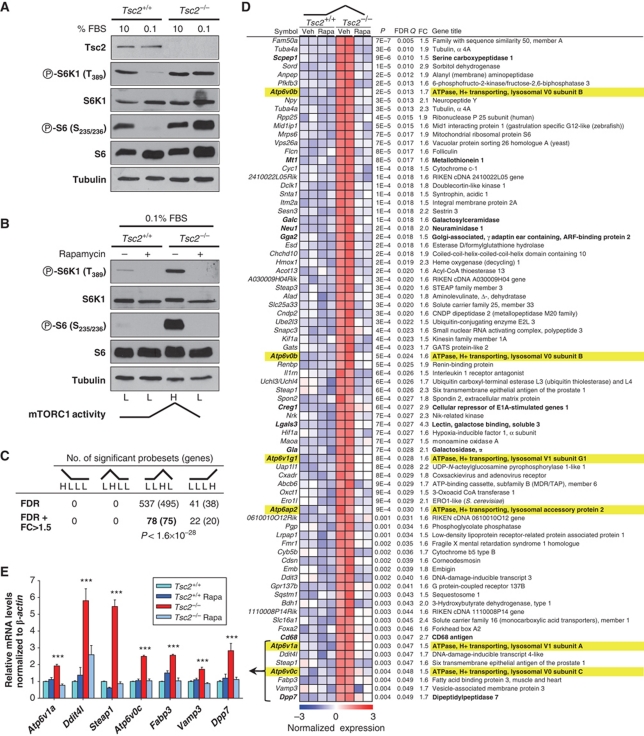
We identified V-ATPase genes in an unbiased screen for genes whose expression was tightly regulated by mTORC1. We used a robust experimental paradigm involving the combination of two interventions, one genetic and one pharmacologic. In Tsc2+/+ immortalized mouse embryo fibroblasts (MEFs), but not Tsc2−/−, mTORC1 is inhibited by serum deprivation (0.1% fetal bovine serum, FBS) (Figure 1A). In Tsc2−/− cells, abnormally increased mTORC1 activity can be corrected by treatment with rapamycin (Figure 1B). By contrast, rapamycin has little effect on mTORC1 in Tsc2+/+ cells in which mTORC1 is already inhibited by low serum (Figure 1B). Thus, under serum-deprived conditions, mTORC1 activity is low in Tsc2+/+ cells (untreated or rapamycin treated), high in Tsc2−/− cells, but lowered by rapamycin; a pattern referred to as a ‘low/low/high/low’ or ‘LLHL’ (Figure 1B). This experimental setup creates an ‘asymmetric’ response to rapamycin (in Tsc2−/− versus Tsc2+/+ cells), which, as illustrated hereafter, can be harnessed to great advantage.
Figure 1.
Identification of mTORC1-regulated genes. Western blot of immortalized MEFs of the indicated genotypes grown for 24 h in the stated amounts of FBS (A), and in the presence of rapamycin (or vehicle) (B). (C) Number of probes (genes) in Affymetrix microarrays corresponding to the indicated patterns (upregulated under one condition compared with the other three by t-test using a Q<0.05 after FDR correction). Data are shown also incorporating a fold change criterion (>1.5-fold; FC>1.5). (D) Heatmap representation of normalized expression values for LLHL probes with a fold change >1.5 (ranked by P-value). Genes encoding V-ATPase subunits and V-ATPase-associated proteins are highlighted in yellow and in bold lysosomal genes are noted. (E) qRT–PCR analysis of genes with the weakest LLHL pattern. Error bars represent s.e.m. (n=4); ***P<0.001.
Using an Affymetrix MOE430A array platform and duplicate samples, genes were identified with an LLHL expression pattern (upregulated in untreated Tsc2−/− cells compared with a baseline formed by Tsc2+/+ cells, untreated or rapamycin treated, and Tsc2−/− rapamycin-treated cells). With a false discovery rate-corrected P-value (FDR Q) cutoff of 0.05 and a threshold of 1.5-fold, we identified 78 probe sets (henceforth referred to as probes). They corresponded to 75 genes (Figure 1C and D).
We hypothesized that if the association of probes with the LLHL pattern was random, there would be similar numbers of probes with the three alternate patterns (HLLL, LHLL and LLLH). Strikingly, however, no probes fitted an HLLL or LHLL pattern, and only 22 probes conformed to an LLLH pattern (Figure 1C). Given these results and assuming a binomial distribution with equal probabilities for all patterns, the probability of finding 78 probes with an LLHL pattern by chance alone was below 1.6 × 10−28.
To validate the microarray findings, qRT–PCR was performed on a subset of genes with the weakest LLHL association (highest P-values). In every instance, the LLHL pattern was confirmed and the fold change was even greater than expected (Figure 1E).
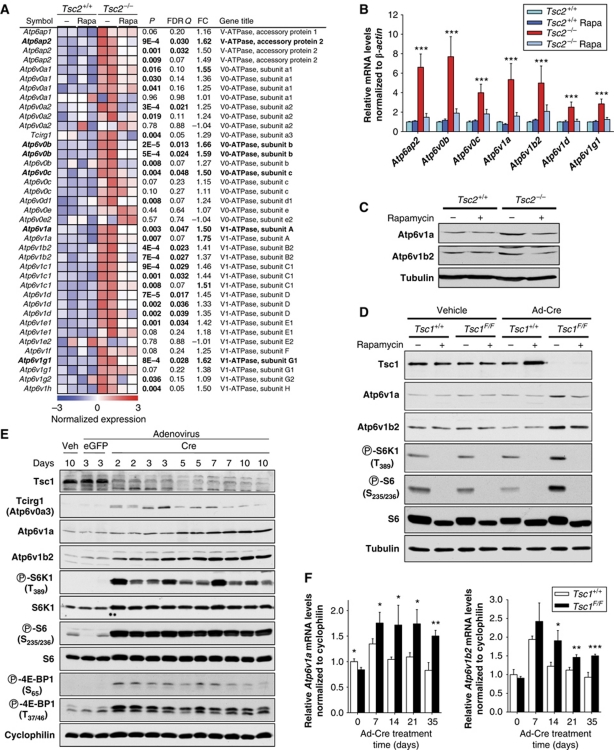
Among the LLHL probes, we found six for V-ATPase genes (Figure 1D; Supplementary Table S1). The probability that there would be six V-ATPase probes among the 78 probes identified by chance alone was <9 × 10−8. Furthermore, an analysis of all the V-ATPase probes in the array showed a striking LLHL pattern for most genes (Figure 2A) and the results were confirmed in a subset of genes by qRT–PCR (Figure 2B) and western blot (Figure 2C).
Figure 2.
mTORC1 regulates V-ATPases. (A) Heatmap representation of normalized expression values for all informative V-ATPase probes in the arrays. qRT–PCR (B) and western blot (C) of V-ATPases in Tsc2+/+ and Tsc2−/− MEFs in 0.1% FBS and treated (or not) with rapamycin for 24 h (n=4). (D) Western blot of Tsc1+/+ and Tsc1F/F primary MEFs exposed (or not) to Ad-Cre and treated (or not) with rapamycin in 0.1% FBS for 24 h. (E) Western blot from liver extracts (assembled from two gels) of mice injected with the indicated adenovirus (or vehicle) and harvested after the stated number of days. (F) qRT–PCR analysis of V-ATPase gene expression in liver extracts from Tsc1+/+ or Tsc1F/F mice injected with Ad-Cre and harvested following the indicated number of days (n=2 per time point). Error bars represent s.e.m.; *P<0.05; **P<0.01; ***P<0.001.
Because Tsc2+/+ and Tsc2−/− cells are also p53−/−, they may have diverged from each other over time, and thus, we examined the role of mTORC1 in V-ATPase gene expression in primary MEFs. MEFs were generated from Tsc1floxP/floxP (Tsc1F/F) (Kwiatkowski et al, 2002) as well as littermate Tsc1+/+ embryos, and exposed to recombinant adenoviruses driving the expression of Cre recombinase (Ad-Cre). Ad-Cre transduction of Tsc1F/F MEFs led to effective recombination of loxP sites (Supplementary Figure S1A), with loss of the Tsc1 protein, activation of mTORC1 (Figure 2D) and V-ATPase upregulation (Figure 2D).
To broaden these observations, we examined publicly available gene expression data sets. Recently, a study was reported in which the effects of rapamycin were evaluated over time in serum-deprived Tsc2−/− and Tsc1−/− immortalized MEFs (Duvel et al, 2010). While in this study rapamycin was administered only to the mutant cells, we observed a previously unrecognized downregulation of V-ATPase expression by rapamycin (Supplementary Figure S2). V-ATPase expression was reduced by 6 h (Supplementary Figure S2) and maximal V-ATPase downregulation was observed at 24 h, the time at which our experiments were conducted (Figure 1D). Similar results were observed in a second publicly available data set of immortalized Tsc2−/− MEFs treated with rapamycin (Cunningham et al, 2007) (Supplementary Figure S3). The analysis of these data sets confirmed our results that mTORC1 regulated V-ATPase expression.
mTORC1 regulation of V-ATPase expression in the mouse
To extend our observations to an in vivo setting, we injected Tsc1F/F mice with Ad-Cre intravenously under conditions known to induce recombination of target genes in hepatocytes (Kucejova et al, 2011). Ad-Cre (but not control Ad-eGFP or saline) led to effective recombination of loxP sites (Supplementary Figure S1B), loss of the Tsc1 protein and robust activation of mTORC1 (Figure 2E). This was accompanied by increased expression of V-ATPase genes at both the mRNA and protein levels (Figure 2E and F; Supplementary Figure S4A). As Ad-Cre injection affected the expression of many housekeeping genes including actin, we used as a reference cyclophilin B, whose expression was seemingly unaffected by Ad-Cre (Figure 2F; Supplementary Figure S4). While control mice were infected also with an adenovirus (Ad-GFP), we could not rule out effects of Cre recombinase beyond loxP site recombination, and additional experiments were performed using Tsc1+/+ mice injected with Ad-Cre as a control (Figure 2F; Supplementary Figure S4B). Independently of the control, Tsc1 loss was accompanied by mTORC1 activation and increased V-ATPase expression.
Next, we explored the regulation of V-ATPases by mTORC1 in a previously reported tumour model. Transgenic mice expressing a constitutively active myristoylated form of Akt1 (myr-Akt) in the prostate develop prostate intraepithelial neoplasia (PIN) and PIN lesions exhibit robust mTORC1 activation (Majumder et al, 2004). Myr-Akt would be expected to inactivate Tsc1/Tsc2 and the PIN phenotype is reversed by treatment with the rapamycin analogue everolimus (also called RAD001) (Majumder et al, 2004). While this represented a very different experimental paradigm and gene expression studies were performed following 12 and 48 h of RAD001 oral administration, a collective analysis showed a similar mTORC1-dependent induction of V-ATPase expression (Supplementary Figure S5). In fact, a statistically significant LLHL pattern was found for 13 probes corresponding to 9 V-ATPase genes. Thus, V-ATPases are regulated by mTORC1 not only in cells in culture, but also in mice in both physiological and pathological states.
Transcriptional regulation of V-ATPases by mTORC1
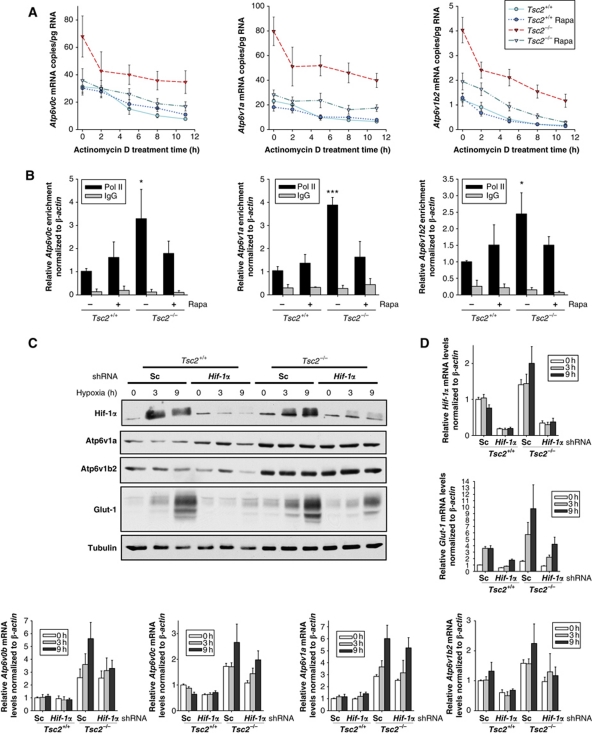
To obtain insight into the mechanism of V-ATPase mRNA upregulation by mTORC1, we examined the relative contribution of mRNA synthesis and stability. While as expected, baseline V-ATPase mRNA levels were higher in Tsc2−/− than Tsc2+/+ cells, mRNA decay rates were very similar (Figure 3A). These data suggested that V-ATPase mRNA levels were upregulated by mTORC1 through a transcriptional mechanism. Consistent with this notion, RNA polymerase II (Pol II) chromatin immunoprecipitation (ChIP) experiments showed Pol II enrichment at V-ATPase coding sequences in Tsc2−/− cells (Supplementary Figure S6). This effect persisted after normalization to β-actin ChIP data suggesting that the effect was not due to a generalized increase in Pol II transcription by mTORC1 (Figure 3B).
Figure 3.
V-ATPases are regulated transcriptionally by mTORC1 independently of Hif-1α. (A) qRT–PCR analysis of V-ATPase mRNA decay rates in the indicated cell types in low serum (n=3). (B) RNA polymerase II (Pol II) occupancy on V-ATPase gene coding sequences compared with IgG control (n=3). Western blotting (C) and qRT–PCR (D) of Tsc2+/+ and Tsc2−/− MEFs stably transduced with a lentiviral construct with a Hif-1α (or a scrambled) shRNA sequence and exposed to hypoxia for the indicated number of hours (n=3). Error bars represent s.e.m.; *P<0.05; ***P<0.001.
We noted that among the 75 genes regulated by mTORC1 was hypoxia-inducible factor 1α (Hif-1α) (Figure 1D), which we previously showed is regulated by Tsc1/Tsc2 and mTORC1 (Brugarolas et al, 2003). Hif-1α is a critical determinant of the activity of the heterodimeric Hif-1 transcription factor (Kaelin, 2008) and a recent ChIP-chip study reported finding Hif-1α on V-ATPase d1 sequences (Xia et al, 2009). Furthermore, several putative hypoxia response elements (HRE) were identified in the promoters of several V-ATPases (Supplementary Table S2). To determine whether Hif-1α contributed to V-ATPase regulation, we examined the effects of both hypoxia and Hif-1α knockdown on V-ATPase expression. As a positive control, the expression of the canonical Hif-1 target Glut-1 was evaluated. Hypoxia stabilized Hif-1α protein and this was accompanied by increased expression of Glut-1 (Figure 3C and D). As expected, the upregulation of Glut-1 by hypoxia was markedly blunted by Hif-1α knockdown (Figure 3C and D). We next examined V-ATPase levels. While in Tsc2+/+ cells, we did not observe an increase in V-ATPase expression following hypoxia (or a consistent downregulation following Hif-1α knockdown), in Tsc2−/− cells there was a trend towards induction by hypoxia and this was somewhat blunted by Hif-1α knockdown, which also, in some cases, downregulated baseline V-ATPase expression levels (Figure 3D). Thus, while Hif-1α may partially contribute to V-ATPase gene regulation in Tsc2−/− cells, Hif-1α does not seem to be the principal regulator of V-ATPase gene expression under these conditions.
mTORC1-dependent V-ATPase regulation requires Tfeb
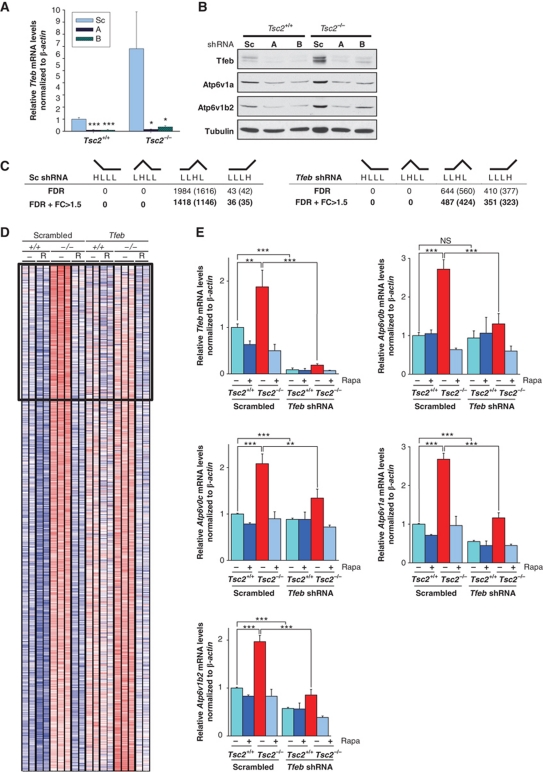
A recent study showed that lysosomal biogenesis is regulated by TFEB (Sardiello et al, 2009), and we noted that among the genes reported to be regulated by TFEB, there were several V-ATPases. Interestingly, the consensus sequence for TFEB (also called Tcfeb in the mouse), which includes the E-box (Sardiello et al, 2009), overlaps with HRE (Wenger et al, 2005) (Supplementary Table S2). We set out to examine whether TFEB may be involved in mTORC1-dependent regulation of V-ATPases. An antibody recognizing TFEB was generated in collaboration with Bethyl, and we evaluated the effects of Tfeb knockdown on V-ATPase expression. Among eight Tfeb shRNAs tested, two were found that substantially lowered Tfeb levels (Figure 4A and B). Tfeb depletion downregulated V-ATPase expression, particularly in Tsc2-deficient cells, in which baseline levels were upregulated (Figure 4B; see also Figure 4E). These data show that Tfeb is required for mTORC1-induced V-ATPase expression.
Figure 4.
V-ATPase regulation by mTORC1 requires Tfeb and Tfeb is an important mediator of the changes in gene expression induced by mTORC1. (A) qRT–PCR of MEFs of the indicated genotypes stably transduced with the indicated shRNA vectors; each Tfeb shRNA (A, B) was compared with the scrambled (Sc) control by an unpaired t-test (n=3). Error bars represent s.e.m.; *P<0.05; ***P<0.001. (B) Western blot of serum-starved MEFs of the indicated genotypes. (C) Number of probes (genes) corresponding to the different patterns upregulated under one condition compared with the other three by t-tests (Q<0.05 after FDR correction) in Illumina microarrays. Data are shown also incorporating a fold change criterion (>1.5-fold; FC>1.5). (D) Heatmap representation of normalized expression values for all 1418 significant LLHL probes with FC>1.5 in scrambled shRNA transduced MEFs compared with Tfeb shRNA transduced MEFs. Box shows probes for which mean expression levels are reduced by >20% following Tfeb knockdown in Tsc2−/− MEFs. (E) qRT–PCR of Tsc2+/+ and Tsc2−/− MEFs stably transduced with the indicated shRNA vectors and treated (or not) with rapamycin (n=3). Error bars represent s.e.m.; NS, non-significant; **P<0.01; ***P<0.001.
Tfeb is an important mediator of mTORC1 changes on gene expression
To determine how broadly TFEB regulated gene expression downstream of mTORC1, we performed gene expression studies in MEFs stably depleted of Tfeb. Having previously used an Affymetrix platform and seeking to determine the robustness of our findings, Illumina microarrays were used. Otherwise, the experimental design was identical as that reported in Figure 1C and D. First, we compared the results in the Tsc2+/+ and Tsc2−/− cells transduced with a scrambled shRNA to those previously obtained with plain Tsc2+/+ and Tsc2−/− cells. While the number of LLHL probes identified with the Illumina chips was much larger than with the Affymetrix arrays, the distribution across patterns was strikingly similar; the majority of probes had an LLHL pattern and no probes were found with a HLLL or LHLL pattern (compare Figures 4C and 1C). Furthermore, among the 75 genes previously identified with a LLHL pattern, a statistically significant LLHL pattern was retained in 48% (Supplementary Figure S7). Considering that the microarray platforms were different, that microarrays were processed at different facilities several years apart, and that stringent criteria were used (FDR Q<0.05), a 48% retention rate was considered satisfactory and suggested that many other genes besides V-ATPases were bona fide targets of mTORC1.
To specifically determine the contribution of Tfeb to mTORC1-induced gene expression and the LLHL pattern, we compared scrambled versus Tfeb shRNA-expressing cells. Quite surprisingly, Tfeb knockdown markedly decreased the number of genes with an LLHL pattern (1146 versus 424; Figure 4C). Furthermore, as visually illustrated in the heatmap (Figure 4D), in 26% of LLHL probes, Tfeb knockdown resulted in a substantial ‘flattening’ of the LLHL pattern (>20% reduction in mean expression levels in Tsc2−/− MEFs by Tfeb knockdown), including several V-ATPases (Supplementary Table S3). LLHL ‘flattening’ was confirmed for several V-ATPase genes by qRT–PCR (Figure 4E). Interestingly, the decrease in the number of genes with an LLHL pattern after Tfeb knockdown was specific for the LLHL pattern. In fact, Tfeb knockdown resulted in an increase in the number of genes with a LLLH pattern (35 versus 323; Figure 4C; Supplementary Table S4). These results suggest that Tfeb is an important effector downstream of mTORC1 involved in the regulation of a substantial number of genes.
Regulation of endocytosis by V-ATPases, TFEB and mTORC1
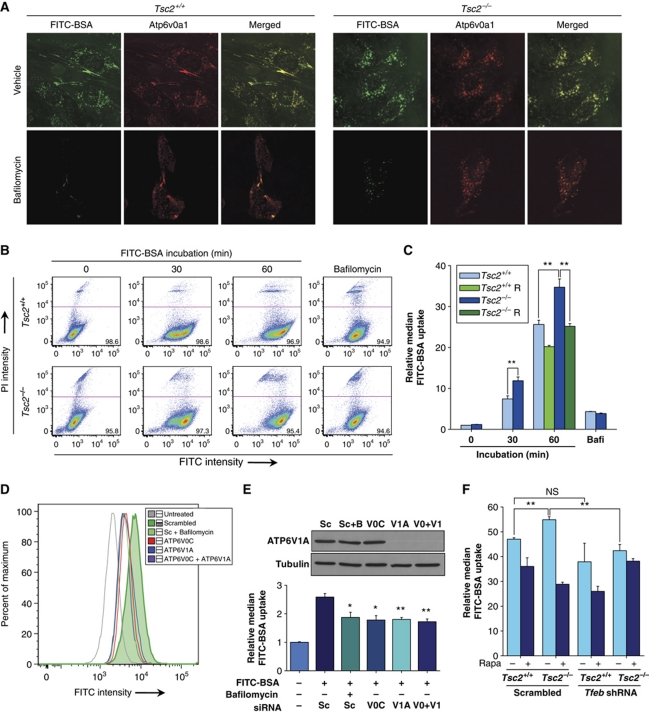
Having determined that mTORC1 regulated V-ATPase expression in cells in culture and in mice and having established that TFEB was required for mTORC1-induced V-ATPase expression, we sought to examine the functional significance of V-ATPase regulation by mTORC1. Because V-ATPases are involved in vesicle trafficking (Marshansky and Futai, 2008), we evaluated the effects of mTORC1 on endocytosis. For these experiments, we used albumin, the most abundant protein in plasma and a protein that is taken up by macrophages and hydrolyzed to amino acids, which are then released in the circulation and are utilized by different cells (Guyton and Hall, 1996). While albumin uptake occurs through different pathways in different cell types (Mayor and Pagano, 2007), albumin is taken up by MEFs through a caveolin-dependent mechanism (Razani et al, 2001). FITC-conjugated bovine serum albumin (FITC-BSA) was efficiently taken up by MEFs, where it was found in punctate structures reminiscent of vesicles, which also contained V-ATPases (Figure 5A). In keeping with the idea that mTORC1 regulates endocytosis, FACS analyses showed higher levels of albumin uptake in Tsc2−/− than Tsc2+/+ cells (Figure 5B and C). In addition, treatment of Tsc2−/− cells with rapamycin downregulated albumin uptake (Figure 5C). Albumin uptake required V-ATPases as determined by experiments using the V-ATPase inhibitor bafilomycin A1 (Bowman and Bowman, 2002) (Figure 5B and C). In addition, albumin uptake was downregulated by siRNA-mediated depletion of essential components of either the V0 or the V1 domain (alone or in combination) in highly transfectable HeLa cervical carcinoma cells (Figure 5D and E).
Figure 5.
mTORC1 regulates endocytosis in a V-ATPase- and Tfeb-dependent manner. (A) Confocal images of serum-starved (0.1% FBS) Tsc2+/+ and Tsc2−/− MEFs incubated with FITC-BSA and evaluated for V-ATPase colocalization following (or not) treatment with the V-ATPase inhibitor bafilomycin A1. (B) FACS scatterplots highlighting live (propidium iodide (PI) negative) Tsc2+/+ and Tsc2−/− MEFs incubated with FITC-BSA for the indicated times; bafilomycin-treated samples shown as a control. (C) Bar graph representation of FITC-BSA incorporation in live cells pretreated (or not) with rapamycin (rapa) by reference to FITC-BSA incorporation in untreated cells (n=4). Representative FACS histogram (D) as well as bar graph representation (n=3) and western blot (E) of HeLa cells transfected with siRNA oligonucleotides targeting the V0C, V1A or both V-ATPase subunits, or a scrambled control (Sc), and incubated for 60 min with FITC-BSA; a bafilomycin A1-treated sample [B] is included as a control. (F) Bar graph representation of FITC-BSA uptake by FACS analysis in the indicated cell types in 0.1% serum and treated (or not) with rapamycin (n=3). Error bars represent s.e.m.; NS, non-significant; *P<0.05; **P<0.01.
Next, we tested whether Tfeb may be similarly involved in endocytosis. Indeed, Tfeb knockdown resulted in a statistically significant decrease in albumin uptake (Figure 5F). The effects of Tfeb knockdown on albumin uptake in Tsc2−/− cells were similar to those achieved by treatment with rapamycin (Figure 5F). Taken together, these data show that mTORC1 promotes endocytosis in a Tfeb- and V-ATPase-dependent manner.
Regulation of Tfeb phosphorylation and subcellular localization by the Tsc1/Tsc2 complex and rapamycin
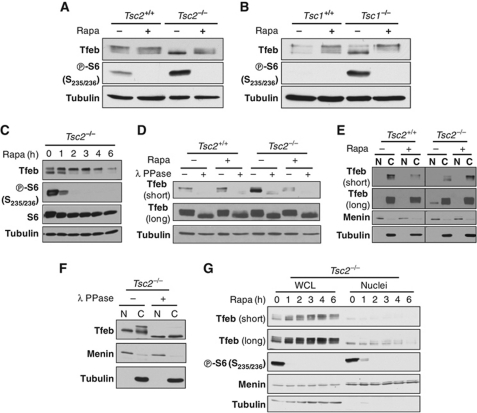
Having established the importance of Tfeb downstream of mTORC1, we explored the mechanism whereby mTORC1 regulated Tfeb. We observed that not only the expression levels of Tfeb appeared to be upregulated by mTORC1, but also that mTORC1 affected Tfeb electrophoretic mobility on SDS–PAGE (Figures 4B and 6A). In Tsc2−/− cells, Tfeb was prominently found in a fast-migrating form(s) (Figures 4B and 6A). Treatment of Tsc2−/− cells with rapamycin shifted Tfeb up giving rise to a pattern similar to that observed in wild-type cells (Figure 6A). Similar results were observed in Tsc1+/+ and Tsc1−/− cells (Figure 6B). To determine how quickly Tfeb mobility was affected by rapamycin, a time course was performed. As shown in Figure 6C, rapamycin treatment for 1 h was sufficient to change the distribution of Tfeb.
Figure 6.
Coordinated regulation of Tfeb phosphorylation and nuclear localization by the Tsc1/Tsc2 complex and rapamycin. (A, B) Western blot of MEFs of the indicated genotypes in 0.1% serum treated or not with rapamycin for 24 h (as in Figure 1B). (C) Western blot of serum-starved (0.1% serum) Tsc2−/− cells treated with rapamycin for the indicated times. (D) Western blot of Tsc2+/+ and Tsc2−/− cells under serum starvation conditions treated (or not) with rapamycin and subsequently incubated, as stated, with λ phosphatase. (E) Western blot of serum-starved Tsc2+/+ and Tsc2−/− cells treated (or not) with rapamycin and subsequently fractionated into nuclear [N] or cytoplasmic [C] fractions. (F) Western blots of nuclear versus cytoplasmic fractions from serum-starved Tsc2−/− cells incubated as indicated with λ phosphatase. (G) Western blot of whole-cell lysates (WCL) or nuclei from serum-starved Tsc2−/− cells treated with rapamycin for the indicated periods of time.
Changes in Tfeb migration may reflect changes in phosphorylation, and phosphoproteomic studies have shown TFEB to be phosphorylated in >10 sites (Dephoure et al, 2008; Mayya et al, 2009). To determine whether Tfeb migration was affected by phosphorylation, we evaluated the effects of λ phosphatase treatment. As shown in Figure 6D, the migration of Tfeb was accelerated by treatment of cell lysates with λ phosphatase. Parenthetically, by comparison to similarly processed samples incubated without λ phosphatase for the same amount of time, phosphatase exposure not only accelerated Tfeb migration, but also led to a downregulation of total Tfeb protein levels (Figure 6D). Notably, even fast-migrating Tfeb in Tsc2−/− cells was subject to a downshift following phosphatase treatment (Figure 6D). These results indicate that even in Tsc2−/− cells, Tfeb retained some level of phosphorylation.
We considered that the changes in Tfeb phosphorylation may be linked to changes in its subcellular localization and performed subcellular fractionation experiments. Whereas in Tsc2+/+ cells, nuclear Tfeb was undetectable, Tfeb was found in nuclear fractions of Tsc2−/− cells (Figure 6E). In the nucleus, Tfeb appeared to be present almost exclusively in a fast-migrating form (Figure 6E). To determine whether nuclear Tfeb retained some level of phosphorylation, nuclear extracts were treated with λ phosphatase. As shown in Figure 6F, this resulted in a further shift, indicating that nuclear Tfeb was also phosphorylated. Since phosphatase treatment collapsed both nuclear and cytosolic Tfeb onto an undistinguishable band, the differences in migration likely reflect differences in phosphorylation.
Next, we sought to determine how rapamycin affected nuclear Tfeb in Tsc2−/− cells. Rapamycin treatment caused a shift up in nuclear Tfeb mobility and a progressive decline in nuclear Tfeb (Figure 6G). Taken together, these data show that the migration and nuclear localization of Tfeb are coordinately regulated by Tsc1/Tsc2 and rapamycin and suggest that mTORC1 controls the phosphorylation state and nuclear localization of Tfeb.
Regulation of TFEB phosphorylation and nuclear localization by mTORC1
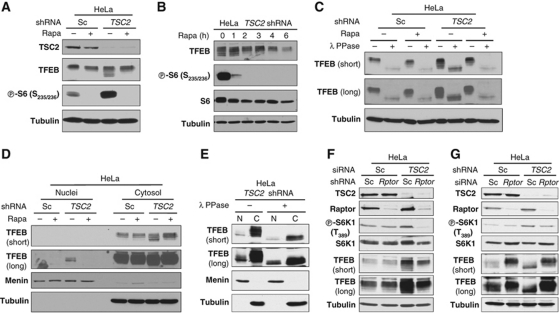
To extend these studies, we examined the regulation of TFEB by mTORC1 in HeLa cells. HeLa cells were transduced with lentiviruses encoding a TSC2 shRNA or a scrambled control and, as for MEFs, cultured for 24 h in 0.1% FBS with or without rapamycin. The results were remarkably similar to those observed in MEFs. TSC2 inactivation led to a downshift in TFEB and this was reverted by rapamycin (Figure 7A and B). As in MEFs, phosphatase treatment accelerated TFEB migration even in cells depleted of TSC2 (Figure 7C). Subcellular fractionation studies showed the presence of fast-migrating TFEB in nuclear fractions of cells depleted of TSC2, but not in controls (Figure 7D) and nuclear TFEB retained some level of phosphorylation (Figure 7E). Thus, the regulation of TFEB phosphorylation and nuclear localization by TSC1/TSC2 and rapamycin was conserved across cell types.
Figure 7.
mTORC1-dependent regulation of TFEB phosphorylation and nuclear localization. (A) Western blot of HeLa cells stably transduced with lentiviruses containing a TSC2 shRNA (or a scrambled control, Sc) and serum-starved (0.1% FBS) ± rapamycin for 24 h. (B) Western blot of serum-starved TSC2-depleted HeLa cells treated with rapamycin for the indicated hours. (C) Western blots from HeLa cells as in (A) incubated, where stated, with λ phosphatase. (D) Western blots of cellular fractions from HeLa cells treated as indicated. (E) Western blot of nuclear [N] and cytoplasmic [C] fractions from TSC2-depleted serum-starved HeLa cells incubated (or not) with λ phosphatase. (F) Western blot of HeLa cells stably transduced with a lentivirus encoding a Raptor shRNA (or a scrambled control, Sc) and subsequently transfected with siRNAs and serum starved. (G) Western blot of HeLa cells stably transduced with lentiviral vectors as indicated and subsequently transfected with siRNA and serum starved. Rptor, Raptor.
To unequivocally establish that TFEB regulation by TSC1/TSC2 and rapamycin was indeed mTORC1 dependent, we inactivated mTORC1 by knocking-down Raptor. HeLa cells were transduced with a Raptor shRNA (or a scrambled control) and, following selection, were subjected to TSC2 depletion using siRNA. As shown in Figure 7F, in cells depleted of Raptor, TSC2 RNAi failed to accelerate TFEB mobility. In a complementary set of experiments, Raptor siRNA (like rapamycin) retarded TFEB mobility in HeLa cells depleted of TSC2 (Figure 7G). Regardless of the means to deplete Raptor, Raptor inactivation prevented the accumulation of faster-migrating TFEB (Figure 7F and G). Taken together, our data indicate that mTORC1 coordinately regulates TFEB phosphorylation and nuclear localization in multiple cell types.
mTORC1-dependent nuclear localization of TFEB requires C-terminal serine-rich motif
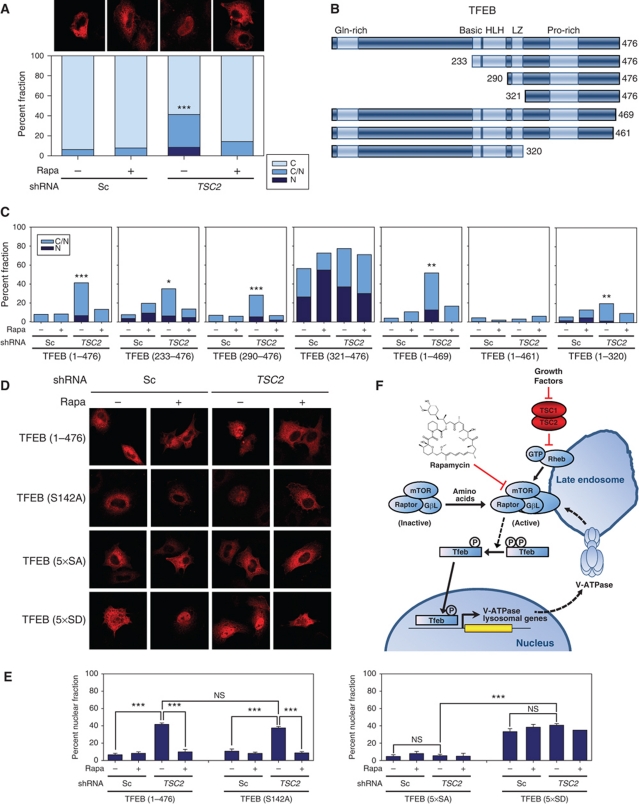
To dissect TFEB regulation by mTORC1, we first identified conditions in which ectopically expressed TFEB behaved like endogenous TFEB. Epitope-tagged TFEB, when expressed at low levels in HeLa cells, preserved its regulation by mTORC1; it was enriched in the nuclei of TSC2-depleted cells and the levels were decreased by rapamycin (Figure 8A). To determine what regions were important for TFEB regulation by mTORC1, we examined a series of deletions (Figure 8B). TFEB233–476 and TFEB290–476 behaved like full-length TFEB, indicating that the N-terminal half of TFEB was dispensable for mTORC1-dependent nuclear localization (Figure 8C). Within this region, mutation of a serine residue (S142) proposed to be phosphorylated by ERK2 and to regulate TFEB nuclear localization (Settembre et al, 2011) had no appreciable effect (Figure 8D and E). Deletion of residues 1–320 resulted in a protein that was enriched in nuclei (Figure 8C). Interestingly, deletion of just 15 amino acids from the C-terminus abrogated mTORC1-dependent nuclear localization (Figure 8C). By contrast, deletion of seven amino acids had no effect (Figure 8C). These data indicated that there were critical residues between 462–469. This corresponded to a serine-rich region (462SSRRSSFS469).
Figure 8.
mTORC1-dependent nuclear TFEB localization requires C-terminal serine-rich motif. (A) Confocal microscopy (representative images and bar graph quantification) of TFEB subcellular localization in HeLa cells stably expressing a scrambled shRNA (Sc) or a shRNA targeting TSC2 transfected with myc-tagged full-length TFEB and incubated for 24 h in 0.1% FBS with or without rapamycin. Percent of cells with cytoplasmic TFEB [C], nuclear [N] or both [C/N]. ‘*’ Illustrates statistically significant differences in nuclear TFEB (N+C/N) between TSC2-depleted HeLa cells by comparison to the other three conditions (n=4). (B) Schematic of full-length TFEB and deletion mutants; HLH, helix-loop-helix; LZ, leucine zipper. (C) Quantification of the subcellular distribution of mutant TFEB, compared with wild-type, in HeLa cells. ‘*’ Illustrates statistically significant differences in nuclear TFEB (N+C/N) between TSC2-depleted HeLa cells by comparison to the other three conditions for each panel (n=3). Confocal microscopy images (D) and bar graph quantification (E) of wild-type or mutant TFEB; 5 × SA, S462/463/466/467/469A; 5 × SD, S462/463/466/467/469D. Error bars represent s.e.m. (n=3). (F) Model of TFEB regulation by mTORC1. NS, non-significant; *P<0.05; **P<0.01; ***P<0.001.
To further evaluate, this serine-rich motif, we mutated all S to A (5 × SA). Interestingly, TFEB5 × SA was largely cytoplasmic and failed to be driven into the nucleus in cells with active mTORC1 (Figure 8D and E). Thus, a serine(s) within this motif is necessary for mTORC1-dependent nuclear TFEB localization. Conversely, mutation of the same residues to aspartic acid resulted in an enrichment of TFEB in the nucleus (Figure 8D and E). TFEB5 × SD localized to the nucleus even in cells with inactive mTORC1. Interestingly, the percentage of cells with TFEB5 × SD in the nucleus was the same as for wild-type TFEB in mTORC1-active cells (Figure 8E; see also Figure 8A). These data show that the presence of phosphomimetic residues in the serine-rich motif obviates the mTORC1 requirement for TFEB nuclear localization. The simplest explanation for these data is that a serine(s) in the C-terminal serine-rich motif is phosphorylated in an mTORC1-dependent manner and that phosphorylation is both necessary and sufficient to drive TFEB into the nucleus (see model in Figure 8F).
Complex TFEB regulation beyond mTORC1 by multiple signals
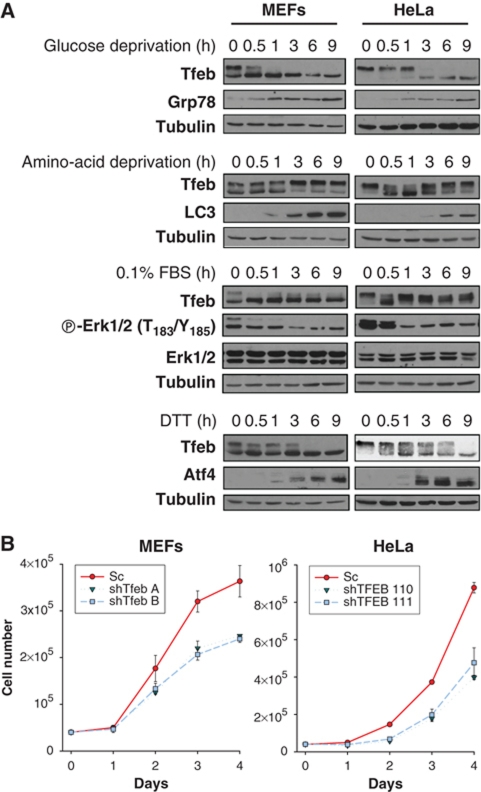
Next, we examined TFEB in wild-type MEFs and HeLa cells grown under standard conditions (10% FBS). TFEB was found in both a slow and fast-migrating form(s) in MEFs, but predominantly in a slow migrating form(s) in HeLa cells (Figure 9A). We evaluated the effects of multiple interventions on TFEB mobility. Glucose withdrawal resulted in a downshift in TFEB in both MEFs and HeLa cells (Figure 9A). Amino acid withdrawal induced acute changes in TFEB migration in HeLa cells and subsequently led to the accumulation of slow migrating TFEB, which was also observed in MEFs (Figure 9A). The mobility of TFEB was also affected by serum deprivation in both MEFs and HeLa cells (Figure 9A). Finally, changes in TFEB mobility were also induced by treatment with a reducing agent, DTT. DTT led to a progressive shift in TFEB to a fast-migrating form(s) in both HeLa and MEFs (Figure 9A).
Figure 9.
Complex TFEB regulation by multiple signals beyond mTORC1. (A) HeLa and MEFs were starved for amino acids, glucose and serum (0.1% FBS), or treated with 10 mM DTT for the indicated times. (B) Graphs depicting cell counts of MEFs and HeLa cells over time (Sc, scrambled). P<10−4 and P<10−7 for Tfeb knockdown MEFs and HeLa cells, respectively, using two-factor ANOVA. Error bars represent s.e.m. (n=3).
Overall, these data show that TFEB is extensively and dynamically regulated by multiple stimuli. While post-translational modifications other than phosphorylation may be involved in the changes observed in TFEB migration, TFEB could be down-shifted by phosphatase treatment in every instance (data not shown). The dynamic changes in TFEB mobility induced by the various interventions could not be explained by their expected effects on mTORC1 suggesting that TFEB regulation involves multiple pathways and is likely to be rather complex. This is consistent with the finding that TFEB is phosphorylated in >10 sites (Dephoure et al, 2008; Mayya et al, 2009; Yu et al, 2011).
Finally, since TFEB is an oncogene (Davis et al, 2003; Kuiper et al, 2003; Srigley and Delahunt, 2009), we tested whether TFEB was implicated in the regulation of cell proliferation. As shown in Figure 9B, stable depletion of TFEB in both MEFs and HeLa cells lowered the rates of cell expansion.
Discussion
Using a robust experimental paradigm, we uncovered a novel regulatory network linking endocytosis to TFEB and mTORC1. An unbiased gene expression analysis coupled with a design amenable to refined statistics led us to identify a connection between mTORC1 and V-ATPase genes. Publicly available data sets confirmed the link and experiments in mice showed that mTORC1 regulated V-ATPase expression also in vivo. A recent study had established that TFEB regulated lysosomal biogenesis (Sardiello et al, 2009) and among the genes regulated by TFEB we found several V-ATPases. These results led us to hypothesize that V-ATPase regulation by mTORC1 may be mediated by TFEB. Indeed, TFEB was necessary for V-ATPase upregulation by mTORC1. mTORC1 regulated TFEB phosphorylation and nuclear localization and a C-terminal serine-rich motif was identified that is essential for mTORC1-dependent TFEB nuclear localization. Our data suggest that mTORC1 induces the phosphorylation of a serine(s) within this motif driving thereby TFEB to the nucleus. TFEB regulates the expression of V-ATPases and other lysosomal genes and TFEB was required for mTORC1-induced endocytosis. To our knowledge this is the first study to show that endogenous TFEB is regulated at the subcellular localization level and that mTORC1 promotes TFEB nuclear localization. These data link an oncogenic transcription factor that is a master regulator of lysosomal biogenesis, TFEB, to mTORC1 and endocytosis.
An increasing amount of evidence implicates TORC1 in endocytosis. mTOR was identified in an RNAi screen for kinases involved in clathrin-mediated endocytosis (Pelkmans et al, 2005) and Tsc2-deficient cells were previously shown to have increased fluid-phase endocytosis (Xiao et al, 1997). In Drosophila melanogaster, a sensitized screen for genes that modified a tissue-specific dTOR overexpression phenotype identified an important regulator of endocytosis, Hsc70-4, a clathrin uncoating factor, and the authors showed that dTOR was involved in fat body endocytosis (Hennig et al, 2006). Interestingly, besides V-ATPases, mTORC1 regulated the expression of multiple lysosomal genes (Figure 1D; see also Supplementary Table S5). These effects may be mediated, at least in part, by TFEB, which regulates lysosome biogenesis (Sardiello et al, 2009). Thus, one mechanism whereby mTORC1 and TFEB may affect endocytosis is by regulating V-ATPase levels and lysosomes.
The lysosome is the final destination not only for endosomal cargo but also for autophagosomes. In response to starvation, mTORC1 is inhibited leading to the formation of autophagosomes that engulf intracellular components for recycling and sustenance (He and Klionsky, 2009; Kroemer et al, 2010). Autophagosomes and their content are delivered to lysosomes, which accumulate in the perinuclear area (Korolchuk et al, 2011). Autophagosomes fuse with lysosomes to generate autolysosomes, a process that rapidly depletes the lysosomal pool (Yu et al, 2010). The degradation of macromolecules and release of their constituents into the cytosol reactivates mTORC1 (Yu et al, 2010). mTORC1 reactivation terminates autophagy and through a poorly understood mechanism repletes the lysosome pool (Yu et al, 2010). However, how the pool of lysosomes is repleted is unknown. Our results suggest that lysosomal reformation in response to autophagy, which is mTORC1 dependent (Yu et al, 2010), may be mediated, at least in part, by TFEB.
mTORC1 coordinately regulates TFEB phosphorylation and nuclear localization. mTORC1 promoted TFEB nuclear localization and this process required a C-terminal serine-rich motif. Serine substitution to non-phosphorylatable residues abrogated TFEB regulation by mTORC1. More importantly, mutation to phosphomimetic amino acids was sufficient to reproduce the effect of mTORC1 on TFEB nuclear localization obviating the need for mTORC1. These data support a model in which mTORC1 (or an effector kinase) directly phosphorylates a serine(s) within the C-terminal serine-rich motif driving thereby TFEB to the nucleus. Consistent with this notion, nuclear TFEB was phosphorylated. This model may seem at odds with the observation that nuclear TFEB is fast-migrating and possibly hypo-phosphorylated by comparison to cytoplasmic (presumably cytosolic) TFEB. However, as our data suggest, nuclear TFEB may be phosphorylated on sites that are not phosphorylated on cytosolic TFEB, and thus no inferences can be drawn about the degree of phosphorylation based on SDS–PAGE migration. Furthermore, while we have identified a critical motif involved in mTORC1 regulation, other sites in TFEB appear to be regulated by mTORC1 (Yu et al, 2011).
Our results further emphasize the link between mTORC1 and the late endosome/lysosome and provide evidence for the existence of bidirectional regulatory loops. mTORC1 localizes to the surface of the late endosome/lysosome in a highly choreographed manner (Korolchuk et al, 2011; Narita et al, 2011). The late endosome/lysosome is necessary for mTORC1 activation (Flinn et al, 2010; Li et al, 2010; Sancak et al, 2010) and mTORC1 is reactivated under conditions of persistent starvation following macromolecule degradation in autolysosomes (Yu et al, 2010). However, not only is mTORC1 downstream of the lysosome, but, inasmuch as mTORC1 regulates TFEB and V-ATPases, mTORC1 is also upstream. mTORC1 reactivation by autolysosomes requires Spinster, a lysosomal efflux sugar transporter. Spinster has been proposed to act by regulating lysosomal pH (Rong et al, 2011), which is controlled primarily by the V-ATPase. Interestingly, not only has luminal pH been implicated in mTORC1 regulation, but changes in cytosolic pH occur in response to alterations in nutrient conditions (Korolchuk et al, 2011) and have been implicated in the regulation of lysosome topology and mTORC1 (Korolchuk et al, 2011).
Recently, TFEB was found to regulate autophagy gene expression (Settembre et al, 2011). Starvation led to ERK2 inactivation, which in turn, decreased TFEB phosphorylation and increased its nuclear localization. This model is in principle compatible with our results. TFEB is phosphorylated in over 10 sites (Dephoure et al, 2008; Mayya et al, 2009; Yu et al, 2011), and mechanisms likely exist that drive TFEB into the nucleus independently of the activation state of mTORC1. However, the studies do differ in one aspect. Whereas mutation of the putative ERK2 phosphorylation site, S142, to A is reported to be sufficient to drive TFEB into the nucleus (Settembre et al, 2011), we find that, at least under low serum conditions, the subcellular localization of TFEB is not appreciably altered by a S142A mutation.
The experimental conditions studied by Settembre et al (2011), which involve evaluating ectopically expressed TFEB in cells in the absence of glucose, amino acids and serum, are quite different from ours. Individual deprivation of glucose, amino acids or serum has profound, dynamic and diverse effects on TFEB (see Figure 9A). Given these observations and the complexity of TFEB phosphorylation (Dephoure et al, 2008; Mayya et al, 2009; Yu et al, 2011), how starvation of all three sources will affect TFEB is unclear. Consistent with this complexity, the changes in TFEB migration observed following deprivation of individual factors could not be explained by the predicted changes on mTORC1 activity. In fact, we speculate that the discovery that mTORC1 regulates TFEB may have been possible only because of the tight experimental system we used.
Besides V-ATPases and lysosomal genes, many other genes downstream of mTORC1 were regulated in a Tfeb-dependent manner. Approximately 25% of genes induced by mTORC1 required Tfeb. Experiments are ongoing to determine how many of these genes are directly regulated by Tfeb. A similar experimental setup previously identified Hif-1α as an mTORC1 effector (Brugarolas et al, 2003), and Hif-1α is an important mediator of a metabolic gene expression programme (Duvel et al, 2010). Along with sterol regulatory element-binding protein-1 (Porstmann et al, 2008), a transcription factor whose nuclear localization is regulated by mTORC1, and Hif-1α, TFEB is an important mediator of mTORC1 effects on gene expression.
The discovery that mTORC1 regulates TFEB has clinical implications. It may explain the expression of melanocyte lineage markers in tumours of patients with TSC (Martignoni et al, 2008), a syndrome arising from mutations in either the TSC1 or TSC2 genes, resulting in constitutive mTORC1 activation (El-Hashemite et al, 2003; Kenerson et al, 2007). Patients with TSC are predisposed to develop renal angiomyolipomas, which express melanocytic markers such as HMB45 and Melan-A (Martignoni et al, 2008), and since expression of these proteins in melanomas is thought to be driven by TFEB (or related family members with whom TFEB can heterodimerize) (Steingrimsson et al, 2004), these markers may be similarly driven by TFEB in renal angiomyolipomas.
Interestingly, TFEB functions as an oncogene in the kidney. The TFEB gene is translocated in a subset of renal tumours where it is subjected to a robust heterologous promoter (Davis et al, 2003; Kuiper et al, 2003). Translocation carcinomas tend also to aberrantly express melanocytic markers (Srigley and Delahunt, 2009), which may be similarly driven by TFEB. TFEB activation may also account for the expression of cathepsin-K, a lysosomal protease that has been recently proposed as a marker for this tumour type (Martignoni et al, 2009). Interestingly, translocation carcinomas are often recognized by immunohistochemical studies showing high levels of nuclear TFEB (Srigley and Delahunt, 2009). Because TFEB nuclear localization is regulated by mTORC1 and as we show, sirolimus excludes TFEB from the nucleus, sirolimus, or its derivatives, temsirolimus and everolimus, may be effective against this tumour type.
While this study focused on a small subset of genes, many genes were identified whose expression was regulated by mTORC1 not only positively but also negatively (Supplementary Figures S8–S10; Supplementary Table S1). Moreover, genes were also found whose expression was regulated by Tsc2 independently of mTORC1 (LLHH and HHLL patterns). As previously reported (Brugarolas et al, 2003), this would point towards mTORC1-independent functions of the Tsc1/Tsc2 complex (Supplementary Figure S8; Supplementary Table S1). In addition, the expression of some genes appeared to be regulated by rapamycin to a significantly greater extent than by Tsc2 (HLHL and LHLH patterns) (Supplementary Figure S8; Supplementary Table S1). Importantly, in all these settings, analyses of alternative patterns showed the associations to be statistically significant (Supplementary Figure S8). Incidentally, among the genes with an HLHL pattern, there was enrichment for genes implicated in glycolysis, pentose phosphate pathway and fatty acid biosynthesis (Supplementary Table S5). These pathways were recently reported to be regulated by mTORC1 (Duvel et al, 2010) and our results suggest that they are affected by rapamycin to a greater extent than by Tsc1/Tsc2.
In summary, our work uncovered a novel regulatory network connecting mTORC1 to a master regulator of lysosome biogenesis, TFEB, which is essential for mTORC1-induced endocytosis.
Materials and methods
Cell culture, drug and hypoxia treatments
MEFs Tsc2+/+; p53−/− (Tsc2+/+) and Tsc2−/−; p53−/− (Tsc2−/−) were a gift of DJ Kwiatkowski (Brigham and Women's Hospital, Harvard Medical School, Boston, MA). Cells were grown in high glucose Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% FBS (HyClone) and 1% (v/v) penicillin/streptomycin (P/S) (Gibco) in a humidified incubator at 37°C and 5% CO2. For experiments, cells were plated to achieve ∼70% confluency at the time of treatment. For low-serum experiments, cells were washed twice in PBS and medium was changed to the same base medium but supplemented with 0.1% FBS and cells were harvested after 24 h. Where indicated, rapamycin (LC Laboratories) was used at 25 nM in methanol and methanol was used as vehicle. Where indicated, cells were treated with actinomycin D (Sigma) at 10 μg/ml, bafilomycin A1 (LC Laboratories) at 100 nM, or DTT (10 mM). For growth curves, cells were plated on day 0, maintained in 10% FBS and triplicates were counted every 24 h. For hypoxia experiments, cells were exposed to 1% O2 and 5% CO2 at 37°C in a hypoxia chamber (Coy Laboratory Products). For starvation time courses, cells were washed with PBS twice and starved of amino acids (amino acid-free media), glucose (glucose-free media) or serum (0.1% FBS) for different amounts of time.
Hif-1α, Tfeb, Raptor and TSC2 shRNAs
Recombinant lentiviruses were generated in HEK293T cells by transient transfection using calcium phosphate with the envelope plasmid pMD2.G (Database ID p486), packaging plasmid psPAX2 (p485) and the specific plasmid of interest. pMD2.G, psPAX2 and pLKO.1 or pGIPZ vectors were transfected at a ratio of 3:8:10. Supernatants were collected for 2 days, passed through a 0.45-μm filter, and used to infect MEFs or HeLa cells for 24 h in medium containing 8 μg/ml polybrene (Sigma). MEFs and HeLa cells were selected with 2 μg/ml puromycin for at least 1 week and HeLa cells were further enriched by FACS. The following vectors were used: pLKO.1 vectors with a Hif-1α shRNA (5′-AGAGGTGGATATGTCTGGG-3′) (p481) generously provided by LB Gardner (Nemetski and Gardner, 2007), a mouse Tfeb shRNA from Open Biosystems (sequence A, TRCN0000085548, 5′-CGGCAGTACTATGACTATGAT-3′) (p633) or a designed Tfeb shRNA (sequence B, 5′-GGCAGTACTATGACTATGATG-3′) (p652), a human TFEB shRNAs from Open Biosystems (sequence 110, TRCN0000013110, 5′-GAGACGAAGGTTCAACATCAA-3′) (p700) or (sequence 111, TRCN0000013111, 5′-GAACAAGTTTGCTGCCCACAT-3′) (p701), a raptor shRNA from Addgene (Addgene plasmid 1858) (p741) or a scrambled sequence (5′-GGGTCTGTATAGGTGGAGA-3′) (p480). pGIPZ vectors containing a TSC2 (p496) or a scrambled (p483) shRNA were from Open Biosystems.
Western blotting and antibodies
Cell lysates and western blotting was performed as previously described (Vega-Rubin-de-Celis et al, 2010), using antibodies from the following sources: S6K1, phospho-S6K1 (T389), S6, phospho-S6 (S235/236), total 4E-BP1, phospho-4E-BP1 (T37/46), phospho-4E-BP1 (S65) from Cell Signaling; HIF-1α, TSC1 and Menin from Bethyl laboratories; TSC2, ATF4 and c-Myc from Santa Cruz; Raptor from Millipore; Erk1/2, phospho-Erk1/2 (T183/Y185) and Tubulin from Sigma; Cyclophilin B from Abcam; GLUT-1 from Novus Biologicals; and Grp78 from BD Biosciences. Atp6v1a and Atp6v1b2 antibodies were a generous gift from D Brown (Massachusetts General Hospital, Boston, MA); and Atp6v0a1 and Atp6v0a3 antibodies were kindly provided by V Marshansky (Massachusetts General Hospital, Boston, MA). Tfeb antibodies were developed in collaboration with Bethyl Laboratories.
For the remaining Materials and methods, please see the Supplementary data section.
Accession numbers
Microarrays were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE27982 and GSE28021.
Supplementary Material
Acknowledgments
We thank Dr WG Kaelin Jr (Dana-Farber Cancer Institute) and the members of the Brugarolas laboratory for helpful discussions; MG Roth, JM Abrams and MA White (UT Southwestern Medical Center) for reading the manuscript; and C Li (Dana-Farber Cancer Institute) for statistical advice. We are indebted to Drs DJ Kwiatkowski (Brigham and Women's Hospital, Harvard Medical School, Boston, MA) for Tsc2-deficient MEFs and Tsc1F/F mice; LB Gardner (New York University) for Hif1-α and scrambled shRNA plasmids; N Esumi (Johns Hopkins University) for TFEB plasmid; and D Brown and V Marshansky (Massachusetts General Hospital, Boston, MA) for V-ATPase antibodies. This work was supported by a fellowship of Excellence (BPOSTDOC06/004) from Generalitat Valenciana (Spain) to SP-L; a fellowship from Fundacion Caja Madrid to SV-R; T32CA124334 and F32CA136087 to TATT; NIBIB EB 05556 to JCS; NIGMS 77253 to DRC; and the following Grants to JB: K08NS051843, RO1CA129387, RSG115739 from the ACS and a BOC 5FY06582 from the March of Dimes Foundation. Microarray samples were processed at the Genomics Shared Resource of the Harold C Simmons Cancer Center supported in part by 1P30CA142543. JB is a Virginia Murchison Linthicum Scholar in Medical Research at UT Southwestern. The content is solely the responsibility of the authors and does not represent official views from any of the granting agencies.
Author contributions: SP-L, SV-RC and JB designed the experiments. SP-L performed experiments, all statistical and microarray analyses and wrote an initial draft of the manuscript. SV-RC conducted confocal microscopy and most TFEB experiments, JCS carried out ChIP assays under the supervision of DRC. NCW was responsible for the mouse experiments. TATT helped with shRNA cloning. LZ was involved in preliminary microarray data characterization. X-JX provided statistical input and revised statistics. JB conceived the study, was responsible for the direction of the project and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT, Eng CH (2008) Mammalian target of rapamycin as a therapeutic target in oncology. Expert Opin Ther Targets 12: 209–222 [DOI] [PubMed] [Google Scholar]
- Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N (2009) Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 296: E592–E602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyenbach KW, Wieczorek H (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol 209: 577–589 [DOI] [PubMed] [Google Scholar]
- Bowman BJ, Bowman EJ (2002) Mutations in subunit C of the vacuolar ATPase confer resistance to bafilomycin and identify a conserved antibiotic binding site. J Biol Chem 277: 3965–3972 [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr (2004) Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18: 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG Jr (2003) TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 4: 147–158 [DOI] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GG, Quilliam LA (2003) Rheb binds TSC2 and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem 278: 32493–32496 [DOI] [PubMed] [Google Scholar]
- Connolly E, Braunstein S, Formenti S, Schneider RJ (2006) Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol 26: 3955–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL (2004) Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev 18: 1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P (2007) mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, Pledger WJ, Cheng JQ (2002) Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem 277: 35364–35370 [DOI] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, Fletcher JA, Fisher DE (2003) Cloning of an alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci USA 100: 6051–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW (2008) Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 22: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, Mackeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hashemite N, Zhang H, Henske EP, Kwiatkowski DJ (2003) Mutation in TSC2 and activation of mammalian target of rapamycin signalling pathway in renal angiomyolipoma. Lancet 361: 1348–1349 [DOI] [PubMed] [Google Scholar]
- Fingar DC, Blenis J (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23: 3151–3171 [DOI] [PubMed] [Google Scholar]
- Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM (2010) The late endosome is essential for mTORC1 signaling. Mol Biol Cell 21: 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8: 917–929 [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Guyton AC, Hall JE (1996) Texbook of Medical Physiology, 9th edn, Philadelphia, PA: WB Saunders Company [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig KM, Colombani J, Neufeld TP (2006) TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol 173: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi I, Kovacevic Z, Lesovoy V, Schmidt-Wolf IG, Barbarash O, Gokmen E, O'Toole T, Lustgarten S, Moore L, Motzer RJ (2007) Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 356: 2271–2281 [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL (2003a) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657 [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL (2003b) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jaeschke A, Hartkamp J, Saitoh M, Roworth W, Nobukuni T, Hodges A, Sampson J, Thomas G, Lamb R (2002) Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J Cell Biol 159: 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr (2008) The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 8: 865–873 [DOI] [PubMed] [Google Scholar]
- Kaper F, Dornhoefer N, Giaccia AJ (2006) Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Res 66: 1561–1569 [DOI] [PubMed] [Google Scholar]
- Kenerson H, Folpe AL, Takayama TK, Yeung RS (2007) Activation of the mTOR pathway in sporadic angiomyolipomas and other perivascular epithelioid cell neoplasms. Hum Pathol 38: 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O'Kane CJ, Deretic V, Rubinsztein DC (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucejova B, Sunny NE, Nguyen AD, Hallac R, Fu X, Pena-Llopis S, Mason RP, Deberardinis RJ, Xie XJ, Debose-Boyd R, Kodibagkar VD, Burgess SC, Brugarolas J (2011) Uncoupling hypoxia signaling from oxygen sensing in the liver results in hypoketotic hypoglycemic death. Oncogene 30: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG (2003) Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet 12: 1661–1669 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, El-Hashemite N, Onda H (2002) A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet 11: 525–534 [DOI] [PubMed] [Google Scholar]
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL (2010) Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 285: 19705–19709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC (2006) Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell 21: 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP (2005) Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121: 179–193 [DOI] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR (2004) mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med 10: 594–601 [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10: 151–162 [DOI] [PubMed] [Google Scholar]
- Marshansky V, Futai M (2008) The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol 20: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, Pan CC, Netto G, Doglioni C, Hes O, Argani P, Chilosi M (2009) Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol 22: 1016–1022 [DOI] [PubMed] [Google Scholar]
- Martignoni G, Pea M, Reghellin D, Zamboni G, Bonetti F (2008) PEComas: the past, the present and the future. Virchows Arch 452: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Pagano RE (2007) Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol 8: 603–612 [DOI] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2: ra46. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grunwald V, Thompson JA, Figlin RA, Hollaender N, Urbanowitz G, Berg WJ, Kay A, Lebwohl D, Ravaud A (2008) Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 372: 449–456 [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavare S, Inoki K, Shimizu S (2011) Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemetski SM, Gardner LB (2007) Hypoxic regulation of Id-1 and activation of the unfolded protein response are aberrant in neuroblastoma. J Biol Chem 282: 240–248 [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436: 78–86 [DOI] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A (2008) SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 8: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T (2002) Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 4: 658–665 [DOI] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, Di Vizio D, Hou H Jr, Kneitz B, Lagaud G, Christ GJ, Edelmann W, Lisanti MP (2001) Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem 276: 38121–38138 [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ (2006) Regulation of the small GTPase Rheb by amino acids. Oncogene 25: 657–664 [DOI] [PubMed] [Google Scholar]
- Rong Y, McPhee C, Deng S, Huang L, Chen L, Liu M, Tracy K, Baehreck EH, Yu L, Lenardo MJ (2011) Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci USA 108: 7826–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J (2004) Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA 101: 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009) A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC (2004) The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6: 91–99 [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG (2005) The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem 280: 18717–18727 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srigley JR, Delahunt B (2009) Uncommon and recently described renal carcinomas. Mod Pathol 22(Suppl 2): S2–S23 [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA (2004) Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38: 365–411 [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003) Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268 [DOI] [PubMed] [Google Scholar]
- Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X (2010) Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry 49: 2491–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G (2005) Integration of oxygen signaling at the consensus HRE. Sci STKE 2005: re12. [DOI] [PubMed] [Google Scholar]
- Wolff NC, Vega-Rubin-de-Celis S, Xie XJ, Castrillon DH, Kabbani W, Brugarolas J (2011) Cell-type-dependent regulation of mTORC1 by REDD1 and the tumor suppressors TSC1/TSC2 and LKB1 in response to hypoxia. Mol Cell Biol 31: 1870–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL (2009) Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA 106: 4260–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GH, Shoarinejad F, Jin F, Golemis EA, Yeung RS (1997) The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem 272: 6097–6100 [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ (2003a) Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest 112: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D (2003b) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5: 578–581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.