Abstract
Within-host competition between parasites is frequently invoked as a major force for parasite evolution, yet quantitative studies on its extent in an organismal group are lacking. Temperate bacteriophages are diverse and abundant parasites of bacteria, distinguished by their ability to enter a facultative dormant state in their host. Bacteria can accumulate multiple phages that may eventually abandon dormancy in response to host stress. Host resources are then converted into phage particles, whose release requires cell death. To study within-host competition between phages, I used the bacterium Escherichia coli and 11 lambdoid phages to construct single and double lysogens. Lysogenic bacterial cultures were then induced and time to host cell lysis and productivity of phages was measured. In double lysogens, this revealed strong competitive interactions as in all cases productivity of at least one phage declined. The outcome of within-host competition was often asymmetrical, and phages were found to vary hierarchically in within-host competitive ability. In double infections, the phage with the shorter lysis time determined the timing of cell lysis, which was associated with a competitive advantage when time differences were large. The results emphasize that within-host competition greatly affects phage fitness and that multiple infections should be considered an integral part of bacteriophage ecology.
Keywords: multiple infection, within-host competition, polylysogeny, evolution of virulence, Escherichia coli, bacteriophage λ
Introduction
The ubiquity of parasites makes multiple infections a common phenomenon. Parasites rarely have a host to exploit for themselves, but share it with other parasite strains or species (Cox, 2001; Read and Taylor, 2001). Understanding the resulting within-host interactions is important, as they are predicted to alter infectious disease dynamics and the parasites evolutionary trajectory (Nowak and May, 1994; May and Nowak, 1995). Furthermore, central evolutionary theories, addressing the evolution of sex, local adaptation and virulence evolution, have been developed in the framework of host–parasite interactions (Hamilton, 1980; Anderson and May, 1982; Thompson, 1994). The influence of multiple infections on these interactions may thus have far-reaching implications.
Bacteriophages (phages) are viruses that infect bacteria. Temperate phages are distinguished by their ability to reproduce by two different transmission modes. After infecting a cell, they either follow the lytic cycle, converting host resources into new viral particles and killing the cell to release them, or they enter lysogeny, integrating their DNA into the genome of the host to become a prophage. Prophages are replicated by bacterial division and are stably maintained over extended periods of time. This changes once host DNA has been damaged. The bacterial stress response and possibly other signals can trigger the induction of the prophage, which then enters lytic development and finally lyses the host cell (Ptashne, 2004; Shkilnyj and Koudelka, 2007; Ghosh et al., 2009).
Lysogenic bacteria constitute a large fraction of microbial communities (Miller and Day, 2008) and a number of observations suggest that they are often multiply infected (Leitet et al., 2006; Williamson et al., 2008). Bacteria are susceptible to many phages, which can integrate into the same host genome if they use different insertion sites (Campbell, 2003). Consequently, polylysogens, cells harboring several different prophages, are found in a variety of bacterial species (Reynolds et al., 1988; Espeland et al., 2004; Asadulghani et al., 2009). Finally, phages contain a vast amount of genetic diversity, to which recombination has contributed an important part, indicating that multiple infections have always played a role in phage biology (Hendrix, 2002).
Superinfection of a lysogen is a constant threat to prophages, because entry of the new phage into the lytic pathway would destroy the resident prophage along with the host (Berngruber et al., 2010). To avoid this threat, prophages must protect their host against superinfection by phages of their own kind. This immunity is hardwired into the genetic circuitry that maintains the lysogenic state and is an essential feature of lysogeny (Lwoff, 1953; Campbell, 2006). In addition, a number of mechanisms allow a prophage to secure a cell against lysis by other, heteroimmune phages. These include the modification of cell surface receptors required for phage adsorption (Matsumoto et al., 1985), premature transcription termination in other phages (Oberto et al., 1989), restriction-modification systems to target DNA of incoming phages (Arber and Dussoix, 1962), increasing the probability that superinfecting phages establish lysogeny (Serra-Moreno et al., 2008), and the induction of host cell death, possibly to prevent the spread of a competing phage (Benzer, 1955).
Bacteriophage–bacteria interactions represent an ideal system for studying the competitive interactions within hosts as well as their effect on parasite fitness. Competition among phages, particularly within the same host, is a largely unexplored area in phage ecological research. This is particularly true for temperate phages. On the one hand, detailed knowledge exists on their molecular genetics; on the other, large-scale surveys suggest that they have an important role in determining microbial diversity and community structure, and direct nutrient fluxes on ecosystem scales (Angly et al., 2006; Suttle, 2007). A link between these two areas of research, that describes how individual interactions among phages shape the phage community, is lacking.
Besides their ecological importance that warrants a closer examination of multiple infections in phages, temperate phages offer several advantages that allow a particularly detailed investigation. First, sequential insertion of phages into the host genome allows a controlled construction of singly and multiply infected cells, so that the effect of the order of infection on success of competing phages can be studied. Second, prophages are almost always single copy, and their lytic development can be triggered externally. This allows the processes of host infection and subsequent within-host growth to be separated, thereby excluding confounding effects of variation in infectious dose on reproductive success (Hochberg, 1998). Third, time to host death (that is, lysis time) can be measured accurately. Although any lytic action eventually kills a cell, lysis time is an important determinant for the speed at which a phage devours a bacterial population and may be interpreted as a component of virulence (Eshelman et al., 2010). Fourth, reproductive success of coinfecting phages can be quantified separately, which overcomes a major obstacle of many experimental studies. Finally, the detailed knowledge on phage and bacterial genetics allows a detailed study of competition in one of the simplest in vivo systems known (Lewontin, 1970).
Whether multiple infections select for more or less virulent parasites is controversial and it has been suggested that it depends critically on the nature of their competitive interactions (Brown et al., 2002). If the reproductive success of a parasite depends directly on its exploitative behavior, multiple infections are expected to favor more virulent parasites, as they use the available resources quicker than their competitor (Frank, 1996). More prudent strategies with a lower virulence may be favored if exploitation of host resources requires an initial investment by the parasites, the benefits of which are then available to all coinfecting parasites (Brown et al., 2002). The availability of such a public good allows ‘cheaters' to persist that do not participate in the collective action and are consequently less virulent. Note that both hypotheses assume a direct link between within-host growth and host mortality, with more vigorous growth causing more damage and a shorter host life span. In phage λ, which is among the phages used in this study, cell lysis is directly controlled by the phage and does not depend on its within-host growth rate (Wang, 2006), hence the relation between lysis timing and competitive success within a single cell is unclear.
In this study, I used 11 different lambdoid phages of Escherichia coli to study within-host competition in doubly infected cells. To this end, I constructed all possible single and double lysogens and induced them under controlled conditions. I measured lysis time in all lysogens and quantified productivity of all phages (Figure 1). Besides gaining detailed information on individual phage interactions, the diversity of phages used allowed me to address questions concerning general patterns of competitive interactions in this group of phages. The results provide evidence that multiple infections have a severe impact on phage fitness, and caution that lysis time is only a weak predictor for the outcome of competition.
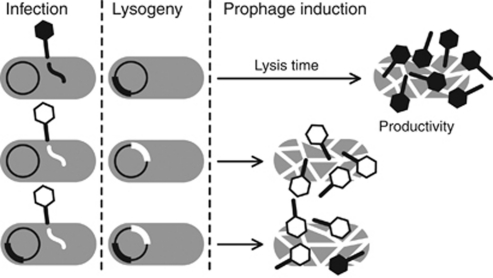
Figure 1.
Life cycle of temperate phages in the context of this study. Upon infection, phages establish lysogeny and reside dormant as prophages in the bacterial chromosome. When induced, they begin lytic development and lyse their host after some time to release virions. In this hypothetical example, the white phage has a shorter lysis time than the black phage and phages differ in productivity. If prophages share a cell and form a double lysogen, both lysis time and productivity may change. In this example, the white phage determines lysis time and competition lowers productivity of both phages.
Materials and methods
Media, reagents and strains
Bacteria were grown in lysogeny broth (LB; 10 gl−1 tryptone, 5 gl−1 yeast extract, 10 gl−1 NaCl). For plates, 1.5% agar was added, for top agar, 0.7%. Bacteria were stored in LB with 15% glycerol at −80 °C. Phages were stored in SMG (0.1 NaCl, 10 m MgSO4, 0.05 Tris (pH 7.5), 0.01% gelatin) at 4 °C (Table 1). Several of the phages that we used belong to a collection of Mexican E. coli phages (Kameyama et al., 1999). Two of our isolates could not be assigned to the original isolates, and were renamed mEpX1 and mEpX2, respectively. Mitomycin C (Sigma, Buchs, Switzerland) at a concentration of 5 μg ml−1 was used for prophage induction.
Table 1. Phages that were used in this study.
| Phage | Immunity group | Receptor specificity |
|---|---|---|
| λ (λPaPa) | XVIII | LamB |
| HK022 | HK022 | FhuA |
| φ80 | φ80 | FhuA and TonB |
| mEp043 | IV | FhuA and TonB |
| mEp213 | IX | FhuA and TonB |
| mEp234 | XVIII | FhuA |
| mEp235 | XVII | FhuA |
| mEp332 | XVIII | OmpC |
| mEp506 | XV | FhuA |
| mEpX1 | mEpX1 | FhuA |
| mEpX2 | mEpX2 | FhuA |
Nomenclature of immunity groups follows Kameyama et al. (1999). Receptor specificities were in agreement with previous results where available.
Construction of single and double lysogens
To obtain single lysogens, phage dilutions were spotted on a lawn of E. coli MG1655 and incubated overnight. Turbid plaques indicated growth of lysogens and bacteria were picked from these plaques and reisolated twice. Lysogens were typed for their immunity group (see below) and the release of phages upon induction was verified. To avoid selecting anomalous lysogens, we isolated several lysogens of every phage and measured their induction curves (see below). A representative lysogen of every phage was selected for all further work.
Double lysogens were constructed by superinfection of single lysogens. Where possible, we controlled for the order of insertion and constructed double lysogens twice, changing the order with which phages were inserted (Figure 2). Double lysogens were typed for their immunities.
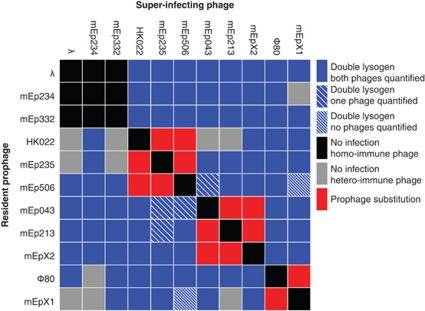
Figure 2.
The outcome of superinfection of 11 different single-lysogens with the same 11 phages.
Typing lysogens with clear-plaque phage mutants
A prophage always immunizes its host against itself and phages of the same immunity group (Table 1). The specific resistance of a lysogen can therefore be used to identify the prophages it harbors. Typing was carried out by cross-streaking a bacterial sample over a streak of a lysogeny-deficient mutant of the respective phage. Continued growth indicated presence of the corresponding immunity and thus presence of the prophage.
Lysogeny-deficient mutants of phages from all immunity groups were obtained by ultraviolet irradiation (200 J m−2). Irradiated lysates were plated on susceptible bacteria and incubated overnight. Clear plaques that indicated lysogeny-deficient mutants were cored, reisolated twice and amplified by confluent lysis.
Prophage induction
Single colonies of lysogens were inoculated in 500 μl of LB and grown overnight in 96-deep well plates at 37 °C in a shaking incubator (400 r.p.m.). To ensure thorough mixing of the culture, a glass bead was added to each well. Plates were covered with breathable film to allow aeration and prevent evaporation and cross-contamination. Overnight cultures were diluted 1/500 in 500 μl pre-warmed LB and incubated for 2 h to bring cultures into exponential growth. A volume of 100 μl was then transferred into an optical 96-well plate and induced with 5 μg ml−1 mitomycin C. Bacterial growth was monitored in plate readers (Spectramax M2e and Spectramax 340PC, Molecular Devices, Sunnyvale, CA, USA) and data on culture density (relative absorbance at 600 nm) were acquired every other minute. After induction has completed, lysates were diluted 1/10 in SMG, treated with 25 μl chloroform, centrifuged at 5000 g for 10 min and stored at 4 °C.
Quantification of phage productivity
Productivity as measured here describes the average number of phages that are released per cell present at the time mitomycin C was added to the culture. It thus integrates over the whole time span during which prophage induction occurs and allows a comparison between single and double lysogens. The titer of free phages in lysates was quantified by spotting a dilution series on double agar plates (Carlson, 2005). Two different phages in a sample were quantified separately by spotting the sample twice on host bacteria with different resistances. These were either knockout strains that lacked a specific receptor (Baba et al., 2006), strains that were transformed with plasmids carrying the repressor gene of a specific phage (see Supplementary Material), or lysogens of a specific phage (Table 2). The latter could not be used in all cases, because certain phages plate inefficiently on some lysogens, which impedes accurate quantification. For the same reason, not all double lysogens were fully analyzed (Figure 2). To obtain an estimate of the average number of phages released per cell, titer data were divided by the average number of bacterial cells present in the culture before induction. This number was estimated once for E. coli MG1655 in 24 independent replicates and used for all lysogens.
Table 2. Bacterial strains and plasmids that were used in this study.
| Bacterial strain/plasmid | Relevant genotype | Phage resistance | Source |
|---|---|---|---|
| MG1655 | — | None | — |
| JW0146 | ΔfhuA766::kan | HK022, Φ80, mEp043, mEp213, mEp234, mEp235, mEpX1, mEpX2, mEp506 | Keio collection (Baba et al. 2006) |
| JW5195 | ΔtonB760::kan | Φ80, mEp043, mEp213 | Keio collection (Baba et al. 2006) |
| JW3996 | ΔlamB732::kan | λ | Keio collection (Baba et al. 2006) |
| JW2203 | ΔompC768::kan | mEp332 | Keio collection (Baba et al. 2006) |
| pλR | pGEM-T Easy vector carrying cI and OR of λ | λ, mEp234, mEp332 | Own construction Promega, Madison, WI, USA |
| pHK022R | As above, but carries cI of HK022 | HK022 | Own construction |
| pΦ80R | As above, but carries cI and OR of Φ80 | Φ80 | Own construction |
Estimation of lysis time
The onset of lysis causes a distinct clearance of the bacterial culture and the time at which turbidity of the culture peaks provides an estimate of lysis time. To this end, R statistical software v2.11.1 (R Development Core Team, 2010) was used to fit a spline to the growth curve that was measured in the plate reader (Supplementary Figure S1, R script available on request).
Data on productivity and lysis time were analyzed using R statistical software. Data on single lysogens were analyzed with one-way analysis of variance and differences between levels were explored with Tukey's range test. Data on double lysogens were analyzed with a two-way analysis of variance (including the factors ‘phage 1' (11 levels) and ‘phage 2' (12 levels, including no phage)). Further comparisons were performed with non-parametric tests (Mann–Whitney U test, Kendall's τ), and where estimates of slopes were required, linear regressions were used.
Results
Construction of single and double lysogens
Using 11 different lambdoid phages, I constructed 44 double lysogens that contained unique combinations of two different phages. In addition, 35 double lysogens were obtained, in which the order of insertion of the phages was reversed. Three causes prevented the construction of 42 double lysogens: (a) the superinfecting phage belonged to the same immunity group as the prophage, (b) the prophage conferred specific resistance to superinfection by a heteroimmune phage, and (c) superinfection caused substitution of the resident prophage (Figure 2).
Order of prophage insertion
I tested whether productivity of a phage depended on the order of insertion into a double lysogen. To this end I correlated data on productivity and lysis time from those double lysogens, which were constructed using two different orders of insertion. Linear regressions revealed good linear fits with slopes close to one for both productivity and lysis time (Supplementary Figure S2; productivity: d.f.=60, r2=0.93, slope=0.97, P<0.0001; lysis time: d.f.=31, r2=0.82, slope=0.91, P<0.0001). λ/mEp213 double lysogens lysed incompletely causing estimates of productivity and lysis time to be highly variable. They were excluded from the regression but kept in the dataset.
Neither productivity nor lysis time depended on the order by which phages were inserted into the bacterial genome. In all likelihood these double lysogens are therefore identical. To avoid pseudoreplication, their data were combined in all subsequent analyses, leaving 44 different double lysogens. Not all lysogens were fully analyzed (Figure 2).
Prophage induction in single lysogens
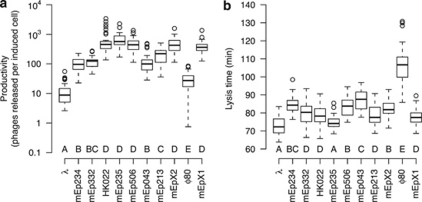
Single lysogens of different phages differed significantly both in their productivity, that is, the amount of phages that were released on average from an induced cell (Figure 3a, F10,440=147.4, P<0.0001), and in their lysis time (Figure 3b, F10,858=204.3, P<0.0001). Across phages, productivity and lysis time did not correlate significantly (Kendall's τ=−0.31, d.f.=10, P=0.22): among different phages, a longer lysis time is not associated with a higher productivity.
Figure 3.
Phage productivity (a) and lysis time (b) in single lysogens. Box plots summarizing baseline data of all 11 phages that were used in this study. Lysogenic cultures of E. coli MG1655 carrying a single prophage were induced with mitomycin C and turbidity was monitored to estimate lysis time. Productivity of different phages was measured as the average number of phages released per cell that was induced. Analysis of variance found significant variation among phages in both data sets. Letters indicate groups of phages that were found to differ significantly according to Tukey's range test.
These data obtained from single lysogens are referred to as baseline productivity and baseline lysis time, respectively, and describe the performance of a phage in the absence of competition. In later analyses, I will compare the performance of phages in double lysogens against their baseline performance.
Prophage induction in double lysogens
Productivity
Most phages failed to maintain their baseline productivity in a double lysogen (median=16% of baseline productivity; Mann–Whitney U test, n=81, U=−1482.5, P<0.0001). The magnitude of this loss varied among lysogens (Supplementary Figure S3) and analysis of variance confirmed that (a) phages differed in their response towards competition (effect size η2 (proportion of total variance explained)=0.52, F10,1655=1422, P<0.0001) and (b) they differed in the effect they had on a competitor (η2=0.21, F11,1655=537, P<0.0001). In addition, a significant interaction indicated that the outcome of competition could not always be predicted by the main effects of the phages alone but depended on their specific combination (η2=0.21, F70,1655=83, P<0.0001). These results are addressed in more detail below. To allow for comparisons among phages, their productivities were normalized and expressed relative to their baseline productivity.
The significant main effects observed above indicated that phages could be described by their competitive abilities. These were summarized in two traits, namely, the change in productivity that a phage experiences in the presence of competitors (‘competitive response') and the effect of the phage on the productivity of its competitors (‘competitive effect'). Both traits were calculated as the geometric mean of the relative productivity in all double lysogens of a particular phage. The traits were found to correlate (n=11, Kendall's τ=−0.67, P=0.003). Phages that responded little to competition exerted a strong effect on others and vice versa (Figure 4a). The results indicated that phages could be ranked according to their competitive ability. Phage λ stood out, as it responded sensitively to competition, yet also affected its competitors. This observation will be addressed later.
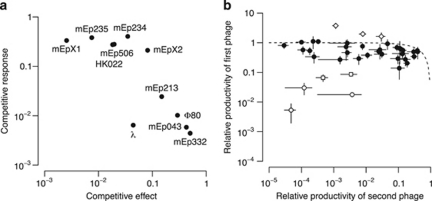
Figure 4.
Within-host competition between lambdoid phages. (a) Parameters describing the competitive ability of each phage. Competitive effect measures the effect of a phage on the productivity of its competitors; higher values indicate less effect. Competitive response measures the response of a phage to competition; lower values indicate a sensitive response. (b) Relative productivity of phages in double lysogens. Every data point corresponds to a unique pair of phages. To order data points and facilitate their comparison, phages with higher relative productivity in a pair were plotted on the y-axis. Error bars show the 95% confidence interval. The dotted line shows the function y=1−x. Points close to the line achieve a combined relative productivity of approximately one. Open symbols are lysogens where phages strongly interfere as defined by a combined relative productivity <0.1 (circles: λ/HK022, λ/mEp213, λ/mEpX1; squares: mEp234/Φ80, mEp506/Φ80), or where a phage in a double lysogen achieves a productivity that is significantly above its baseline productivity (diamonds: λ/mEpX2, mEp043/mEp234, mEp213/mEp234).
The relation between the relative productivities of two phages in the same lysogen is shown in Figure 4b. There was not a single case in which both phages maintained their productivity at the baseline level. Overall, the combined relative productivities within a double lysogen do not sum up to one (median=0.56; Mann–Whitney U test, n=44, U=−288, P<0.0001), which indicates that the loss in productivity cannot be explained by a sharing of resources alone. In the majority of cases, however, combined relative productivities were close to one and phages either suffered symmetrically from competition, or one of the phages was disproportionally affected (closed symbols in Figure 4b). In a minority of cases patterns deviated (open symbols in Figure 4b) and a phage benefited from the presence of a second phage (this was phage mEp234 in two of three cases) or both phages suffered high losses (as defined by a combined relative productivity <0.1). Phages λ or Φ80 were involved in all of the latter cases.
To summarize, analysis of competitive interactions that unfold upon prophage induction in double lysogens presented the following pattern: phage productivity changes substantially compared with single lysogens and at least one phage suffers a decrease in productivity. The magnitude of this loss is determined by both phages and allows describing their competitive abilities by competitive response and effect. These two traits are correlated and suggest the existence of a hierarchy of competitive abilities. The outcome of competition is further influenced by an interaction term that depends on the specific combination of two phages. In some cases, phages mutually interfere or one of the phages gains a net benefit.
Lysis time
Lysis time varied substantially among double lysogens (Supplementary Figure S4) and followed a pattern that was qualitatively similar to the one found for productivity: analysis of variance revealed significant effects of both phages (effect size η2 (proportion of total variance explained)=0.14, F10,2996=109.3, P<0.0001; η2=0.13, F11,2996=93.6, P<0.0001) and a significant interaction term (η2=0.37, F71,2996=41.8, P<0.0001). Lysis time in a double lysogen depended on both participating phages, and was further influenced by a combination-specific factor.
If both phages had operated independently during prophage induction, one would expect lysis time to be determined by the faster phage. Our data did not contradict this hypothesis (Figure 5a, Mann–Whitney U test, n=40, U=14.5, P=0.85, median difference=0.1 min) but did clearly reject a null hypothesis that lysis time in a double lysogen will be the average of the lysis times of the individual phages (Mann–Whitney U test, n=40, U=−312.5, P<0.0001, median difference=−4.3 min). The results remained qualitatively similar once phage Φ80 was removed from the dataset.
Figure 5.
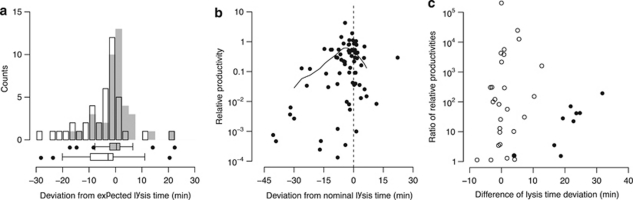
Shifts in lysis time and relation to productivity and competitive success. (a) Difference of lysis time of double lysogens from expectation under two hypotheses: (i) lysis time is determined by the faster phage in a double lysogen (gray bars), and (ii) lysis time is close to the average of the baseline lysis times of both phages in a double lysogen (open bars). (b) Phage relative productivity in relation to deviation from baseline lysis time. The solid curve is a spline that was fitted to the data. The dashed line indicates no deviation. (c) Competitive success of a phage in a double lysogen plotted against its deviation from baseline lysis time relative to that of its competitor (positive values indicate its baseline lysis time is closer than the competitor's). Closed circles are double lysogens with Φ80; open circles are all other double lysogens.
Relationship between productivity and lysis time
Across all double lysogens that were tested, productivity of a phage declined the further the lysis time of a double lysogen deviated from the baseline lysis time (Figure 5b). Beyond a difference of a few minutes, this decline became substantial. There was no evidence that the direction of the deviation mattered, but the data covered mainly lysis times shorter than baseline lysis times.
On the basis of the preceding observations (earlier lysis time dominates in double lysogens, productivity declines with deviation from baseline lysis time), I asked whether the relative deviation of two phages from their baseline lysis time was associated with a competitive advantage of the phage whose baseline lysis time was closer to the lysis time of the double lysogen. To test this, I calculated two values for all double lysogens: the ratio of the relative productivities of both phages and the difference of their deviations from lysis time (Figure 5c). These two values did not significantly correlate when all phages were included (Kendall's τ=0.02, n=37, P=0.87), but did so for double lysogens that contained phage Φ80 (Kendall's τ=0.67, n=9, P=0.013). This phage has a particularly long baseline lysis time (Figure 3b) and hence might be the only case where shifts in lysis time were substantial enough to affect productivity.
Cases of mutual interference
When both phages suffered a severe reduction in productivity, lysis was often observed to begin very early or proceed slowly. Double lysogens that included Φ80 had often lysis times below the baseline lysis time of both phages (Supplementary Figure S4). The two largest drops (14.0 min and 8.3 min below the shorter lysis time) occurred in those double lysogens, in which both phages also suffered strong reductions in reproductive output (open squares in Figure 4b). Double lysogens of λ together with HK022, mEp213 and mEpX1 showed a strong reduction in productivity of both phages (open circles in Figure 4b). This phenomenon was due to incomplete lysis of the bacterial culture. Although both phages lysed their host efficiently as single lysogens, lysis began very early in double lysogens but then proceeded slowly (Supplementary Figure S5).
Discussion
Patterns of competitive interactions among lambdoid phages
Multiple infections are predicted to be important determinants of infectious disease dynamics and pathogen evolution. The genetic and physiological factors that determine the outcome of within-host competition are therefore of critical importance, yet they remain poorly understood. Empirical studies are typically limited to small numbers of parasite genotypes, and it is unclear whether the observed results can be generalized. I have studied within-host competition among 11 temperate phages of E. coli, which has allowed me to make inferences about the general rules that govern these interactions. Multiple infections in temperate phages have received some attention and the fitness consequences for individual phages have been addressed (Boyd and Bidwell, 1962; Serra-Moreno et al., 2008), but never in the context of prophage induction. Recently, it has been shown that prophage induction can occur in response to changes in bacterial density and it has been proposed that it is more than an adaptation to escape from a cell bound to die (Ghosh et al., 2009). The ability of prophages to respond to changes in transmission opportunities emphasizes the importance of prophage induction in the life cycle of temperate phages.
The results herein emphasize the importance of multiple infections for natural phage populations, as they clearly show that lytic productivity of prophages declines, often substantially, when a host is shared between phages. Within-host competitive ability varied considerably among phages, which caused competition outcomes to be often asymmetric. Because resources in a bacterial cell are limited, exploitation competition is bound to occur. This was supported by the observation that the sum of relative phage productivities in double lysogens did rarely exceed, and was often closed to one (Figure 4b). This upper limit was frequently not reached, however, which suggests that a second type of competition was operating, interference competition, which caused a net reduction of total productivity. The reason for this interference remains to be elucidated, but it is conceivable that crosstalk between two simultaneously executed lytic programs within the same cell may hinder efficient resource depletion.
It will be important to show whether the estimates obtained here can predict total phage fitness. Prophage induction covers only one part of the phage life cycle. A second key step is the infection of a cell, and it has been shown here and elsewhere (Boyd and Bidwell, 1962; Serra-Moreno et al., 2008) that the order of arrival is of critical importance.
Competition has an important role in structuring communities. In this study, I found that the outcome of competition was explained to a large degree (73% of the variance of productivity) by main effects, and hence depended mainly on the characteristics of the participating phages and less on their specific combination (Goldberg, 1996). This allowed me to describe phages by their individual competitive abilities, which are competitive effect (the ability to suppress competitors) and competitive response (resistance of a phage toward suppression), two measures that are correlated.
The observation that phages can be ranked according to their within-host competitive abilities raises the question whether this ranking extends to their complete life cycle, and therefore compromises genetic diversity in phage populations if competitively superior phages dominate. I suggest three possibilities that can explain phage coexistence. First, I observed that phage productivity did not only depend on main effects, but to a smaller extent also on the specific combination of the participating phages (21% of the variance of productivity). This may cause competitive abilities to be partly intransitive, and thus prevent the fixation of a superior competitor (Kerr et al., 2002). Second, natural environments are more variable than the assay conditions of this study. For example, it is known that lambdoid prophages vary both in their sensitivity toward an inducing signal and in their response towards different signals (Łoś et al., 2009; Refardt and Rainey 2010). Finally, it is also conceivable that competitive ability depends on the host genotype (de Roode et al., 2004).
Competition and the evolution of lysis time
In double lysogens of lambdoid phages, the faster phage dictated lysis time. This finding differs from a previous study, where no consistent directional shift was found when λS holin alleles conferring different lysis times were coexpressed together with the λS holin wild-type allele (Raab et al., 1988). It may be that dominance of a shorter lysis time requires differences beyond the holin gene itself. Evidence that the observed shift was associated with a competitive advantage of phages with a short baseline lysis time was weak however. Although productivity declined with deviations from baseline lysis time, this did not translate into a clear competitive benefit for phages with a short lysis time. Most shifts in lysis time were in the order of minutes (lower quartile −8.1 min, upper quartile 0.7 min), and because phages accumulate linearly over time in a cell (Wang, 2006), this scale is too small to explain the observed changes in productivity over orders of magnitude. Only Φ80, which has a comparatively long lysis time, exhibits a decline in productivity that appears at least partially to be caused by a shortening of its lysis time (Inokuchi and Ozeki, 1970). It is interesting to speculate whether the lysis times of the phages in this study have already been shortened by natural selection in response to within-host competition and whether the remaining differences are therefore too small to confer a measurable competitive advantage.
If the timing of cell lysis of phage λ is extended beyond its normal lysis time, burst size can be more than doubled, yet comes at the cost of a longer generation time. Because generation time and burst size both contribute to fitness, it has been proposed that an intermediate lysis time optimizes phage fitness (Wang, 2006). At the same time, it has been noted that this optimization might fail in the presence of competing phages. My data confirm that multiple infections indeed can thwart the attempt of a phage to optimize lysis time. In addition, there is tentative evidence that competition can select phages with a shorter lysis time, yet it is also evident that lysis time is not the sole determinant of competitive success.
I therefore propose that competitive success largely depends on the ability to monopolize cellular resources. A phage that dominates intracellular processes is competitively superior. Deviations from baseline lysis time then reflect the degree by which the developmental program of a phage is distorted and thus indicate competitive inferiority. This would explain why productivity of a phage decreased both when the lysis time of a double lysogen was shorter or longer than its baseline lysis time.
Mutual interference and parasitism
While the majority of competitions followed a pattern, in which at least one of the competitors maintained productivity close to the baseline level, a number of phage pairs (all of which involved either phage λ or Φ80) appeared to strongly mutually interfere with each other's lytic development, which caused a substantial loss of productivity for both phages. In the case of λ, this corroborates similar phenomena involving lambdoid and T-even phages, which are mediated by the Rex system of λ (Benzer, 1955; Toothman and Herskowitz, 1980; Parma et al., 1992). Such strong interference competition (spite), in which the harm inflicted to a competitor comes at a considerable cost to oneself, can be favored by kin selection if it benefits a third, closely related party (West and Gardner, 2010). This reasoning has been put forward to explain the maintenance of bacteriocin production in bacteria, in which toxin release is lethal to the producer (Inglis et al., 2009). In this study, it may be advantageous because it prevents the invasion of foreign phages into a population of λ lysogens (Parma et al., 1992).
In three double lysogens, one phage increased its productivity above the baseline level, and productivity of the second phage was reduced. The underlying mechanism of this parasitic interaction is not known, yet it is interesting to note that mEp234 benefits from coinfection with mEp043 and mEp213, two phages with weak competitive effect. mEp234 has a low lytic efficiency as single lysogen (D Refardt, unpublished results) and while it may dominate competition intracellularly, it must rely on the lytic action of its competitor to finally release its virions.
Conclusion
Lytic productivity of lambdoid prophages was strongly altered in the presence of competitors. In several cases, their productivity was nearly nullified. Together with the emerging evidence that polylysogeny is a common phenomenon in natural bacterial populations, my study suggests that multiple infections must be taken into account if phage population dynamics are to be understood.
Acknowledgments
I thank I Wurmitzer for help in the laboratory, J Engelstädter, O Silander and T Stadler for help with perl and R scripts, S Alizon, TF Cooper, Y Ulrich and two anonymous reviewers for constructive comments on the manuscript, ME Gottesman, JW Little and I-N Wang for sharing phages, and S Bonhoeffer for all his support. This study is part of the project ‘Genetic Diversity of Hosts and Parasites' (grant no. 0-21248-08), funded by the Competence Center Environment and Sustainability of the ETH (http://www.cces.ethz.ch/).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards R, Carlson C, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage λ. J Mol Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, et al. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. 2006Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection Mol Syst Biol 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S. Fine structure of a genetic region in bacteriophage. Proc Natl Acad Sci USA. 1955;41:344–354. doi: 10.1073/pnas.41.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berngruber TW, Weissing FJ, Gandon S. Superinfection inhibition and the evolution of viral latency. J Virol. 2010;84:10200–10208. doi: 10.1128/JVI.00865-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JSK, Bidwell DE. Superinfection of lysogenic strains of Salmonella typhimurium Q1: prophage substitution and double lysogenization. J Gen Microbiol. 1962;29:659–686. doi: 10.1099/00221287-29-4-659. [DOI] [PubMed] [Google Scholar]
- Brown SP, Hochberg ME, Grenfell BT. Does multiple infection select for raised virulence. Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. [DOI] [PubMed] [Google Scholar]
- Campbell A. Prophage insertion sites. Res Microbiol. 2003;154:277–282. doi: 10.1016/S0923-2508(03)00071-8. [DOI] [PubMed] [Google Scholar]
- Campbell A.2006General aspects of lysogenyIn: Calendar RL (ed).The Bacteriophages2nd ednOxford University Press: New York, NY; 66–73. [Google Scholar]
- Carlson K.2005Working with bacteriophages: common techniques and methodological approachesIn: Kutter E, Sulakvelidze A (eds).Bacteriophages: Biology and applications CRC Press: Boca Raton, FL; 429–484. [Google Scholar]
- Cox FEG. Concomitant infections, parasites and immune responses. Parasitology. 2001;122:S23–S38. doi: 10.1017/s003118200001698x. [DOI] [PubMed] [Google Scholar]
- de Roode JC, Culleton R, Cheesman SJ, Carter R, Read AF. Host heterogeneity is a determinant of competitive exclusion or coexistence in genetically diverse malaria infections. Proc R Soc B. 2004;271:1073–1080. doi: 10.1098/rspb.2004.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshelman CM, Vouk R, Stewart JL, Halsne E, Lindsey HA, Schneider S, et al. Unrestricted migration favours virulent pathogens in experimental metapopulations: Evolutionary genetics of a rapacious life history. Phil Trans R Soc B. 2010;365:2503–2513. doi: 10.1098/rstb.2010.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeland EM, Lipp EK, Huq A, Colwell RR. Polylysogeny and prophage induction by secondary infection in Vibrio cholerae. Environ Microbiol. 2004;6:760–763. doi: 10.1111/j.1462-2920.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Quart Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Roy K, Williamson KE, Srinivasiah S, Wommack K, Radosevich M. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl Environ Microbiol. 2009;75:7142–7152. doi: 10.1128/AEM.00950-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg DE. Competitive ability: definitions, contingency and correlated traits. Phil Trans R Soc B. 1996;351:1377–1385. [Google Scholar]
- Hamilton WD. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Hochberg ME. Establishing genetic correlations involving parasite virulence. Evolution. 1998;52:1865–1868. doi: 10.1111/j.1558-5646.1998.tb02266.x. [DOI] [PubMed] [Google Scholar]
- Inglis RF, Gardner A, Cornelis P, Buckling A. Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2009;106:5703–5707. doi: 10.1073/pnas.0810850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi H, Ozeki H. Phenotypic mixing between bacteriophage Φ80 and λ in vitro and in vivo. Virology. 1970;41:701–710. doi: 10.1016/0042-6822(70)90434-4. [DOI] [PubMed] [Google Scholar]
- Kameyama L, Fernandez L, Calderon J, Ortiz-Rojas A, Patterson TA. Characterization of wild lambdoid bacteriophages: Detection of a wide distribution of phage immunity groups and identification of a Nus-dependent, nonlambdoid phage group. Virology. 1999;263:100–111. doi: 10.1006/viro.1999.9888. [DOI] [PubMed] [Google Scholar]
- Kerr B, Riley M, Feldman M, Bohannan B. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Leitet C, Riemann L, Hagström Å. Plasmids and prophages in Baltic Sea bacterioplankton isolates. J Mar Biol Assoc UK. 2006;86:567–575. [Google Scholar]
- Lewontin RC. The units of selection. Annu Rev Ecol Syst. 1970;1:1–18. [Google Scholar]
- Łoś JM, Łoś M, Węgrzyn G, Węgrzyn A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb Pathog. 2009;47:289–298. doi: 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Lwoff A. Lysogeny. Bacteriol Rev. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Ichikawa N, Tanaka S, Morita T, Matsuhiro A. Molecular cloning of Φ80 adsorption-inhibiting cor gene. Jpn J Genet. 1985;60:475–483. [Google Scholar]
- May RM, Nowak MA. Coinfection and the evolution of parasite virulence. Proc R Soc B. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- Miller RV, Day MJ.2008Contribution of lysogeny, pseudolysogeny, and starvation to phage ecologyIn: Abedon ST (ed).Bacteriophage Ecology: Population Growth, Evolution and Impact of Bacterial Viruses Cambridge University Press: Cambridge, UK; 114–143. [Google Scholar]
- Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc R Soc B. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- Oberto J, Weisberg RA, Gottesman ME. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989;207:675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- Parma DH, Snyder M, Sobolevski S, Nawroz M, Brody E, Gold L. The Rex system of bacteriophage λ: tolerance and altruistic cell death. Genes Dev. 1992;6:497–510. doi: 10.1101/gad.6.3.497. [DOI] [PubMed] [Google Scholar]
- Ptashne M. A Genetic Switch: Phage Lambda Revisited. Third. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, NY; 2004. [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing: Vienna, Austria; 2010. R: A language and environment for statistical computing. [Google Scholar]
- Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence of translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- Refardt D, Rainey PB. Tuning a genetic switch: experimental evolution and natural variation of prophage induction. Evolution. 2010;64:1086–1097. doi: 10.1111/j.1558-5646.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- Reynolds RB, Reddy A, Thorne CB. Five unique temperate phages from a polylysogenic strain of Bacillus thuringiensis subsp. aizawai. J Gen Microbiol. 1988;134:1577–1585. doi: 10.1099/00221287-134-6-1577. [DOI] [PubMed] [Google Scholar]
- Serra-Moreno R, Jofre J, Muniesa M. The CI repressors of Shiga toxin-converting prophages are involved in coinfection of Escherichia coli strains, which causes a down regulation in the production of Shiga toxin 2. J Bacteriol. 2008;190:4722–4735. doi: 10.1128/JB.00069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkilnyj P, Koudelka GB. Effect of salt shock on stability of λimm434 lysogens. J Bacteriol. 2007;189:3115–3123. doi: 10.1128/JB.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Thompson JN. The Coevolutionary Process. University of Chicago Press: Chicago, IL; 1994. [Google Scholar]
- Toothman P, Herskowitz I. Rex-dependent exclusion of lambdoid phages. I. Prophage requirements for exclusion. Virology. 1980;102:133–146. doi: 10.1016/0042-6822(80)90076-8. [DOI] [PubMed] [Google Scholar]
- Wang I. Lysis timing and bacteriophage fitness. Genetics. 2006;172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Gardner A. Altruism, spite, and greenbeards. Science. 2010;327:1341–1344. doi: 10.1126/science.1178332. [DOI] [PubMed] [Google Scholar]
- Williamson KE, Schnitker J, Radosevich M, Smith D, Wommack K. Cultivation-based assessment of lysogeny among soil bacteria. Microbial Ecol. 2008;56:437–447. doi: 10.1007/s00248-008-9362-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.