Abstract
Herbivorous reptiles depend on complex gut microbial communities to effectively degrade dietary polysaccharides. The composition of these fermentative communities may vary based on dietary differences. To explore the role of diet in shaping gut microbial communities, we evaluated the fecal samples from two related host species—the algae-consuming marine iguana (Amblyrhynchus cristatus) and land iguanas (LI) (genus Conolophus) that consume terrestrial vegetation. Marine and LI fecal samples were collected from different islands in the Galápagos archipelago. High-throughput 16S rRNA-based pyrosequencing was used to provide a comparative analysis of fecal microbial diversity. At the phylum level, the fecal microbial community in iguanas was predominated by Firmicutes (69.5±7.9%) and Bacteroidetes (6.2±2.8%), as well as unclassified Bacteria (20.6±8.6%), suggesting that a large portion of iguana fecal microbiota is novel and could be involved in currently unknown functions. Host species differed in the abundance of specific bacterial groups. Bacteroides spp., Lachnospiraceae and Clostridiaceae were significantly more abundant in the marine iguanas (MI) (P-value>1E−9). In contrast, Ruminococcaceae were present at >5-fold higher abundance in the LI than MI (P-value>6E−14). Archaea were only detected in the LI. The number of operational taxonomic units (OTUs) in the LI (356–896 OTUs) was >2-fold higher than in the MI (112–567 OTUs), and this increase in OTU diversity could be related to the complexity of the resident bacterial population and their gene repertoire required to breakdown the recalcitrant polysaccharides prevalent in terrestrial plants. Our findings suggest that dietary differences contribute to gut microbial community differentiation in herbivorous lizards. Most importantly, this study provides a better understanding of the microbial diversity in the iguana gut; therefore facilitating future efforts to discover novel bacterial-associated enzymes that can effectively breakdown a wide variety of complex polysaccharides.
Keywords: Galápagos iguanas, fecal microbiota, 16S rRNA-based pyrosequencing, dietary differences
Introduction
Herbivory involves subsistence on the fibrous parts of plants, for example, leaves, stems and buds (Parra, 1978) and the subsequent breakdown of the complex carbohydrates (that is, celluloses and hemicelluloses) that are present in plant cell walls. Presently, 118 families of glycoside hydrolases have been identified that assist herbivores with the hydrolysis and degradation of plant cell walls (http://www.cazy.org/Glycoside-Hydrolases.html). Vertebrate hosts lack an extensive diversity of these endogenous glycoside hydrolases, and therefore rely on their endosymbiotic relationship with the gut microbiota to effectively hydrolyze complex carbohydrates or polysaccharides (Davison and Blaxter, 2005; Mackie et al., 2008). For example, in mammalian systems, hindgut and subsequently foregut fermentation has evolved in herbivorous hosts (Hume and Warner, 1980; Mackie et al., 2008). These adaptations were probably a result of enhancing the efficiency and roles of bacterial fermentation in the gut. This in turn provides an intuitive indication of the importance of bacterial community in breaking down indigestible polysaccharides and its subsequent provision of nutrients and energy to the host.
Compared with mammals, herbivory in extant reptilian hosts has not been commonly observed. It is estimated that 80% of mammalian species are herbivorous (Stevens and Hume, 1995), whereas only less than 2% of reptiles of >7800 recognized species of squamate reptiles follow the herbivorous dietary regimen (Espinoza et al., 2004). In general, reptilian herbivory has been observed in a few turtles, 40 species of tortoises (King, 1996; Stevens and Hume, 1998) and about 50 species of lizards belonging to the infraorder Iguania (Pough, 1973; Troyer, 1983). The paucity of herbivorous reptiles has been linked to ectothermy and the limitations of small body size, which does not allow maximal gut capacity to digest the plant materials (Mackie et al., 2004). However, recent studies have shown that reptiles have also evolved over time to adapt to herbivory, for example, by having slower metabolic rates that facilitate increased gut transit time and by behavioral adaptations that maintain body temperature comparable to that of a mammalian host (Zimmerman and Tracy, 1989; King, 1996). Similar to the mammalian system, these adaptations possibly occurred to facilitate the role of gut microbiota in breaking down recalcitrant dietary polysaccharides. To illustrate, herbivorous iguanas can digest 54% of cell wall constituents and supply 30%–40% of their energy needs from hindgut fermentation (McBee and McBee, 1982), implicating a symbiotic gut microbiota capable of effectively degrading complex polysaccharides. However, unlike the mammalian gut microbiota, very few studies have been carried out to provide insights on the phylogenetic diversity present in the herbivorous lizard gut microbiota.
Among herbivorous lizards, the marine iguanas (MI) (Amblyrynchus cristatus) and land iguanas (LI) (Conolophus subscristatus and C. pallidus) on the Galápagos Islands constitute one of the most unique pairs of Iguanidae to study and compare their gut microbiota. The common ancestor for both iguanas, the Ctenosaur, was thought to have drifted on rafts of vegetation from Central or South America to the Galápagos Islands (Cogger and Zweifel, 1998), and subsequently differentiated into two separate genera 10–20 million years ago (Rassmann, 1997) before the age of the current islands. Marine iguanas are the only iguanas that have adapted to feed primarily on soft macrophytic algae growing in the intertidal and subtidal zones (Shepherd and Hawkes, 2005). In contrast, LI feed on terrestrial flora like Opuntia cactus pads, lantana flowers and cordia plants (Christian et al., 1984), and have a qualitatively more similar diet to Galápagos giant tortoises (GT) (Geochelone nigra) than to MI. This interesting difference in the iguanas' diet was also observed by Darwin during his visit to the islands in 1835, where he wrote in his diary—‘I opened the stomach of several (marine iguanas), and in each case found it largely distended with minced seaweed of that kind which grows in thin foliaceous expansions of a bright-green or dull-red color…I opened the stomach of several (land iguanas), and found them full of vegetable fibres and leaves of different trees, especially of a species of acacia.' (Darwin, 1835).
For most dietary regimens, the acquisition of a new diet is a fundamental driver for the evolution of new species and hence differentiation in the gut microbiota (Ley et al., 2008a). Given the vastly different dietary intake of the marine and LI, it is therefore hypothesized that both iguanas would have distinct differences in their gut microbiota. To address this hypothesis, we sampled feces from a total of 47 MI and LI endemic to Galápagos Islands, as well as two outgroups, GT and green iguanas (GI) (Iguana iguana). Genomic DNA was extracted from these samples for high-throughput 16S rRNA-based pyrosequencing to provide a comprehensive examination of the microbial diversity found in the fecal surrogates. Comparative analyses were subsequently carried out to evaluate the extent of similarity and dissimilarity between the microbial diversity of the herbivorous lizards. This study serves to provide insight on phylogenetic composition and abundances of gut microbiota in herbivorous lizards and to examine the microbial diversity in lizards with different dietary preferences.
Materials and methods
Sampling site and iguana characteristics
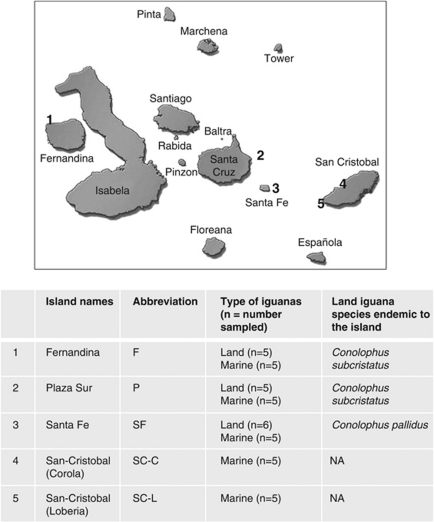
A field trip was conducted to the Galápagos archipelago with sampling trips by boat to the islands of Fernandina (latitude −0.30273, longitude −91.6436), Plaza Sur (latitude −0.58408, longitude −90.1094), Santa Fe (latitude −0.79986, longitude −90.0875) and two locations on San Cristobal (San Cristobal-Carola: latitude −0.89122, longitude −89.6121; San Cristobal-Loberia: latitude −0.92708, longitude −89.6122) during August-September 2009 (Figure 1). The sampling period coincided with the cool and dry season, characterized by relatively cold waters (18–20 °C) generated by the Humboldt Current that are nutrient-rich (Constant, 2000). Marine iguanas (A. cristatus) were found on the coastal margins of all sampling sites, whereas LI (C. subcristatus and C. pallidus) were found on Fernandina, Plaza Sur and Santa Fe, but not San Cristobal. It is well documented that MI typically gather in large groups, dive underwater to forage for sea algae, and bask under the sun to thermoregulate their body temperature (Boersma, 1982; Trillmich and Trillmich, 1986; Wikelski and Trillmich, 1994). In contrast, the LI are typically solitary (Snell et al., 1984), maintain a more constant body temperature under the sun, and consume mainly the cactus pads found in the arid littoral zone (Christian et al., 1984).
Figure 1.
Sampling sites and collection details. Fecal samples from LI and MI were collected in Fernandina, Plaza Sur, Santa Fe and San–Cristobal of Galápagos Islands. NA denotes that LI are no longer extant on San–Cristobal.
Fecal sampling from outgroup reptilian herbivores
Another group of iguanidae herbivores, GI (Iguana iguana) from El Salvador, Central America was selected as one outgroup from a different geographical location. A second outgroup, Galápagos GT (Geochelone nigra) residing in San Cristobal was also chosen as an outgroup for comparison with marine and LI based on the same geographical location but different order of reptilian herbivore. Fecal samples were obtained from GI (n=2) imported from El Salvador and also from Galápagos GT residing in San Cristobal–Galapaguera (n=4).
Fecal sampling
A total of 47 fecal samples were collected based on sampling procedure described previously (Mackie et al., 2004). In brief, LI were captured by noose and stimulated around the anal vent to elicit defecation. Fresh feces from all other reptiles were collected at the time of defecation by scooping the feces into a sterile 15-ml centrifuge tube. Fecal samples were stored at 4 °C during the entire field trip, and then at −20 °C in laboratory before DNA extraction. All procedures were non-invasive and conducted in accordance with guidelines from the American Society of Icthyologists and Herpetologists, approved by the Charles Darwin Research Station and Galápagos National Park (PNG Autorization de Proyecto PC-21-06 Ext 01-09). Field work and laboratory research was covered under University of Illinois IACUC #09041. Iguana fecal samples were exported to the University of Illinois with permission from the Galápagos National Park under CITES Permit No. 007-09/PNG.
DNA extraction
Genomic DNA from feces was extracted using the UltraClean Soil DNA Isolation kit (MoBio, Carlsbad, CA, USA). Slight modifications to the manufacturer's protocol were made to ensure a representative extraction of bacterial and archaea populations present in the samples. In brief, lysozyme and achromopeptidase were added to the extraction buffer, and the sample mixture was incubated at 37 °C for 1 h before DNA extraction. Concentration of genomic DNA was then measured with Qubit fluorometer (Invitrogen, Carlsbad, CA, USA).
Barcoded PCR and 454 pyrosequencing
Samples for 454 FLX pyrosequencing were amplified with universal forward 519F (5′-Fusion A-Barcode -CAGCMGCCGCGGTAATWC-3′) and reverse 926R (5′-Fusion B-Barcode- CCGTCAATTCMTTTRAGTT-3′) primer pairs (http://www.roche.com). PCR reaction mixtures comprised 100 ng of genomic DNA, 25 μl of Premix F (Epicentre Biotechnologies, Madison, WI, USA), 200 n (each) of forward and reverse primers, 0.5 U of Ex Taq DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan), and the volume added up to 50 μl with molecular-biology grade water. PCR with 30 cycles of thermal program (denaturation, 95 °C for 30 s; annealing, 55 °C for 45 s; and extension, 72 oC for 60 s) was carried out. All amplicons were gel-excised, concentrated and purified with Wizard DNA purification kit (Promega, Madison, WI, USA). The concentrations were then measured by Qubit fluorometer (Invitrogen).
Pyrotag handling and analysis
454 pyrosequencing was carried out on 454 FLX Titanium (454 Life Sciences—a Roche Company, Branford, CT, USA). The paired-end pyrosequencing services were provided by Roy J Carver Biotechnology Center, University of Illinois. A total of 248 594 16S rRNA sequences (also referred to as 16S pyrotags) were obtained from 454 Titanium pyrosequencing run. The 16S pyrotags were sorted based on their respective barcodes to form a total of 47 pyrotag libraries representing the collected fecal samples. Raw sequence reads were checked for their quality to minimize the effects of random sequencing errors. Briefly, the quality check included the elimination of sequences that did not perfectly match the proximal PCR primer, and that with short sequencing length (<150 nt). All 16S pyrotags were then removed of their primers, barcodes and adaptor sequences, and had an average read length of 370 nt after trimming (Supplementary Table S1). 16S pyrotags identified with reverse orientation were also reverse complemented on RDP Pipeline Initial Process (Cole et al., 2009). Processed pyrotags were then aligned based on RDP Infernal, which allowed secondary structure alignment (Cole et al., 2009). The aligned pyrotags were visually checked with Jalview: http://www.jalview.org/Web_Installers/install.htm, and manual adjustments were performed to improve the alignment whenever necessary.
Taxonomical classification and statistical analysis
RDP Classifier was used for taxonomical assignments of the aligned 16S pyrotags at 95% confidence level (Cole et al., 2009). MEGAN was used to illustrate and compare the relative abundance of an identified taxonomy on a heat map (Huson et al., 2007). Primer-E worksheets that detailed the presence and absence, as well as the percentage abundances of individual bacterial genera were collated, and subsequently performed with multidimensional scaling plot (MDS) on Primer-E software (http://www.primer-e.com/). A Jaccard dissimilarity matrix was calculated using the ‘vegdist' function in the Vegan package in R (https://r-forge.r-project.org/projects/vegan/) on the genus-level community matrix. This matrix was then used to calculate the average community dissimilarity among each host species pair. In addition, RDP Lib Compare was used to estimate the probability of observing abundance difference in a given phylogenetic taxon (Wang et al., 2007). All other statistical comparison of numerical means was carried out by t-test (Microcal Origin, Northampton, MA, USA).
Rarefaction curves
Aligned sequences for each sample were generated with their individual cluster files based on the RDP pyrosequencing pipeline. The cluster files were in turn used to generate rarefaction curves that defined the number of operational taxonomic units (OTUs) defined at 97% similarity level with respect to the total number of pyrotags read (Supplementary Figure S1). Regression analysis was also carried out on Sigma Plot to fit the rarefaction curves into double rectangular hyperbola curve models (Supplementary Table S2). On the basis of the regression curves, the number of OTUs (97% gene similarity) identified based on 6000 pyrotags were noted for comparison of microbial richness.
Results
Microbial community in herbivorous reptiles
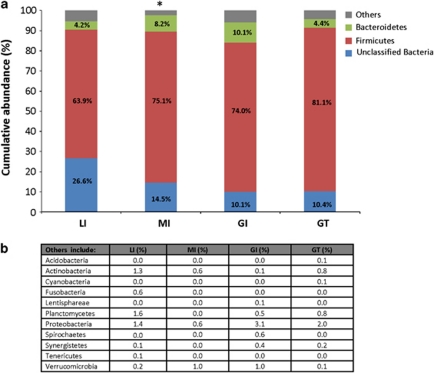
Microbial communities in all examined herbivorous reptiles were dominated by Bacteria at abundances ranging from 99.8 to 100% of the total bacterial community. Archaea were only detected in LI, GI and GT at relatively low abundance, ranging from 0.1 to 0.2% of total microbial community. In contrast, archaeal populations could not be detected at this sequencing depth in feces of MI. Within the bacterial population, Firmicutes were the predominant phylum in the fecal microbiota, and were present with an overall abundance >63.9% of the total microbial community (Figure 2). Besides Firmicutes, unclassified Bacteria also accounted for a moderate proportion of the total microbial community, and were present at 26.6, 14.5, 10.1 and 10.4% in the LI, MI, GI and GT, respectively. All examined reptilian populations had Bacteroidetes in varying abundances, ranging from 4.2 to 10.1% of the total microbial community. The remaining fecal microbiota of the examined reptiles comprised low abundance phyla (<3.1% of total microbial community) like Planctomycetes, Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Spirochaetes, Cyanobacteria and Synergistetes, which were not consistently present in all of the different host populations (Figure 2b).
Figure 2.
Bacterial distribution evaluated at the phylum taxonomical level. (a) The gut microbiota of herbivorous reptiles was predominated by Firmicutes, unclassified Bacteria and Bacteroidetes. (b) The low-abundance phyla were grouped together as ‘others', and their specific abundances were listed. *denotes that Archaea were detected in all herbivorous reptiles, at abundance ranging from 0.1 to 0.2% of total microbial community, except in MI. LI, land iguanas; MI, marine iguanas; GI, green iguanas; GT, giant tortoises.
Clustering differences in bacterial lineages and their abundances
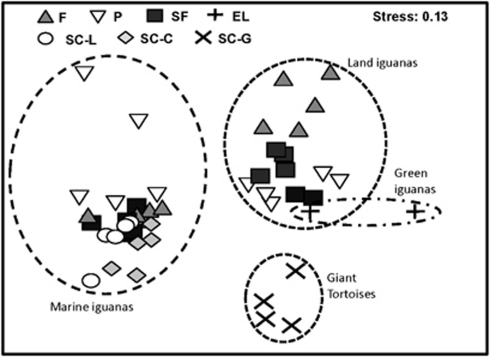
In Figure 3, the occurrence and relative abundance of bacterial lineages detected in the reptilian hosts were evaluated for their clustering differences. On the basis of the multidimensional scaling plot (MDS), the MI, LI and GT populations clustered separately, whereas the LI and GI populations were spatially clustered together (Stress=0.13). A further statistical analysis of the Jaccard dissimilarities validated that the gut microbiota is significantly different among the host species (Supplementary Figure S2, Permutation Manova, F=13.64, P-value <0.001). In particular, the seaweed-consuming MI population only shared 44–49% similarity to the LI and GI populations that consumed terrestrial flora (Supplementary Figure S2).
Figure 3.
Host-specific clustering differences in bacterial lineages. The abundance and occurrence of bacterial lineages present in feces of each individual reptile were compared and plotted on multidimensional scaling plot (MDS). Apparent clustering was observed in relation to host species (circled in dashed lines), regardless of the sampling location. Abbreviations denote the sampling locations, that is, F, Fernandina; P, Plaza Sur; SF, Santa Fe; EL, El-Salvador; SC-L, San Cristobal-Loberia; SC-C, San Cristobal-Corola ; SC-G, San Cristobal-–Galapaguera.
Comparison of bacterial groups among herbivorous reptiles
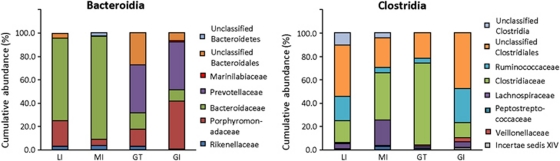
Further evaluation at finer taxonomical level was carried out to determine the distribution of microorganisms in reptiles residing on different islands. The two predominant phyla (that is, Bacteroidetes and Firmicutes) present in the reptilian fecal microbiota were comprised of the classes Flavobacteria, Sphingobacteria, Bacteroidia, Bacilli, Clostridia and Erysipelotrichi (Supplementary Figure S3). Among them, classes Bacteroidia and Clostridia were consistently present in all examined feces at relatively higher abundances. A further evaluation of these two classes showed that Bacteroidia in LI and MI fecal microbiota is comprised mainly of the family Bacteroidaceae, and in abundance that was significantly higher than that present in GT and GI (Figure 4, RDP Lib Compare, P-value >1E−9). The abundance of Bacteroidaceae, which was primarily made up of genus Bacteroides, was also significantly higher in MI than LI (RDP Lib Compare, P-value>6E−14). In contrast, the predominant bacterial family in GT and GI fecal microbiota was Prevotellaceae, and Prevotellaceae was undetected in the fecal microbiota of Galápagos LI and MI (Figure 4). A similar evaluation of the class Clostridia showed that 21.6–47.9% of the Clostridia were unclassified Clostridiales. The class Clostridia also included Clostridiaceae, which further comprised classified and unclassified Clostridiaceae 1 subfamily, as well as genera Sarcina and Clostridium. Together, the family Clostridiaceae was present at significantly higher abundance in the fecal microbiota of MI and GT as compared with LI and GI (Figure 4, RDP Lib Compare, P-value>6E−14). Among the different host populations, Lachnospiraceae was also relatively predominant in the MI fecal microbiota (RDP Lib Compare, P-value>1.5E−8). In contrast, Ruminococcaceae was present in >5-fold higher abundance in the fecal microbiota of LI and GI relative to MI and GT (RDP Lib Compare, P-value>6E−14).
Figure 4.
Bacterial distribution evaluated at a finer taxonomical level. The classes Bacteroidia and Clostridia were consistently present at high abundances in all herbivorous reptiles. A further evaluation within Bacteroidia and Clostridia showed differences in the profiles and abundance of bacterial groups among the herbivorous reptiles.
Comparison of archaeal groups among herbivorous reptiles
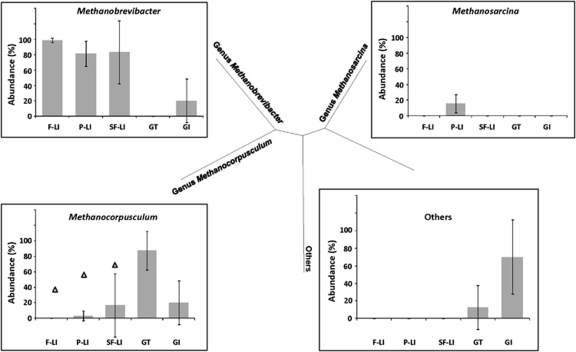
Archaeal populations were not detected in any of the studied MI fecal samples. Although archaeal population was detected in the feces of LI, GT and GI, it was dominated by a few genera. Genus Methanobrevibacter was ubiquitously detected in fecal microbiota of all LI and GI, and Methanobrevibacter was present at >81.6% of total archaeal community in the LI (Figure 5). The presence of other archaeal genera could only be found in some LI individuals at some of the islands. For example, the genus Methanosarcina was found in only four of the five sampled LI in Plaza Sur at an average abundance of 15.5±11.7% of total archaeal population, but was absent in all other reptiles and at all other sites. In addition, genus Methanocorpusculum was present in one of the LI sampled from Fernandina (0.01%±0.03%), Plaza Sur (2.9%±6.4%) and Santa Fe (16.7%±40.8%), respectively (Figure 5). Similarly, Methanocorpusculum was detected in one of the two GI from El Salvador, and at a higher abundance (20.0%±28.3%) than LI. In contrast, Methanocorpusculum was ubiquitously present in all examined GT, and constitutes the most abundant archaeal group (87.5%±25.0%) in the fecal microbiota of GT. Uncultivated Archaea were also present in the GT and GI feces, at average abundances of 12.5±25% and 70±42.4%, respectively (Figure 5).
Figure 5.
Archaeal population detected in fecal microbiota. No Archaea were detected in MI. The genus Methanobrevibacter, Methanosarcina and Methanocorpusculum were detected in the herbivorous reptiles that consumed terrestrial plant material. Triangles denote that only one LI at each location, respectively, was shown to harbor Methanocorpusculum. F-LI, land iguanas in Fernandina; P-LI, land iguanas in Plaza Sur; SF-LI land iguanas in Santa Fe.
Host-specific differences in microbial richness
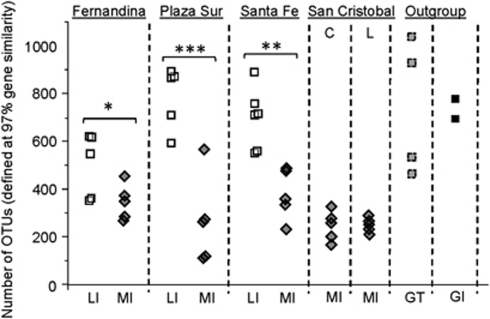
Microbial richness was defined based on the number of OTUs (OTUs at 97% gene similarity) identified at 6000 16S pyrotags (Supplementary Table S2). Figure 6 shows that the number of OTUs identified in the MI fecal microbiota ranged from 112 to 567 OTUs, and the microbial richness in MI was generally >2-fold lower than in LI, GT and GI. To further illustrate, microbial richness in LI fecal microbiota was significantly higher than in MI residing on Fernandina (P-value=0.054), Plaza Sur (P-value=8.73E−4) and Santa Fe (P-value=0.002), respectively. Similarly, microbial richness in GI and GT fecal microbiota was significantly higher than in the MI population (P-value>3.246E−5). Microbial richness among the LI, GI and GT was, however, not significantly different from each other (P-value>0.45).
Figure 6.
Comparison of microbial richness among the herbivorous reptiles. Microbial richness was defined based on number of OTUs (defined at 97% gene similarity). *denotes fairly significant difference evaluated at 90% confidence interval (CI), **significant at 95% CI, ***highly significant at 99% CI.
Discussion
Compared with mammalian hosts, herbivorous iguanas rely on external heat to maintain body temperature. As such, their diurnal body temperatures fluctuate according to ambient temperature, and have a shorter period to maintain at a level optimal for anaerobic gut fermentation. Nevertheless, iguanas are efficient herbivores and achieve digestion of 54% of cell wall constituents to supply 30–40% of the host-energy needs (McBee and McBee, 1982; Christian et al., 1984; Troyer, 1984; Van Marken Lichtenbelt, 1992), suggesting efficient hydrolytic and fermentative processes despite these limitations. Very little is known about the gut microbiota that contributes to effective hydrolysis and fermentation in the herbivorous iguanas. Past studies have relied on microscopic examinations, cultivation based enumeration and molecular fingerprinting methods to identify that microorganisms indeed contribute to the breakdown of complex dietary polysaccharides through hindgut fermentation (McBee and McBee, 1982; Mackie et al., 2004). Preliminary studies based on small full-length 16S rRNA libraries showed that the fecal microbiota of both marine and land iguanas were dominated by Firmicutes belonging to the genus Clostridium (Mackie et al., 2008; Nelson et al., 2010). The current study builds upon existing knowledge, and serves to provide a more detailed taxonomic examination of the gut microbiota in LI and MI residing in the Galápagos archipelago, as well as herbivorous GT found on the same archipelago and GI from Central America.
Our 16S rRNA-based pyrosequencing revealed that at the phyla level, the microbial community of herbivorous reptiles, regardless of the host species and diet, was predominated by Firmicutes and Bacteroidetes. This broad microbial distribution of major phyla is similar to those found in mammals (Eckburg et al., 2005; Ley et al., 2008b) and also to the single report for a captive carnivorous snake, the Burmese python (Costello et al., 2010), suggesting that Firmicutes and Bacteroidetes account for important and core roles in the host metabolism (Turnbaugh et al., 2006, 2008). For example, cellulose, hemicelluloses and pectin can first be hydrolyzed by Firmicutes and Bacteroidetes to their constituent hexoses and pentoses that in turn are converted by mixed-acid fermentation pathways with pyruvate as the central intermediate, and then into short-chain fatty acids (that is, acetate, propionate and butyrate) that contribute to the energy requirements of the host. Past studies further demonstrated that acetate, propionate and butyrate could be measured at high concentrations in the feces of both LI and MI (Mackie et al., 2004).
In addition to Firmicutes and Bacteroidetes, a significant proportion of the fecal microbiota in the Galápagos iguanas, as well as in the GI and GT was unclassified Bacteria. A portion of the 16S pyrotags was also not assigned by RDP Classifier to the respective taxonomical level at high confidence level (Supplementary Table S3), in turn suggesting that a large group of bacteria present in iguana fecal microbiota was novel and could be involved in currently unknown functions. A previous attempt to isolate novel bacteria from Galápagos marine iguana feces resulted in a range of Clostridia isolated from media containing different carbohydrate sources, and these isolates await full identification and characterization (Mackie, unpublished data). In a separate enrichment culture that actively degraded agar by rapid liquefaction, repeated attempts to isolate pure cultures of the agar-degraders failed, suggesting novel metabolic and functional pathways, operating synergistically, that require further elucidation before subsequent isolation attempts (Nelson et al., 2010).
Although our current study used short-length pyrosequencing and was not able to identify the bacterial groups at species level, deep sequencing still enabled quantitative comparison of the gut microbiota. For example, a comparison of the relative abundances of fecal microbiota showed that seaweed-consuming MI were separated from all other terrestrial flora-consuming lizards (Figure 3), even though marine and LI have been shown to have speciated from a common ancestor. An additional evaluation revealed differences in the abundances of some bacterial groups among the LI and MI. For example, Lachnospiraceae and Clostridiaceae were significantly more abundant in the MI (Figure 4). In contrast, Ruminococcaceae were present at a higher abundance in the LI (Figure 4). These findings, along with those from other studies (Ley et al., 2008a, 2008b), suggest that diet is a strong factor shaping gut microbiota composition. Therefore, we hypothesize that the observed differences in bacterial groups can possibly be explained by the contrasting diet consumed by the two groups of iguanas.
Besides differences in the Firmicutes, there were also observed differences in the abundance of Bacteroides spp. in both LI and MI. MI are the only iguanas that have adapted to feed primarily on soft macrophytic algae, which contain a range of polysaccharides, many with sulfated sugar moieties that are absent in terrestrial plants eaten by LI. In the intestinal tract, Bacteroides spp. are versatile in hydrolyzing a broad array of polysaccharides (Shah and Gharbia, 1993), and a recent study has demonstrated that B. plebieus contains beta-agarases and beta-porphyranases capable of breaking down algal polysaccharides such as agar, carrageenan, laminarin and porphyrans (Hehemann et al., 2010). Given that MI consume seaweed as its only diet since speciation 10–20 M years ago, the higher abundance of Bacteroides spp. could potentially be driven by the need to hydrolyze algal polysaccharides. In contrast, LI consume a diet that is relatively harder to degrade because of the composition of the terrestrial plant material that is lignified and higher in cellulose and hemicellulose content. Our findings revealed that the number of OTUs in the LI were consistently higher than in the MI (Figure 6). This increase in the OTU numbers could be related to the need for a diverse gene repertoire to effectively hydrolyze and ferment the recalcitrant plant cell-wall polysaccharides prevalent in the diet of LI. This observation is in agreement with past observations made in herbivores, which harbored higher bacterial diversity compared with carnivores and omnivores (Ley et al., 2008a).
In addition, Galápagos LI tend to have a larger body size compared with MI, in turn maximizing the gut capacity and lengthening diet-retention time. Typical values for retention time in the intestinal tract of LI, as well as MI, is ∼7–10d (Troyer, 1984; Wikelski et al., 1993), which facilitates longer contact time between gut microbial symbionts and insoluble substrates and contributes to high digestive efficiency in herbivorous iguanas. LI had a ubiquitous presence of hydrogen-using Methanobrevibacter spp. in their fecal microbiota (Figure 5). In addition, LI residing on Plaza Sur also had Methanosarcina spp., although at a lower abundance than genus Methanobrevibacter. Besides the LI, the presence of Methanocorpusculum is also observed to be ubiquitously associated with GT that consumed terrestrial plants (Figure 5). In contrast, no archaeal population was detected in the MI, further suggesting that host species and dietary differences can differentiate the gut microbiota.
It is worth noting that in the only report of methane production from fecal material collected from a few terrestrial iguanas kept in zoos, methane production rates were 322 nmol g-1 per hour for the GI (Iguana iguana) and 275 nmol g-1 per hour for the Bahamas iguana (I. delicatissima) (Hackstein and Van Alen, 1996). These production rates ranked within the vertebrates with higher methane production rates and are similar to those reported for ruminants, further reiterating an active methanogenic archaeal population in the iguanas. In an effort to explain the reduced abundance of methanogenic archaea in MI, alternative pathways for terminal carbon and electron flow were investigated (Supplementary Text 1). We used primers targeting the APS reductase (apsA) gene to detect sulfate-reducing bacteria as many of the algal polysaccharides are sulfated. A number of sequences were recovered from both iguana species that had closest BLAST hits to Desulfovibrio spp. (D. termitidis, D. vulgaris, D. desufuricans and D. pigra). In addition, the formyltetrahydrofolate synthase (fthfs) gene was used as evidence for the acetogenic pathway, and BLAST results showed similarity to Ruminococcus productus, Clostridium thermaceticum and uncultured bacteria. The detection of these bacterial groups may likely explain for the reduced abundance of methanogenic archaea in MI, although the quantitative significance of these alternate pathways for terminal carbon and electron disposal remain to be further elucidated.
Interestingly, our pyrosequencing data suggested subtle variations in the gut microbiota of the same host species residing in different islands. For example, a weak clustering effect based on the locations of the LI individuals could be observed. A comparison of the LI found on different islands showed that LI residing in Fernandina had a lower microbial richness compared with those in Plaza Sur and Santa Fe (Figure 6, P-value=0.01 and 0.03, respectively). Likewise, MI found in San Cristobal has a lower number of OTUs compared with those in Fernandina and Santa Fe (Figure 6, P-value=0.01). Biogeographical patterns in the Salmonella population of Galápagos iguanas have been previously reported (Wheeler et al., 2011), and it is likely that such biogeographical patterns extend to the entire intestinal/fecal microbiota. Conclusive evidence to confirm biogeographical population patterns of gut microbiota will be further analyzed in future studies.
In summary, we performed 16S rRNA-based pyrosequencing on fecal microbiota sampled from Galápagos iguanas and other reptiles, and provide a first detailed account of the microbial community inhabiting the gut of herbivorous reptiles. Although the microbial profile is primarily predominated by Firmicutes and Bacteroidetes, and resembled the gut microbiota of other herbivorous hosts, a large portion of the bacteria in the herbivorous reptiles remain unclassified and yet to be cultured. In addition, the microbial community and richness are intricately linked to the host species and their dietary preferences. With a better understanding of the microbial diversity in herbivorous reptiles, future studies would aim to elucidate their functional roles through metagenomics and metatranscriptomics. It is anticipated that such effort would facilitate possible discoveries of novel enzymes that can breakdown a wide variety of polysaccharides, as well as microbial-associated pathways that help herbivorous hosts overcome plant-defense mechanisms.
Acknowledgments
We would like to thank M Wikelski for long-term support and concepts, as well as access to infrastructure on the islands (CDRS and GNP) that made the study possible. We thank Augusto G Haz Beltran and Lenin Cruz Beldon for assistance with the field work and sample collection. We also like to thank the Charles Darwin Research Station, Director Washington Tapia and the Galápagos National Parks for professional and logistical assistance in conducting field work and obtaining all the necessary permits for collection and export of fecal samples. PYH was supported in part by funds from USDA-NRI (1-489318-231000-191100). EW was supported by the US Environmental Protection Agency Science to Achieve Results Fellowship program. This publication was developed with partial support from a STAR Research Assistance Agreement No. 91684301-1 awarded by the U.S. Environmental Protection Agency. It has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Boersma PD.1982The benefits of sleeping aggregations in marine iguanas, Amblyrhynchus cristatusIn: Burghardt GM, Rand AS (eds).Iguanas of the World: Their Behavior, Ecology and Conservation Noyes Publications: New Jersey; 292–299. [Google Scholar]
- Christian KA, Tracy CR, Porter WP. Diet, digestion, and food preferences of Galapagos land iguanas. Herpetologica. 1984;40:205–212. [Google Scholar]
- Cogger HG, Zweifel RG.1998Encyclopedia of Reptiles and Amphibians2nd edn.Academic Press: San Diego [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant P.2000The Galapagos Islands4th edn.Odyssey Publications: Hong Kong [Google Scholar]
- Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1835. In: Browne J, Neve M (eds).The Voyage of the Beagle: Charles Darwin's Journal of Researches Penguin: London, UK; (abridged edition, 1989). [Google Scholar]
- Davison A, Blaxter M. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol Biol Evol. 2005;22:1273–1284. doi: 10.1093/molbev/msi107. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (New York, NY) 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza RE, Wiens JJ, Tracy CR. Recurrent evolution of herbivory in small, cold-climate lizards: breaking the ecophysiological rules of reptilian herbivory. Proc Natl Acad Sci. 2004;101:16819–16824. doi: 10.1073/pnas.0401226101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein JHP, Van Alen T. Fecal methanogens and vertebrate evolution. Evolution. 1996;50:559–572. doi: 10.1111/j.1558-5646.1996.tb03868.x. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hume ID, Warner ACI.1980Evolution of microbial digestion in mammalsIn: Ruckebusch Y, Thivend P (eds).Digestive Physiology and Metabolism in Ruminants MTD Press: Lancaster, UK; 665–684. [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. Reptiles and Herbivory. Chapman and Hall: New York; 1996. [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science (New York, NY) 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie RI, Nelson DM, Wheeler E, Wikelski M, Cann IK.2008Fermentative digestion in herbivorous lizards: Bacterial population analysis in the intestinal tract of free-living land (Conolophus pallidus) and marine iguanas (Amblyrhynchus cristatus) on the Galapagos archipelagoIn: Morris S, Vosloo A (eds).Molecules to Migration: the Pressures of Life Medimond Publishing Company: Bologna, Italy; 193–202. [Google Scholar]
- Mackie RI, Rycyk M, Ruemmler RL, Aminov RI, Wikelski M. Biochemical and microbiological evidence for fermentative digestion in free-living land iguanas (Conolophus pallidus) and marine iguanas (Amblyrhynchus cristatus) on the Galapagos archipelago. Physiol Biochem Zool. 2004;77:127–138. doi: 10.1086/383498. [DOI] [PubMed] [Google Scholar]
- McBee RH, McBee VH.1982The hindgut fermentation in the green iguana, Iguana iguanaIn: Burghardt GM, Rand AS (eds).Iguanas of the World: Their Behavior, Ecology and Conservation Noyes Publications: New Jersey; 77–83. [Google Scholar]
- Nelson DM, Cann IK, Altermann E, Mackie RI. Phylogenetic evidence for lateral gene transfer in the intestine of marine iguanas. PLoS One. 2010;5:e10785. doi: 10.1371/journal.pone.0010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra R.1978Comparison of foregut and hindgut fermentation in herbivoresIn: Montgomery GG (ed).The Ecology of Arboreal Folivores Smithsonian Institution Press: Washington, D.C.205–229. [Google Scholar]
- Pough FH. Lizard energetics and diet. Ecology. 1973;54:837–844. [Google Scholar]
- Rassmann K. Evolutionary age of the Galapagos iguanas predates the age of the present Galapagos islands. Mol Phylogenet Evol. 1997;7:158–172. doi: 10.1006/mpev.1996.0386. [DOI] [PubMed] [Google Scholar]
- Shah HN, Gharbia SE. Ecophysiology and taxonomy of Bacteroides and related taxa. Clin Infect Dis. 1993;16 (Suppl 4:S160–S167. doi: 10.1093/clinids/16.supplement_4.s160. [DOI] [PubMed] [Google Scholar]
- Shepherd SA, Hawkes MW. Algal food preferences and seasonal foraging strategy of the marine iguana, Amblyrhynchus cristatus, on Santa Cruz, Galapagos. Bulletin Marine Sci. 2005;77:51–72. [Google Scholar]
- Snell HL, Snell HM, Tracy CR. Variation among populations of Galapagos land iguanas (Conolophus): contrasts of phylogeny and ecology. Bio J Linnean Soc. 1984;21:185–207. [Google Scholar]
- Stevens CE, Hume ID.1995Comparative Physiology of the Vertebrate Digestive System2nd edn.Cambridge University Press: Cambridge [Google Scholar]
- Stevens CE, Hume ID. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev. 1998;78:393–427. doi: 10.1152/physrev.1998.78.2.393. [DOI] [PubMed] [Google Scholar]
- Trillmich KGK, Trillmich F. Foraging strategies of the marine iguana, Amblyrhynchus cristatus. Behav Ecol Sociobiol. 1986;18:259–266. [Google Scholar]
- Troyer K. The biology of Iguanine lizards: Present status and future directions. Herpetologica. 1983;39:317–328. [Google Scholar]
- Troyer K. Structure and function of the digestive tract of a herbivorous lizard Iguana iguana. Physiol Zool. 1984;57:1–8. [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Van Marken Lichtenbelt WD. Digestion in an ectothermic herbivore, the green iguana (Iguana iguana): Effect of food composition and body temperature. Physiol Zool. 1992;65:649–673. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler E, Cann IKO, Mackie RI. Genomic fingerprinting and serotyping of Salmonella from Galapagos iguanas demonstrates island differences in strain diversity. Environ Microbiol Rep. 2011;3:166–173. doi: 10.1111/j.1758-2229.2010.00203.x. [DOI] [PubMed] [Google Scholar]
- Wikelski M, Gall B, Trillmich F. Ontogenetic changes in food intake and digestion rate of the herbivorous marine iguana (Amblyrhynchus cristatus, Bell) Oecologia. 1993;94:373–379. doi: 10.1007/BF00317112. [DOI] [PubMed] [Google Scholar]
- Wikelski M, Trillmich F. Foraging strategies of the Galapagos marine iguana (Amblyrhynchus cristatus): Adapting behavioral rules to ontogenetic size change. Behaviour. 1994;128:255–279. [Google Scholar]
- Zimmerman LC, Tracy CR. Interactions between the environment and ectothermy and herbivory in reptiles. Physiol Zool. 1989;62:374–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.