Abstract
Infectious diseases are an enormous burden to global health and since drug discovery is costly, those infectious diseases that affect the developing world are often not pursued by commercial drug-discovery efforts. Therefore, pragmatic means by which new therapeutics can be discovered are needed. One such approach is target repurposing, where pathogen targets are matched with homologous human targets that have been pursued for drug discovery for other indications. In many cases, the medicinal chemistry, structural biology and biochemistry knowledge around these human targets can be directly repurposed to launch and accelerate new drug-discovery efforts against the pathogen targets. This article describes the overarching strategy of target repurposing as a tool for initiating and prosecuting neglected disease drug-discovery programs, highlighting this approach with three case studies.
Infectious diseases are the biggest cause of human death and disability [101]. The WHO has reported that nearly 400 million years of healthy life were lost to infections in 2004 – twice the number due to any other cause and five times the number due to cancer. Despite the acute need for new drugs, there are many hurdles to overcome to make such anti-infective medications a reality. Drug discovery and development is expensive, and much of the work has to be done in technology-rich laboratories and clinics. It typically costs hundreds of millions of dollars and takes over a decade to advance from invention to market [1]. Drug discovery and development is also risky, in that as few as 5% of candidate drugs that enter clinical trials achieve approval and clinical use. Failure rates for anti-infective drugs exceed 70% in clinical trials [2]. For any indication, even drug candidates with good efficacy and safety may still be abandoned if they fall too far behind the launch of competitor drugs onto the market, or if there is little expectation of improvement in the standard of care at the time of launch [3]. While many important contributions to drug discovery are made from academic and government laboratories, the bulk of the expense (and risk) in taking an unproven compound through development is largely borne by companies competing for a share of the US$600 billion global market for pharmaceuticals. The commercial value of this market is centered in North America, Europe and Japan.
The WHO also reports that TB, malaria, and a group of other tropical diseases are among the most prevalent of these infections [102]. Several of these tropical diseases are summarized in Table 1, sorted in order of disability-adjusted life years (DALYs), a metric of global burden of disease that describes the impact of a specific condition on quality and length of life. To provide a frame of reference, also included in the table are two conditions (lung and prostate cancer) that attract significant research and development resources for delivery to patients in the developed world.
Table 1.
Summary of the impact of the top causes of death and disability, with a primary focus on neglected tropical diseases.
| Disease | DALYs† (millions) | Approximate numbers of candidates‡ | |||
|---|---|---|---|---|---|
| PCD | Phase I | Phase II | Phase III | ||
| Lower respiratory infections | 94 | 6‡ | 0‡ | 1‡ | 0‡ |
| HIV/AIDS | 59 | 81‡ | 19‡ | 59‡ | 8‡ |
| Tuberculosis | 34.7 | 23 | 2 | 5 | 0 |
| Malaria | 34.6 | 9 | 0 | 5 | 3 |
| Leishmaniases | 2.3 | 6 | 0 | 1 | 1 |
| Schistosomiasis | 2.1 | 0 | 0 | 0 | 0 |
| African trypanosomiasis | 1.5 | 3 | 0 | 1 | 0 |
| Chagas disease | 0.7 | 1 | 0 | 0 | 0 |
| Lung cancer | 11.2 | 28 | 10 | 30 | 9 |
| Prostate cancer | 1.6 | 34 | 11 | 30 | 5 |
Some of these conditions, such as respiratory infections, are often manageable with existing drugs and supportive care. However, the lack of access to these drugs and care has resulted in these diseases being a persistent cause of death and disability in impoverished populations. Improvements in treatment availability should be a priority for these illnesses. Conversely, there are other infectious diseases for which new drug discovery is needed to achieve improved outcomes. Drivers for new drug discovery include known drug resistance (malaria and TB), reliance on a single treatment and the consequence if resistance were to develop against this treatment (schistosomiasis), inadequate drug safety (African trypanosomiasis), and inadequate drug efficacy (Chagas disease and visceral leishmaniasis).
The disproportionate impact of R&D costs on neglected tropical disease drug discovery
These factors contribute to two different worlds of drug discovery. Diseases that are leading causes of mortality and morbidity in Western societies may be targeted with dozens, or even hundreds, of discovery projects and drug candidates. In contrast, some of the global infectious diseases are targeted by only a handful of drug candidates. Even the strongest of the infectious disease pipelines has only a fifth the number of candidates as for individual cancer indications, and many have only one, or none (Table 1). For example, in comparing human African trypanosomiasis (HAT) (1.5 million DALYs) and prostate cancer (1.6 million DALYs), one can see that while there are approximately 80 candidate compounds ranging from preclinical development through Phase II clinical trials for prostate cancer, there are only four for HAT. Considering the failure rates typical in drug discovery it is clear that there are too few initiatives to expect success against the global infectious diseases [103]. This consequence of the two worlds of drug discovery is illustrated by the observation that of 1393 new medicines that reached the market between 1975 and 2000, only 1% was directed at malaria, TB or tropical diseases [4].
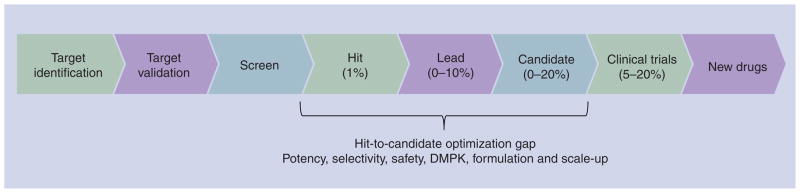
A preclinical optimization gap further restricts drug-discovery success. Irrespective of the disease target, in order to be considered a candidate drug a molecule must typically be effective in disease models, have appropriate stability and tissue penetration adequate to achieve therapeutic levels to patients, have low toxicity, and be suitable for cost-effective manufacturing. Molecules identified from screening almost never have these collective properties. Instead, suitable drug candidates are invented through the optimization of stability, solubility, potency, selectivity, pharma-cokinetics, pharmacodynamics and toxicity of compounds obtained from screening. This optimization costs millions of dollars and has, historically, been carried out in drug companies. It requires expertise in medicinal and formulation chemistry, pharmacology and toxicology, plus the synthesis of large quantities of the chemical compounds of interest and extensive in vivo experimentation. Teams of chemists work with pharmacologists and toxicologists to design and synthesize variations of active molecules in an effort to achieve optimal activity. Even with strong teams only a tiny fraction (<0.1%) of molecules identified in early stages of drug discovery can be optimized into compounds that merit advancement to clinical trials. This results in an optimization gap from screen to candidate that claims the great majority of early-stage discovery projects (Figure 1). Typically, optimization projects for malaria, TB and other tropical diseases can only afford to employ just one or two chemists [5], a number that represents a quarter or less of the chemistry support typically provided to non-tropical disease projects in companies. This makes success even less likely and the timelines longer.
Figure 1. Location of the gap in optimization resource and expertise in neglected tropical disease drug discovery.
Percentages of compounds proceeding to the next step are shown in parentheses.
A further challenge to any drug-discovery program is the assumption that a proposed therapeutic target is ‘druggable’, meaning that is can be manipulated for therapeutic effect by drug-like molecules [6]. Genome sequencing and biochemistry efforts have uncovered many pathogen-specific enzyme targets that could be essential to parasite survival [7–9]. This would seem highly desirable from a drug-discovery perspective, as the presumed challenges of attaining selectivity for the pathogen target over host targets would be reduced or eliminated. However, not all proposed therapeutic targets are drug-gable. Target families proven to be druggable in successful human drug-discovery programs should have reduced risk that the parasite target will not be druggable.
In summary, in order to improve drug pipelines for neglected tropical diseases it will be necessary to overcome the enormous challenges inherent in drug discovery (and exacerbated in the resource-poor area of neglected tropical disease drug discovery). In particular, approaches to drug discovery in this field must come up with ways to facilitate the bridging of the optimization gap that has impeded the advancement of compounds from screen to drug [5]. One of these approaches can be target repurposing.
Target repurposing
Target repurposing exploits the facts that a) many drugs bind specific proteins and b) industry discovery is protein target focused. Evolution has resulted in similar protein designs between organisms, often with conserved features of binding and active sites. As a result, drug-like chemicals can often bind proteins that are structurally related to the targets to which these chemicals were originally designed to bind. If the related protein is itself a potential drug target, then this crossbinding can guide repositioning of a discovery program from one disease to another. Genomes of many pathogens have now been fully sequenced, permitting the prediction and confirmation of parasite protein sequences, and prioritization of putative targets based on sequence similarity to human targets. The pharmaceutical industry has produced hundreds of thousands of drug-like compounds against several thousand drug targets and many of these programs include compounds that have successfully passed the initial pharmacology and toxicology tests associated with candidate optimization. While not all drug-gable human drug targets are present in parasitic pathogens, use of these compounds and knowledge for those targets that do overlap is a proven strategy that can enable a new drug-discovery program to quickly obtain drug candidates.
A concern inherent in the target repurposing approach is the risk that compounds derived from medicinal chemistry programs against human targets may have toxic effects mediated through the same or related human targets. While drugs developed for use in developed countries may have side effects that are considered acceptable because they can be readily managed in a strong supportive care setting, use of these same products could be severely problematic in regions that lack easy access to supportive care. Nonetheless, given the acute (and sometimes fatal) pathology of some parasitic diseases, some off-target effects may prove to be acceptable risks. For example, the repurposing of trypanosomal phosphodies-terases (PDEs) represent an ongoing approach for discovery of drugs for African sleeping sickness and Chagas disease [10–12], two indications for which drugs are either highly toxic, or of modest efficacy. The most closely homologous human enzyme is PDE4, inhibition of which has been linked to emesis. If achieving selectivity between host and pathogen targets proves to be impossible, then one must thus consider whether such a side effect profile is acceptable given the current state of the therapeutics for these diseases.
The identification of pathogen proteins related to known drug targets can be aided by databases such as TDR Targets DB [104]. The availability of these resources make it possible to consider a comprehensive repositioning of existing drug-discovery expertise against pathogens causing malaria, TB and the other tropical diseases defined by the WHO as most in need of new drug treatments. The scene has, thus, been set to permit an integration of past research investments in drug discovery with major unmet needs in global health.
We review three examples that illustrate application of the target repurposing approach to bring new therapeutics into clinical research and practice.
HIV protease inhibitors
A particularly striking example of how the repositioning of chemistry expertise can favorably impact drug discovery was the rapid development of treatments for HIV infection following the sequencing of the virus genome in 1985. The rapid identification of clinically suitable anti-HIV protease inhibitors in the 1990s was built on prior chemistry expertise gained with human aspartic proteases. This approach of ‘repurposing’ discovery chemistry expedited the invention of inhibitors with drug-like potency, selectivity and safety. It helped to de-risk these projects and deliver drug candidates for AIDS just ten years after the determination of the HIV genome.
The first step was to recognize the presence of candidate drug targets in the HIV genome, a task made possible by extensive investment in HIV genome sequencing and cellular biology. One of the candidate targets identified was an aspartic protease predicted to share a common biochemical mechanism with a family of human proteases that had already been targeted for drug discovery. Analysis of the HIV genome revealed a protein with a short motif of amino acids known to be a common feature of aspartic acid proteases. The prediction that HIV utilized an aspartic protease in its life cycle was confirmed by genetic studies showing that conversion of the active site aspartic acid to an asparagine resulted in deficits in the proteolytic processing of HIV pre-proteins [13]. This also resulted in a block to the production of infectious virus. Subsequent determination of the x-ray crystallographic structure of the HIV protease confirmed the prediction that it was a homolog of known aspartic proteases, raising the possibility that anti-HIV drug discovery could be facilitated with knowledge from members of this enzyme family that had previously been targeted by medicinal chemistry [14,15].
One of the best-studied human aspartic acid proteases at the time was renin, an enzyme that triggers a cascade of reactions that result in an elevation of blood pressure. Drugs acting on a downstream enzyme in this cascade, angioten-sin-converting enzyme, had already become well established as safe and effective treatments for hypertension. Seeing the success of angiotensin-converting enzyme inhibitors, numerous companies had explored targeting renin as a further means to control blood pressure. However, while many renin inhibitors had been found, none had the desired combination of oral bioavail-ability and selectivity. It seemed the medicinal chemistry attack on renin was a dead end.
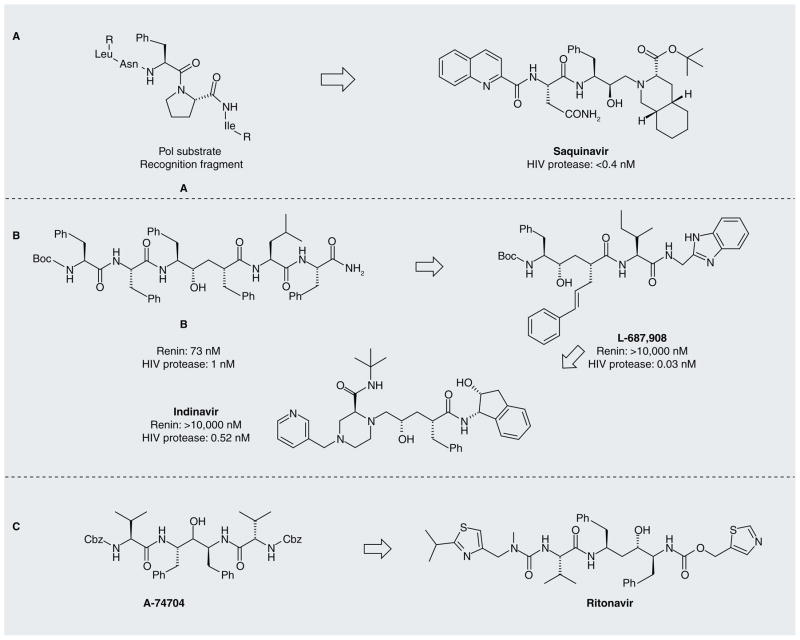
The speed with which HIV spread in the USA and other countries fostered a strong mobilization of drug-discovery interest. It was soon realized that some inhibitors of renin and other human aspartic proteases could also inhibit the HIV protease [16]. This group took an approach to optimize potency and selectivity of transition-state mimetics by exploiting differences between human and HIV protein substrates near the site of cleavage [17]. Human substrates of aspartic proteases are nearly devoid of proline residues adjacent to the cleavage site, while many of the HIV substrates, such as the Pol protein precursor, are enriched for proline residues. Inhibitor analogs could be made more selective for the HIV protease by incorporating features of a proline side chain in the position occupied by proline in authentic substrates. This work led to saquinavir, the first protease inhibitor approved by the US FDA for treatment of HIV infection (Figure 2A). This 1995 product approval came just 10 years after the initial sequencing of the HIV genome.
Figure 2.
Evolution of HIV-1 protease inhibitors.
Several other groups jump-started their HIV drug-discovery programs by screening collections of renin inhibitors. At Merck, Sharp & Dohme this screen led to the early identification of potent inhibitors that could block HIV production in cells [18]. However, these early compounds had poor solubility that precluded their usefulness as drugs. Several additional rounds of medicinal chemistry were required to achieve a potent, selective and orally active drug with pharmacokinetics suitable for the clinic (Figure 2B) [19].
A third approach to facilitate discovery of potent and selective drugs against HIV exploited the fact that the active site of aspartic proteases lies at the interface of two domains [20]. In the human aspartic proteases these domains are non-identical, resulting in a nonsymmetrical active site. In contrast, the HIV protease is a homodimer of two identical single-domain subunits, resulting in a symmetrical active site that has different binding properties to the human enzymes. Such a site can be targeted by ligands that have a twofold axis of symmetry, while the human aspartic proteases will not recognize such ligands. This work led to highly selective inhibitors but these initial compounds had poor oral bioavailability. This problem was then targeted by optimization efforts, resulting in ritonavir (Figure 2C) [21], which was approved by the FDA in 1996.
Therefore, the early invention of HIV protease inhibitors was aided by the knowledge of medicinal chemistry and enzyme mechanisms that had been gained with other aspartic protease targets, in particular renin. This contributed to the rapid progression of protease-inhibitor drug candidates to the clinic, and the creation of a rich pipeline for drugs to treat HIV infection. Several additional benefits came from this broad mobilization of medicinal chemistry against the HIV protease. One was the rapid delivery of multiple different products to the market. In the next few years it was found that combinations of these products were particularly effective at blunting the ability of the virus to escape from inhibition. A second, unexpected benefit was the finding that one of the inhibitors, ritonavir, was highly effective at preventing the biotransformation of other prote-ase inhibitors by cytochrome P450–3A4 [22]. This provided significant plasma concentration levels of each inhibitor without affecting the plasma concentrations of ritonavir, enhancing the therapeutic benefit of such drug cocktails. This has led to widespread use of ritonavir as a potentiator of other HIV protease inhibitors due to its favorable influence on their systemic exposure.
Eflornithine
One of two front-line treatments for HAT, eflornithine is a suicide inhibitor of ornithine decarboxylase that was initially studied as a human cancer therapeutic. The drug interferes with polyamine biosynthetic pathways that are involved in the generation of small-amine intermediates that are incorporated into nucleic acid and amino acid synthesis: typically spermine, spermidine and putrescine. The rate-limiting step of this reaction sequence is catalyzed by ornithine decarboxylase, and the ornithine analog α-difluoromethylornithine (eflornithine) is a suicide inhibitor of this enzyme (for a review of polyamine synthesis inhibition as a therapeutic approach, see [23]). Unfortunately, the drug was found to have poor efficacy in cancer and the clinical development was stopped. However, it was recognized by others that trypanosomes utilize a homologous ornithine decarboxylase enzyme. This led to the hypothesis that eflor-nithine might interrupt polyamine synthesis in the parasite, and be useful as a trypanocidal drug. This hypothesis was confirmed in cellular and in mouse infection experiments [24], and the mechanism of action was subsequently supported by x-ray crystallographic analysis [25]. The compound was shown to clear Trypanosoma bru-cei gambiense infections in humans [26], although the drug is not as effective against the more virulent rhodesiense strain. This is thought to be due primarily to the more rapid regeneration of ornithine decarboxylase in Trypanosoma bru-cei rhodesiense, providing this strain with a means by which to overcome drug treatment [27]. Eflornithine remains one of the two front-line therapeutics for stage II T.b. gambiense infections, and is the most recently approved drug for this disease (1990). From a target-repurposing perspective, the difference in efficacy between pathogen strains would not have been obvious based on sequence homology and thus the eflornithine case underscores a further need to understand the molecular biology of the parasite targets, in addition to their structural homology to human targets. It is worth noting that, in a second repurposing of this drug, eflornithine has also been approved as a topical agent for treatment of female facial hirsutism [28], also by modulating polyamine biosynthesis.
N-myrisoyl transferase
A key post-translational modification of proteins is catalyzed by a myristoyl-CoA-protein N-myristoyltransferase (NMT). This enzyme transfers a molecule of myrisitc acid to N-terminal glycine residues, resulting in membrane targeting of the modified protein [29]. Inhibitors of this target class have been explored as therapeutics for cancer and fungal diseases [30,31]. The essentiality of the homologous enzyme in T. brucei (TbNMT) has been demonstrated via RNA interference [32], and the TbNMT enzyme displays 55% identity to human NMT2. This suggested that TbNMT was a tractable drug target for HAT.
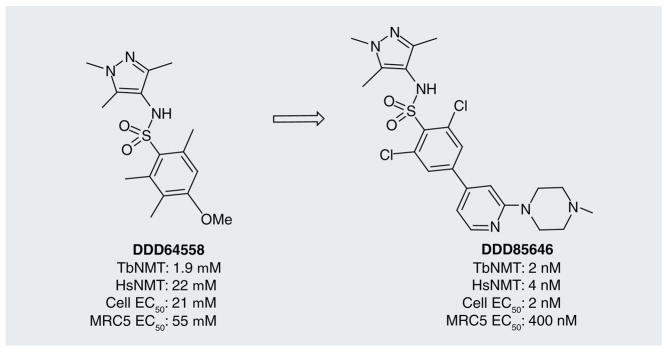
Despite the existence of chemical matter against the human homologue, Frearson et al. elected to perform a high-throughput screen of 62,000 compounds against TbNMT, resulting in a 2-μM hit compound DDD64558 (Figure 3) [33], which had modest selectivity over the human homolog. The optimization process (generating over 200 analogs) resulted in a 1000-fold improvement in potency, providing DDD65646. Although this optimized compound was nonselective over human NMT, it showed high activity against T. brucei cells with greater than 200-fold selectivity over human cells and in vivo activity, clearing acute mouse infections of T.b. rhodesiense and extending survival of infected animals. The mechanism of action was confirmed as involving TbNMT based on reduction of [3H]-myristoylated proteins, and rescue of trypanosomes by overexpres-sion of TbNMT. Furthermore, binding to the homologous Leishmania major enzyme LmNMT (95% identity in the binding site region) was confirmed crystallographically.
Figure 3.
The optimized TbNMT inhibitor DDD65656 resulting from the initial high-throughput screening hit DDD64558.
Notably, while the optimization of the compounds has led to a much-reduced selectivity of the drug for TbNMT over the host enzyme, the cellular selectivity is relatively good. Thus, while further improvements are needed for this series of compounds to achieve CNS exposure and a greater selectivity over the human NMT, this case study is a promising success illustrating target repurposing.
Resources for target repurposing
The target repurposing approach is strongly enabled by a number of existing resources. The identification of target homologs in pathogens, and candidate compounds for testing, is aided by the availability of annotated pathogen genome databases, such as PlasmoDB [34], pathogen target bioinformatics resources, such as TDR Targets [35], public data repositories of screening data, such as PubChem [105], Collaborative Drug Discovery [106], ChEMBL (formerly StARLite) [107], and BindingDB [108], and structural biology resources, such as the Protein Data Bank [109]. The implementation of chemical validation studies is aided by purchasable compound collections, such as the Library of Pharmacologically Active Compounds (Sigma–Aldrich) and the NIH Clinical Collection [110].
Future perspective
As illustrated through the examples in this review, repositioning of molecules and target knowledge from existing drug-discovery programs can facilitate rapid and cost-effective advancement towards invention of new therapeutics for emerging and neglected diseases (Figure 1). This repositioning can also lower barriers for commercial stakeholders to participate infectious disease drug discovery through the leveraging of compounds (and expertise) from their past research investments. Extension of target repurposing should help the neglected tropical disease drug-discovery effort benefit from the extensive knowledge derived from industrial drug-discovery efforts, for which the research investment of the US pharmaceutical industry alone was $67.4 billion in 2010 [111].
However, despite the identification of numerous target matches, which could be used to drive repurposing of drug discovery for neglected diseases, progress has still been slow. Indeed, two of the specific examples were cited above are two decades old. Why has this approach not been more widely exploited?
We believe one of the major, as yet unfilled gaps, is the lack of validation evidence, especially pharmacological validation. To fill this gap there is a growing number of projects applying existing compounds as toolkits with which to assess the tractability of target homologs for disruption of pathogen viability (e.g., with PDEs [36,37], kinases [38,39] and other targets that have a rich medicinal chemistry history). The validation of targets and pathways with small-molecule agents, in concert with genetic evidence, can provide a higher level of certainty regarding the likelihood of these targets’ being converted to fruitful therapeutic approaches.
Some additional resources could be highly beneficial to support these efforts. For example, Chong and Sullivan calculate that there are approximately 8900 unique drug molecules that are either in clinical use or progressed through clinical trials that represent potentially strong ligand-repurposing starting points [40]. Citing their own hosted Clinical Compound Repository at Johns Hopkins University (USA) as a model, the authors propose that a modest investment of public–private funding (<$10 million) in an expanded collection that contains these 8900 compounds could strongly enable target repurposing by enabling screening campaigns against this collection. In addition, availability of these compounds as singletons to test specific hypotheses for target repurposing, could essentially bypass the time-consuming effort of de novo synthesis that is required to benchmark human-targeted compounds against homologous pathogen targets. As a result of these synthetic challenges, access to small molecules is frequently a limiting step to launching such initiatives.
Another highly valuable addition to the public domain drug-discovery efforts would be ready access to core chemistry, molecular modeling, drug-metabolism profiling and pharmacokinetic resources. This could be similar to that which is already in place for x-ray crystallography (e.g., the Structural Genomics Consortium). In such a model, a queue of synthesis programs is pursued upon request, with the goal to provide a key resource free of charge to initiate target-repurposing programs. The output of this core resource could contain key analogs for screening, small-scale resynthesis of known active agents, and in vitro profiling of physicochemical and metabolic properties that can be directive of optimization efforts. The data generated from such experiments can strongly inform new projects against emerging parasite targets, and provide justification in these programs for further investment from funding agencies. Importantly, such a core resource could provide pivotal guidance to investigators who are entering into the early stages of drug target validation and optimization. Notably, the National Institutes of Health have implemented the Therapeutics for Rare and Neglected Disease program, the goal of which is to provide the rare and neglected disease research community access to drug-discovery expertise and resource.
Conclusion
The process for target repurposing can be implemented in a variety of ways, but they all rely on the following premises: first, parasites express essential targets that have human homologs; second, some fraction of these human homologs has been pursued by the drug-discovery industry, and, therefore, lead matter must exist; third, assessment of this lead matter against parasite homologs should uncover chemical matter that can serve as tools to validate the target as an antiparasitic approach, and as leads for further optimization towards leads and clinical candidates. This approach can seed new drug-discovery and/or development programs against the parasite targets without costly high-throughput screening campaigns, and with reduced risk of chemotype attrition due to poor physicochemical properties.
In applying these premises, as typified by the few examples described in this article, it is clear that a target-repurposing approach can be a faster, more cost-effective method for drug discovery over existing ‘traditional’ de novo drug-discovery approaches.
Executive summary.
Although there is a significant impact on global health, neglected diseases are those that affect the poorest parts of the world, with little research and development effort expended on the discovery of new drugs.
The high cost and attrition rate of new drug-discovery approaches further restrict the pace of neglected tropical disease drug discovery.
Many pathogen genomes have been elucidated, enabling bioinformatic matching of targets between pathogens and mammals, enabling the knowledge and compounds for these targets to be repurposed for anti-infective agents. This is referred to as ‘target repurposing’.
Three programs are highlighted for their varying levels of success in applying the concepts of target repurposing: HIV protease inhibitors, eflornithine and N-myrtistoyl transferases.
Key Terms
- Disability-adjusted life year
Metric developed by the WHO that describes global disease burden by combining years of life lost due to death, and years of life lost due to less-than-full health
- Human African trypanosomiasis
The disease has had a health impact of 1.5 million disability-adjusted life years, approximately equivalent to prostate cancer (1.6 million disability-adjusted life years), yet has a small fraction of new drugs in any stage of discovery and development
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
Support from the National Institutes of Health (R01 AI082577) is gratefully acknowledged. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
- 1.Dimasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22(2):151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 2.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 3.Gudiksen M, Fleming E, Furstenthal L, Ma P. What drives success for specialty pharmaceuticals? Nat Rev Drug Discov. 2008;7(7):563–567. doi: 10.1038/nrd2594. [DOI] [PubMed] [Google Scholar]
- 4.Trouiller P, Olliaro P, Torreele E, Orbinski J, Laing R, Ford N. Drug development for neglected diseases: a deficient market and a public-health policy failure. Lancet. 2002;359(9324):2188–2194. doi: 10.1016/S0140-6736(02)09096-7. [DOI] [PubMed] [Google Scholar]
- 5.Nwaka S, Hudson A. Innovative lead discovery strategies for tropical diseases. Nat Rev Drug Discov. 2006;5(11):941–955. doi: 10.1038/nrd2144. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 7.Gull K. The cell biology of parasitism in Trypanosoma brucei: insights and drug targets from genomic approaches? Curr Pharm Des. 2002;8(4):241–256. doi: 10.2174/1381612023396212. [DOI] [PubMed] [Google Scholar]
- 8.Lakhdar-Ghazal F, Blonski C, Willson M, Michels P, Perie J. Glycolysis and proteases as targets for the design of new anti-trypanosome drugs. Curr Top Med Chem. 2002;2(5):439–456. doi: 10.2174/1568026024607472. [DOI] [PubMed] [Google Scholar]
- 9.Fairlamb AH. Metabolic pathway analysis in trypanosomes and malaria parasites. Philos Trans R Soc Lond B Biol Sci. 2002;357(1417):101–107. doi: 10.1098/rstb.2001.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King-Keller S, Li M, Smith A, et al. Chemical validation of phosphodiesterase C as a chemotherapeutic target in Trypanosoma cruzi, the etiological agent of Chagas’ disease. Antimicrob Agents Chemother. 2010;54:3738–3745. doi: 10.1128/AAC.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laxman S, Beavo JA. Cyclic nucleotide signaling mechanisms in trypanosomes: possible targets for therapeutic agents. Mol Interventions. 2007;7:203–215. doi: 10.1124/mi.7.4.7. [DOI] [PubMed] [Google Scholar]
- 12.Oberholzer M, Marti G, Baresic M, Kunz S, Hemphill A, Seebeck T. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 2007;21:720–731. doi: 10.1096/fj.06-6818com. [DOI] [PubMed] [Google Scholar]
- 13.Kohl NE, Emini EA, Schleif WA, et al. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navia MA, Fitzgerald PM, Mckeever BM, et al. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- 15.Blundell T, Pearl L. Retroviral proteinases. A second front against AIDS. Nature. 1989;337(6208):596–597. doi: 10.1038/337596a0. [DOI] [PubMed] [Google Scholar]
- 16.Richards AD, Roberts R, Dunn BM, Graves MC, Kay J. Effective blocking of HIV-1 proteinase activity by characteristic inhibitors of aspartic proteinases. FEBS Lett. 1989;247(1):113–117. doi: 10.1016/0014-5793(89)81251-7. [DOI] [PubMed] [Google Scholar]
- 17.Roberts NA, Martin JA, Kinchington D, et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248(4953):358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 18.Vacca JP, Guare JP, Desolms SJ, et al. L-687,908, a potent hydroxyethylene-containing HIV protease inhibitor. J Med Chem. 1991;34(3):1225–1228. doi: 10.1021/jm00107a050. [DOI] [PubMed] [Google Scholar]
- 19.Dorsey BD, Levin RB, Mcdaniel SL, et al. L-735,524: the design of a potent and orally bioavailable HIV protease inhibitor. J Med Chem. 1994;37(21):3443–3451. doi: 10.1021/jm00047a001. [DOI] [PubMed] [Google Scholar]
- 20.Erickson J, Neidhart DJ, Vandrie J, et al. Design, activity, and 2.8 Å crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science. 1990;249(4968):527–533. doi: 10.1126/science.2200122. [DOI] [PubMed] [Google Scholar]
- 21.Kempf DJ, Marsh KC, Denissen JF, et al. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92(7):2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J Pharmacol Exp Ther. 1996;277(1):423–431. [PubMed] [Google Scholar]
- 23.Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Ann Rev Pharmacol Toxicol. 1995;35(1):55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 24.Bacchi C, Nathan H, Hutner S, Mccann P, Sjoerdsma A. Polyamine metabolism: a potential therapeutic target in trypanosomes. Science. 1980;210(4467):332–334. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- 25.Grishin NV, Osterman AL, Brooks HB, Phillips MA, Goldsmith EJ. X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with α-difluoromethylornithine. Biochemistry. 1999;38(46):15174–15184. doi: 10.1021/bi9915115. [DOI] [PubMed] [Google Scholar]
- 26.Kuzoe FA. Current situation of African trypanosomiasis. Acta Trop. 1993;54(3–4):153–162. doi: 10.1016/0001-706x(93)90089-t. [DOI] [PubMed] [Google Scholar]
- 27.Iten M, Mett H, Evans A, Enyaru JCK, Brun R, Kaminsky R. Alterations in ornithine decarboxylase characteristics account for tolerance of Trypanosoma brucei rhodesiense to D,L-α-difluoromethylornithine. Antimicrob Agents Chemother. 1997;41:1922–1925. doi: 10.1128/aac.41.9.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra B, Noveck R, Behr D, Palmisano M. Percutaneous absorption and pharmacokinetics of eflornithine HCl 13.9% cream in women with unwanted facial hair. J Clin Pharmacol. 2001;41(9):972–978. doi: 10.1177/00912700122010951. [DOI] [PubMed] [Google Scholar]
- 29.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276(43):39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 30.Selvakumar P, Lakshmikuttyamma A, Shrivastav A, Das SB, Dimmock JR, Sharma RK. Potential role of N-myristoyltransferase in cancer. Prog Lipid Res. 2007;46(1):1–36. doi: 10.1016/j.plipres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Georgopapadakou NH. Antifungals targeted to protein modification: focus on protein N-myristoyltransferase. Expert Opin Investig Drugs. 2002;11(8):1117–1125. doi: 10.1517/13543784.11.8.1117. [DOI] [PubMed] [Google Scholar]
- 32.Price HP, Menon MR, Panethymitaki C, Goulding D, Mckean PG, Smith DF. Myristoyl-CoA:protein N-myristoyl-transferase, an essential enzyme and potential drug target in kinetoplastid parasites. J Biol Chem. 2003;278(9):7206–7214. doi: 10.1074/jbc.M211391200. [DOI] [PubMed] [Google Scholar]
- 33.Frearson JA, Brand S, Mcelroy SP, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464(7289):728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aurrecoechea C, Brestelli J, Brunk BP, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37(Database issue):D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguero F, Al-Lazikani B, Aslett M, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7(11):900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoraghi R, Seebeck T. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc Natl Acad Sci USA. 2002;99(7):4343–4348. doi: 10.1073/pnas.062716599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rascon A, Soderling SH, Schaefer JB, Beavo JA. Cloning and characterization of a cAMP-specific phosphodiesterase (TbPDE2B) from Trypanosoma brucei. Proc Natl Acad Sci USA. 2002;99(7):4714–4719. doi: 10.1073/pnas.002031599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahia D, Oliveira LM, Lima FM, et al. The TryPIKinome of five human pathogenic trypanosomatids: Trypanosoma brucei, Trypanosoma cruzi, Leishmania major, Leishmania braziliensis and Leishmania infantum – new tools for designing specific inhibitors. Biochem Biophys Res Commun. 2009;390(3):963–970. doi: 10.1016/j.bbrc.2009.10.086. [DOI] [PubMed] [Google Scholar]
- 39.Brown JR, Auger KR. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007;448(7154):645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
Websites
- 101.The global burden of disease, 2004 update. www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html.
- 102.WHO metrics: disability-adjusted live year (DALY) www.who.int/healthinfo/global_burden_disease/metrics_daly/en.
- 103.Closing the global health innovation gap. BIO Ventures for Global Health. http://www.bvgh.org/LinkClick.aspx?fileticket=3An6aKB2z6Y%3D&tabid=79.
- 104.TDR Targets. www.tdrtargets.org.
- 105.PubChem. http://pubchem.ncbi.nlm.nih.gov.
- 106.Collaborative drug discovery. www.collaborativedrug.com.
- 107.CHemBL. www.ebi.ac.uk/chemb.
- 108.BindingDB. www.bindingdb.org.
- 109.Protein Data Bank. www.pdb.org.
- 110.NIH Clinical Collection. www.nihclinicalcollection.com.
- 111.PhRMA. R&D Investment by US biopharmaceutical companies reached record levels in 2010. www.phrma.org/media/releases/rd-investment-us-biopharmaceutical-companies-reached-record-levels-2010.
- 112.Pharmaprojects database. www.pharmaprojects.com.
- 113.ClinicalTrials.gov. http://clinicaltrials.gov.