Abstract
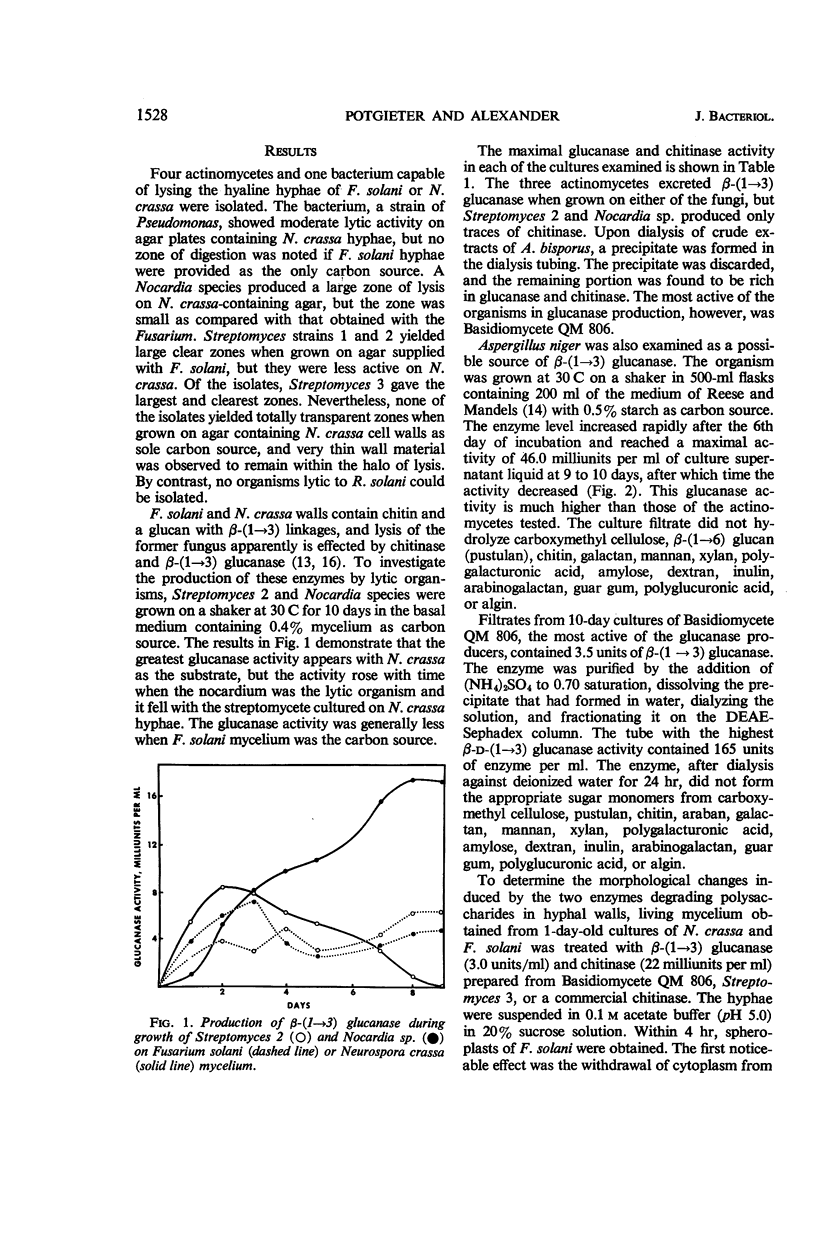
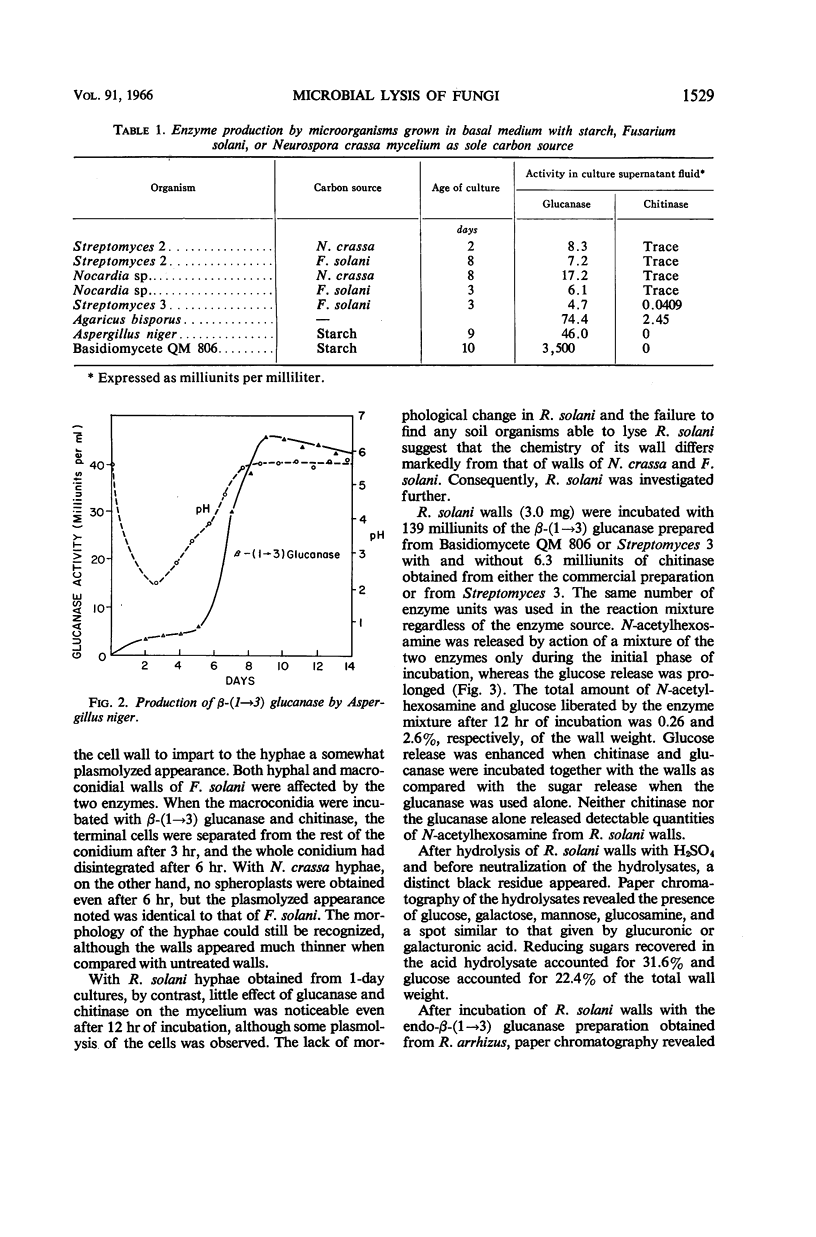
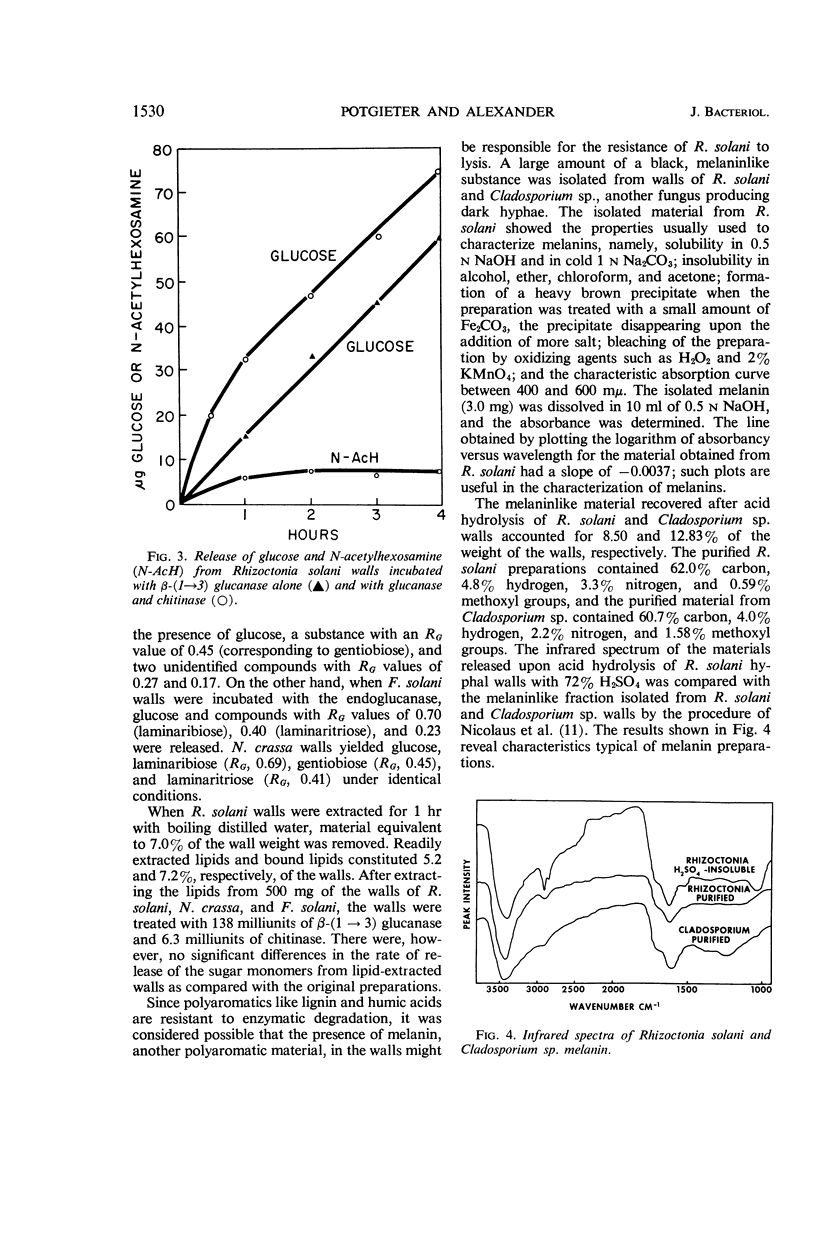
Potgieter, H. J. (Cornell University, Ithaca, N.Y.), and M. Alexander. Susceptibility and resistance of several fungi to microbial lysis. J. Bacteriol. 91:1526–1532. 1966.—Strains of Streptomyces, Nocardia, and Pseudomonas capable of lysing hyphae of Fusarium solani or Neurospora crassa were obtained by selective culture, but attempts to isolate an organism lysing Rhizoctonia solani failed. When provided with F. solani or N. crassa as carbon sources, the actinomycetes tested produced β-(1 → 3) glucanase and chitinase. A mixture containing purified chitinase and β-(1 → 3) glucanase induced spheroplast formation in F. solani, caused some morphological changes in N. crassa, but had almost no effect on R. solani hyphae. The polysaccharides in R. solani walls, which contain a large amount of glucose as well as galactose, mannose, and glucosamine, were not hydrolyzed appreciably by the two enzymes. Laminaribiose and laminaritriose were released by enzymatic hydrolysis of F. solani and N. crassa walls, and gentiobiose was liberated from R. solani and N. crassa walls. Melaninlike materials were found in R. solani walls, accounting for 8.50% of the wall weight. A role for melanin in protecting hyphae from microbial lysis is suggested.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al-Doory Y., Larsh H. W. Quantitative Studies of Total Lipids of Pathogenic Fungi. Appl Microbiol. 1962 Nov;10(6):492–495. doi: 10.1128/am.10.6.492-495.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BACHMANN B. J., BONNER D. M. Protoplasts from Neurospora crassa. J Bacteriol. 1959 Oct;78:550–556. doi: 10.1128/jb.78.4.550-556.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTNICKI-GARCIA S., REYES E. CHEMISTRY OF SPORE WALL DIFFERENTIATION IN MUCOR ROUXII. Arch Biochem Biophys. 1964 Oct;108:125–133. doi: 10.1016/0003-9861(64)90363-7. [DOI] [PubMed] [Google Scholar]
- BONNER T. G., DUNCANA Infra-red spectra of some melanins. Nature. 1962 Jun 16;194:1078–1079. doi: 10.1038/1941078a0. [DOI] [PubMed] [Google Scholar]
- CARLISLE D. B. Softening chitin for histology. Nature. 1960 Sep 24;187:1132–1133. doi: 10.1038/1871132b0. [DOI] [PubMed] [Google Scholar]
- HORIKOSHI K., ARIMA K. X-ray diffraction patterns of the cell wall of Aspergillus oryzae. Biochim Biophys Acta. 1962 Feb 26;57:392–394. doi: 10.1016/0006-3002(62)91140-x. [DOI] [PubMed] [Google Scholar]
- Nicolaus R. A., Piattelli M., Fattorusso E. The structure of melanins and melanogenesis. IV. On some natural melanins. Tetrahedron. 1964 May;20(5):1163–1172. doi: 10.1016/s0040-4020(01)98983-5. [DOI] [PubMed] [Google Scholar]
- POTGIETER H. J., ALEXANDER M. POLYSACCHARIDE COMPONENTS OF NEUROSPORA CRASSA HYPHAL WALLS. Can J Microbiol. 1965 Feb;11:122–125. doi: 10.1139/m65-017. [DOI] [PubMed] [Google Scholar]
- REESE E. T., MANDELS M. Beta-D-1, 3 Glucanases in fungi. Can J Microbiol. 1959 Apr;5(2):173–185. doi: 10.1139/m59-022. [DOI] [PubMed] [Google Scholar]
- REISSIG J. L., STORMINGER J. L., LELOIR L. F. A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem. 1955 Dec;217(2):959–966. [PubMed] [Google Scholar]
- SMOGYI M. Notes on sugar determination. J Biol Chem. 1952 Mar;195(1):19–23. [PubMed] [Google Scholar]
- SWAN G. A. Chemical structure of melanins. Ann N Y Acad Sci. 1963 Feb 15;100:1005–1019. [PubMed] [Google Scholar]
- Skujins J. J., Potgieter H. J., Alexander M. Dissolution of fungal cell walls by a streptomycete chitinase and beta-(1-3) glucanase. Arch Biochem Biophys. 1965 Aug;111(2):358–364. doi: 10.1016/0003-9861(65)90197-9. [DOI] [PubMed] [Google Scholar]



