Summary
Assembly of heptameric Sm protein rings on snRNAs (Sm cores), essential for snRNP function, is mediated by the SMN complex. Specific Sm core assembly depends on Sm proteins and snRNA recognition by SMN/Gemin2- and Gemin5-containing subunits, respectively. The mechanism by which the Sm proteins are gathered and illicit Sm core assembly is prevented is unknown. Here, we describe the 2.5 Å crystal structure of Gemin2 bound to SmD1/D2/F/E/G pentamer and SMN’s Gemin2-binding domain, a key assembly intermediate. Remarkably, through its extended conformation, Gemin2 wraps around the crescent-shaped pentamer, interacting with all five Sm proteins and gripping its bottom- and top-sides and outer perimeter. It further reaches into its RNA-binding pocket, preventing it from binding RNA. Interestingly, SMN-Gemin2 interaction is abrogated by an SMA (spinal muscular atrophy)-causing mutation in an SMN helix that mediates Gemin2 binding. These findings provide mechanistic insights into SMN complex function, linking snRNP biogenesis and SMA pathogenesis.
Introduction
Small nuclear ribonucleoprotein particles (snRNPs) are a major class of non-coding RNA-protein complexes that play key roles in post-transcriptional gene expression, including pre-mRNA splicing and suppression of premature termination (Kaida et al., 2010; Staley and Guthrie, 1998; Wahl et al., 2009). Each snRNP consists of one ~100–200 nucleotide small nuclear RNA (snRNA), called U1, U2, U4, U5, U11, U12, and U4atac, and a heptameric ring of Sm proteins (B/B′, D1, D2, D3, E, F, and G) that surrounds the snRNA’s Sm site (Sm core), as well as several proteins specific to each U snRNA (Kambach et al., 1999; Newman and Nagai, 2010; Patel and Steitz, 2003; Pomeranz Krummel et al., 2009; Weber et al., 2010; Will and Luhrmann, 2001). Sm cores are essential for the function, stability and nuclear localization of snRNPs, and their assembly is a key step in snRNP biogenesis (Mattaj et al., 1993; Will and Luhrmann, 2001). In vitro, purified Sm proteins can spontaneously form Sm cores on any RNA or oligoribonucleotide that contains a sequence resembling an Sm site, AUUUUUG or AUUUGUG (Raker et al., 1999). This assembly occurs stepwise from three Sm heteromeric subcore complexes, SmD1/D2, SmF/E/G and SmB/D3. SmD1/D2 and SmF/E/G first associate, forming a pentameric subcore that avidly binds RNA and this subsequently recruits SmB/D3, completing the formation of a highly stable Sm core (Raker et al., 1996). However, as sequences on which Sm proteins have the propensity to assemble do not uniquely define snRNAs (Pellizzoni et al., 2002b), in cells potentially deleterious illicit Sm core assembly is prevented by the SMN complex, a molecular assembly machine that confers the necessary stringent specificity, ensuring Sm core assembly only on snRNAs (Fischer et al., 1997; Liu et al., 1997; Meister et al., 2001a; Pellizzoni et al., 2002b).
The SMN complex is comprised of SMN, Gemins 2–8 and Unrip (Baccon et al., 2002; Carissimi et al., 2005; Carissimi et al., 2006; Charroux et al., 1999; Charroux et al., 2000; Grimmler et al., 2005; Gubitz et al., 2002; Liu et al., 1997; Pellizzoni et al., 2002a). Recent findings showed that the active SMN complex, comprised of all of its known components, is made up of distinct subunits (Battle et al., 2007; Carissimi et al., 2005; Carissimi et al., 2006; Chari et al., 2008; Yong et al., 2010). The Sm proteins are recognized by a subunit that includes SMN and Gemin2 (Chari et al., 2008; Yong et al., 2010). The specificity for snRNAs is determined separately by Gemin5 (Battle et al., 2006; Lau et al., 2009), which recognizes a large ~50–60 nucleotide structure, called the snRNP code, that includes the Sm site and an adjacent 3′-terminal stem-loop structure found in all the pre-snRNAs and distinguishing them from other classes of RNAs (Golembe et al., 2005; Yong et al., 2004a; Yong et al., 2010). Additional subunits, one containing Gemins 6/7/8 and Unrip that can interact with SMN/Gemin2, and another comprised of the putative RNA helicase Gemin3 that can interact with Gemin4 and Gemin5, are also required for Sm core assembly in complex eukaryotes but their specific functions are not yet known (Battle et al., 2007; Carissimi et al., 2006; Yong et al., 2010). Despite significant advances, essential details on the process by which specific Sm core assembly is achieved in cells remain to be defined and a lack of atomic resolution structure of SMN complex components has limited progress on its mechanism and function. To date, only the structures of one domain in SMN, the Tudor domain (Selenko et al., 2001; Sprangers et al., 2003), and of a Gemins 6/7 heterodimer (Ma et al., 2005) have been described.
Here, we identified Gemin2 as the protein that binds a pentamer of Sm proteins comprised of SmD1/D2 and SmF/E/G. We determined the crystal structure of this complex bound to SMN’s Gemin2 binding domain to 2.5 Å, providing important mechanistic insights for SMN complex function and on Sm core assembly. An additional dimension of interest in the SMN complex and the snRNP assembly pathway comes from the fact that reduced levels of functional SMN, due to protein deficiency (>97% of the cases) or loss of function mutations, cause spinal muscular atrophy (SMA), a common motor neuron degenerative disease and a leading hereditary cause of infant mortality (Lefebvre et al., 1995; Talbot and Davies, 2001; Wirth et al., 2006). Information from the structure we determined explains the molecular basis of an SMA-causing patient mutation in SMN, linking a defect in Gemin2-mediated Sm pentamer recruitment to SMA.
Results
Gemin2 binds a pentamer of Sm proteins, D1, D2, E, F and G
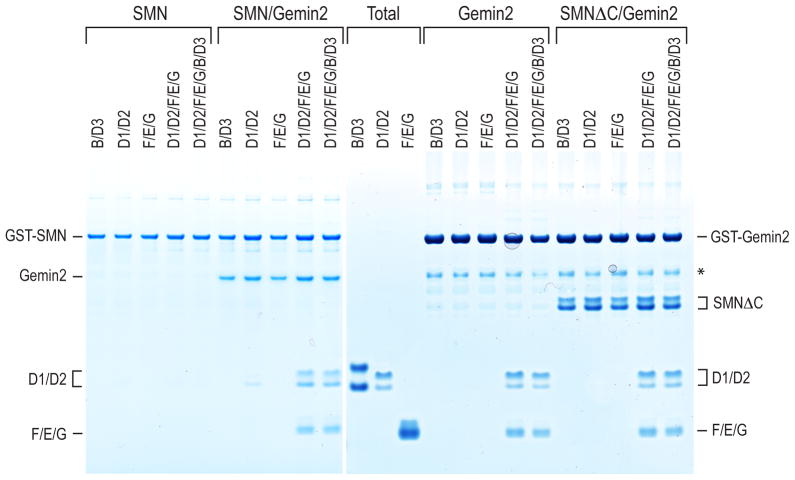
Previous studies identified subunits of the SMN complex, including a key intermediate containing SMN, Gemin2 and a subset of Sm proteins in human cells (Battle et al., 2007; Carissimi et al., 2005; Carissimi et al., 2006; Chari et al., 2008; Yong et al., 2010). SMN and Gemin2, also known as SMN interacting protein 1 (SIP1) (Liu et al., 1997), exist as a heterodimer that further oligomerizes via SMN’s C-terminal YG domain (Pellizzoni et al., 1999). To define interactions of SMN and Gemin2 with Sm proteins, we performed in vitro binding experiments using recombinant SMN, Gemin2 with all the Sm proteins, co-expressed according to their known heteromeric interactions, SmD1/D2, SmF/E/G, and SmB/D3 (Kambach et al., 1999; Raker et al., 1996). Binding assays with the purified proteins showed that SMN/Gemin2 bound to SmD1/D2 and SmF/E/G together, but little or no binding was detected to either of these alone (Figure 1), suggesting that SMN/Gemin2 binds to a pentamer that these di- and tri-heteromeric Sm protein complexes can form (Raker et al., 1996). No binding to SmB/D3 was detected under these conditions. SmB/D3 was previously shown to interact with SMN, dependent on post-translational arginine methylation of their RG-rich domains (Brahms et al., 2001; Friesen and Dreyfuss, 2000; Friesen et al., 2001a), which for efficiency of expression were not included in our constructs. Surprisingly, deletions of most of SMN, leaving only SMN’s Gemin2 binding domain (SMNGe2BD; SMN residues 26–62) (Liu et al., 1997; Wang and Dreyfuss, 2001), showed similar Sm pentamer binding, indicating that most of SMN is not necessary for this interaction (Figure 1 and data not shown). Indeed, Gemin2 alone could bind the Sm pentamer with similar efficiency to that of SMN/Gemin2 (Figure 1).
Figure 1. Gemin2 binds the Sm pentamer, D1, D2, E, F, and G.
Recombinant GST-SMN or GST-SMN/Gemin2 was used for the binding to recombinant Sm proteins (left panel). Similarly in a separate experiment, recombinant GST-Gemin2 or GST-Gemin2 with C-terminus truncated SMN (residues 1–194) was used for the binding to Sm proteins (right panel). Either individual subcore complex (SmD1/D2, SmF/E/G or SmB/D3), or the five (SmD1/D2/F/E/G) or all seven Sm proteins (SmD1/D2/F/E/G/B/D3) were used in binding. For efficient expression, SmB/D3 proteins do not contain their C-terminal RG-rich domain. The asterisk indicates degradation products of GST-Gemin2. The total panel shows 5% of the proteins used for binding.
Determination of the crystal structure of Gemin2-SMNGe2BD-Sm pentamer
The structure of an SMN complex intermediate that contains the majority of the Sm proteins is critical for understanding the Sm core assembly process and we therefore set out crystallization trials of SMN/Gemin2 with SmD1/D2 and SmF/E/G. Attempts to obtain crystals containing full length SMN were not successful. However, a complex containing Gemin2, SMNGe2BD and the Sm pentamer migrated as a single peak in gel filtration and yielded well diffracting crystals. The structure of human SmD1/D2 dimer was previously determined to 2.5 Å (Kambach et al., 1999). In addition, the structures of human SmB/D3 as well as bacterial and archeal Sm-like proteins were determined to high resolution (Collins et al., 2003; Kambach et al., 1999; Sauter et al., 2003; Toro et al., 2001), and we used them to build structure models of SmF, E, and G. Using SmD1/D2 and SmF/E/G models for molecular replacement in combination with single-wavelength anomalous diffraction (SAD) phasing from selenomethionine (Se-met) labeling in Gemin2 and SmF, E, G, we solved the structure of this seven-component complex to 2.5 Å resolution (Table 1).
Table 1.
Data Collection and Refinement Statistics
| Data Collection | ||
|---|---|---|
| Data set | Native | Se-Met derivative |
| Wavelength (Å) | 0.99993 | 0.9791 |
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) | a = 82.83 | a = 82.90 |
| b = 84.60 | b = 86.56 | |
| c = 104.66 | c = 105.03 | |
| Highest resolution (Å) a*, b*, and c* | 2.4, 3.2, 2.6 | 3.0, 4.0, 3.0 |
| Unique reflections | 19831 | 11688 |
| Completeness (%) | 99.3 (98.6)a | 98.1 (98.3)d |
| 76.0 (20.0)b | 75.2 (25.1)e | |
| Rsym(%) | 0.124 (0.284)c | 0.182(0.217)f |
| Mean I/σ | 13.7 (3.8) | 7.6 (4.3) |
| Redundancy | 6.3 (3.8) | 7.7 (5.1) |
| Refinement Statistics | ||
| Resolution range for refinement (Å) | 40-2.5 | |
| R factor (%) | 25.7 | |
| Rfree factor (%) | 33.1 | |
| Number of reflections | 19831 | |
| Number of protein atoms | 4554 | |
| Number of water molecules | 33 | |
| RMSD bond lengths (Å) | 0.014 | |
| RMSD bond angles (°) | 1.64 | |
| Ramachandran plot (%)Favored, additional allowed, disallowed | 92.3, 6.4, 1.3 | |
For resolution range of 40-3.2 Å, the number in parenthesis corresponds to the highest resolution shell of 3.35-3.20 Å.
For resolution range of 40-2.5 Å, the number in parenthesis corresponds to the highest resolution shell of 2.62-2.50 Å. Before ellipsoidal truncation of the data, the completeness is 98.8% (95.2% in the highest resolution shell).
The number in parenthesis corresponds to the highest resolution shell of 3.35-3.20 Å, which was unaffected by ellipsoidal truncation. The Rsym of the resolution range 3.2-2.4 Å was not obtained due to anisotropic diffraction. We performed ellipsoidal truncation on the merged dataset.
For resolution range of 40-4.0 Å, the number in parenthesis corresponds to the highest resolution shell of 4.2-4.0 Å.
For resolution range of 40-3.0 Å, the number in parenthesis corresponds to the highest resolution shell of 3.16-3.00 Å. Before ellipsoidal truncation of the data, the completeness is 98.1% (84.7% in the highest resolution shell).
The number in parenthesis corresponds to the highest resolution shell of 4.2-4.0 Å, which was unaffected by ellipsoidal truncation. The Rsym of the resolution range 4.0-3.0 Å was not obtained due to anisotropic diffraction. We performed ellipsoidal truncation on the merged dataset.
Rsym = Σ|I-<I>|/ΣI, where I is the observed intensity and <I> is the average intensity from multiple measurements.
R factor = Σ|Fobs-Fcalc|/ΣFobs,
5.1% of the truncated dataset was excluded from the refinement to calculate Rfree.
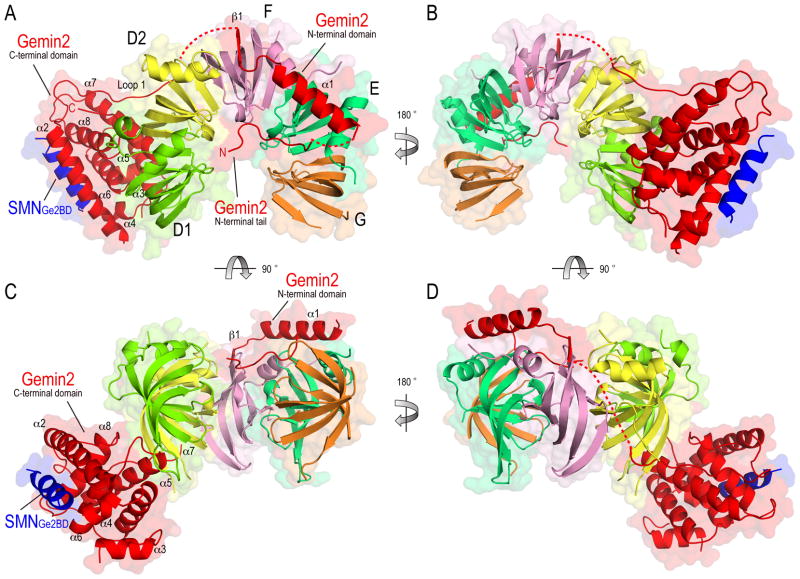
The final refined model of Gemin2-SMNGe2BD-Sm pentamer (PDB ID code 3S6N) is shown in Figure 2. Overall, the five Sm proteins in the complex are arranged as a crescent-shaped 5/7th of a ring. The arrangement of the Sm proteins in the pentamer, as SmD1, D2, F, E, and G, clockwise from a top view (Figure 2A), is the same as in Sm cores visualized in structures of U1 snRNP (Pomeranz Krummel et al., 2009; Weber et al., 2010) (Figure S1A). Consistent with the binding experiments (Figure 1), Gemin2 contacts the Sm pentamer. Gemin2 has an extended conformation, wrapping around the Sm pentamer and interacting with both SmD1/D2 and SmF/E/G via two distinct structural domains. Gemin2’s C-terminal domain (residues 100–280) interacts with both SmD1/D2 and with SMNGe2BD on opposite distal surfaces, while its N-terminal domain (residues 1–69) interacts with SmF/E/G. SMNGe2BD comprises a single helix (residues 37–51) that does not contact the Sm pentamer (Figure 2).
Figure 2. Overall structure of the Gemin2-SMNGe2BD-Sm pentamer complex.
Four rotational views of the ribbon diagram of the complex structure (PDB ID code 3S6N) are shown with the surface representation. The five Sm proteins, SmD1, D2, F, E, and G are colored in lime, lemon, pink, dark green and orange, respectively. Gemin2 and SMNGe2BD peptide are colored in red and blue, respectively. Gemin2’s domains and secondary structures are labeled. In panel (A), the five Sm proteins are arranged in the clockwise order of SmD1, D2, F, E and G to form a 5/7 of doughnut shape with their canonical N-terminal helices facing out. The structure contains visible structures of SmD1 (residues 1–81), SmD2 (residues 23–76 and 90–116), SmF (residues 3–76), SmE (residues 14–90), SmG (residues 14–51 and 56–72), Gemin2 (residues 22–31, 47–69, 83–123, 134–149 and 179–276) and SMN (residues 37–51). The disordered loop connecting the N-terminal tail and α1 of Gemin2 is shown in a red dashed line in panel (A). The disordered loop connecting the N- and C-terminal domains of Gemin2 are also shown in red dashed lines in panels (A), (B) and (D). The figures are prepared using PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC). The rotation of the views is indicated by arrow and the degree of the rotation.
Structure of the Sm proteins in the Gemin2-SMNGe2BD-Sm pentamer
Each of the Sm proteins in the pentamer has a canonical Sm fold, consisting of an N-terminal α-helix followed by a strongly bent five-stranded anti-parallel β-sheet with loops connecting each of these segments (Kambach et al., 1999) (Figure S1A). In addition to the 2.5 Å SmD1/D2 structure (Kambach et al., 1999), two recently published structures of assembled human Sm cores, a 5.5 Å structure of U1 snRNP reconstituted from recombinant proteins (Pomeranz Krummel et al., 2009) and a 4.4 Å structure of lightly protease-digested native U1 snRNP purified from HeLa cells (Weber et al., 2010), provided useful references against which the structure of the Sm proteins in our complex could be compared. The SmD1/D2 structure in our complex is very similar to the 2.5 Å dimer structure (rmsd of 0.39 Å for all the equivalent main chain atoms), and the Cα trace for SmF/E/G is very similar to that of the corresponding proteins in the U1 snRNP at 4.4 Å (rmsd 1.16 Å), demonstrating an overall similarity of Sm proteins’ folds and arrangement in all contexts (Figure S1B).
Our structure revealed several differences and details on Sm protein organizations that were not observed in previous structures. In our complex, more residues in SmD2 are visible and certain loop regions and C-terminal tail of SmD2 and SmD1 adopt slightly different conformations (Figure S1B), likely due to their contact with SmF/E/G as well as with Gemin2. Our structure also provides a view of the intermolecular interactions between neighboring Sm proteins in the pentamer, including the interactions within the SmF/E/G subcore complex as well as at the SmD2-SmF interface that connects the two Sm subcore complexes (see details in Figure S1).
Notably, the Gemin2-bound Sm pentamer has a narrower conformation compared to that in the assembled, snRNA-bound Sm core of U1 snRNP (Weber et al., 2010). While individual Sm proteins in the two structures show little deviations (rmsd of 0.67, 0.89, 0.71, 0.79, and 0.90 Å for SmD1, D2, F, E, and G, respectively), comparing the entire Sm pentamer from the two structures shows significant differences. As shown in Figure S2A, when the Cα of SmD1/D2 in the two structures are superimposed (0.78 Å rmsd), SmF/E/G comparison gives a 5.81 Å rmsd on average (3.13, 5.90, and 8.07 Å rmsd for SmF, E and G, respectively). Similarly, when SmF/E/G coordinates are aligned (1.16 Å rmsd), SmD1/D2 deviates by 4.70 Å (5.85 and 3.05 Å rmsd for SmD1 and D2, respectively) (Figure S2B). As a result, the width of the opening between SmD1 and SmG in Gemin2-bound Sm pentamer is smaller than in the assembled Sm core. For example the distance between the most conserved residues of the pentamer, SmD1 Asn37 to SmG Asn39 is 27.4 Å in our structure vs. 31.2 Å in the U1 snRNP structure (Figure S2C). There is also a smaller angle between SmD1 Asn37, SmF Asn41 and SmG Asn39 (64.9° vs. 76.2°) (Figure S2C). Consequently, the space between SmD1 and SmG is not sufficient for SmB/D3 to fit in and the pentamer’s RNA-binding pocket could not accommodate the snRNA’s Sm site (Figure S2D and see below in Figure 4D), at least not in the conformation it has based on the U1 snRNP model (Weber et al., 2010). These findings suggest that while the interfaces within each of the Sm subcore complexes, SmD1/D2 and SmF/E/G, are rigid, there is considerable angular flexibility at the SmD2-SmF interface that may be utilized during Sm core assembly.
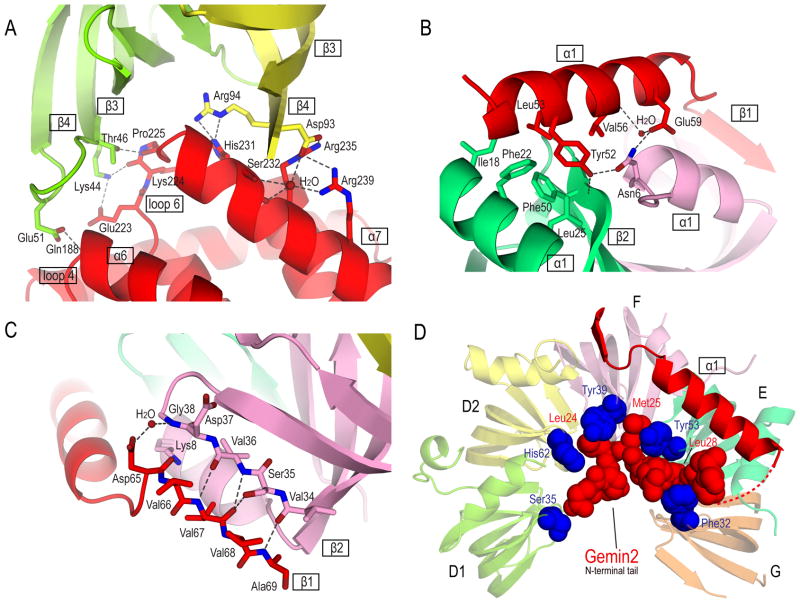
Figure 4. Detailed views of Gemin2 structure and its interactions with the Sm pentamer.
Ribbons are colored as in Figure 2. Amino acid residues involved in the interactions as described in the text are shown in sticks (A–C) or spheres (D). Side chain carbon atoms are labeled in the same color as their respective protein chains. Oxygen and nitrogen are colored in red and blue, respectively. Potential hydrogen-bonds and salt bridges are shown as dashed lines. Water molecules are shown as red spheres.
(A) Interactions between Gemin2’s C-terminal domain and SmD1/D2.
(B) Interactions between α1 of Gemin2 and SmF/E.
(C) Interactions between β1 of Gemin2 and β2 of SmF.
(D) Gemin2’s N-terminal tail inserts into the Sm pentamer’s RNA-binding pocket and interacts with the five Sm proteins. The five amino acids shown in blue spheres, Ser35, His62, Tyr39, Tyr53 and Phe32 in Sm D1, D2, F, E and G, respectively, are involved in the interactions with RNA by stacking nucleotide bases from the top. Gemin2’s Leu24, Met25, and Leu28 block the RNA nucleotide binding pockets of SmF, E, and G, respectively.
Structure of Gemin2 and its interactions with the Sm pentamer
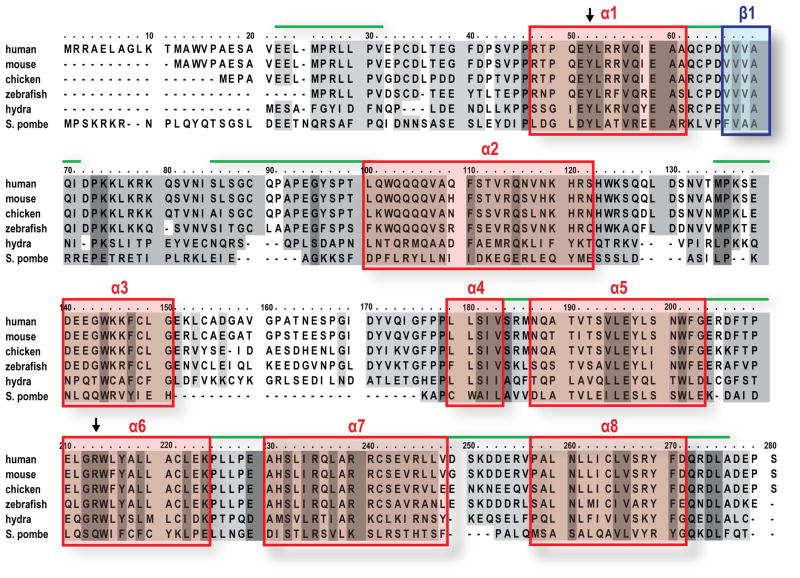
The contact between Gemin2 and the Sm pentamer is remarkably extensive, encompassing a large combined buried area (total 5130 Å2). Gemin2’s C- (residues 100–280) and N-terminal (residues 1–69) domains interact with SmD1/D2 and SmF/E, respectively. A relatively unstructured loop 1 (residues 70–99) connects these two domains and surrounds the perimeter of SmD2/F. Additionally Gemin2’s N-terminal tail (residues 1–46) extends into the inner RNA-binding pocket of the Sm pentamer (Figure 2). These extensive contacts between Gemin2 and the Sm pentamer, supported by high quality electron density maps (Figure S3), reveal how Gemin2 could stabilize the pentamer, which by itself is relatively unstable (Raker et al., 1996), and is consistent with Gemin2’s strong binding to the pentamer compared to either SmD1/D2 or SmF/E/G alone (Figure 1). Multiple sequence alignment of Gemin2 orthologs from divergent eukaryotic organisms shows a high degree of amino acid sequence as well as secondary structure conservation (Figure 3). In accord with Gemin2’s structure, the highest conservation is observed for α1 and β1 in the N-terminal domain and the helices of the C-terminal domain.
Figure 3. Multiple sequence alignment of Gemin2 orthologs from diverse organisms.
The sequence alignment was performed using the Praline program (Simossis and Heringa, 2005). Human, Homo sapiens (accession number NP_003607); mouse, Mus musculus (NP_079932); chicken, Gallus gallus (NP_989530); zebrafish, Danio rerio (NP_001017608); hydra, Hydra magnipapillata (XP_002160350); S. pombe, Schizosaccharomyces pombe (CAB88094). Conservation of sequence is represented based on the BLOSUM score matrix. Partially conserved residues (conservation score 4–7) are highlighted as light gray; highly conserved residues (conservation score 8–10) are highlighted as dark gray. Gemin2’s secondary structure, as seen in the crystal, is indicated with red boxes for α-helices, blue box for the β-sheet and green lines for the loops. Positions of the mutations (Tyr52 and Arg213) are indicated by arrows.
Gemin2’s C-terminal domain consists of seven α-helices (Figure 2). As shown in a model in Figure 4A that is supported by the electron density map in Figure S3A, helices α5–8 (residues 187–271) form an anti-parallel four-helix bundle, and α2 (residues 100–122) is a long orthogonal helix packed against α6 and α8. Between the two well-structured segments, there are two short helices (α3-α4) and two invisible loops: one between α2 and α3 and the other between α3 and α4. The four-helix bundle and α2 account for the entire binding surface to SmD1/D2 as well as most of the binding surface to SMNGe2BD. The interactions between Gemin2 and SmD1/D2 mainly involve polar networks between loop 4, 6 of Gemin2 and β3-β4 of SmD1, as well as between α7 of Gemin2 and β3-β4 of SmD2. The main chain of Gln188 (loop 4) and Pro225 (loop 6) in Gemin2 form hydrogen-bonds with Glu51 and Thr46 in SmD1, respectively. In addition, the side chain of Glu223 and carbonyl group of Lys224 from loop 6 of Gemin2 form salt bridges with Lys44 of SmD1. Ser232, Arg235 and Arg239, which are highly conserved among Gemin2 orthologs, make extensive hydrophilic network with a water molecule and Asp93 from SmD2. Moreover, His231 of Gemin2 forms hydrogen-bonds with Arg94 of SmD2 (Figure 4A).
Connecting the N- and C-terminal domains of Gemin2 is loop 1 (residues 70–99), whose second half (residues 83–99) interacts with SmD2. Although its first half (residues 70–82) is invisible in our structure, its trajectory and length suggest that this segment loosely wraps around the perimeter of SmD2/F (Figure 2). The amino acid sequence of this loop varies among Gemin2 orthologs of divergent organisms, however, its length in all exceeds 22 residues (Figure 3), which is sufficient to cover the distance between the N- and C-terminal domains of Gemin2.
Gemin2’s N-terminal domain contains a single α-helix (α1), which mainly contacts SmF/E, followed by a short sequence (VVVA), which forms a β-strand that pairs with the β-sheet of SmF (Figures 4B, 4C, S3B and S3C). α1 of Gemin2 contains a highly conserved YLxxVxxE motif (Figure 3), of which the conserved residues Tyr52, Leu53 and Val56 form a hydrophobic patch and contact the hydrophobic surface formed by Ile18, Phe22, Leu25 and Phe50 of SmE (Figure 4B). In addition, the interactions between Gemin2’s α1 and SmF/E are substantiated through an extensive hydrogen-bonding network involving the side chains of Tyr52 and Glu59 in Gemin2, side chain of Asn6 in SmF and main chain amide group of Phe50 in SmE. This network is further extended to the main chain carbonyl group of Val56 in Gemin2’s α1 via a well-defined water molecule (Figure 4B). Consistently, Gemin2’s interacting residues in SmE (Ile18, Phe22, Leu25, and Phe50) and in SmF (Asn6) are also highly conserved (Weber et al., 2010). These findings indicate a functional significance of these interactions, which is further supported by mutagenesis studies on one of the critical residues, Tyr52 (see below in Figure 5). The short β-strand following α1 of Gemin2 pairs with the second half of β2 of SmF through anti-parallel β sheet interactions (Figure 4C). These interactions are further enhanced by additional hydrogen-bondings: between the main chain of Val67 of Gemin2 and the side chain of Ser35 of SmF, between carbonyl oxygen of Asp65 in Gemin2 and the side chain of Lys8 in SmF, and between the side chain of Asp65 in Gemin2 and main chain of Gly38 in SmF mediated by a water molecule (Figure 4C).
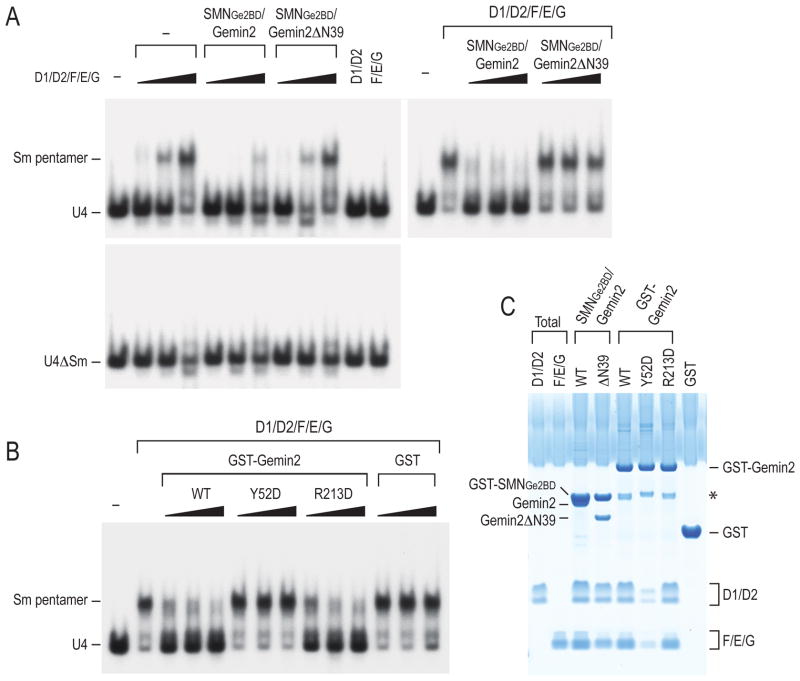
Figure 5. Gemin2 prevents the Sm pentamer from binding RNA.
(A) Gemin2 dose-dependently inhibits the Sm pentamer from binding RNA and its N-terminus is critical for this inhibition. Gemin2 or Gemin2ΔN39 (1 μg) co-purified with GST-SMNGe2BD was pre-incubated with increasing amounts of the five Sm proteins (D1/D2/F/E/G; 10–50 ng of each) and then assayed for binding to [32P]-α-UTP labeled U4 or U4ΔSm snRNA. SmD1/D2 or SmF/E/G was used as controls (left panel). In a parallel experiment, SmD1/D2/F/E/G was pre-incubated with increasing amounts (1–9 μg) of Gemin2 or Gemin2ΔN39 that were co-purified with GST-SMNGe2BD and assayed for RNA-binding. The positions of the free RNA and the Sm pentamers assembled on the RNA are indicated.
(B) The effects of wild type or various mutant Gemin2 proteins on the Sm pentamer’s binding to RNA. Increasing amounts (1–9 μg) of GST, GST-Gemin2 wild type (WT), or various GST tagged Gemin2 mutants (Y52D and R213D) were pre-incubated with SmD1/D2/F/E/G and assayed for binding to U4 snRNA as in (A).
(C) Protein binding between wild type or various Gemin2 mutants and the Sm pentamer. Recombinant wild type and mutant Gemin2 proteins (2.5–5 μg) used in (A) and (B) were assayed for binding to SmD1/D2/F/E/G (1 μg). The total panel shows 5% of the proteins used for binding.
The RNA-interacting residues of Sm proteins are highly conserved among archaeal and eukaryotic Sm and Sm-like proteins. The crystal structures of two archaeal Sm protein homo-heptamers complexed with oligo(U) RNA reveal how each Sm protein contacts each uracil (Thore et al., 2003; Toro et al., 2001). Based on this insight as well as biochemical studies, Weber and colleagues provided a similar interaction model, in which each Sm protein provides one aromatic residue on the top and one positively charged residue at the bottom to sandwich each base while simultaneously using the most conserved residue among Sm proteins, Asn, to form hydrogen-bonds with the base from the side (Weber et al., 2010). Strikingly, however, in our structure the N-terminal tail of Gemin2 occludes the RNA binding surface of the Sm pentamer and extensively overlaps with the positions of the U1 snRNA’s Sm-site in the pocket. Gemin2’s N-terminal residues 22–31 extend into the center of the Sm pentamer’s RNA-binding pocket, contacting most of the base-binding sites. Specifically, the electron density map shows that Gemin2’s Leu24, Met25 and Leu28 are right at the base-binding pocket (Figure 4D and S3D). This result suggests that Gemin2 would interfere with the Sm pentamer’s binding to snRNA.
Gemin2 inhibits the Sm pentamer from binding RNA
To test the effect of Gemin2 on the Sm pentamer’s snRNA binding as predicted from the structure, we measured the binding of the Sm pentamer to [32P]-α-UTP labeled U4 or U4ΔSm snRNA in the absence or presence of Gemin2 and its various mutants by electrophoretic mobility shift (Raker et al., 1999). The Sm pentamer bound efficiently to U4 but not to U4ΔSm, indicating that it forms an RNP dependent on the nucleotides present in an Sm site (Figure 5A). Importantly, Gemin2 inhibited the pentamer’s binding to U4 in a dose-dependent manner (Figure 5A and 5B). This inhibition is dependent on direct binding of Gemin2 to the pentamer, as the mutation of a highly conserved Gemin2 residue Tyr52 (Y52D) (Figures 3 and 4B) impairs Gemin2-pentamer interaction (Figure 5C) and also fails to inhibit the pentamer’s U4 binding (Figure 5B). In contrast, another Gemin2 mutation, R213D, which does not affect Gemin2’s binding to the pentamer (Figure 5C), maintains the ability to inhibit the pentamer’s RNA binding similarly to wild type Gemin2 (Figure 5B). The R213D mutation impairs SMN-Gemin2 interaction (see below in Figure 6), suggesting that SMN is not required for Gemin2 to inhibit the pentamer’s RNA binding. However, deletion of Gemin2’s N-terminal tail up to α1, Gemin2ΔN39, while maintaining the ability to bind the Sm pentamer (Figure 5C), had little or no effect on the pentamer’s binding to the RNA (Figure 5A). Thus, Gemin2’s N-terminal tail, which is inserted into the pentamer’s RNA-binding pocket, plays an important role in preventing the Sm pentamer from binding RNAs.
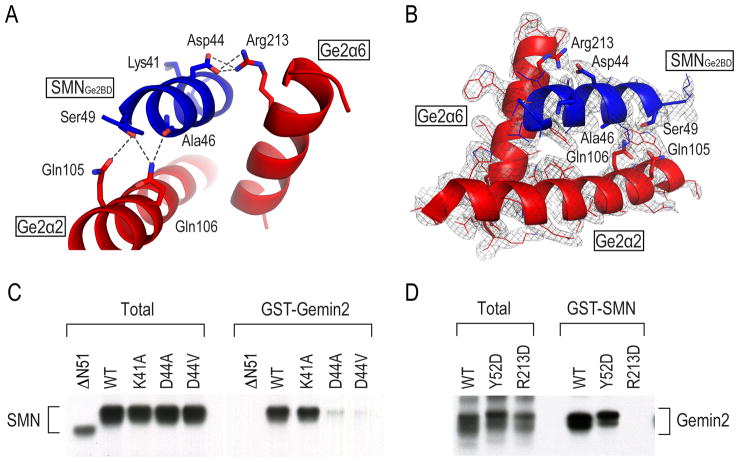
Figure 6. Critical residues involved in SMNGe2BD-Gemin2 interaction and an SMA-causing patient mutation in SMN abrogates its interaction with Gemin2.
(A) A detailed view of the SMNGe2BD-Gemin2 polar interactions. Ribbons are colored as in Figure 2. Amino acid residues, side chain carbon atoms, oxygen and nitrogen, and potential hydrogen-bonds and salt bridges are labeled as in Figure 4. Asp44 (D44) of SMN is mutated in an SMA patient. Lys41 of SMN is not involved in interactions with Gemin2 and is therefore used as a control for the binding experiment in (C). Additional hydrophobic interactions, involving Leu39, Tyr43, Val47 and Phe50 in SMN interacting with a hydrophobic pocket in Gemin2, are shown in Figure S4.
(B) Electron density map of an additional view of the interaction regions between Gemin2 and SMNGe2BD. SigmaA-weighted 2Fo-Fc electron density map (gray mesh) is contoured at 1.2σ. The color schemes are the same as in (A). Protein models are shown in cartoon representation with contacting residues in sticks.
(C) The SMA patient mutation, D44V, abrogates SMN-Gemin2 interaction. In vitro translated [35S]-Met labeled wild type SMN and the indicated mutant proteins were incubated with recombinant GST-Gemin2. SMNΔN51 lacks the Gemin2 binding domain (Wang and Dreyfuss, 2001) and is used as a negative control. The total panel shows 5% of the proteins used for the binding.
(D) Reciprocal binding reactions were performed as in (C). In vitro translated [35S]-Met labeled wild type Gemin2 or mutants (Y52D and R213D) were incubated with recombinant GST-SMN that was immobilized on beads.
SMN-Gemin2 interaction explains an SMA-causing SMN mutation
As shown by two different angles of the view of the structure and with electron density map (Figure 6A and 6B), the interaction of SMNGe2BD with Gemin2 is mediated by a single helix (SMN residues 37–51) adjacent and anti-parallel to the α2 in Gemin2’s C-terminal domain, on the opposite side of this domain’s SmD1/D2 binding surface (Figure 2). The SMN helix contacts α2, α6, and α8 of Gemin2 via hydrophobic (Figure S4) and polar interactions (Figure 6A and 6B). For example, the main chain carbonyl oxygen of Ala46 and the side chain of Ser49 in SMN form hydrogen-bonds with the side chains of Gln105 and Gln106 in Gemin2 (Figure 6A). In addition, the side chain of Arg213 (R213) in Gemin2 forms salt bridge interactions with that of SMN’s Asp44 (D44) (Figure 6A). This interaction is of particular interest, as D44V mutation in SMN is an SMA-causing patient mutation (Sun et al., 2005). To test the importance of this interaction, we determined the effect of mutations in either R213 or D44 on full-length SMN and Gemin2 binding. As shown in Figure 6C, D44V as well as D44A abrogated Gemin2 binding, consistent with previous observations that also demonstrated that SMN D44V decreases the protein’s snRNP assembly activity (Ogawa et al., 2007). In contrast, mutation of a nearby residue that according to the structure is not involved in SMN-Gemin2 interaction, K41A, had no effect. Reciprocally, and as expected from the structure, mutating R213 in Gemin2, R213D, abrogated its binding to SMN (Figure 6D). On the other hand, Gemin2’s Sm pentamer binding-defective mutant, Y52D (Figure 5C), showed no difference in binding to SMN (Figure 6D). Together, these findings highlight the importance of SMN’s D44 and Gemin2’s R213 in bridging SMN-Gemin2 interaction, providing a structural basis of the D44V patient mutation at an atomic resolution.
Discussion
The structure and biochemical experiments provide mechanistic insights into the process by which the SMN complex assembles Sm cores and a structural basis for understanding the effect of an SMA-causing SMN mutation. Sm core assembly is a remarkable architectural feat requiring the seven Sm proteins to be brought together and form a ring around the pre-snRNAs’ Sm site, a short nucleotide sequence present also in numerous other RNAs (Wahl et al., 2009; Will and Luhrmann, 2001). To accomplish this, the SMN complex must gather the Sm proteins, inhibit their propensity for illicit Sm core assembly on unintended RNAs until a pre-snRNA joins (Neuenkirchen et al., 2008; Yong et al., 2004b). Our findings demonstrate that Gemin2 serves as the arm of the SMN complex that gathers five out of the seven Sm proteins, holding them as a pentamer poised for Sm core assembly and at the same time preventing them from binding RNAs. The structure explains how this is accomplished, revealing Gemin2 to be a key factor in snRNP biogenesis. Gemin2, through its extended conformation and remarkably extensive interactions with all five Sm proteins, grips the pentamer from its bottom and top sides, and from its outer parameter and inner pocket. Though its specific function was not previously known, Gemin2 has been shown to have a role in Sm core assembly (Feng et al., 2005; Ogawa et al., 2007; Shpargel and Matera, 2005). Consistent with this, the ubiquitously expressed Gemin2 is essential for viability of all eukaryotic organisms (Jablonka et al., 2002; Owen et al., 2000; Paushkin et al., 2000). Notably, Gemin2 gene deletion in the mouse causes embryonic lethality, at an even earlier stage than SMN gene deletion (Jablonka et al., 2002; Schrank et al., 1997). Furthermore, Gemin2’s sequence and domain structure are more phylogenetically conserved than that of all other SMN complex components, including SMN (Cauchi, 2010).
Our findings indicate that the N-terminal tail of Gemin2, particularly residues 22–31, plays a role in inhibiting the pentamer from binding RNA as it occupies the pentamer’s RNA-binding pocket. Furthermore, several residues in this part of Gemin2, including Met25 and Leu28 interact with the residues in the Sm proteins that are involved in binding Sm site nucleotides and are positioned in a way that would hinder RNA binding (Figure 4D). Interestingly, these residues are conserved in Gemin2 orthologs from divergent organisms or are substituted by residues that are compatible with having the same activity, suggesting that this is a conserved function of Gemin2. The pentamer’s narrower conformation in the Gemin2-bound state compared to that in the assembled Sm core would be expected to also restrict access of RNAs to the binding pocket. However, as the structure of an Sm pentamer alone is unknown, it is not possible to determine if Gemin2 binding plays a role in inducing or stabilizing the narrower conformation. Recent studies have shown that pICln, a protein that can bind Sm proteins and inhibit their interaction with snRNA (Friesen et al., 2001b; Pesiridis et al., 2009; Pu et al., 1999), can bind at the SmD1-G opening, forming a closed hetero-hexameric ring that cannot bind snRNA (Chari et al., 2008). A complex suggested to represent a downstream intermediate, comprised of Drosophila C-terminal deleted SMN, Gemin2 and the Sm pentamer, which by electron microscopy shows a similar overall morphology to that of our structure, has also been described (Chari et al., 2008). Our data demonstrate that Gemin2 can bind the Sm pentamer and prevent it from binding snRNAs independent of pICln. Thus, there are at least two mechanisms of pentamer inhibition that are not incompatible and could occur sequentially, first by pICln, and subsequently by Gemin2. However, as pICln is not obligatory for Gemin2-pentamer association, it is also possible that the pentamer binds directly to Gemin2, which links the pentamer to SMN.
For the subsequent steps of Sm core assembly to occur, after pre-snRNA is brought in by Gemin5, Gemin2’s N-terminus, possibly up to α1, would need to be displaced from the Sm pentamer’s RNA-binding pocket to allow the pre-snRNA to bind. The observation that Gemin2ΔN39 can bind the pentamer, suggests that such a displacement would not have the undesirable effect of dissociating the pentamer from the SMN complex. The SmD1-G opening and the Sm site-binding pocket would also need to be widened, utilizing the SmD2-F interface as a hinge. How these structural transitions are effected remains to be determined. Completion of Sm core assembly requires several additional steps and ATP hydrolysis, involving additional proteins about which little structural information is available. In complex eukaryotes, access of RNA to the inhibited intermediate, comprised minimally of SMN/Gemin2-Sm pentamer, is likely to be limited to only bona fide RNA substrates, pre-snRNAs, delivered by Gemin5 (Yong et al., 2010). While it is clear that SMN is oligomeric in cells (Wan et al., 2008), the number of SMN subunits in a complex is unknown, bringing the possibility that it serves as a scaffold for more than one Gemin2-Sm pentamer forming on the same complex simultaneously. SMN determines the capacity of Sm core assembly (Wan et al., 2005) and its oligomerization is particularly important for this function as it serves to recruit essential components for this process. SmB/D3 association with the SMN complex is mediated at least in part by their direct interaction with SMN, which depends on SMN’s oligomerization via its C-terminal YG-rich domains (residues 268–279) (Pellizzoni et al., 1999) and in which the Tudor domain (residues 91–142) plays a role by binding to RG tails of SmB/D3 (Brahms et al., 2001; Sprangers et al., 2003), an interaction that is strongly enhanced by arginine methylation that is carried out by the methylosome/PRMT5 (Brahms et al., 2001; Friesen et al., 2001a; Friesen et al., 2001b; Meister et al., 2001b). There is evidence that an additional subunit that includes Gemins 6/7/8 and Unrip can also associate with SMN/Gemin2 and Sm proteins (Carissimi et al., 2006; Yong et al., 2010). Interestingly, Gemins 6 and 7 form a heterodimer and both have Sm folds and it has therefore been suggested that they might bind the pentamer in the same position where SmB/D3 bind, potentially forming a closed heptameric ring intermediate (Ma et al., 2005). This could further help maintain the pentamer’s association with SMN/Gemin2, together with Gemin8 and Unrip. The function of Gemins 3 and 4, which exist as a dimer and associate with Gemin5, is not known, but the presence of a DEAD box domain in Gemin3 suggests that it may function as an RNA helicase and may be the source of the ATPase activity on which the assembly reaction depends. With the available structure of the key intermediate we describe here, several aspects of the mechanism and regulation of the SMN-Gemins complex as a molecular assembly machine for snRNP biogenesis can now be readily addressed.
The structure further explains why D44V of SMN is an SMA-causing mutation. In the vast majority of SMA patients, the disease results from reduced levels of the SMN protein rather than from nonsense mutations (Wirth et al., 2006). We suggest that D44V is a loss of function mutation because it decreases the ability of SMN bearing this mutation to bind Gemin2 and thus impairs the SMN complex’s capacity to recruit the Sm pentamer for snRNP assembly. These findings thus further link SMN’s function in snRNP biogenesis to SMA. Further atomic level structural information could suggest approaches to enhance SMN-Gemin2 interaction as a potential therapy for SMA.
Experimental Procedures
Plasmid construction and protein expression and purification
All of the plasmids used in the studies contain human cDNAs. Full-length SmD1 and SmD2 were constructed in a single pCDFDuet vector (Novagen) with N-terminal His(6)-tag followed by Tobacco Etch Virus (TEV) cleavage site (His6-Tev) fused to SmD2. Full-length SmF and SmE were also constructed in a single pCDFDuet vector, with N-terminal His6-Tev fused to SmE. Full-length SmG was made in pET28 vector (Novagen) with His6-Tev at the N-terminus. Full-length Gemin2 was made in pCDF vector (Novagen) with His6-Tev at its N-terminus. SMNGe2BD, containing SMN residues 26–62, was fused with an N-terminal GST tag in pET42 vector (Novagen). Mutants of Gemin2 or SMN were created from the plasmids containing wild type Gemin2 or SMN cDNAs using the QuikChange site-directed mutagenesis kit (Stratagene). SmD1/D2 was purified by Ni-column first, followed by TEV protease cleavage, secondary pass of Ni-column, cation exchange, and gel filtration chromatography. SmF/E and SmG were co-expressed and purified by a similar procedure except that anion exchange was used instead. Gemin2 and SMNGe2BD were co-expressed and purified by glutathione affinity chromatography, followed by TEV protease cleavage, Ni-column, and anion exchange chromatography. To make the heptamer of the Gemin2-SMNGe2BD-Sm pentamer complex, equal molar amount of the SmD1/D2, SmF/E/G, and Gemin2/SMNGe2BD complexes were mixed in gel filtration buffer (20 mM Tris-HCl, pH8.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP [tris(2-carboxyethyl)phosphine]) supplemented with 0.5 M NaCl, and subjected to HiLoad superdex200 gel filtration chromatography. The fractions containing all seven components were checked by SDS-PAGE, pooled and concentrated to 7–11 mg/ml, and used for crystallization studies.
Selenomethionine (Se-Met) incorporation was performed on the subcore complexe SmF/E/G and Gemin2/SMNGe2BD by adapting the methionine pathway inhibition method (Van Duyne et al., 1993). Cell culture and protein purifications were performed as described above, except that cells were cultured in M9 minimal medium containing amino-acid supplement (Lys, Phe, Thr to final concentration of 100 mg/L, Ile, Leu, and Val to 50 mg/L, and Se-Met to 80 mg/L) for 15 min before protein induction with 1 mM IPTG. The Se-Met-labeled SmF/E/G and Gemin2/SMNGe2BD were mixed with native SmD1/D2 in equal molar stoichiometry for gel filtration as described above. The Se-Met-labeled heptamer was concentrated to about 8 mg/ml for crystallization studies.
For GST-Gemin2 and mutant protein expression and purification, GST fusion proteins were produced from BL21(DE3) E. coli cells containing the expression plasmids in pGEX-4T-1 vector according to the manufacturer’s suggestions (GE Healthcare) with few modifications (Extended Experimental Procedures).
In vitro transcription and translation of SMN and Gemin2
Various Myc-tagged SMN (wild type, K41A, D44A and D44V mutants) and Gemin2 (wild type, Y52D and R213D mutants) constructs were in vitro transcribed and translated with [35S]-Met labeling using the TNT Quick Coupled Transcription/Translation System according to the manufacturer’s instructions (Promega).
In vitro protein binding assays
Protein binding assays were performed according to the manufacturer’s instructions (GE Healthcare) with few modifications (Extended Experimental Procedures). Briefly, 1 μg of GST fusion proteins was immobilized on 25 μl glutathione-Sepharose beads and incubated with 10 μl of the in vitro transcribed and translated [35S]-Met labeled proteins in binding buffer (50 mM Tris, 200 mM NaCl, 0.2 mM EDTA, 0.05% NP-40, 2 mM DTT and protease inhibitors). Bound proteins were resolved by SDS-PAGE and detected by autoradiography. Alternatively, immobilized GST fusion proteins were incubated with 3 μg of purified recombinant Sm proteins in buffer containing 20 mM Tris-HCl, pH8.0, 250 mM NaCl, 2.5 mM MgCl2 and 0.02% Triton X-100. Bound proteins were resolved by SDS-PAGE and visualized by SimplyBlue staining (Invitrogen).
Crystallization, data collection, and structure determination
Human Gemin2-SMNGe2BD-Sm pentamer complex crystals were grown in 1% PEG8000, 100 mM Tris-HCl, pH7.6-8.2 by hanging-drop vapor diffusion method at 20°C within a couple of days. They form in space group P212121, with a = 82.8 Å, b = 84.6 Å, and c = 104.7 Å. Each asymmetric unit contains one Gemin2-SMNGe2BD-Sm pentamer complex. The crystals were cryo-protected by gradually transferring from reservoir solution containing 10% to 40% PEG400, and frozen in liquid nitrogen. Se-Met labeled complex crystals were grown under similar conditions. The X-ray diffraction datasets of native and Se-Met derivative complex crystals were collected at the Advanced Light Source (ALS, Berkeley, CA) beamlines 8.2.1 and 8.2.2. Data were processed by HKL2000 (Otwinowski and Minor, 1997). Initially, the dataset obtained from the native crystals could only be truncated to reach the maximal resolution of about 3.2 Å. The subcore complex of SmD1/D2 inside the heptamer complex was readily located by molecular replacement with the 2.5 Å crystal structure (PDB ID code 1B34) as the search model. The components of SmF, E, and G were located by iterative homolog model building and molecular replacement searching in combination with SmD1/D2. Multi-wavelength anomalous dispersion (MAD) datasets were collected at different wavelengths on several Se-Met derivative crystals, due to severe X-ray decay of a single crystal. Nevertheless, the best Se-Met derivative crystal dataset collected at 0.9796 Å was of high quality and could be used as single-wavelength anomalous dispersion (SAD) in combination with molecular replacement by the SmD1/D2/F/E/G model for phase improvement by PHASER (McCoy et al., 2007) from CCP4 suite (Potterton et al., 2003). In this way, most of the helices of Gemin2 and SMNGe2BD were located. Since the diffraction of the crystals was severely anisotropic, the native dataset was reprocessed and truncated ellipsoidally followed by anisoscaling (Strong et al., 2006). This extended the resolutions to 2.4 Å and 2.6 Å in the directions of a* and c*, respectively, while the resolution in the direction of b* still remained at 3.2 Å. After this processing, the resulting electron density maps for many side chains were improved. The models were gradually improved by cycles of manual rebuilding in Coot (Emsley and Cowtan, 2004) using combinations of methods, including density modification, CNS simulated annealing (Brunger et al., 1998), REFMAC refinement (Winn et al., 2001), and PHASER molecular replacement plus SAD (McCoy et al., 2007). Eleven Se-Met sites were found in the SAD Se-Met dataset. Eight of them were from SmF/E/G and all matched the locations of the atoms S from Met. Three of them were from Gemin2, which provided guidance for the assignment of Gemin2 sequence. In the final stage of model refinement, TLS followed by restrained refinement, as implemented in REFMAC, was used. The final model (PDB ID code 3S6N ) contains SmD1 (residues 1–81), SmD2 (residues 23–76 and 90–116), SmF (residues 3–76), SmE (residues 14–90), SmG (residues 14–51 and 56–72), Gemin2 (residues 22–31, 47–69, 83–123, 134–149, and 179–276), and SMN (residues 37–51). The higher than average R-free of the structure model (Table 1) is due to severe anisotropic diffraction (Strong et al., 2006) and a few regions having relatively low quality electron density, which cannot be confidently assigned in the model. Ramachandran plot by MOLPROBITY (Davis et al., 2007) shows 92.3% of the dihedral angles in favored region, 6.4% in additional allowed region, and 1.3% (7 out of 545) in disallowed region (Table 1). All the 7 outliers are located in the loop regions with relatively poor electron density. Only the regions supported by high quality electron density maps are presented and discussed in detail (Figure S3 and 6B).
In vitro RNA binding assays
[32P]-α-UTP labeled U4 or U4ΔSm snRNA was produced by in vitro transcription as previously described (Pellizzoni et al., 2002b). Binding of Sm pentamers to the radio-labeled snRNAs was performed in buffer containing 20 mM HEPES, pH8.0, 70 mM KCl, 2.5 mM MgCl2, 0.5 mM EDTA and 0.01% Triton X-100. Various amounts of Sm proteins (10–50 ng of each) were used for the binding to snRNAs. Where indicated, 1 μg of recombinant Gemin2 or its deletion mutant (Gemin2ΔN39) was pre-incubated with the Sm pentamer for 10 min before RNA was added for binding. After 30 min binding at 25°C, 2 M urea (final concentration) was added to the reaction mixture and the RNPs were analyzed by 6% native gel electrophoresis. For the RNA binding experiments using Gemin2 or its mutants (Y52D and R213D), 50 ng of each Sm protein was pre-incubated with various amounts of Gemin2 or the mutant proteins (1–9 μg).
Supplementary Material
Acknowledgments
We thank Corie Ralston and Christy Bertoldo for their assistance with X-ray data collection at the Advanced Light Source (ALS, Berkeley, CA) synchrotron beamlines 8.2.1 and 8.2.2. We thank Emily Anna Bridges for help with bacterial cell culture. We are grateful to the members of our laboratory for helpful discussions and comments on this manuscript. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J Biol Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Wang J, Dreyfuss G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J Biol Chem. 2007;282:27953–27959. doi: 10.1074/jbc.M702317200. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carissimi C, Baccon J, Straccia M, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 2005;579:2348–2354. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Carissimi C, Saieva L, Baccon J, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J Biol Chem. 2006;281:8126–8134. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- Cauchi RJ. SMN and Gemins: ‘We are family’ … or are we?: insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays. 2010 doi: 10.1002/bies.201000088. [DOI] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, Cubeddu L, Naidoo N, Harrop SJ, Kornfeld GD, Dawes IW, Curmi PM, Mabbutt BC. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J Biol Chem. 2003;278:17291–17298. doi: 10.1074/jbc.M211826200. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feng W, Gubitz AK, Wan L, Battle DJ, Dostie J, Golembe TJ, Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum Mol Genet. 2005;14:1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001a;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001b;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Otter S, Peter C, Muller F, Chari A, Fischer U. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum Mol Genet. 2005;14:3099–3111. doi: 10.1093/hmg/ddi343. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- Jablonka S, Holtmann B, Meister G, Bandilla M, Rossoll W, Fischer U, Sendtner M. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc Natl Acad Sci U S A. 2002;99:10126–10131. doi: 10.1073/pnas.152318699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Ma Y, Dostie J, Dreyfuss G, Van Duyne GD. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure. 2005;13:883–892. doi: 10.1016/j.str.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Boelens W, Izaurralde E, Jarmolowski A, Kambach C. Nucleocytoplasmic transport and snRNP assembly. Mol Biol Rep. 1993;18:79–83. doi: 10.1007/BF00986760. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001a;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001b;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Ogawa C, Usui K, Aoki M, Ito F, Itoh M, Kai C, Kanamori-Katayama M, Hayashizaki Y, Suzuki H. Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J Biol Chem. 2007;282:11122–11134. doi: 10.1074/jbc.M609297200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Owen N, Doe CL, Mellor J, Davies KE. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum Mol Genet. 2000;9:675–684. doi: 10.1093/hmg/9.5.675. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Paushkin S, Charroux B, Abel L, Perkinson RA, Pellizzoni L, Dreyfuss G. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J Biol Chem. 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem. 2002a;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002b;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Pesiridis GS, Diamond E, Van Duyne GD. Role of pICLn in methylation of Sm proteins by PRMT5. J Biol Chem. 2009;284:21347–21359. doi: 10.1074/jbc.M109.015578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol Cell Biol. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Grimmler M, Schwarzer V, Schoenen F, Fischer U, Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- Thore S, Mayer C, Sauter C, Weeks S, Suck D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J Biol Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Ottinger E, Cho S, Dreyfuss G. Inactivation of the SMN complex by oxidative stress. Mol Cell. 2008;31:244–254. doi: 10.1016/j.molcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dreyfuss G. Characterization of functional domains of the SMN protein in vivo. J Biol Chem. 2001;276:45387–45393. doi: 10.1074/jbc.M105059200. [DOI] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–4184. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin Pediatr Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol. 2004a;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004b;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.